Clonal Associations among Staphylococcus aureus Isolates from Various Sites of Infection (original) (raw)
Abstract
A molecular epidemiological analysis was undertaken to identify lineages of Staphylococcus aureus that may be disproportionately associated with infection. Pulsed-field gel electrophoresis analysis of 405 S. aureus clinical isolates collected from various infection types and geographic locations was performed. Five distinct S. aureus lineages (SALs 1, 2, 4, 5, and 6) were identified, which accounted for 19.01, 9.14, 22.72, 10.12, and 4.69% of isolates, respectively. In addition, 85 lineages which occurred with frequencies of <2.5% were identified and were termed “sporadic.” The most prevalent lineage was methicillin-resistant S. aureus (SAL 4). The second most prevalent lineage, SAL 1, was also isolated at a high frequency from the anterior nares of healthy volunteers, suggesting that its prevalence among clinical isolates may be a consequence of high carriage rates in humans. Gene-specific PCR was carried out to detect genes for a number of staphylococcal virulence traits. tst and cna were found to be significantly associated with prevalent lineages compared to sporadic lineages. When specific infection sites were examined, SAL 4 was significantly associated with respiratory tract infection, while SAL 2 was enriched among blood isolates. SAL 1 and SAL 5 were clonally related to SALs shown by others to be widespread in the clinical isolate population. We conclude from this study that at least five phylogenetic lineages of S. aureus are highly prevalent and widely distributed among clinical isolates. The traits that confer on these lineages a propensity to infect may suggest novel approaches to antistaphylococcal therapy.
Staphylococcus aureus is an important opportunistic pathogen, causing a variety of hospital- and community-acquired infections. Recent reports of the National Nosocomial Infections Surveillance System ranked S. aureus as a leading cause of hospital-acquired bacteremia, pneumonia, and surgical wound infection (7). S. aureus acquires antibiotic resistance with remarkable proficiency, and strains for which vancomycin is the only effective therapeutic agent have emerged. The recently reported reduced susceptibility to vancomycin highlights the importance of understanding the molecular epidemiology of S. aureus infection and identifying new therapeutic targets (17, 46).
Bacterial population analyses indicate that phylogenetic lineages are not always randomly distributed within clinical isolate populations (24, 30–34, 49). In the S. aureus species, discrete lineages or subtypes which exist due to strong selective pressures imposed by antibiotic use and due to other factors that have not been clearly defined can be identified. For example, the majority of methicillin-resistant S. aureus (MRSA) strains expanded clonally and globally upon acquisition of the 30-kb mec determinant (24). Only recently has evidence showing that horizontal transfer resulted in the spread of this determinant to other phylogenetic lineages emerged (3, 24, 31). A large-scale study of the genetic structure of the S. aureus population involving multilocus enzyme electrophoresis (MLEE) analysis of 2,077 clinical and environmental isolates revealed that 81% of isolates were confined to five electrophoretic types (ETs). One ET was methicillin resistant, while another included the majority of toxic shock syndrome toxin 1 (TSST-1)-producing strains (34). The latter, termed ET41, was determined to be the S. aureus clone responsible for the majority of epidemiologically unrelated cases of menstrual toxic shock syndrome (TSS) (33). No obvious selection criteria accounted for the occurrence of three additional ETs, which together represented 37% of isolates. Another clinically important phylogenetic subset of the S. aureus species are the phage type 95 isolates which increased in frequency in Danish hospitals from 3.8% of all isolates in 1997 to 19.3% of all isolates in 1993 (41). Molecular epidemiological analysis of representative phage type 95 isolates showed that they were indistinguishable by both pulsed-field gel electrophoresis (PFGE) and MLEE, indicating that they were clonal in origin (41). The same PFGE pattern was observed among outbreak strains in the United States, indicating that this clone is widely disseminated among S. aureus clinical isolates (1, 6, 45). The genetic and molecular basis for the overrepresentation of distinct lineages of S. aureus, except perhaps for the methicillin-resistant lineage, remains unknown.
In previous studies, we analyzed the genomic DNA fingerprints of S. aureus ocular infection isolates derived from epidemiologically unrelated patients at three clinical centers in the United States (5). Five distinct lineages were observed, which accounted for 58% of the isolates. One of the lineages accounted for almost 26% of the isolates, while another was mecA+ (5). In that study, it was not determined whether specific lineages were prevalent because of a tropism for ocular tissues or were prevalent due to a disproportionate association with S. aureus infection regardless of anatomical site. The purpose of this study is to determine whether lineages of S. aureus that are disproportionately associated with infection at all sites occur in the clinical isolate population, and if so, to begin to identify the phenotypic and genotypic traits that account for their overrepresentation among clinical isolates.
MATERIALS AND METHODS
Bacterial strains.
S. aureus clinical isolates were collected from patients with bloodstream, catheter tip, bone or joint, respiratory tract, ocular, soft tissue, wound, and skin infections. The geographic locations from which the isolates were derived were Arkansas, Ohio, Massachusetts, Illinois, California, Louisiana, Florida, Pennsylvania, Oklahoma, Nebraska, Texas, Munich (Germany), and London (United Kingdom). Isolates were also collected from 10 additional randomly selected sites through The Surveillance Network, an Internet-based network of clinical microbiology laboratories in the United States administered through MRL Pharmaceutical Services Inc. ET41 strains (34), Minn 8 and KD222, were obtained as kind gifts from Patrick Schlievert, University of Minnesota School of Medicine. Phage type 95 strains, 896-A-SC-02 and 145A-259, which were isolated from a contaminated anesthetic outbreak in the United States (1, 6, 34, 45), were kind gifts from Robert Arbeit, Veterans Affairs Medical Center, Boston, Mass. Upon receipt, species identification of isolates was confirmed by appearance on mannitol salt agar, and then isolates were stored frozen at −70°C in 25% (vol/vol) glycerol–brain heart infusion.
Genomic DNA fingerprinting by PFGE.
Genomic DNA fingerprinting by PFGE was performed as previously described (29), except that lysostaphin (50 μg/ml) was added to the lysis solution for the preparation of genomic DNA. Isolates with similar banding patterns and no more than three band differences were considered clonally related and were designated as an S. aureus lineage (SAL) (45). Isolates with no more than four band differences were considered subtypes of a given SAL. Once isolates were recognized as having identical or similar banding patterns, a second gel was run containing all isolates from the same group to verify lineage relationships.
Antibiotic susceptibility testing.
Isolates were tested by the agar disk diffusion method using BBL Sensi-Discs and NCCLS interpretation tables. Antimicrobial agents tested included cefazolin, ciprofloxacin, clindamycin, erythromycin, oxacillin, penicillin, trimethoprim-sulfamethoxazole (TMP-SXT), and vancomycin. In addition to the agar disk diffusion method, the broth microdilution method was used to test for reduced susceptibility to vancomycin (46).
Genotypic characterization of prevalent lineages by PCR.
The genetic determinants for the following virulence traits were detected in whole-cell lysates of S. aureus isolates using oligonucleotide primers (Table 1) derived from published sequences: (i) collagen binding protein (cna) (37), (ii) TSST-1 (tst) (4), (iii) fibronectin binding protein A (fnbA) (44), (iv) fibronectin binding protein B (fnbB) (19), (v) alpha-hemolysin (hla) (35), (vi) beta-hemolysin (hlb) (9), (vii) clumping factor (clfA) (28), and (viii) methicillin resistance (mecA) (13). PCR was performed exactly as described previously (5).
TABLE 1.
Primer sequences for amplification of virulence genes
| Gene | Primer sequence |
|---|---|
| mecA | 5′ AAC AGG TGA ATT ATT AGC ACT TGT AAG 3′ |
| 5′ ATT GCT GTT AAT ATT TTT TGA GTT GAA 3′ | |
| hla | 5′ GGT TTA GCC TGG CCT TC 3′ |
| 5′ CAT CAC GAA CTC GTT CG 3′ | |
| hlb | 5′ GCC AAA GCC GAA TCT AAG 3′ |
| 5′ CGC ATA TAC ATC CCA TGG C 3′ | |
| fnbA | 5′ GCG GAG ATC AAA GAC AA 3′ |
| 5′ CCA TCT ATA GCT GTG TGG 3′ | |
| fnbB | 5′ GGA GAA GGA ATT AAG GCG 3′ |
| 5′ GCC GTC GCC TTG AGC GT 3′ | |
| clf | 5′ CGA TTG GCG TGG CTT CAG 3′ |
| 5′ GCC AGT AGC CAA TGT CAC 3′ | |
| tst | 5′ AAG CCC TTT GTT GCT TGC G 3′ |
| 5′ ATC GAA CTT TGG CCC ATA CTT T 3′ |
Capsular polysaccharide serotyping.
Capsule typing was performed by Jean C. Lee, Channing Laboratory, Harvard University, Boston, Mass.
Statistical analysis.
Pearson's chi-square (χ2) and Fisher's exact tests were used to determine the significance of frequency data. Bonferroni's correction for multiple comparisons was applied where multiple tests were performed, thereby reducing the nominal P value for statistical significance to 0.025. Otherwise, the nominal P value for statistical significance was 0.05.
RESULTS
Clonal associations among S. aureus clinical isolates.
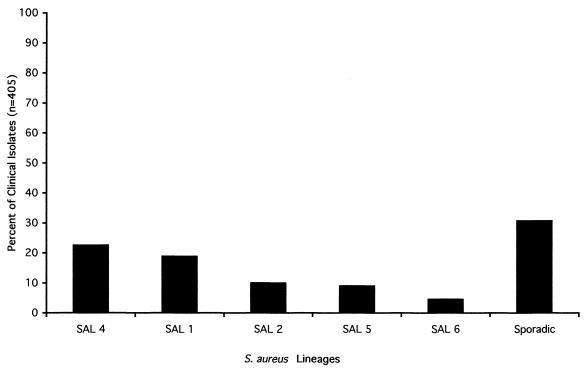
Genomic DNA fingerprint patterns for 405 epidemiologically unrelated human S. aureus clinical isolates collected from various infection sites and geographical locations were generated by PFGE analysis (Fig. 1). A total of 91 distinct genomic DNA fingerprint patterns or SALs were observed. Eighteen SALs occurred more than once with frequencies ranging from 2 to 92 isolates. Of these, the most prevalent lineages were SAL 1 (Fig. 2) and SAL 4 (Fig. 3), which accounted for 19.01 and 22.72% of the isolates, respectively. SAL 5 (Fig. 4) accounted for 10.12% of the isolates, while SAL 2 and SAL 6 accounted for 9.14 and 4.69% of isolates, respectively (Fig. 5). Cumulatively, SALs 1, 2, 4, 5, and 6 accounted for 65.68% of all isolates. Isolates comprising the five most prevalent lineages were collected from at least 10 of the 23 collection sites, suggesting a broad geographic distribution for these lineages. An additional lineage, SAL 3, accounted for 3.47% of isolates and was considered borderline prevalent (Fig. 1). The remaining 85 lineages were termed “sporadic” and occurred with frequencies of 2.5% or less. However, the vast majority of sporadic lineages (81 of 85) occurred with frequencies of <1%.
FIG. 1.
Relative distribution of SALs identified following PFGE analysis of S. aureus clinical isolates from various sources. Sporadic SALs were those which occurred at a frequency of <2.5% of all isolates.
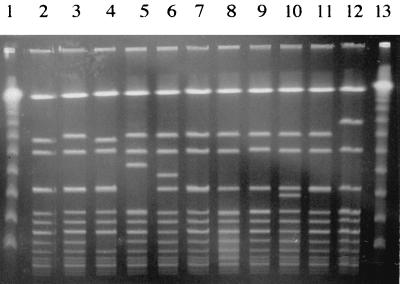
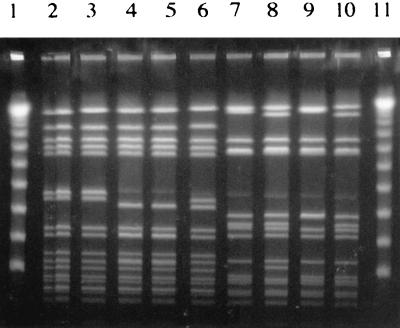
FIG. 2.
Genomic DNA fingerprint patterns of SAL 1 clinical isolates analyzed by PFGE following _Sma_I digestion of chromosomal DNA. Lanes 1 and 13, lambda ladder; lanes 2 and 3, respiratory tract isolates; lanes 4 and 5, blood isolates; lanes 6 and 7, catheter tip isolates; lanes 8 and 9, bone or joint isolates; lanes 10 and 11, ET41 strains KD222 and Minn 8, respectively; lane 12, agr group III isolate (16, 27).
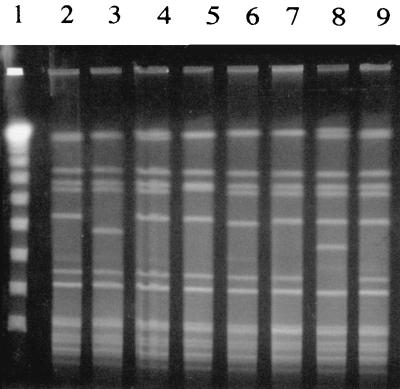
FIG. 3.
Genomic DNA fingerprint patterns of SAL 4 clinical isolates analyzed by PFGE following _Sma_I digestion of chromosomal DNA. Lanes 1, lambda ladder; lanes 2 and 4, respiratory tract isolates; lane 3, bone or joint isolates; lanes 5 and 6, blood isolates; lane 7, ocular isolate; lanes 8 and 9, catheter tip isolates.
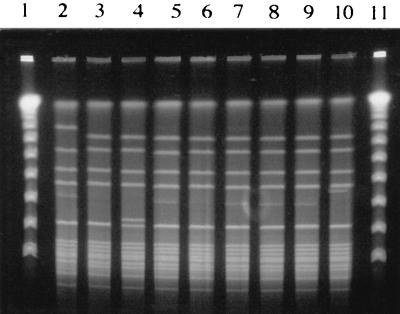
FIG. 4.
Genomic DNA fingerprint patterns of SAL 5 clinical isolates analyzed by PFGE following _Sma_I digestion of chromosomal DNA. Lanes 1 and 11, lambda ladder; lanes 2 and 5, bone or joint isolates; lane 3, ocular isolate; lanes 4 and 6, respiratory tract isolates; lane 7, blood isolate; lane 8, catheter tip isolate; lanes 9 and 10, phage type 95 isolates 145A-259 and 896A-SC-02, respectively.
FIG. 5.
Genomic DNA fingerprint patterns of SAL 2 (lanes 7 to 10) and SAL 6 (lanes 2 to 6) clinical isolates analyzed by PFGE following _Sma_I digestion of chromosomal DNA. Lanes 1 and 11, lambda ladder; lanes 2 and 5, bone or joint isolates; lane 3, respiratory tract isolate; lane 4, blood isolate; lane 6, cellulitis isolate; lane 7, bone or joint isolate; lane 8, blood isolate; lane 9, catheter tip isolate; lane 10, respiratory tract isolate.
Antibiotic susceptibility profiles of prevalent SALs.
To determine the influence of antibiotic resistance on the expansion of the prevalent SALs identified in this study, antibiotic susceptibility testing was performed by the agar disk diffusion and broth microdilution methods. The majority of all isolates comprising both prevalent and sporadic lineages were Penr, which is consistent with the observation of others that now more than 90% of S. aureus infection isolates are resistant to penicillin (47). SAL 4 isolates were uniformly oxacillin resistant and widely resistant to all other antibiotics tested, with the exception of vancomycin and TMP-SXT. This finding suggests that antibiotic selection pressure was a major factor contributing to the expansion of SAL 4. Oxacillin resistance and/or the methicillin resistance genetic determinant was also associated with individual isolates of lineages other than SAL 4 (SAL 1, SAL 5, and SAL 6 and eight sporadic lineages), supporting the suggestion that mec has spread horizontally within the S. aureus species (3, 31). SALs 1, 2, 5, and 6 were largely, though not uniformly, susceptible in antibiograms (Table 2), suggesting that factors other than antibiotic selection pressure influenced the expansion of these lineages. As would be anticipated, sporadic isolates showed variable antibiotic susceptibility patterns. It is noteworthy that all isolates tested in this study were vancomycin susceptible as determined by both the broth microdilution and agar disk diffusion methods.
TABLE 2.
Antibiotic susceptibility profiles for prevalent and sporadic SALs
| Antibiotica | No. of isolates with indicated profile for SALb: | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 (n = 17) | 2 (n = 4) | 4 (n = 8) | 5 (n = 11) | 6 (n = 10) | Sporadic (n = 12) | |||||||||||||
| S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | |
| Cefazolin | 17 | 0 | 0 | 4 | 0 | 0 | 1 | 0 | 7 | 11 | 0 | 0 | 9 | 0 | 1 | 11 | 0 | 1 |
| Ciprofloxacin | 14 | 2 | 1 | 4 | 0 | 0 | 2 | 0 | 6 | 10 | 1 | 0 | 10 | 0 | 0 | 11 | 0 | 1 |
| Clindamycin | 12 | 5 | 0 | 4 | 0 | 0 | 2 | 0 | 6 | 11 | 0 | 0 | ND | ND | ND | 5 | 4 | 3 |
| Erythromycin | 10 | 4 | 3 | 3 | 1 | 0 | 1 | 0 | 7 | 5 | 0 | 6 | 5 | 0 | 5 | 4 | 5 | 3 |
| Oxacillin | 17 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 8 | 8 | 3 | 0 | 8 | 1 | 1 | 10 | 0 | 2 |
| Penicillin | 2 | 0 | 15 | 0 | 0 | 4 | 0 | 0 | 8 | 2 | 0 | 9 | 0 | 0 | 10 | 2 | 0 | 10 |
| TMP-SXT | 15 | 1 | 1 | 4 | 0 | 0 | 6 | 0 | 2 | 11 | 0 | 0 | ND | ND | ND | 10 | 0 | 2 |
| Vancomycin | 17 | 0 | 0 | 4 | 0 | 0 | 8 | 0 | 0 | 11 | 0 | 0 | 10 | 0 | 0 | 12 | 0 | 0 |
Infection site specificity of prevalent SALs.
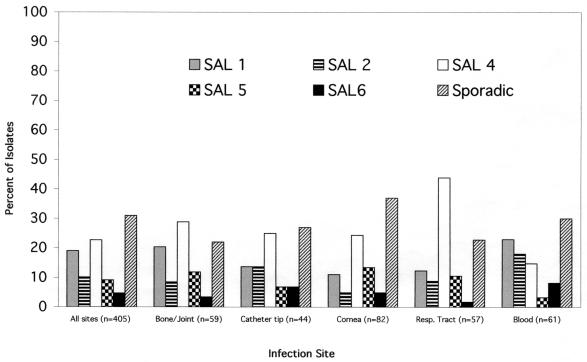
Pathogenic S. aureus strains have the capacity to colonize and establish infection in a remarkably wide range of body sites including blood, indwelling biomaterials, mucosal surfaces, bone, and vitreous and other tissues. Little is known of the basis for tropism for S. aureus for specific infection sites. Therefore, it was of interest to determine whether the prevalent SALs identified in this study show significant associations with specific infection sites. Figure 6 shows the distribution of SAL 1, 2, 4, 5, and 6 and sporadic isolates at sites of infection frequently associated with S. aureus infection and their distribution from all sites. The distribution of prevalent and sporadic lineages among isolates collected from bone or joint, catheter tip, or corneal sites of infection did not differ significantly from their distribution from all sites (P > 0.05, χ2). This suggests that the lineages identified in this study do not possess a tropism for these specific sites of infection. In contrast, the distribution of lineages derived from the respiratory tract differed significantly from that for all sites (P = 0.0046, χ2). This difference was primarily attributable to an approximately twofold enrichment in SAL 4 isolates collected from the respiratory tract compared with all sites of infection (43.86 versus 19.25%; P < 0.0001 [χ2], nominal P = 0.025). Among blood isolates, an enrichment of SAL 2 isolates was observed (18.03 versus 8.72%) which also was borderline statistically significant (P = 0.026 [χ2], nominal P = 0.025).
FIG. 6.
Relative distributions of prevalent and sporadic SALs at anatomical sites frequently associated with S. aureus infection and at all sites. Resp., respiratory.
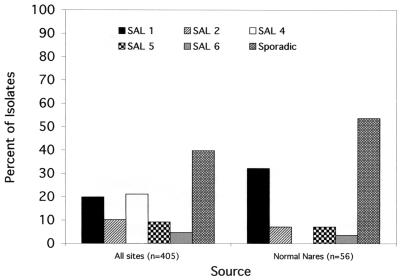
Lineage distribution among normal flora.
Indigenous flora represent an important reservoir for disease causing S. aureus in humans. To determine whether the distribution of SALs among clinical isolates is related to their distribution among normal flora, PFGE analysis was performed on 55 S. aureus isolates collected from the anterior nares of healthy volunteers. As shown in Fig. 7, a significantly different profile of lineage distribution was observed for normal flora isolates compared with clinical isolates. SAL 1 was enriched among normal flora (32.14 versus 19.01%; P = 0.022, compared to a nominal P value of 0.025), while SAL 4 isolates were not represented among the normal flora isolates (P < 0.0001). Interestingly, a number of SALs (SALs 9, 11, and 21) were significantly associated with normal flora but occurred infrequently among clinical isolates (P ≤ 0.017, Fisher's exact test), suggesting that they have a reduced propensity to cause disease compared to that of disease-associated SALs.
FIG. 7.
Frequency and distribution of SALs most commonly associated with normal flora isolates compared with their frequency and distribution among all clinical isolates.
Genotypic characterization of SALs prevalent among S. aureus clinical isolates.
The disproportionately large number of infections associated with SALs 1 to 6 (65.7%, 266 of 405) suggests that they possess unique combinations of genes that confer an enhanced propensity to cause infection. It is known that S. aureus expresses more than 30 secreted and cell surface proteins, many of which have been cloned, sequenced, and ascribed potential roles in pathogenesis such as (i) attachment, (ii) evasion of host defenses, or (iii) tissue invasion-penetration (39). To begin an investigation into the specific factors that cause SALs 1 to 6 to predominate among clinical isolates, PCR was employed to identify genetic determinants for known staphylococcal virulence traits among prevalent and sporadic lineages. The results of this analysis are shown in Table 3. Certain traits were found associated with all SALs, regardless of prevalence. For example, a positive PCR signal for hla (alpha-hemolysin), fnbA (fibronectin binding protein A), and clfA (clumping factor) was observed in 100, 89.7, and 96.2% of all isolates, respectively, regardless of lineage identity. In contrast, tst and cna were very significantly associated with prevalent lineages compared with sporadic lineages (P < 0.0001; nominal P = 0.025), suggesting a potentially important role for these proteins in the virulence of the organism. However, in neither case were these determinants uniformly associated with all prevalent lineages, but rather both demonstrated obvious association with specific lineages. For example, a positive PCR signal for cna was associated with 93.6, 100, and 100% of SAL 1, SAL 5, and SAL 6 isolates, respectively; 8.7% of SAL 2 isolates; and 0% of SAL 4 isolates. Similarly, tst was strongly associated with SAL 1 isolates (95.4%), weakly associated with SAL 4 (7.7%) and SAL 5 (4.0%) isolates, and not at all associated with SAL 2 or SAL 6 isolates. Most striking was the complete lack of positive signal for tst among sporadic lineages. The genetic determinant for beta-hemolysin, hlb, showed a moderate degree of lineage specificity among the prevalent lineages, being more closely associated with SAL 4 and SAL 6 (65.9 and 70.5%, respectively) than with SAL 1, SAL 2, and SAL 5 (9.6, 0, and 0%, respectively). However, despite a strong association between hlb and two of the prevalent lineages, there was no significant difference in the frequency of this determinant among prevalent and sporadic lineages (35 versus 46.2%; P = 0.128, Fisher's exact test). A positive PCR signal for fnbB was observed among 22.7% of isolates comprising sporadic lineages; however, of the prevalent SALs, fnbB was associated solely with SAL 2.
TABLE 3.
Occurrence and frequency of potential virulence genes and mecA among prevalent and sporadic SALs as determined by PCR analysis
| Group tested | % Positive for gene (no. of positive isolates/no. of isolates tested) | |||||||
|---|---|---|---|---|---|---|---|---|
| mecA | tst | cna | fnbA | fnbB | hla | hlb | clfA | |
| SAL | ||||||||
| SAL 1 | 5.5 (2/37) | 95.4 (42/44) | 93.6 (44/47) | 85.7 (24/28) | 0 (0/36) | 100 (21/21) | 9.6 (3/31) | 100 (5/5) |
| SAL 2 | 0 (0/21) | 0 (0/7) | 8.7 (2/23) | 90 (9/10) | 95.2 (20/21) | 100 (8/8) | 0 (0/18) | 100 (2/2) |
| SAL 4 | 90.2 (37/41) | 7.7 (2/26) | 0 (0/47) | 96 (24/25) | 0 (0/39) | 100 (30/30) | 65.9 (31/47) | 100 (15/15) |
| SAL 5 | 10.5 (2/19) | 4 (1/25) | 100 (27/27) | 85.7 (12/14) | 0 (0/23) | 100 (20/20) | 0 (0/20) | 100 (6/6) |
| SAL 6 | 0 (0/9) | 0 (0/11) | 100 (17/17) | 100 (8/8) | 0 (0/16) | 100 (10/10) | 70.5 (12/17) | 100 (4/4) |
| Total prevalent lineages | 32.2 (41/127) | 39.8 (45/113) | 55.9 (90/161) | 91 (77/85) | 14.8 (20/135) | 100 (88/88) | 34.5 (46/133) | 100 (32/32) |
| Sporadic lineages | 22.2 (14/63) | 0 (0/38) | 25 (15/60) | 86.6 (39/45) | 22.7 (10/44) | 100 (31/31) | 38.1 (21/55) | 90.4 (19/21) |
| All isolates | 27.9 (55/197) | 28.6 (45/157) | 44.9 (107/237) | 89.7 (122/136) | 20.1 (38/189) | 100 (128/128) | 38.8 (77/200) | 96.2 (51/53) |
| Frequency analysis result (Fisher's exact test; prevalent vs sporadic) | 0.175 | <0.0001 | <0.0001 | 0.556 | 0.247 | 0.000 | 0.738 | 0.152 |
Capsular polysaccharide analysis of SALs 1 to 6 and sporadic isolates.
It is now well established that CP5 and CP8 strains cause the majority (70 to 80%) of all S. aureus infections (20). In this study, 75.4% of S. aureus clinical isolates were CP5 or CP8, which is consistent with rates reported in the literature (Table 4) (20). However, when the distributions of CP5, CP8, and nontypeable isolates among disease-prevalent and sporadic lineages were compared, significant differences were observed. For example, prevalent lineages were significantly enriched in CP8 isolates (65 versus 22.3%, P < 0.0001, χ2) compared with sporadic lineages, while the sporadic lineages were significantly enriched in nontypeable isolates (50.7 versus 7.0%, P < 0.0001, χ2). The proportions of isolates designated CP5 did not differ among prevalent and sporadic lineages (28 versus 26.8%, P = 1.0, χ2). Our findings that a majority of prevalent lineages are enriched in CP8 strains and that sporadic lineages are enriched in nontypeable strains are consistent with the hypothesis that SAL 1 to SAL 6 cause the majority of S. aureus infections.
TABLE 4.
Distribution of CP5, CP8, and nontypeable strains among prevalent and sporadic SALs
| Group tested | % of strains with characteristic (no. with characteristic/total no. tested) | ||
|---|---|---|---|
| CP5 | CP8 | Nontypeable | |
| SAL | |||
| SAL 1 | 0 (0/31) | 93.5 (29/31) | 6.4 (2/31) |
| SAL 2 | 0 (0/12) | 91.6 (11/12) | 8.3 (1/12) |
| SAL 4 | 96.5 (28/29) | 3.4 (1/29) | 0 (0/29) |
| SAL 5 | 0 (0/19) | 100 (19/19) | 0 (0/19) |
| SAL 6 | 0 (0/9) | 55.5 (5/9) | 44.4 (4/9) |
| Prevalent lineages | 28 (28/100) | 65 (65/100) | 7.0 (7/100) |
| Sporadic lineages | 26.8 (18/67) | 22.3 (15/67) | 50.7 (34/67) |
| All isolates | 27.5 (46/167) | 47.9 (80/167) | 24.5 (41/167) |
| Frequency analysis result (prevalent vs sporadic)a | 1.000 | <0.0001 | <0.0001 |
SAL 1 and SAL 5 are phylogenetically related to known virulent lineages.
To determine whether SALs 1 to 6 are genetically related to lineages previously documented to be prevalent among clinical isolates, the genomic DNA fingerprints of prevalent SALs were compared with those for isolates representing ET41 (33) and the phage type 95 clones (1, 6, 41). PFGE analysis revealed that SAL 1 shares an identical _Sma_I digestion pattern with the ET41 isolate Minn 8 and differs by only one band from the ET41 isolate KD222, indicating that SAL 1 is clonally related to the predominant lineages associated with cases of menstrual TSS (Fig. 2, lanes 10 and 11). Furthermore, the outbreak isolates 896-A-SC-02 and 145A-259 shared identical _Sma_I digestion patterns with SAL 5 (Fig. 4, lanes 9 and 10). This evidence indicates that at least two of the prevalent lineages identified, SAL 1 and SAL 5, are genetically related to SALs which were documented by others to be widespread in the S. aureus clinical isolate population. Interestingly, SAL 1 was also found to be clonally related to isolates comprising the recently identified S. aureus agr group III, which secretes a type III quorum-sensing octapeptide (Fig. 2, lane 12) (16, 27).
DISCUSSION
S. aureus is a highly versatile organism with the capacity to colonize and establish infections in a wide range of body sites. Little is known of the basis for the tropism of S. aureus for specific infection sites. However, there is evidence to suggest that certain SALs have a strong association with specific tissues. Musser et al. reported an MLEE clone of S. aureus, termed ET41, which accounted for 88% of cases of urogenital TSS but which occurred in the urogenital tract of 28% of healthy carriers (33). Why ET41 is involved with the majority of cases of menstrual TSS is not known. However, it was suggested that ET41 is highly adapted to the cervicovaginal tract and that as a consequence the probability of this lineage causing infection in a milieu disposed toward TSS is greater than that for other clones. In our study, SAL 1 was found to have the same _Sma_I banding pattern by PFGE analysis as that of ET41 (Fig. 2), indicating that they are the same lineage. Furthermore, SAL 1 was found to be strongly associated with the mucosal surface of the anterior nares in healthy volunteers. These data suggest that SAL 1-ET41 is adapted to at least two mucosal surfaces in healthy humans, the anterior nares and the cervicovaginal mucosa. Our finding that SAL 1 is the second most common lineage found among clinical isolates, accounting for 19.01% of all infections, may be a consequence of its prevalence on mucosal surfaces, which are the primary source of S. aureus for infection at other sites (22, 23). However, an enhanced virulence capacity for SAL 1 over those of other lineages cannot be ruled out for all infection sites.
A significant percentage (43%) of the respiratory tract infection isolates typed in this study were multiply antibiotic-resistant SAL 4. Since the majority of cases of staphylococcal respiratory tract infection are nosocomial in origin, occurring primarily in an elderly population with underlying infection and a history of prior antibiotic therapy (15, 18, 40, 48), the finding that a large proportion of infection isolates from the respiratory tract are multiply antibiotic resistant is not surprising. However, why the majority of these infections are caused by SAL 4 is unclear. SAL 4 may possess a specific tropism for tissues of the respiratory tract, or SAL 4 may superinfect patient surfaces as a result of the antibiotic elimination of competing commensal flora. While there is now strong evidence to suggest that the mec determinant is harbored by multiple divergent phylogenetic lineages (3, 31), it is also the case that more than half of MRSA isolates are of a single clone which is widespread and common in the United States (31, 34). Our studies provide support for the idea that mecA can be associated with divergent lineages but that a single lineage, designated here as SAL 4, remains the predominant cause of all MRSA infections. The relationship between SAL 4 and previously recognized prominent MRSA lineages will be determined in follow-up studies. Interestingly two SAL 1 isolates also carried the mecA gene. The ability of SAL 1 to acquire methicillin resistance together with the widespread distribution of this lineage in both disease-related and colonizing strains could pose a significant threat of the emergence of a new MRSA clone which may already be highly adapted to the human host. Other associations between infection sites and specific SALs that showed borderline statistical significance were noted. For example, compared with all sources, blood isolates were enriched approximately twofold with SAL 2 (18 versus 8.7%, P = 0.026, χ2; nominal P = 0.025). The basis for this enrichment is unclear. However, recent reports have highlighted a potentially important role for fibronectin binding proteins A and B in the attachment of S. aureus to endothelial cells (38). The unique presence of both fnbA and fnbB in most SAL 2 strains may explain the almost twofold enrichment of SAL 2 among blood isolates.
PCR was used to begin to identify the specific factors that may contribute to the prevalence of SAL 1 to SAL 6 among clinical isolates. Certain traits, such as the genetic determinants for fibronectin binding protein A (fnbA), clumping factor (clfA), and alpha-hemolysin (hla), were present in all strains tested regardless of SAL, suggesting an important role for these conserved elements in the survival of S. aureus. Other traits, such as tst, cna, and hlb, are known to be associated with mobile genetic elements and were found in this study to be associated with certain lineages and not with others, suggesting limited horizontal transfer among lineages (9, 10, 14, 25, 26). For example, cna was present in almost all SAL 1, SAL 5, and SAL 6 isolates but was completely absent from SAL 2 and SAL 4 isolates. Gillaspy et al. (14) suggest that all strains of S. aureus possess the integration site for cna. It would be of interest to determine whether this is in fact the case for SAL 2 and SAL 4 strains. Interestingly, a highly significant association was observed between possession of cna and prevalent lineages (P < 0.0001), suggesting an important role for collagen binding adhesin in the expansion of virulent clones. Allelic replacement experiments have shown a role for collagen binding adhesin in the virulence of S. aureus in a mouse model of septic arthritis (36). The specific role of collagen binding adhesin in establishing S. aureus infection at other sites requires further attention. Other investigators have reported a strong correlation between capsular type 8 strains and the cna gene (42). This is consistent with our observation that cna is associated with only CP8 lineages (SALs 1, 5, and 6) and is absent in CP5 lineages (SAL 4).
The genetic determinant for TSST-1 (tst) was largely clustered within SAL 1, with a low incidence in SAL 4 and SAL 5, and was not present at all among sporadic isolates. This contrasts with results of previous studies which indicate that the genetic determinant for TSST-1 is associated with a diversity of genetic backgrounds within the S. aureus species (33, 34). This discrepancy may be due to allelic variations in the tst gene, which have been previously documented (33) and which may not be detectable by PCR analysis. This result highlights a limitation of the present study, which is that detection of genetic determinants by PCR may be limited by primer specificity for individual alleles. Therefore, care has to be taken in the interpretation of patterns of virulence traits as determined by PCR. However, because of its specificity PCR may highlight subtle allelic variations in virulence genes which play a role in the expansion of certain prevalent lineages.
It is now well established that the majority of S. aureus infections are caused by strains reactive to either anti-CP8 or anti-CP5 capsular antibodies (2, 20), and the results of this study are consistent with these data. However, while CP5 and CP8 lineages are readily delineated from each other, certain lineages include CP5 or CP8 strains together with nontypeable strains. This observation suggests that structural alterations in operons encoding capsular type occurred in these lineages. Recombinations among capsular biosynthetic operons resulting in intralineage shifting in capsular polysaccharide serotype have been identified before in human pathogens (8). Since antistaphylococcal vaccines consisting of CP5 and CP8 are currently under investigation for their protective efficacy against S. aureus infection (12), alterations in the capsule structure of lineages prevalent among clinical isolates would have serious implications for the long-term efficacy of such vaccines. Further studies monitoring serotype stability among prevalent lineages would provide a basis to evaluate the potential utility of serotype-specific vaccines.
Taken together, the results of these studies provide strong evidence for the existence of five phylogenetic lineages of S. aureus which are highly prevalent and widely distributed among clinical isolates. These studies support the concept that the basic unit of bacterial pathogenicity is the clone or lineage that expands due to the possession of unique combinations of virulence genes (11). Such clones are likely to be characterized by the production of virulence factors that enhance colonization, persistence and invasion at the infecting site, as well as factors that permit their widespread dissemination and evasion of host responses. The molecular epidemiology of pathogenic SALs has received little attention. Prospective multicenter studies that rigorously control for repeated isolation of samples from the same patient, multifocal infections with the same strain, and outbreak strains at a single location will help to elucidate the mechanisms by which certain clones outcompete other clones, disseminate widely, and display enhanced virulence. Such studies may reveal novel approaches to infectious disease control.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants EY 10867 (to M.C.B.) and EY 11648 (to M.S.G.) and by Research to Prevent Blindness, Inc.
REFERENCES
- 1.Arbeit R D. Laboratory procedures for epidemiologic analysis. Edinburgh, United Kingdom: Churchill Livingstone, Ltd.; 1997. [Google Scholar]
- 2.Arbeit R D, Karakawa W W, Vann W F, Robbins J B. Predominance of two newly described capsular polysaccharide types among clinical isolates of Staphylococcus aureus. Diagn Microbiol Infect Dis. 1984;2:85–91. doi: 10.1016/0732-8893(84)90002-6. [DOI] [PubMed] [Google Scholar]
- 3.Archer G L, Niemeyer D M, Thanassi J A, Pucci M J. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob Agents Chemother. 1994;38:447–454. doi: 10.1128/aac.38.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blomster-Hautamaa D A, Kreiswirth B N, Kornblum J S, Novick R P, Schlievert P M. The nucleotide and partial amino acid sequence of toxic shock syndrome toxin-1. J Biol Chem. 1986;261:15783–15786. [PubMed] [Google Scholar]
- 5.Booth M C, Hatter K L, Miller D, Davis J, Kowalski R, Parke D W, Chodosh J, Jett B D, Callegan M C, Penland R, Gilmore M S. Molecular epidemiology of Staphylococcus aureus and Enterococcus faecalis in endophthalmitis. Infect Immun. 1998;66:356–360. doi: 10.1128/iai.66.1.356-360.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control. Postsurgical infections associated with an extrinsically contaminated intravenous anesthetic agent—California, Illinois, Maine, Michigan. Atlanta, Ga: Centers for Disease Control; 1990. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. National nosocomial infections surveillance (NNIS) system report. Data summary from January 1999-May 1999. Atlanta, Ga: Centers for Disease Control and Prevention; 1999. [Google Scholar]
- 8.Coffey T J, Enright M C, Daniels M, Morona R, Hryniewicz W, Paton J C, Spratt B G. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol. 1998;27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 9.Coleman D, Knights J, Russell R, Shanley D, Birkbeck T H, Dougan G, Charles I. Insertional inactivation of the Staphylococcus aureus β-toxin by bacteriophage φ13 occurs by site and orientation specific integration of the φ13 genome. Mol Microbiol. 1991;5:933–939. doi: 10.1111/j.1365-2958.1991.tb00768.x. [DOI] [PubMed] [Google Scholar]
- 10.Coleman D C, Sullivan D J, Russell R, Arbuthnott J P, Carey B F, Pomeroy H M. Staphylococcus aureus bacteriophages mediating the simultaneous lysogenic conversion of β-lysin, staphylokinase and enterotoxin A: molecular mechanism of triple conversion. J Mol Microbiol. 1989;135:1679–1697. doi: 10.1099/00221287-135-6-1679. [DOI] [PubMed] [Google Scholar]
- 11.Falkow S. What is a pathogen? ASM News. 1997;63:359–370. [Google Scholar]
- 12.Fattom A I, Sarwar J, Ortiz A, Naso R. A Staphylococcus aureus capsular polysaccharide (CP) vaccine and CP-specific antibodies protect mice against bacterial challenge. Infect Immun. 1996;64:1659–1665. doi: 10.1128/iai.64.5.1659-1665.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geha D J, Uhl J R, Gustaferro C A, Persing D H. Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J Clin Microbiol. 1994;32:1768–1772. doi: 10.1128/jcm.32.7.1768-1772.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillaspy A F, Patti J M, Pratt F L, Iandolo J J, Smeltzer M S. The Staphylococcus aureus collagen adhesin-encoding gene (cna) is within a discrete genetic element. Gene. 1997;196:239–248. doi: 10.1016/s0378-1119(97)00256-4. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez C, Rubio M, Romero-Vivas J, Picazo J J. Bacteremic pneumonia due to Staphylococcus aureus: a comparison of disease caused by methicillin-resistant and methicillin-susceptible organisms. Clin Infect Dis. 1999;29:1171–1177. doi: 10.1086/313440. [DOI] [PubMed] [Google Scholar]
- 16.Guangyong J, Beavis R, Novick R P. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 17.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover F C. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–146. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 18.Iwahara T, Ichiyama S, Nada T, Shimokata K, Nakashima N. Clinical and epidemiologic investigations of nosocomial pulmonary infections caused by methicillin-resistant Staphylococcus aureus. Chest. 1994;105:826–831. doi: 10.1378/chest.105.3.826. [DOI] [PubMed] [Google Scholar]
- 19.Jonsson K, Signas C, Muller H P, Lindberg M. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur J Biochem. 1991;202:1041–1048. doi: 10.1111/j.1432-1033.1991.tb16468.x. [DOI] [PubMed] [Google Scholar]
- 20.Karakawa W W, Sutton A, Schneerson R, Karpas A, Vann W F. Capsular antibodies induce type-specific phagocytosis of capsulated Staphylococcus aureus by human polymorphonuclear leukocytes. Infect Immun. 1988;56:1090–1095. doi: 10.1128/iai.56.5.1090-1095.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karch H, Heesemann J, Laufs R, O'Brien A D, Tacket C O, Levine M M. A plasmid of the enterohemorrhagic Escherichia coli O157:H7 is required for expression of a new fimbrial antigen for adhesin to epithelial cells. Infect Immun. 1987;55:455–461. doi: 10.1128/iai.55.2.455-461.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev. 1997;10:505–520. doi: 10.1128/cmr.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kluytmans J A, Mouton J W, Ijzerman E P, Vandenbroucke-Grauls C M, Maat A W, Wagenvoort J H, Verbrugh H A. Nasal carriage of Staphylococcus aureus as a major risk factor for wound infections after cardiac surgery. J Infect Dis. 1995;171:216–219. doi: 10.1093/infdis/171.1.216. [DOI] [PubMed] [Google Scholar]
- 24.Kreiswirth B, Kornblum J, Arbeit R D, Eisner W, Maslow J N, McGeer A, Low D E, Novick R P. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science. 1993;259:227–230. doi: 10.1126/science.8093647. [DOI] [PubMed] [Google Scholar]
- 25.Kreiswirth B N, Projan S J, Schlievert P M, Novick R P. Toxic shock syndrome toxin-1 is encoded by a variable genetic element. Rev Infect Dis. 1989;11:S83–S89. doi: 10.1093/clinids/11.supplement_1.s83. [DOI] [PubMed] [Google Scholar]
- 26.Lee C Y, Iandolo J J. Mechanism of bacteriophage conversion of lipase activity in Staphylococcus aureus. J Bacteriol. 1985;164:288–293. doi: 10.1128/jb.164.1.288-293.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayville P, Guangyong J, Beavis R, Hongmei Y, Goger M, Novick R P, Muir T W. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc Natl Acad Sci USA. 1999;96:1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDevitt D, Francois P, Vaudaux P, Foster T J. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol Microbiol. 1994;11:237–248. doi: 10.1111/j.1365-2958.1994.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 29.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcal isolates using restriction fragments with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musser J M, Bemis D A, Ishikawa H, Selander R K. Clonal diversity and host distribution in Bordetella bronchiseptica. J Bacteriol. 1987;169:2793–2803. doi: 10.1128/jb.169.6.2793-2803.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Musser J M, Kapur V. Clonal analysis of methicillin-resistant Staphylococcus aureus strains from intercontinental sources: association of the mec gene with divergent phylogenetic lineages implies dissemination by horizontal transfer and recombination. J Clin Microbiol. 1992;30:2058–2063. doi: 10.1128/jcm.30.8.2058-2063.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Musser J M, Mattingly S J, Quentin R, Goudeau A, Selander R K. Identification of a high-virulence clone of type III Streptococcus agalactiae (group B Streptococcus) causing invasive neonatal disease. Proc Natl Acad Sci USA. 1989;86:4731–4735. doi: 10.1073/pnas.86.12.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Musser J M, Schlievert P M, Chow A W, Ewan P, Kreiswirth B N, Rosdahl V T, Naidu A S, Witte W, Selander R K. A single clone of Staphylococcus aureus causes the majority of cases of toxic shock syndrome. Proc Natl Acad Sci USA. 1990;87:225–229. doi: 10.1073/pnas.87.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Musser J M, Selander R K. Genetic analysis of natural populations of Staphylococcus aureus. New York, N.Y: VCH Publishers; 1990. [Google Scholar]
- 35.O'Reilly M, Kreiswirth B, Foster T J. Cryptic α-toxin gene in toxic shock syndrome septicaemia strains of Staphylococcus aureus. Mol Microbiol. 1990;4:1947–1955. doi: 10.1111/j.1365-2958.1990.tb02044.x. [DOI] [PubMed] [Google Scholar]
- 36.Patti J M, Bremell T, Krajewska-Pietrasik D, Abdelnour A, Tarkowski A, Ryden C, Hook M. The Staphylococcus aureus collagen adhesin is a virulence determinant in experimental septic arthritis. Infect Immun. 1994;62:152–161. doi: 10.1128/iai.62.1.152-161.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patti J M, Jonsson H, Guss B, Switalski L M, Wiberg K, Lindberg M, Hook M. Molecular characterization and expression of a gene encoding a Staphylococcus aureus collagen adhesin. J Biol Chem. 1992;267:4766–4772. [PubMed] [Google Scholar]
- 38.Peacock S J, Foster T J, Cameron B J, Berendt A R. Bacterial fibronectin-binding proteins and endothelial cell surface fibronectin may mediate adherence of Staphylococcus aureus to resting human endothelial cells. Microbiology. 1999;145:3477–3486. doi: 10.1099/00221287-145-12-3477. [DOI] [PubMed] [Google Scholar]
- 39.Projan S J, Novick R P. The molecular basis of pathogenicity. Edinburgh, United Kingdom: Churchill Livingstone, Ltd.; 1997. [Google Scholar]
- 40.Rikitomi N, Nagatake T, Sakamoto T, Matsumoto K. The role of MRSA (methicillin-resistant Staphylococcus aureus) adherence and colonization in the upper respiratory tract of geriatric patients in nosocomial pulmonary infections. Mol Immunol. 1994;38:607–614. doi: 10.1111/j.1348-0421.1994.tb01830.x. [DOI] [PubMed] [Google Scholar]
- 41.Rosdahl V T, Witte W, Musser M, Jarlov J O. Staphylococcus aureus strains of type 95. Spread of a single clone. Epidemiol Infect. 1994;113:463–470. doi: 10.1017/s0950268800068473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryding U, Flock J I, Flock M, Soderquist B, Christensson B. Expression of collagen-binding protein and types 5 and 8 polysaccharide in clinical isolates of Staphylococcus aureus. J Infect Dis. 1997;176:1096–1099. doi: 10.1086/516520. [DOI] [PubMed] [Google Scholar]
- 43.Selander R K, Caugant D A, Whittam T S. Genetic structure and variation in natural populations of Escherichia coli. Washington, D.C.: American Society for Microbiology; 1987. [Google Scholar]
- 44.Signands C, Raucci G, Joensson K, Lindgren P E, Anantharamaiah G M, Höök M, Lindberg M. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc Natl Acad Sci USA. 1989;86:699–703. doi: 10.1073/pnas.86.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tenover F C, Arbeit R, Archer G, Biddle J, Byrne S, Goering R, Hancock G, Hebert G A, Hill B, Hollis R, Jarvis W R, Kreiswirth B, Eisner W, Maslow J, McDougal L K, Miller J M, Mulligan M, Pfaller M A. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J Clin Microbiol. 1994;32:407–415. doi: 10.1128/jcm.32.2.407-415.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tenover F C, Lancaster M V, Hill B C, Steward C D, Stocker S A, Hancock G A, O'Hara C M, Clark N C, Hiramatsu K. Characterization of staphylococci with reduced susceptibilities to vancomycin and other glycopeptides. J Clin Microbiol. 1998;36:1020–1027. doi: 10.1128/jcm.36.4.1020-1027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thornsberry C. Trends in antimicrobial resistance among today's bacterial pathogens. Pharmacotherapy. 1995;15:3S. [PubMed] [Google Scholar]
- 48.Watanakunakorn C. Bacteremic Staphylococcus aureus pneumonia. Scand J Infect Dis. 1987;19:623–627. doi: 10.3109/00365548709117196. [DOI] [PubMed] [Google Scholar]
- 49.Whittam T S, Wachsmuth I K, Wilson R A. Genetic evidence of clonal descent of Escherichia coli O157:H7 associated with hemorrhagic colitis and hemolytic uremic syndrome. J Infect Dis. 1988;157:1124–1133. doi: 10.1093/infdis/157.6.1124. [DOI] [PubMed] [Google Scholar]