Mutational Disruption of Plasma Membrane Trafficking of Saccharomyces cerevisiae Yor1p, a Homologue of Mammalian Multidrug Resistance Protein (original) (raw)
Abstract
The ATP binding cassette (ABC) transporter protein Yor1p was identified on the basis of its ability to elevate oligomycin resistance when it was overproduced from a high-copy-number plasmid. Analysis of the predicted amino acid sequence of Yor1p indicated that this protein was a new member of a subfamily of ABC transporter proteins defined by the multidrug resistance protein (MRP). In this work, Yor1p is demonstrated to localize to the Saccharomyces cerevisiae plasma membrane by both indirect immunofluorescence and biochemical fractionation studies. Several mutations were generated in the amino-terminal nucleotide binding domain (NBD1) of Yor1p to test if the high degree of sequence conservation in this region of the protein was important for function. Deletion of a phenylalanine residue at Yor1p position 670 led to a mutant protein that appeared to be retained in the endoplasmic reticulum (ER) and that was unstable. As shown by others, deletion of the analogous residue from a second mammalian MRP family member, the cystic fibrosis transmembrane conductance regulator (CFTR), also led to retention of this normally plasma membrane-localized protein in the ER. Changes in the spacing between or the sequences flanking functional motifs of Yor1p NBD1 led to defective trafficking or decreased activity of the mutant proteins. Analyses of the degradation of wild-type and ΔF670 Yor1p indicated that the half-life of ΔF670 Yor1p was dramatically shortened. While the vacuole was the primary site for turnover of wild-type Yor1p, degradation of ΔF670 Yor1p was found to be more complex with both proteasomal and vacuolar contributions.
Multiple-drug resistance has often been linked with increased expression of ATP-binding cassette (ABC) transporter proteins that act as multispecific drug efflux pumps (see reference 15 for a review). The first known ABC transporter protein mediating multidrug resistance was human MDR1. MDR1 is localized to the plasma membranes of cells and is overexpressed in an array of multidrug-resistant cell lines and tumors (reviewed in reference 57). Biochemical experiments indicate that MDR1 transports compounds that typically have not been modified by the cell (reviewed in reference 16).
More recently, a second ABC transporter has been found in multidrug-resistant lung tumor cells (8). This ABC transporter protein, designated multidrug resistance protein (MRP), has several properties that make it distinct from MDR1. First, MRP transports modified substrates, such as glutathione and glucuronide conjugates (28, 36, 39, 45). Second, while both MDR1 and MRP possess a repeating structure of a set of transmembrane domains followed by the characteristic ABC transporter nucleotide binding domain, MRP has an additional set of transmembrane domains at its amino terminus (2). Finally, nucleotide binding domain 1 (NBD1) of MRP exhibits a characteristic spacing of functional motifs and high sequence similarity that serves to define a group of ABC transporter proteins referred to as the MRP family (8).
Like all known ABC transporters, NBD1 of the MRP family contains a Walker A, LSGGQ, and Walker B element (26, 64). The identical spacing of these elements in NBD1 of the MRP family serves to define this class of ABC transporter proteins (8). A second conserved feature in the MRP family NBD1 region is the presence of a phenylalanine residue between the Walker A and LSGGQ motifs. This phenylalanine was shown to be precisely deleted from an MRP family member, the human cystic fibrosis (CF) transmembrane conductance regulator (CFTR), in 60% of patients with CF (62). Wild-type CFTR must arrive at the plasma membrane to function normally, and deletion of this phenylalanine residue (ΔF508) causes the resulting mutant protein to be retained in the endoplasmic reticulum (ER) (7), where it is degraded by the proteasome (29, 65).
Saccharomyces cerevisiae has also been found to contain a group of ABC transporter proteins showing the characteristic structural features indicative of the MRP family. Ycf1p was the first member of the S. cerevisiae MRP family and was identified by its important role in cadmium tolerance (60). A second member of the S. cerevisiae MRP family was cloned as a key determinant of oligomycin resistance (9, 33). This ABC transporter protein was designated Yor1p (stands for yeast oligomycin resistance protein).
In this work, we localize Yor1p to the plasma membrane in cells. Analyses of wild-type and mutant forms of Yor1p suggest that the trafficking and turnover of this yeast protein have remarkable similarity to those of human CFTR. As seen for the major disease-associated allele of CFTR (ΔF508), degradation of an analogous form of Yor1p involves multiple proteolytic systems.
MATERIALS AND METHODS
Yeast strains and media.
The S. cerevisiae strains used are listed in Table 1. Yeast transformation was performed by the lithium acetate method as described previously (27). Standard yeast media containing supplements appropriate for growth of auxotrophic strains (56) were employed for growth of cells. Selection with the drug oligomycin was performed as described previously (33). Gradient plates were prepared by pouring 25 ml of YPGE medium (56) containing the desired maximum concentration of oligomycin into plates resting at an incline. Once this drug-containing medium had solidified, plates were laid flat and overlaid with YPGE medium (25 ml). Relative resistance levels were then assessed by performing a spot test assay (69) along the gradient.
TABLE 1.
S. cerevisiae strains used
| Strain designation | Genotype | Source or reference |
|---|---|---|
| SEY6210 | MATα leu2-3,112 ura3-52 lys2-801 trp1-Δ901 his3-Δ200 suc2-Δ9 Mel− | Scott Emr |
| DKY7 | MATα leu2-3,112 ura3-52 lys2-801 trp1-Δ901 his3-Δ200 suc2-Δ9 Mel− yor1-1::hisG | 33 |
| DKY14 | MATα leu2-3,112 ura3-52 lys2-801 trp1-Δ901 his3-Δ200 suc2-Δ9 Mel− yor1-1::hisG pep4::LEU2 | This study |
| DKY15 | MATα leu2-3,112 ura3-52 lys2-801 trp1-Δ901 his3-Δ200 suc2-Δ9 Mel− yor1-1::hisG ubc7::HIS3 | This study |
| DKY16 | MATα leu2-3,112 ura3-52 lys2-801 trp1-Δ901 his3-Δ200 suc2-Δ9 Mel− pre1 pre2 yor1-1::hisG | This study |
| CTY252 | MATa ura3 leu2 sec12-3 | Rob Piper |
| EAE18 | MATa ura3 leu2 sec12-3 yor1-1::hisG | This study |
| TVY1 | MATα leu2-3,112 ura3-52 lys2-801 trp1-Δ901 his3-Δ200 suc2-Δ9 Mel− pep4::LEU2 | Scott Emr |
| WCG4-11/21a | MATa leu2-3,112 ura3 his3-11,15 pre1-1 pre2-1 | Dieter Wolf |
Plasmids.
The low-copy-number plasmid containing the YOR1 gene was constructed in several steps. First, a 1.4-kb _Sac_II/_Sna_BI fragment from YOR1 was cloned into pBluescript KSII(−) (cut with _Sac_II and _Eco_RV). The _Hin_dIII site that lies 3′ to the insert was then removed by cleaving the sequence with _Hin_dIII and _Xho_I and treating it with the Klenow fragment and deoxynucleoside triphosphates, followed by religation to make plasmid pDK42. The _Sac_II/_Kpn_I fragment from pDK42 was then cloned into pRS316 that had been cut with the same enzymes, resulting in plasmid pDK58. The 5′ end of the gene was then cloned into pDK58 as a _Sac_I/_Sac_II fragment, giving rise to the full-length gene in pRS316 (pDK59). The template for site-directed mutagenesis (pSM111) contained the _Hin_dIII/_Spe_I fragment from nucleotides +1826 to +2913 relative to translation start site (which contains the first nucleotide binding domain) in plasmid pBCSK+ (Stratagene).
Site-directed mutagenesis.
Mutations in the first nucleotide binding domain of YOR1 were introduced by a PCR-based strategy (54). Briefly, mutagenic primers were annealed to the pSM111 template along with the T7 primer. The mutagenic primers were as follows: ΔF670, TCT TTA TTG AAT GGg GAt CC TAT GAT GTT ATC TCT TAC AG; K715M, CCT GGC TAA ATT GAT tCt gGC CaT TTG ACC ACC AGA TAA AGT AAT ACC; K715Q, CCT GGC TAA ATT GAT ACG TGC ttg TTG ACC ACC AGA TAA AGT AAT ACC; and insA652 (alanine insert at position 652), CCA TGG ATA ACC ACA CAT TAA agc TAA GTC CCC GTT GAC TTC AAC C. Nucleotides that differ from those of the wild type are indicated in lowercase letters. A second PCR used a YOR1 primer (+2090 to +2110) and the T3 primer. The products of each of the mutagenic reactions and the overlapping product were gel purified, combined, and used for a second PCR with the T3 and T7 primers. The products of the second PCR were cloned back into pSM111 as either a _Hin_dIII/_Spe_1 fragment (in the case of the alanine insertion) or an _Nsi_1/_Spe_I fragment (with the K715M, K715Q, and ΔF670 mutations). Following verification of mutations by restriction mapping, plasmids were sequenced to ensure that no additional errors had been introduced. DNA fragments containing the mutations were then placed in the context of the wild-type gene.
Production of rabbit anti-Yor1p antiserum.
Antisera directed against either the amino-terminal 110 amino acids or the 80 carboxy-terminal amino acids were used in this study. The production of the C-terminal antiserum has been described elsewhere (33). A fusion protein between glutathione _S_-transferase (GST) and the amino-terminal 110 amino acids of Yor1p was constructed in plasmid pGEX-KG (17). PCR was used to amplify the region and introduce _Eco_RI and _Hin_dIII sites at amino acids 1 and 110, respectively. This allowed the fragment to be cloned into the expression vector cleaved at these same sites. The resulting plasmid was designated pDK40 and sequenced to ensure that no errors had been introduced. The GST-Yor1p fusion protein was purified by standard methods and used to immunize rabbits as described previously (20, 33). Both the N-terminal and C-terminal antisera were affinity purified with Aminolink Plus (Pierce) matrix coupled to appropriate GST-Yor1p fusion proteins by the manufacturer’s protocol.
Immunoblotting.
Cells were grown in complete synthetic medium minus uracil to an optical density at 600 nm (OD600) of 0.3 to 0.5, harvested by centrifugation, resuspended in sorbitol breaking buffer (0.3 M sorbitol, 0.1 M NaCl, 5 mM MgCl2, 10 mM Tris [pH 7.5], 1× Boehringer Mannheim complete protease inhibitors), and lysed with glass beads, and extracts were cleared with a low-speed spin at 4°C. Protein concentration was measured in 1% sodium dodecyl sulfate (SDS) by the method of Lowry et al. (40). Protein extracts were solubilized in an equal volume of sample buffer (40 mM Tris-HCl [pH 6.8], 8 M urea, 15% SDS, 0.1 mM EDTA, 1% β-mercaptoethanol, 0.01% bromophenol blue) and heated to 37°C for 20 min. Equal amounts of protein were subjected to electrophoresis on SDS-polyacrylamide gels, transferred to nitrocellulose membranes, and probed with the antibody indicated below. For immunodetection of Yor1p, the C-terminal antibody was used for all Western blot and indirect immunofluorescence analyses and the N-terminal antibody was employed for all immunoprecipitation experiments. Immunoreactive material was detected with a donkey anti-rabbit immunoglobulin conjugated to horseradish peroxidase (Amersham) and by chemiluminescence (Pierce).
Immunoprecipitation, pulse-labeling, and chase.
Labeling and immunoprecipitations were performed essentially as described in reference 11 with the following modifications. Cells were grown overnight in complete synthetic medium to an OD600 of 0.3 to 0.5. Cells lacking a temperature-sensitive allele were then harvested, resuspended in fresh prewarmed medium without methionine to an OD600 of 2, and incubated for 10 min with gentle shaking. Cells were metabolically labeled by adding 15 μCi of Express protein labeling mix (New England Nuclear) per OD600 unit of cells and shaking at 30°C for 10 min. A 100× chase solution containing 100 mM ammonium sulfate, 0.3% l-cysteine, and 0.4% l-methionine was added to the culture following labeling, and aliquots (5 × 107 cells) were removed. The sec12-3 cells were grown at 23°C and shifted to 37°C for 10 min to impose the sec12-3 block or left at 23°C to permit continued Sec12-3p function. Following the temperature shift, cells were labeled at either the permissive or the restrictive temperature for 10 min and chased as described above. Time point aliquots were transferred to chilled tubes, and sodium azide was added to a final concentration of 10 mM. Cells were washed once with 10 mM sodium azide and resuspended in 200 μl of lysis buffer (15 mM Tris [pH 7.6], 1 mM EDTA, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1× Boehringer Mannheim complete protease inhibitors). Glass beads were added, and cells were lysed by vigorously vortexing them three times for 1 min each time, with 1-min cooling intervals. SDS was then added to 1%, and two more 1-min vortexing rounds were performed. The resulting protein extracts were diluted with 5 volumes of immunoprecipitation (IP) buffer (15 mM Tris [pH 7.6], 150 mM NaCl, 1% Triton X-100, 2 mM sodium azide) and cleared by centrifugation. Polyclonal anti-Yor1p (1:100 dilution of the anti-N-terminal antibody) was added to the cleared lysate and incubated at 4°C in an end-over-end rotator for 1 h. Following this incubation, preswollen protein A-Sepharose beads (Sigma) were added and incubated end-over-end overnight at 4°C. Immune complexes bound to protein A-Sepharose beads were washed three times with IP buffer containing 0.1% SDS and two times with detergent-free wash buffer (10 mM Tris [pH 7.6], 50 mM NaCl, 2 mM sodium azide), resuspended in 40 μl of sample buffer, and heated to 37°C for 20 min. Twenty microliters of each sample was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) (7.5% acrylamide), fixed in 7.5% acetic acid, soaked in 1 M sodium salicylate (pH 6.0) for 30 min, dried, and subjected to autoradiography or quantitated by phosphorimaging (Packard Cyclone).
Cell fractionation and sucrose gradient density centrifugation.
Cellular membranes were resolved by sucrose gradient density centrifugation essentially as described in references 35 and 53. Cells (100 ml) were grown to an OD600 of 0.3 to 0.5, sodium azide and potassium fluoride were added to 10 mM, and then the cells were briefly chilled on ice and harvested. Cells were then washed once in 10 mM sodium azide–10 mM potassium fluoride–5 mM Tris (pH 7.6) and resuspended in 0.5 ml of STE10 plus protease inhibitors (10% sucrose, 10 mM Tris [pH 7.6], 10 mM EDTA, 1× Boehringer Mannheim complete protease inhibitors). Glass beads were added, and the suspension was vortexed for 2 min. An additional 1 ml of STE10 plus protease inhibitors was then added and vortexed, and the supernatant was transferred to a new tube. This lysate was clarified by centrifugation for 3 min at 300 × g. Three hundred microliters of this cleared supernatant was then loaded onto a linear sucrose gradient prepared by sequentially layering STE solutions containing 10, 20, 35, or 60% sucrose to form a 5-ml step gradient. When sucrose gradients were centrifuged in the presence of Mg2+, we used the same protocol except that the sucrose solutions were prepared in TM buffer (10 mM Tris [pH 7.6], 1 mM MgCl2). Each gradient was then tipped to a horizontal position and allowed to diffuse for 5 h. The gradients were then returned to an upright position, loaded with extract, and subjected to 14 h of centrifugation in a SW55.1 rotor (Beckman) at 50,000 rpm and 4°C. Fractions (380 μl) were collected from the top of the gradient, and proteins were precipitated with trichloroacetic acid. Protein samples were neutralized with the addition of 10 μl of 1 M Tris base and resuspended by heating them in 50 μl of sample buffer at 37°C for 30 min. The protein composition of each fraction was then analyzed by SDS-PAGE followed by Western blotting with appropriate antibodies.
Indirect immunofluorescence.
Immunofluorescence experiments were performed essentially as described previously (51, 67). Cells (50 ml) were grown to an OD600 of approximately 0.5 in either yeast extract-peptone-dextrose or selective medium, at which point 5.8 ml of 37% formaldehyde was added directly to the culture. The culture was then shaken at 30°C for an additional 30 min. Cells were then harvested by centrifugation, resuspended in paraformaldehyde fixative (51) lacking MgCl2, and gently shaken overnight at room temperature. Fixed cells were washed four times in 1 ml of sorbitol buffer (1.2 M sorbitol, 50 mM sodium phosphate [pH 6.5]) with 1% β-mercaptoethanol and resuspended in 1 ml of sorbitol buffer minus β-mercaptoethanol. Spheroplasting was performed by adding 20 μl of a 1-mg/ml solution of oxylyticase (Enzogenetics) and shaking the suspension for 20 min at 30°C. Spheroplasted cells were then washed three times in sorbitol buffer and permeabilized by adding SDS to 0.05% and incubating the cells at 30°C for 5 min. Permeabilized cells were then washed three times with sorbitol buffer, adsorbed to polyethylenimine-coated slides, and exposed to antibodies. Yor1p was visualized by decoration with an affinity-purified anti-Yor1p (C-terminal) antibody and an anti-rabbit fluorescein isothiocyanate-conjugated goat antibody (Organon Teknika). Confocal microscopy was performed by standard techniques at the University of Iowa Electron Microscopy Center with a Bio-Rad confocal imaging system fitted to a conventional microscope with a 100× lens objective. Images were filtered by Kalman averaging and merged. Images were obtained and processed identically.
RESULTS
Wild-type Yor1p localizes to the plasma membrane by indirect immunofluorescence.
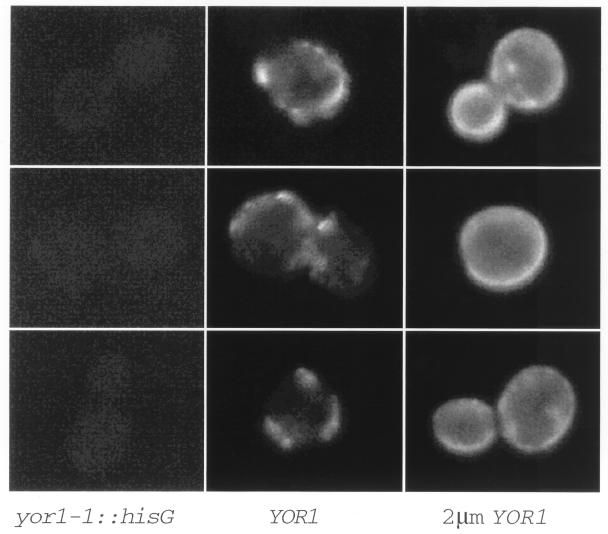
We have previously described the isolation of the YOR1 gene on the basis of its important role in mediating oligomycin resistance (33). From the striking sequence similarity between Yor1p and other ABC transporter proteins of the MRP subfamily, we speculated that Yor1p might mediate oligomycin resistance through action as a multidrug transporter protein. To gain insight into the possible mechanism of Yor1p function, the subcellular location of the protein was determined. Indirect immunofluorescence experiments were carried out with affinity-purified antibodies that recognize the carboxy terminus of Yor1p (33). Immunofluorescence was first performed on strain SEY6210 (YOR1) and its isogenic yor1-1::hisG derivative DKY7. As can be seen in Fig. 1, the wild-type strain showed a highly punctate staining pattern around the periphery of the cell similar to that seen for other plasma membrane-targeted ABC transporter proteins (11, 49). No specific immunofluorescence was visible in the yor1-1::hisG mutant cells, confirming that the staining pattern in wild-type cells was specific for Yor1p. Overproduction of Yor1p from a high-copy-number plasmid carrying the wild-type gene led to a dramatic increase in the level of plasma membrane staining by the anti-Yor1p antibody. This result is consistent with the increased presence of Yor1p at the plasma membrane, which correlates with the increased oligomycin resistance seen in these cells (33). We interpret these data as providing evidence for the plasma membrane being the functional site for Yor1p activity.
FIG. 1.
Yor1p localization by indirect immunofluorescence. Isogenic strains with various levels of the YOR1 gene were prepared for indirect immunofluorescence essentially as described previously (67). Strains carried the yor1-1::hisG allele (left panels), had a single copy of YOR1 (middle panels), or were transformed with the wild-type YOR1 gene carried on a 2 μm plasmid (right panels). Cells were labeled with affinity-purified rabbit anti-Yor1p C-terminal antibody, followed by incubation with fluorescein isothiocyanate-conjugated goat anti-rabbit antibody.
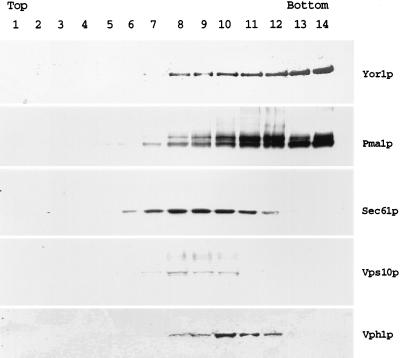
To confirm the plasma membrane localization of Yor1p indicated by the indirect immunofluorescence experiments, a wild-type cell lysate was fractionated on a sucrose gradient. The distributions of Yor1p and several marker proteins of known subcellular locations were compared by Western blot analysis with appropriate antisera (Fig. 2). Yor1p was enriched in the highest-density fractions of the sucrose gradient in a manner similar to that of the plasma membrane marker Pma1p (55). The distribution of the ER membrane protein Sec61p (59) was distinctly different from that of Yor1p and was most highly enriched in the lower-density fractions of the gradient. Markers for the Golgi membrane, Vps10p (also called Pep1p) (43), and the vacuolar membrane, Vph1p (42), were also enriched in the lower-density regions of the gradient. This sucrose gradient analysis provides strong support for the belief that Yor1p is localized to the plasma membrane.
FIG. 2.
Biochemical fractionation of Yor1p. Wild-type cells were lysed with glass beads and fractionated over a sucrose gradient. Aliquots of each sucrose gradient fraction were precipitated with trichloroacetic acid, resuspended in Laemmli buffer, and electrophoresed by SDS-PAGE. The proteins were then transferred to nitrocellulose, and the resulting blot was probed with the indicated antisera. The relative positions of the light (top) and heavy (bottom) fractions of the sucrose gradient are indicated on the figure. Antisera employed listed on the right-hand side are as follows: Yor1p is the affinity-purified rabbit anti-Yor1p antibody used in Fig. 1, Pma1p corresponds to the plasma membrane ATPase protein (55), Sec61p is an integral membrane subunit of the translocon in the ER (59), Vps10p (also called Pep1p) is the Golgi apparatus- or prevacuole-localized carboxypeptidase Y (CPY) receptor (43), and Vph1p is the 100-kDa subunit of the vacuolar ATPase (42).
Mutagenesis of the first nucleotide binding domain of Yor1p.
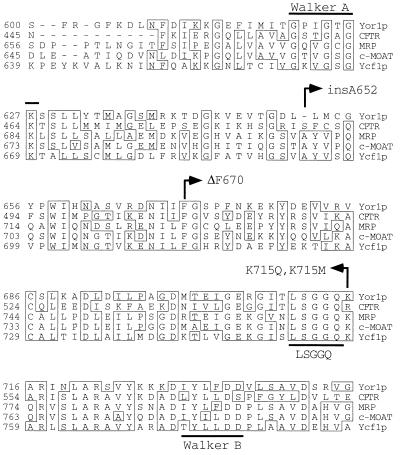
ABC transporters are defined by their characteristic nucleotide binding domains. Like other nucleotide binding proteins, ABC transporters contain Walker A and Walker B motifs (64). However, the sequence similarity of ABC transporters extends over the entire nucleotide binding domain and is further defined by the presence of a short peptide sequence (LSGGQ) that lies between the Walker A and B elements (26). While the spacing between these three functional motifs is similar in all ABC transporter proteins, different families of these related proteins have been identified based on variations in the lengths of sequences separating the Walker A, Walker B, and LSGGQ motifs. One of these groups has been referred to as the MRP family since the sequence of this ABC transporter protein originally served to define this strictly conserved spacing (8). Sequence analysis of Yor1p indicated that this protein was also a member of the MRP family of ABC transporters, and an alignment showing the conserved spacing and sequences of several members of this group is shown in Fig. 3.
FIG. 3.
Alignment of NBD1 regions from MRP family members. An alignment of the NBD1 segments from each of the indicated ABC transporter proteins is shown. The numbers refer to the respective positions along the polypeptide chain, and residues that are identical in at least three of the five selected proteins are boxed. The conserved structural motifs present in all ABC transporters are indicated by the heavy lines, and the sites of mutations in Yor1p NBD1 are denoted by arrows.
To explore the functional significance of the conserved spacing and primary sequence shared by Yor1p and the MRP family, several different site-directed mutations were produced in the NBD1 region of the YOR1 gene. The phenylalanine residue at position 670 in Yor1p corresponds to a phenylalanine that is conserved in all the MRP family members. Removal of this phenylalanine residue from CFTR was previously shown to eliminate normal plasma membrane targeting of this protein and to cause the resulting ΔF508 mutant of CFTR to be retained in the ER (7). The potential functional role of the corresponding phenylalanine residue in Yor1p was evaluated by constructing a ΔF670 allele of YOR1.
Three other mutant versions of Yor1p were generated. While NBD1 of Yor1p clearly shows the striking sequence identity of the MRP family, computer analysis of this region in Yor1p indicates that a single amino acid gap must be introduced into the Yor1p sequence in order to maintain the alignment with the sequences of other family members (33). To examine the functional relevance of this one amino acid gap, an alanine was inserted into the predicted position of the gap. Additionally, experiments with both CFTR and Ycf1p have suggested that changing the basic residue immediately following the LSGGQ motif in these two proteins to methionine or glutamine produces a derivative with increased function (47, 61, 67). The lysine at Yor1p position 715 was changed to either methionine or glutamine to determine if these alterations could increase the function of Yor1p.
Functions and expression of mutant Yor1p derivatives.
Each mutant form of Yor1p was analyzed for its relative ability to complement the oligomycin-hypersensitive phenotype of a strain lacking a functional YOR1 structural gene. Mutant YOR1 genes were introduced on low-copy-number plasmids into a strain carrying the yor1-1::hisG allele and analyzed by spot test assay on plates containing a gradient of oligomycin (Fig. 4). The ΔF670 YOR1 allele was also introduced on a high-copy-number plasmid to determine if residual function could be detected when the mutant protein was overproduced.
FIG. 4.
Oligomycin resistance phenotypes of Yor1p and mutant variants. DKY7 (yor1-1::hisG) cells were transformed with low-copy-number plasmids bearing the genes that express the indicated forms of Yor1p or with the low-copy-number vector (pRS316). Transformants were grown to an _A_600 of approximately 1, and 1,000 cells were placed on YPGE medium containing a gradient of oligomycin (indicated by the wedge of increasing height). The plate was incubated at 30°C and photographed.
None of the mutant Yor1p derivatives were able to confer oligomycin resistance to a level approaching that of the wild-type protein. The ΔF670 Yor1p form was the most highly defective and exhibited oligomycin tolerance that was indistinguishable from that of yor1-1::hisG cells carrying the vector alone, irrespective of the presence of this mutant allele on a low- or high-copy-number plasmid. The insertion of alanine at position 652 (insA652) severely reduced the function of the resulting mutant Yor1p. Finally, the two replacements of the basic residue downstream of the LSGGQ motif (K715M and K715Q) produced forms of Yor1p that were highly defective in the ability to confer oligomycin resistance.
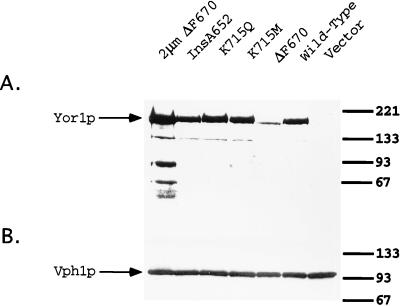
To ensure that the observed changes in oligomycin tolerance were due to alterations in protein function rather than in amount, steady-state levels of each mutant protein were compared to those of the wild-type by Western blot analysis (Fig. 5). All the mutant Yor1p forms were produced at levels comparable to those of the wild-type protein with the exception of ΔF670 Yor1p. This mutant derivative was consistently detected at a reduced steady-state level relative to that of the wild-type protein, suggesting that this lesion reduced the stability of Yor1p. Even when ΔF670 Yor1p was overproduced from a 2μm plasmid, the functional defect of this mutant protein was not suppressed. Since the levels of the ΔF670 Yor1p form produced from the 2μm plasmid are even higher than those of the wild-type protein, the functional defect caused by the ΔF670 lesion cannot be solely due to the decreased levels of the steady-state protein.
FIG. 5.
Steady-state levels of mutant Yor1p derivatives. Whole-cell extracts were prepared from DKY7 (yor1-1::hisG) cells expressing the indicated forms of Yor1p from low-copy-number plasmids. Protein (75 μg) was electrophoresed by SDS-PAGE, transferred to nitrocellulose, and blotted with anti-Yor1p (A) or anti-Vph1p (B) as a loading control.
Localization of mutant Yor1p derivatives.
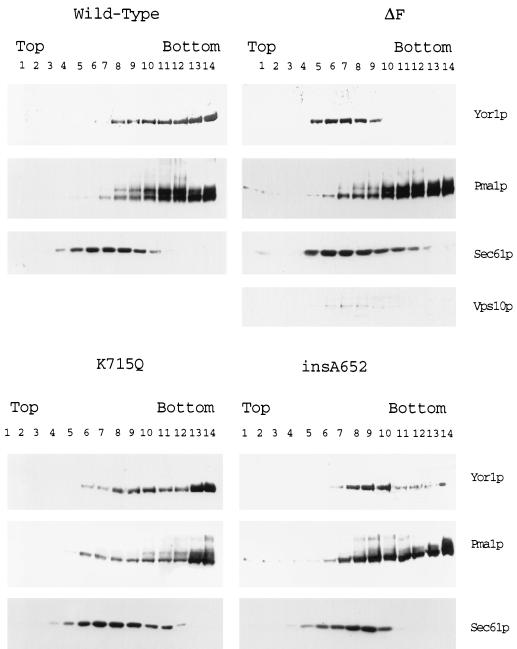
The Western blot data above confirmed that the observed defects seen in these mutant forms of Yor1p were not due to an effect on the levels of synthesis of these proteins. The localization of the mutant transporter proteins was next assessed by biochemical fractionation on sucrose gradients. The positions of the various forms of Yor1p and marker proteins of known subcellular distribution were determined by Western blotting (Fig. 6).
FIG. 6.
Subcellular fractionation of Yor1p NBD1 mutants. Cells lacking a chromosomal YOR1 locus (DKY7) were transformed with low-copy-number plasmids bearing the genes that express the indicated forms of Yor1p. Lysates were prepared and centrifuged through sucrose gradients as described in the legend to Fig. 2. Aliquots of each sucrose gradient were then subjected to Western blotting with the affinity-purified anti-Yor1p antibody (Yor1p) or the rabbit anti-Pma1p (Pma1p), rabbit anti-Vps10p (Vps10p), or rabbit anti-Sec61p (Sec61p) antiserum as listed on the right-hand side of each panel. The orientation of the sucrose gradient fractions is indicated as in Fig. 2.
The first mutant Yor1p form examined was the ΔF670 derivative. ΔF670 Yor1p was easily detectable in the sucrose gradient fractions and showed a distinct pattern of enrichment relative to that of the wild-type protein. ΔF670 Yor1p was found to enrich in the center of the sucrose gradient, while Yor1p was concentrated towards the most dense regions of the gradient. When the same gradient fractions were probed with the anti-Pma1p antiserum, the characteristic enrichment of this integral plasma membrane protein was seen in the most dense fractions of the gradient. However, unlike the wild-type protein, ΔF670 Yor1p was seen to enrich in the fractions of the gradients that contained the ER marker Sec61p and the Golgi or endosomal marker Vps10p. These data are consistent with the hypothesis that ΔF670 Yor1p fails to normally exit the ER during its biosynthesis and that it fails to reach the plasma membrane. This aberrant location of ΔF670 Yor1p is likely to at least contribute to the functional defect of this protein.
The K715Q and insA652 Yor1p forms were also analyzed by Western blotting of sucrose gradient fractions. Both of these mutant proteins exhibited a fractionation pattern intermediate between those of the wild-type and ΔF670 forms of Yor1p. The fractionation behavior of the insA652 form of Yor1p was closer to that of ΔF670 Yor1p, as the majority of the insA652 derivative was present in the center of the gradient. However, unlike ΔF670 Yor1p, detectable levels of the insA652 protein were found associated with the most dense sucrose gradient fractions. K715Q Yor1p was clearly present in the densest fractions of the gradient but was also seen to modestly accumulate in the center of the gradient. Since the trafficking defect of K715Q Yor1p is less pronounced than that of the insA652 form, we ascribe the large reduction in function seen in cells expressing K715Q Yor1p to a reduction in the activity of this protein. These data suggest that both the K715Q and the insA652 Yor1p have defects of varying severities in their ability to move out of the ER. However, defective trafficking is most likely to explain the defect in the insA652 allele while a decrease in activity of the K715Q protein seems more consistent with the behavior of this mutant Yor1p derivative.
The stability of ΔF670 Yor1p is reduced relative to that of the wild-type protein.
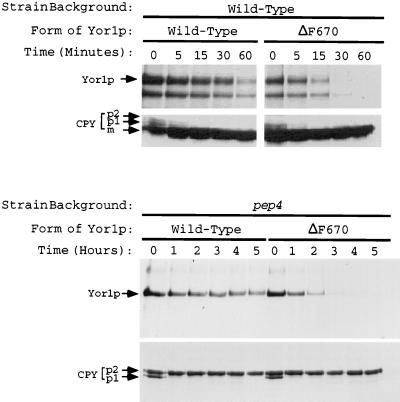
The reduced steady-state levels of ΔF670 Yor1p suggested that this mutant derivative might be less stable than the corresponding wild-type protein. To directly test this possibility, pulse-chase analysis was carried out on strains expressing either the wild-type or ΔF670 form of Yor1p. Cells were labeled with [35S]methionine and then incubated in a large excess of unlabeled amino acids. Aliquots were withdrawn at various times, and Yor1p was recovered by immunoprecipitation. Immunoprecipitated material was resolved by SDS-PAGE, followed by autoradiography or phosphorimaging (Fig. 7).
FIG. 7.
Vacuolar proteases influence the turnover of both wild-type and ΔF670 Yor1p. Isogenic yor1-1::hisG strains that either contained (top panels) or lacked (bottom panels) the PEP4 locus were transformed with low-copy-number plasmids bearing the gene that expresses the wild-type or ΔF670 form of Yor1p. Selected transformants were then analyzed by pulse-chase analysis followed by immunoprecipitation with the anti-Yor1p antiserum or an anti-CPY polyclonal antibody (from R. Piper). Levels of immunoprecipitated proteins were quantitated by phosphorimaging. The time scale for the pulse-chase in the PEP4 background was in minutes, while that for the pulse-chase with the pep4 strain was in hours. The position of Yor1p and the positions of the various forms of CPY (58) are denoted by arrows.
The wild-type protein was degraded with a half-life of approximately 42 min, while the half-life of ΔF670 Yor1p was only 15 min, demonstrating the markedly increased rate of turnover of this mutant form of Yor1p. This result indicates that ΔF670 Yor1p has two defects relative to the wild-type protein: altered subcellular localization and decreased stability. The two forms of immunoprecipitable Yor1p seen in Fig. 7 (top panels) are believed to be due to cleavage by vacuolar proteases during sample processing since the lower form of Yor1p does not appear in pep4 cells (see below) and does not display a precursor-product relationship over time.
In analyses of other plasma membrane-localized ABC transporter proteins, Ste6p and Pdr5p, it was found that these proteins were delivered to the vacuole for degradation after their residence in the plasma membrane (3, 11, 35). To examine the involvement of vacuolar proteases in the turnover of Yor1p and mutant derivatives, a pep4 strain was employed. Loss of the PEP4 gene strongly depresses maturation of vacuolar proteases and stabilizes proteins that are normally degraded in the vacuole (1, 31). The levels of degradation of wild-type and ΔF670 Yor1p were compared in isogenic wild-type and pep4 cells by pulse-chase analysis.
Loss of the PEP4 gene led to a large increase in the stability of wild-type Yor1p (Fig. 7). The half-life of Yor1p increased from 42 min in a PEP4 background to 284 min in the absence of Pep4p. This result clearly implicated normal vacuolar protease levels in the turnover of Yor1p. The behavior of ΔF670 Yor1p in response to the presence of the PEP4 gene was also examined. The half-life of ΔF670 Yor1p was increased from 15 to 58 min when the PEP4 gene was deleted. While the turnover of both the wild-type protein and ΔF670 Yor1p was reduced in the pep4 background, the half-lives of these two ABC transporters were still very different. Even in the absence of vacuolar proteases, ΔF670 Yor1p exhibited a half-life that was only 20% that of the wild-type protein in the same genetic background. However, the half-life of ΔF670 Yor1p was longer in a pep4 mutant strain than in the isogenic PEP4 cells. This result was unexpected, as we hypothesized that ΔF670 Yor1p was trapped in the ER and expected it to be turned over by the ER degradation system, as was shown for other defective proteins retained in this organelle (13, 18, 24, 68). Typically, loss of vacuolar proteases does not affect turnover of proteins that are targets for degradation in the ER (13, 19).
To further assess if ΔF670 Yor1p was both retained and degraded in the ER, we carried out two additional experiments. First, additional sucrose gradient analyses were performed under different conditions to ascertain that ΔF670 Yor1p was localized in the ER. Second, a cell containing a temperature-sensitive allele of the SEC12 gene was used to demonstrate that ΔF670 Yor1p was degraded even if ER-to-Golgi apparatus transport was arrested.
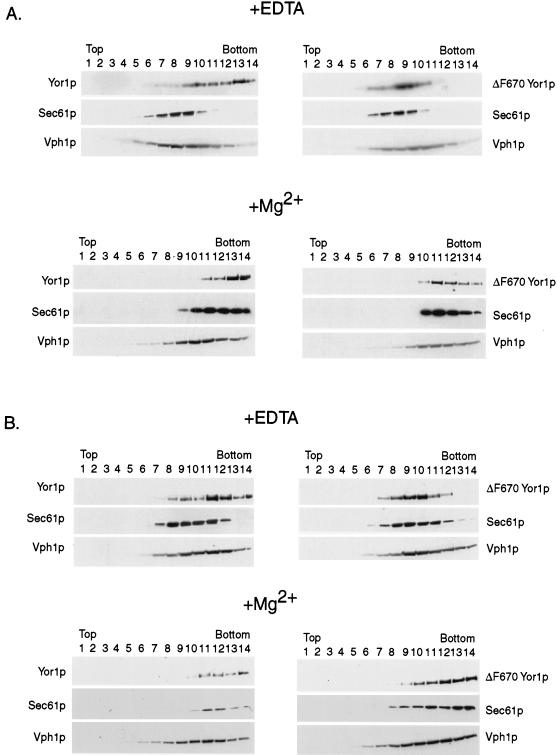
Roberg et al. (53) demonstrated that ER membranes from S. cerevisiae exhibit different densities in sucrose gradients depending on the Mg2+ concentration in these gradients. If Mg2+ is included in the sucrose gradient, then ribosomes remain docked to the ER and the resulting complex exhibits a higher density than if ribosomes are removed by EDTA chelation of Mg2+. Extracts were prepared from wild-type and pep4 cells expressing either wild-type or ΔF670 Yor1p. This analysis was performed on pep4 cells to ensure that if a fraction of ΔF670 Yor1p arrived at the vacuole, it would not be degraded by vacuolar proteases. These extracts were then resolved on sucrose gradients either in the presence or in the absence of Mg2+ as described previously (53). Aliquots of each fraction were collected and analyzed by Western blotting with antibodies directed against Yor1p, Sec61p, and Vph1p.
In cells containing an intact PEP4 gene (Fig. 8), the fractionation profiles of both wild-type and ΔF670 Yor1p, Sec61p, and Vph1p were the same as previously discussed (Fig. 2 and 6). However, when Mg2+ was included in the gradient, the peaks of immunoreactivity of both ΔF670 Yor1p and Sec61p were dramatically shifted to the denser fractions of the gradient. The new peaks of these two proteins were now present in fractions previously found to be enriched in the plasma membrane protein Pma1p. The Western blot profiles of wild-type Yor1p and Vph1p were slightly compressed towards the fractions of higher density, since both of these integral membrane proteins are likely to have some amount of ER precursor that will be shifted to the high-density fractions upon inclusion of Mg2+.
FIG. 8.
Mg2+-dependent fractionation of ΔF670 Yor1p in PEP4 or pep4 cells. Lysates were prepared from PEP4 (A) or pep4 (B) cells expressing either wild-type or ΔF670 Yor1p. These lysates were subjected to sucrose gradient centrifugation in the presence or absence of Mg2+ as described previously (53). Aliquots of the gradient were then analyzed by Western blotting (see Materials and Methods) for the presence of either form of Yor1p and for Sec61p or Vph1p.
This same fractionation protocol was carried out on a pep4 mutant strain expressing either wild-type or ΔF670 Yor1p. Loss of the PEP4 gene did not affect the fractionation profile of either form of Yor1p (Fig. 8). This finding provides strong evidence against the notion that ΔF670 Yor1p reaches the vacuole, where it can be acted on by vacuolar proteases. Since inactivation of the vacuolar proteases failed to allow ΔF670 Yor1p to be detected in vacuole-enriched fractions, we argue that ΔF670 Yor1p is unlikely to be degraded in this organelle. This observation is consistent with the idea that ΔF670 Yor1p is degraded in the ER by the proteasome.
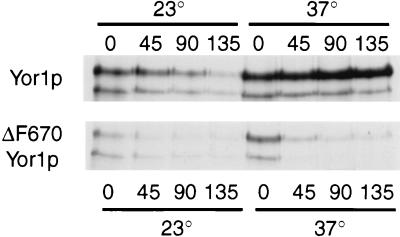
To examine whether ΔF670 Yor1p is a target of the ER degradation system, we prepared a strain carrying a sec12-3 temperature-sensitive mutation that expressed either the wild-type or the ΔF670 form of Yor1p. Sec12p is required to form vesicles from the ER membrane during ER-to-Golgi apparatus transport (46). The sec12-3 strain expressing these two forms of Yor1p was then analyzed by a pulse-chase immunoprecipitation experiment at the permissive and restrictive temperatures to assess the stability of wild-type and ΔF670 Yor1p (Fig. 9).
FIG. 9.
Effect of loss of Sec12p function on turnover of wild-type and ΔF670 Yor1p. A strain containing the temperature-sensitive sec12-3 allele and lacking the YOR1 gene was constructed by one-step gene disruption of the YOR1 locus to produce EAE18. Low-copy-number plasmids bearing the gene that expresses either the wild-type or ΔF670 form of Yor1p were then introduced into this gene background. Turnover of these two forms of Yor1p was then assessed by pulse-chase immunoprecipitation analysis at the permissive (23°C) and restrictive (37°C) temperatures. The numbers refer to times in minutes after chase addition.
The pulse-chase immunoprecipitation demonstrated that the stability of wild-type Yor1p increased upon imposition of the sec12-3 block. This finding is consistent with the idea that wild-type Yor1p must leave the ER to be degraded in a Pep4p-dependent manner. In opposition to the stabilization seen for wild-type Yor1p after the temperature shift of the sec12-3 strain, ΔF670 Yor1p was degraded with similar kinetics at both the permissive and restrictive temperatures. This finding indicates that inactivation of the SEC12 gene product failed to stabilize ΔF670 Yor1p and supports the hypothesis that this mutant protein is degraded at the level of the ER. However, final confirmation that ΔF670 Yor1p is trapped in the ER awaits detection of this mutant protein by microscopic techniques which have not yet been successful (data not shown). To directly explore the contribution of the ER degradation system to the turnover of ΔF670 Yor1p, we used mutant backgrounds that were defective in this proteolytic machinery.
Proteasome-mediated turnover of ΔF670 Yor1p.
The simplest explanation for the observed stabilization of ΔF670 Yor1p by a pep4 mutation is a direct role for vacuolar proteases in the turnover of this protein. However, it is also possible that the pep4 defect acts indirectly to deplete a component required for normal proteasome-mediated turnover of proteins trapped in the ER. A candidate for such an indirect action is depletion of ubiquitin, as the vacuole is an important site for degradation of ubiquitinated proteins that arrive here after endocytosis from the plasma membrane (reviewed in reference 23). The absence of degradation has the potential to affect ubiquitin metabolism in a fashion that may depress degradation of ΔF670 Yor1p.
To determine if changes in ubiquitin levels could reverse the apparent stabilization of ΔF670 Yor1p that was observed in the pep4 mutant background, we transiently increased ubiquitin levels in pep4 cells and examined the effect on the stability of ΔF670 Yor1p. This was accomplished through use of a _CUP1_-ubiquitin fusion gene that produces high levels of ubiquitin when cells are exposed to copper (12). Strains lacking pep4 and expressing either wild-type or ΔF670 Yor1p from low-copy-number plasmids were transformed with a plasmid carrying a _CUP1_-ubiquitin fusion gene. Transformants were grown in the presence or absence of copper and evaluated for stability of Yor1p by pulse-chase analysis.
Induction of the _CUP1_-ubiquitin fusion gene by copper caused a dramatic destabilization of ΔF670 Yor1p in the pep4 mutant strain (Fig. 8). The half-lives of ΔF670 Yor1p were 64 and 22 min when the protein was evaluated in the absence and presence of copper, respectively. Turnover of wild-type Yor1p was not affected by addition of copper. Treatment of cells lacking the _CUP1_-ubiquitin fusion gene with copper failed to influence the turnover of either form of Yor1p (data not shown).
This experiment supported the idea that ΔF670 but not wild-type Yor1p was subject to ubiquitin-dependent protein turnover. Ubiquitin-dependent degradation involves conjugation of a ubiquitin moiety to a target protein (reviewed in reference 63). This reaction requires the participation of ubiquitin-conjugating enzymes termed Ubc proteins in S. cerevisiae (25). The ubiquitin peptide is transferred to the target protein, which in turn is subjected to degradation by the proteasome (25). We tested the involvement of a Ubc enzyme (Ubc7p) and the proteasome in the turnover of ΔF670 Yor1p by using mutant strains defective in these loci. A ubc7 mutant was selected, as other studies have shown that loss of this gene influences the turnover of other proteins that are degraded in the ER (5, 24). A yor1-1::hisG ubc7 strain was constructed and transformed with low-copy-number plasmids expressing either wild-type or ΔF670 Yor1p. The stability of Yor1p was examined by pulse-chase analysis (Fig. 10).
FIG. 10.
ΔF670 but not wild-type Yor1p is responsive to changes in ubiquitin metabolism. (A) A strain lacking the PEP4 gene was transformed with a 2 μm plasmid containing a CUP1-UBI1 gene fusion along with low-copy-number plasmids bearing the gene for either wild-type or ΔF670 Yor1p. Levels of immunoprecipitable Yor1p were determined by pulse-chase analysis either in the absence (open symbols) or the presence (filled symbols) of copper sulfate to induce ubiquitin expression. The percentage of Yor1p remaining after chase was plotted as a function of time. The half-life (T1/2) of each immunoprecipitable Yor1p form is shown on the right-hand side in minutes. (B) An isogenic pair of yor1-1::hisG cells either lacking (ubc7) or containing (UBC7) an intact copy of chromosomal UBC7 was transformed with low-copy-number plasmids bearing the gene for wild-type or ΔF670 Yor1p. Yor1p stability was analyzed by pulse-chase analysis as described above. The filled symbols indicate the presence of UBC7, while the open symbols correspond to a strain carrying a ubc7-Δ1::HIS3 allele (32). The half-lives (in minutes) of the Yor1p forms are shown in the column on the right.
The turnover of wild-type Yor1p was not affected by loss of UBC7. The wild-type Yor1p half-life was 42 min in the UBC7 background and 50 min in the ubc7 strain. However, the turnover of ΔF670 Yor1p was depressed upon loss of Ubc7p, changing from 15 min in a UBC7 strain to 37 min in a ubc7 cell. These data are consistent with a role for Ubc7p in the ubiquitin-dependent turnover of ΔF670 Yor1p.
Along with that of Ubc7p, we examined the contribution of the proteasome to the degradation of ΔF670 Yor1p. A yor1-1::hisG pre1-1 pre2-2 strain was constructed, and the levels of stability of wild-type and ΔF670 Yor1p were assessed by pulse-chase analysis as described above (Fig. 11). Low-copy-number plasmids bearing PRE1 and PRE2 were also introduced into this background in order to restore proteasome function (21, 22, 68).
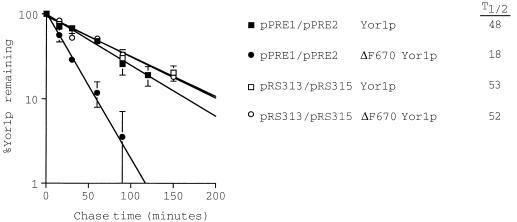
FIG. 11.
A functional proteasome is required for rapid degradation of ΔF670 Yor1p. A yor1-1::hisG mutant cell lacking normal proteasome function (pre1-1 pre2-2) was transformed with low-copy-number plasmids bearing the gene for either the wild-type or ΔF670 form of Yor1p. Low-copy-number plasmids carrying PRE1 and PRE2 (pPRE1/pPRE2) or the vector plasmids alone (pRS313/pRS315) were introduced into these transformants to examine the consequence of varying the activity of the proteasome on turnover of these two forms of Yor1p. Appropriate transformants were analyzed for stability of wild-type or ΔF670 Yor1p by pulse-chase analysis as described above. The calculated half-lives (T1/2; in minutes) are on the right.
Loss of proteasome activity increased the half-life of ΔF670 Yor1p to a value indistinguishable from that of the wild-type protein (52 versus 53 min). Restoring the PRE1 and PRE2 genes decreased the stability of ΔF670 Yor1p to 18 min but had no significant effect on the degradation of wild-type Yor1p. These data are consistent with the turnover of ΔF670 Yor1p occurring through the ER degradation pathway in a fashion similar to that described for 3-hydroxy-3-methylglutaryl-coenzyme A reductase (18) and carboxypeptidase Y mutants (24).
DISCUSSION
Polarized epithelial cells express both MRP and a closely related protein designated the multispecific organic anion transporter (c-MOAT) (48). In these polarized cells, MRP and c-MOAT are localized to the basolateral and apical membranes, respectively (reviewed in reference 34). Analyses of the complement of ABC transporters found in the S. cerevisiae genome have shown that a family of MRP-related transporters, including Yor1p and Ycf1p, is present in this organism (10, 44). This work has identified Yor1p as a plasma membrane-targeted transporter. Previous studies have defined Ycf1p as a component of the vacuolar membrane (37, 67). Thus, S. cerevisiae expresses a family of MRP-related ABC transporters and these transporters are destined for delivery to different membrane locations in the cell. These results illustrate the conserved physiology of the MRP family of proteins between animal cells and S. cerevisiae, supporting the notion that analysis of this fungal MRP family will provide valuable insight into the function of the cognate mammalian proteins.
One of the most intensively studied integral membrane proteins with respect to its intracellular trafficking is the human CFTR (52), associated with the disease CF. Most CFTR alleles associated with CF, including the most common CF-associated allele, ΔF508, have the effect of causing CFTR to be trapped in the ER (62, 66). We probed NBD1 of Yor1p by mutagenesis to determine if this segment of the protein was similarly important in the biogenesis of this closely related yeast ABC transporter protein.
The ΔF670 allele of Yor1p produced a transporter protein that exhibited trafficking behavior striking in its similarity to that of the ΔF508 CFTR. These experiments suggest that ΔF670 Yor1p is trapped in the ER in a fashion analogous to that of ΔF508 CFTR (7). At least two explanations can be proposed to explain the consequences of the loss of this NBD1 phenylalanine codon: (i) important primary sequence information is deleted and (ii) spatial relationships are disturbed in the deletion mutants. The finding that the spacing of the functional motifs in Yor1p NBD1 is shorter by 1 amino acid than in the other members of the MRP family (33) suggested that the spacing within this region might be flexible. To investigate this possibility, we inserted an alanine residue at the gap in Yor1p NBD1 to convert the spacing of this protein to that seen for the other MRP members. This mutant exhibited trafficking behavior that was reminiscent of that of the wild-type CFTR. CFTR matures very slowly in the ER, with approximately 75% of the protein never reaching the plasma membrane (41). Analysis of the insA652 Yor1p derivative suggests that this mutant protein is much slower to leave the ER than wild-type Yor1p but that a small amount is capable of reaching the plasma membrane. This is seen both in the immunoreactive insA652 Yor1p being in the densest fractions of the sucrose gradient and in the weak but reproducible oligomycin resistance conferred by this mutant factor. In contrast, ΔF670 Yor1p could not be detected in the dense sucrose gradient fractions and failed to complement the oligomycin hypersensitivity of a yor1 strain.
One of the most surprising features of ΔF670 Yor1p was the stabilization of this protein that occurred upon loss of the PEP4 gene. Vacuolar proteases have not been observed to affect the stability of other integral membrane proteins that are degraded in the ER (14, 19, 38, 50). The rescue of this apparent stabilization by overexpression of ubiquitin has at least two possible explanations. First, it is possible that ΔF670 Yor1p turnover is especially sensitive to levels of ubiquitin that may be slightly reduced in pep4 mutant strains. Elevation of ubiquitin levels may replenish whatever ubiquitin pool has been depleted. Second, vacuolar protease function may be required to produce an activity important in ΔF670 Yor1p turnover at the ER. This possibility has precedent, as degradation of ΔF508 CFTR has been found to involve several distinct proteolytic systems (29). Overproduction of ubiquitin might enhance proteasome function and allow accelerated turnover of ΔF670 Yor1p by this proteolytic system, although under normal conditions, the proteasome would be one of several contributors to the degradation of ΔF670 Yor1p. The recent demonstration that proteins can leave the yeast vacuole (6) is consistent with the notion that the vacuole may provide protein maturation as well as degradation function. Experiments are under way to further investigate the role of pep4 on ΔF670 Yor1p turnover.
A final issue concerning the degradation of ΔF670 Yor1p is the precise localization of this factor. While our data support the belief that this mutant protein is retained in the ER, we cannot eliminate the possibility that ΔF670 Yor1p is found in intracellular locations in a Sec12p-independent fashion, where it is then degraded. Indirect immunofluorescence has failed to detect ΔF670 Yor1p (data not shown), although this protein can easily be assayed by other immunological methods (Fig. 5 and 7). We are now constructing green fluorescent protein fusions to ΔF670 Yor1p to directly visualize the subcellular distribution of this protein. ΔF508 CFTR has recently been demonstrated to localize to unusual pericentriolar structures in cells unable to fully degrade this mutant protein (30). It will be important to determine if ΔF670 Yor1p is also found in these aggresome structures.
Substitutions in the basic residue immediately following the LSGGQ motif in the MRP family members CFTR and Ycf1p have been found to have dramatic effects on the function of the resulting mutant proteins. A Ste6p-CFTR chimera containing CFTR NBD1, which failed to function in the presence of the ΔF508 form of NBD1, regained activity when either Q or M replaced the arginine downstream of the NBD1 LSGGQ (61). A Ycf1p derivative containing a mutation analogous to ΔF508 (Ycf1p ΔF713), did not respond to Q or M replacement of the lysine following its NBD1 LSGGQ (67). Interestingly, an otherwise wild-type Ycf1p containing the K758Q or K758M lesion exhibited markedly increased function (67). These data from analyses of other MRP family members led us to expect that analogous replacements in Yor1p would elevate its biological function. However, both Yor1p K715Q and K715M are defective in function relative to that of the wild-type protein and K715Q Yor1p exhibits a modest defect in normal plasma membrane fractionation as evidenced by an elevated level of Yor1p immunoreactivity in the region of the sucrose gradient corresponding to the ER. Taken together, these data are consistent with the basic residue following the NBD1 LSGGQ motif being a key determinant in the folding and transport of the MRP family of ABC transporters. In certain instances (CFTR, Ycf1p), alteration of this residue can make the protein fold or be transported more rapidly, while in other instances (Yor1p), alterations of this position depress the ability to fold and/or transport.
Irrespective of the exact explanation behind the observed defects elicited by the NBD1 mutations that we have generated in Yor1p, alterations in this domain of Yor1p have dramatic effects on the function of the resulting mutant transporter protein. This sensitivity can be contrasted with the resistance of the a-factor transporter Ste6p to changes in function caused by alterations in the primary sequence of its NBD1 region (4). Two different single amino acid deletions in Ste6p NBD1 were found to have no detectable effect on the function of the resulting mutant protein. This comparison serves to illustrate the importance of the conserved spacing and high sequence conservation in the MRP family of proteins compared to that in other ABC transporter proteins of similar overall structure.
With Yor1p and CFTR, alterations in the NBD1 region often lead to a defect in transport from the ER. In the case of the ΔF alleles of CFTR or Yor1p, the resulting trafficking defect also results in enhanced proteolysis of the mutant protein. Both of the ΔF variants become substrates for ubiquitin-dependent proteolysis catalyzed by the proteasome. The finding of the similarity in molecular phenotype exhibited by ΔF508 CFTR and ΔF670 Yor1p provides the opportunity for genetic analysis of the mechanisms underlying the cell biology of the intracellular handling of this important class of ABC transporters.
ACKNOWLEDGMENTS
We thank Rob Piper, Mark Stamnes, Scott Emr, Mark Hochstrasser, Dieter Wolf, Karl Kuchler, Chris Kaiser, and Ralf Kölling for discussions and materials. Antibodies were provided by André Goffeau, Tom Rapaport, Colin Stirling, Rob Piper, and Mark Rose. The ubc7 disruption plasmid was provided by Manfred Koegl and Stephan Jentsch. Thanks go to Rob Piper for a critical reading of the manuscript.
This work was supported in part by NIH grants GM49825 (W.S.M.-R.) and DK25295 (University of Iowa Diabetes and Endocrinology Research Center). W.S.M.-R. is an established investigator of the American Heart Association.
REFERENCES
- 1.Ammerer G, Hunter C, Rothman J H, Saari G C, Valls L A, Stevens T H. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol Cell Biol. 1986;6:2490–2499. doi: 10.1128/mcb.6.7.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakos E, Hegedus T, Hollo Z, Welker E, Tusnady G E, Zaman G J R, Flens M J, Varadi A, Sarkadi B. Membrane topology and glycosylation of the human multidrug resistance-associated protein. J Biol Chem. 1996;271:12322–12326. doi: 10.1074/jbc.271.21.12322. [DOI] [PubMed] [Google Scholar]
- 3.Berkower C, Loayza D, Michaelis S. Metabolic instability and constitutive endocytosis of STE6, the a-factor transporter of Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1185–1198. doi: 10.1091/mbc.5.11.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkower C, Michaelis S. Mutational analysis of the yeast a-factor transporter STE6, a member of the ATP binding cassette protein superfamily. EMBO J. 1991;10:3777–3785. doi: 10.1002/j.1460-2075.1991.tb04947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- 6.Bryant N J, Piper R C, Weisman L S, Stevens T H. Retrograde traffic out of the yeast vacuole to the TGN occurs via the prevacuolar/endosomal compartment. J Cell Biol. 1998;142:651–663. doi: 10.1083/jcb.142.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng S H, Gregory R J, Marshall J, Paul S, Souza D W, White G A, O’Riordan C R, Smith A E. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- 8.Cole S P C, Bhardwaj G, Gerlach J H, Mackie J E, Grant C E, Almquist K C, Stewart A J, Kurz E U, Duncan A M V, Deeley R G. Overexpression of a transporter gene in multidrug-resistant human lung cancer cell line. Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 9.Cui Z, Hirata D, Tsuchiya E, Osada H, Miyakawa T. The multidrug resistance-associated protein (MRP) subfamily (Yrs1/Yor1) of Saccharomyces cerevisiae is important for the tolerance to a broad range of organic anions. J Biol Chem. 1996;271:14712–14716. doi: 10.1074/jbc.271.25.14712. [DOI] [PubMed] [Google Scholar]
- 10.Decottignies A, Goffeau A. Complete inventory of the yeast ABC proteins. Nat Genet. 1997;15:137–145. doi: 10.1038/ng0297-137. [DOI] [PubMed] [Google Scholar]
- 11.Egner R, Mahé Y, Pandjaitan R, Kuchler K. Endocytosis and vacuolar degradation of the plasma membrane-localized Pdr5 ATP-binding cassette multidrug transporter in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:5879–5887. doi: 10.1128/mcb.15.11.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellison M J, Hochstrasser M. Epitope-tagged ubiquitin. J Biol Chem. 1991;266:21150–21157. [PubMed] [Google Scholar]
- 13.Finger A, Knop M, Wolf D H. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur J Biochem. 1993;218:565–574. doi: 10.1111/j.1432-1033.1993.tb18410.x. [DOI] [PubMed] [Google Scholar]
- 14.Galan J-M, Cantegrit B, Garnier C, Namy O, Haguenauer-Tsapis R. ER degradation of a mutant yeast plasma membrane protein by the ubiquitin-proteasome pathway. FASEB J. 1998;12:315–323. doi: 10.1096/fasebj.12.3.315. [DOI] [PubMed] [Google Scholar]
- 15.Gottesman M M, Hrycyna C A, Schoenlein P V, Germann U A, Pastan I. Genetic analysis of the multidrug transporter. Annu Rev Genet. 1995;29:607–649. doi: 10.1146/annurev.ge.29.120195.003135. [DOI] [PubMed] [Google Scholar]
- 16.Gottesman M M, Pastan I. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu Rev Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- 17.Guan K-L, Dixon J E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 18.Hampton R, Gardner R G, Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hampton R Y, Rine J. Regulated degradation of HMG-CoA reductase, an integral membrane protein of the endoplasmic reticulum, in yeast. J Cell Biol. 1994;125:299–312. doi: 10.1083/jcb.125.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 21.Heinemeyer W, Gruhler A, Mohrle V, Mahe Y, Wolf D H. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chymotryptic activity and degradation to ubiquitinated proteins. J Biol Chem. 1993;268:5115–5120. [PubMed] [Google Scholar]
- 22.Heinemeyer W, Kleinschmidt J A, Saidowsky J, Escher C, Wolf D H. Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress-induced proteolysis and uncover its necessity for cell survival. EMBO J. 1991;10:555–562. doi: 10.1002/j.1460-2075.1991.tb07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hicke L. Ubiquitin-dependent internalization and down-regulation of plasma membrane proteins. FASEB J. 1997;11:1215–1226. doi: 10.1096/fasebj.11.14.9409540. [DOI] [PubMed] [Google Scholar]
- 24.Hiller M M, Finger A, Schweiger M, Wolf D H. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 25.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 26.Hyde S C, Emsley P, Hartshorn M J, Mimmack M M, Gileadi U, Pearce S R, Gallagher M P, Gill D R, Hubbard R E, Higgins C F. Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature. 1990;346:362–365. doi: 10.1038/346362a0. [DOI] [PubMed] [Google Scholar]
- 27.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jedlitschky G, Leier I, Buchholz U, Center M, Keppler D. ATP-dependent transport of glutathione S-conjugates by the multidrug resistance-associated protein. Cancer Res. 1994;54:4833–4836. [PubMed] [Google Scholar]
- 29.Jensen T J, Loo M A, Pind S, Williams D B, Goldberg A, Riordan J R. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 30.Johnston J A, Ward C L, Kopito R R. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones E W, Zubenko G S, Parker R R. PEP4 gene function is required for expression of several vacuolar hydrolases in Saccharomyces cerevisiae. Genetics. 1982;102:665–667. doi: 10.1093/genetics/102.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jungmann J, Reins H-A, Schobert C, Jentsch S. Resistance to cadmium mediated by ubiquitin-dependent proteolysis. Nature. 1993;361:369–371. doi: 10.1038/361369a0. [DOI] [PubMed] [Google Scholar]
- 33.Katzmann D J, Hallstrom T C, Voet M, Wysock W, Golin J, Volckaert G, Moye-Rowley W S. Expression of an ATP-binding cassette transporter-encoding gene (YOR1) is required for oligomycin resistance in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6875–6883. doi: 10.1128/mcb.15.12.6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keppler D, Konig J. Expression and localization of the conjugate export pump encoded by the MRP2 (cMRP/cMOAT) gene in liver. FASEB J. 1997;11:509–516. doi: 10.1096/fasebj.11.7.9212074. [DOI] [PubMed] [Google Scholar]
- 35.Kolling R, Hollenberg C P. The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 1994;13:3261–3271. doi: 10.1002/j.1460-2075.1994.tb06627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leier I, Jedlitschky G, Buchholz U, Cole S P C, Deeley R G, Keppler D. The MRP gene encodes an ATP-dependent export pump for leukotriene C4 and structurally related conjugates. J Biol Chem. 1994;45:27807–27810. [PubMed] [Google Scholar]
- 37.Li Z-S, Szczypka M, Lu Y-P, Thiele D J, Rea P A. The yeast cadmium factor protein (YCF1) is a vacuolar glutathione S-conjugate pump. J Biol Chem. 1996;271:6509–6517. doi: 10.1074/jbc.271.11.6509. [DOI] [PubMed] [Google Scholar]
- 38.Loayza D, Tam A, Schmidt W K, Michaelis S. Ste6p mutants defective in exit from the endoplasmic reticulum reveal aspects of an ER quality control pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:2767–2784. doi: 10.1091/mbc.9.10.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loe D W, Stewart R K, Massey T E, Deeley R G, Cole S P C. ATP-dependent transport of aflatoxin B1 and its glutathione conjugates by the product of the multidrug resistance protein (MRP) gene. Mol Pharmacol. 1997;51:1034–1041. doi: 10.1124/mol.51.6.1034. [DOI] [PubMed] [Google Scholar]
- 40.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 41.Lukacs G L, Mohamed A, Kartner N, Chang X-B, Riordan J R, Grinstein S. Conformational maturation of CFTR but not its mutant counterpart (ΔF508) occurs in the endoplasmic reticulum and requires ATP. EMBO J. 1994;13:6076–6086. doi: 10.1002/j.1460-2075.1994.tb06954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manolson M F, Proteau D, Preston R A, Stenbit A, Roberts B T, Hoyt M A, Preuss D, Mulholland J, Botstein D, Jones E W. The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H+-ATPase. J Biol Chem. 1992;267:14294–14303. [PubMed] [Google Scholar]
- 43.Marcusson E G, Horazdovsky B F, Cereghino J L, Gharakhanian E, Emr S D. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- 44.Michaelis S, Berkower C. Sequence comparison of yeast ATP-binding cassette transporter proteins. Cold Spring Harbor Symp Quant Biol. 1995;60:291–307. doi: 10.1101/sqb.1995.060.01.034. [DOI] [PubMed] [Google Scholar]
- 45.Muller M, Meijer C, Zaman G J R, Borst P, Scheper R J, Mulder N H, de Vries E G E, Jansen P L M. Overexpression of the gene encoding the multidrug resistance-associated protein results in increased ATP-dependent glutathione S-conjugate transport. Proc Natl Acad Sci USA. 1994;91:13033–13037. doi: 10.1073/pnas.91.26.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakano A, Brada D, Schekman R. A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J Cell Biol. 1988;107:851–863. doi: 10.1083/jcb.107.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paddon C, Loayza D, Vangelista L, Solari R, Michaelis S. Analysis of the localization of STE6/CFTR chimeras in a Saccharomyces cerevisiae model for the cystic fibrosis defect CFTRΔF508. Mol Microbiol. 1996;19:1007–1017. doi: 10.1046/j.1365-2958.1996.444973.x. [DOI] [PubMed] [Google Scholar]
- 48.Paulusma C C, Bosma P J, Zaman G J R, Bakker C T M, Otter M, Scheffer G L, Scheper R J, Borst P, Oude Elferink R P J. Congenital jaundice in rats with a mutation in a multidrug resistance-associated protein gene. Science. 1996;271:1126–1128. doi: 10.1126/science.271.5252.1126. [DOI] [PubMed] [Google Scholar]
- 49.Piper P, Mahé Y, Thompson S, Pandjaitan R, Holyoak C, Egner R, Mühlbauer M, Coote P, Kuchler K. The Pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J. 1998;17:4257–4265. doi: 10.1093/emboj/17.15.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plemper R K, Egner R, Kuchler K, Wolf D H. Endoplasmic reticulum degradation of a mutated ATP-binding cassette transporter Pdr5 proceeds in a concerted action of Sec61 and the proteasome. J Biol Chem. 1998;273:32848–32856. doi: 10.1074/jbc.273.49.32848. [DOI] [PubMed] [Google Scholar]
- 51.Pringle J R, Adams A E M, Drubin D G, Haarer B K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 52.Riordan J, Rommens J M, Kerem B-S, Alon N, Rozmahel R, Grzelczack Z, Zielenski J, Lok W, Plavsic N, Chou J-L, Drumm M L, Iannuzzi M C, Collins F S, Tsui L-C. Identification of the cystic fibrosis gene: cloning and characterization of the cDNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 53.Roberg K J, Rowley N, Kaiser C. Physiological regulation of membrane protein sorting late in the secretory pathway of Saccharomyces cerevisiae. J Cell Biol. 1997;137:1469–1482. doi: 10.1083/jcb.137.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 55.Serrano R, Keilland-Brandt M C, Fink G R. Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+ and Ca2+-ATPases. Nature. 1986;319:689–693. doi: 10.1038/319689a0. [DOI] [PubMed] [Google Scholar]
- 56.Sherman F, Fink G, Hicks J. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1979. [Google Scholar]
- 57.Simon S M, Schindler M. Cell biological mechanisms of multidrug resistance in tumors. Proc Natl Acad Sci USA. 1994;91:3497–3504. doi: 10.1073/pnas.91.9.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stevens T H, Esmon B, Schekman R. Early stages in the yeast secretory pathway are required for the transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- 59.Stirling C J, Rothblatt J, Hosobuchi M, Deshaies R, Schekman R. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol Biol Cell. 1992;3:129–142. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szczypka M S, Wemmie J A, Moye-Rowley W S, Thiele D J. A yeast metal resistance protein similar to human CFTR and multidrug resistance-associated protein. J Biol Chem. 1994;269:22853–22857. [PubMed] [Google Scholar]
- 61.Teem J L, Berger H A, Ostedgaard L S, Rich D P, Tsui L-C, Welsh M J. Identification of revertants for the cystic fibrosis ΔF508 mutation using STE6-CFTR chimeras in yeast. Cell. 1993;73:335–346. doi: 10.1016/0092-8674(93)90233-g. [DOI] [PubMed] [Google Scholar]
- 62.Tsui L-C. The spectrum of cystic fibrosis mutations. Trends Genet. 1992;8:392–398. doi: 10.1016/0168-9525(92)90301-j. [DOI] [PubMed] [Google Scholar]
- 63.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 64.Walker J E, Saraste M, Runswick M J, Gay N. Distantly related sequences in the alpha and beta subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;8:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ward C L, Omura S, Kopito R R. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 66.Welsh M J, Smith A E. Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell. 1993;73:1251–1254. doi: 10.1016/0092-8674(93)90353-r. [DOI] [PubMed] [Google Scholar]
- 67.Wemmie J A, Moye-Rowley W S. Mutational analysis of the Saccharomyces cerevisiae ATP-binding cassette transporter protein Ycf1p. Mol Microbiol. 1997;25:683–694. doi: 10.1046/j.1365-2958.1997.5061868.x. [DOI] [PubMed] [Google Scholar]
- 68.Werner E D, Brodsky J L, McCracken A A. Proteasome-dependent endoplasmic reticulum degradation: an unconventional route to a familiar fate. Proc Natl Acad Sci USA. 1996;93:13797–13801. doi: 10.1073/pnas.93.24.13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu A, Wemmie J A, Edgington N P, Goebl M, Guevara J L, Moye-Rowley W S. Yeast bZIP proteins mediate pleiotropic drug and metal resistance. J Biol Chem. 1993;268:18850–18858. [PubMed] [Google Scholar]