Saccharomyces cerevisiae Mid2p Is a Potential Cell Wall Stress Sensor and Upstream Activator of the PKC1-MPK1 Cell Integrity Pathway (original) (raw)
Abstract
The MID2 gene of Saccharomyces cerevisiae encodes a protein with structural features indicative of a plasma membrane-associated cell wall sensor. MID2 was isolated as a multicopy activator of the Skn7p transcription factor. Deletion of MID2 causes resistance to calcofluor white, diminished production of stress-induced cell wall chitin under a variety of conditions, and changes in growth rate and viability in a number of different cell wall biosynthesis mutants. Overexpression of MID2 causes hyperaccumulation of chitin and increased sensitivity to calcofluor white. α-Factor hypersensitivity of _mid2_Δ mutants can be suppressed by overexpression of upstream elements of the cell integrity pathway, including PKC1, RHO1, WSC1, and WSC2. Mid2p and Wsc1p appear to have overlapping roles in maintaining cell integrity since _mid2Δ wsc1_Δ mutants are inviable on medium that does not contain osmotic support. A role for MID2 in the cell integrity pathway is further supported by the finding that MID2 is required for induction of Mpk1p tyrosine phosphorylation during exposure to α-factor, calcofluor white, or high temperature. Our data are consistent with a role for Mid2p in sensing cell wall stress and in activation of a response that includes both increased chitin synthesis and the Mpk1p mitogen-activated protein kinase cell integrity pathway. In addition, we have identified an open reading frame, MTL1, which encodes a protein with both structural and functional similarity to Mid2p.
The cell wall is an essential organelle in fungal species. In Saccharomyces cerevisiae it is composed of four polysaccharide polymer classes: β-1,3-glucan, β-1,6-glucan, mannan, and chitin. The functions provided by the yeast cell wall include the determination of cell shape, protection of osmotic integrity, and scaffolding for extracellular proteins important for nutrient uptake and agglutination between mating partners. Recent research has emphasized the dynamic nature of this structure (see references 10 and 37 for reviews), which undergoes major changes in shape and composition during the vegetative budding cycle and the alternative developmental pathways of mating and sporulation.
Stress can lead to alteration of polymer levels in the yeast cell wall. This effect is perhaps best documented for chitin. Schekman and Brawley (44) noted that during shmoo formation, a process during which the cell wall is rapidly remodeled, additional chitin is synthesized and deposited at the base and neck region of the mating projection. Roncero and Duran (43) noted that exposure to calcofluor white, a substance which binds primarily chitin in the yeast cell wall, interferes with proper wall synthesis and induces elevated chitin synthesis in vivo. Recently, Popolo et al. (39) and Ram et al. (41) observed that mutational disruption of cell wall biosynthesis caused increased chitin deposition. This finding led to the proposal that alterations in cell wall assembly cause yeast to engage a compensation mechanism that includes increased chitin synthesis.
The PKC1-MPK1 signal transduction pathway plays an essential role in maintaining the integrity of the cell wall during both mating and vegetative growth. Igual et al. (20) showed that at least part of this influence on cell wall construction is the result of control of transcription of a variety of genes involved in cell wall biosynthesis. Pkc1p, a serine/threonine protein kinase (28), serves to stimulate a mitogen-activated protein (MAP) kinase cascade comprised of Bck1p (Slk1p) (11, 26, 29), Mkk1p/Mkk2p (21), and Mpk1p (Slt2p) (27). Mutation of components in the PKC1-MPK1 pathway have a range of effects, such as cell lysis, caffeine sensitivity, cell cycle progression defects, and defective cytoskeletal organization. The molecular basis of yeast Pkc1p stimulation is not yet fully understood; however, the GTP-bound form of the small G-protein, Rho1p, has been shown to physically associate with Pkc1p, resulting in Pkc1p activation (14, 24, 35).
Studies of the extracellular matrix of mammalian cells, a structure analogous to the yeast cell wall, have revealed a class of protein receptors known as integrins. Integrins possess a large extracellular domain, a single membrane-spanning region, and a short cytoplasmic domain. Activation of protein kinases and small GTP-binding proteins such as RhoAp by integrins affects cell adhesion, cellular ion levels, and polarized growth. Recently, Bickle et al. (3) have proposed that disturbances in the cell wall cause activation of the Rho1p GTPase via the Rom2p exchange factor in a manner analogous to integrin signaling. Although the proteome of S. cerevisiae does not include integrin homologs, there are a number of cell surface proteins topologically resembling integrins that could potentially carry out equivalent extracellular sensing/intracellular signaling processes. These proteins, usually type I in orientation, contain large extracellular regions rich in serine and threonine residues, single transmembrane domains, and relatively small cytoplasmic regions.
WSC1 (HCS77/SLG1) encodes a protein with sensor/signaler-like characteristics that has recently been identified as an upstream activator of Pkc1p (19, 22, 47). Three homologous proteins, encoded by the WSC2, WSC3, and WSC4 genes, appear to have overlapping activity with Wsc1p since their deletion can increase the severity of _wsc1_Δ phenotypes (47). _wsc_Δ mutants display characteristics of cells with decreased Pkc1p activity, namely, reduced growth rate and sorbitol-suppressible, temperature-dependent cell lysis. Overexpression of genes presumably downstream of the WSC family, such as RHO1 and PKC1, can suppress some of the effects of WSC gene deletions. The mechanism by which Wsc proteins stimulate Pkc1p activity is not yet known, but it has been suggested that Wsc1p might somehow regulate Rho1p activity and thereby affect downstream targets of Rho1p such as Pkc1p (2).
Mid2p, although not a member of the WSC family, is another potential extracellular sensor that has been identified as a participant in a number of cellular processes. In wild-type MATa cells, transcription of MID2 increases severalfold in response to α-factor and cells lacking Mid2p die during exposure to α-factor (36). Multicopy MID2 has been found to suppress a variety of mutant phenotypes, including the temperature sensitivity of _mpt5_Δ mutants (46), growth in profilin (_pfy1_Δ)-deficient cells (32), and temperature-sensitive growth in _cik1_Δ and _kar3_Δ mutants (31). Additionally, KAI1, an internal fragment of MID2 was identified as a multicopy inhibitor of excessive protein kinase A (TPK1) activity (12).
We identified MID2 as a high-copy-number activator of the Skn7p transcription factor. A relationship between Mid2p and the cell wall is suggested by a number of genetic interactions between MID2 and cell wall biosynthesis genes. Alteration of MID2 gene dosage affects stress-related cell wall chitin deposition, suggesting that MID2 is partly required for induction of cell wall stress-induced chitin synthesis. Furthermore, genetic interactions between MID2 and elements of the PKC1-MPK1 pathway, as well as a requirement for MID2 during induction of Mpk1p tyrosine phosphorylation under a variety of stress conditions, together suggest a role for Mid2p upstream of the PKC1-MPK1 cell wall integrity pathway.
MATERIALS AND METHODS
Plasmids, strains, and gene deletion constructs.
Oligonucleotides used in this study are listed in Table 1. Yeast strains used in this study are listed in Table 2. The MID2 locus, contained within a 2.45-kb _Nhe_I-_Xho_I genomic DNA fragment, was subcloned into pBluescript II (pBSII) SK+ at compatible _Xba_I and _Sal_I restriction sites. A 2.5-kb _Kpn_I-_Sst_I fragment containing MID2 was excised from this plasmid and then inserted into pRS316, pRS425, and pRS426 at corresponding _Kpn_I-_Sst_I sites in the polylinker. Deletion of the entire MID2 open reading frame (ORF) was accomplished by integration of a mid2Δ::KANMX2 cassette (48). mid2Δ::KANMX2 was generated by using the oligonucleotides mid2Δup and mid2Δdown. Correct insertion of the cassette and deletion of the reading frame was confirmed by PCR analysis of yeast colonies using the oligonucleotides mid2Δtest and KANMX2 internal. Deletion of WSC1 was accomplished by replacement of the WSC1 reading frame with a wsc1Δ::KANMX2 cassette generated by the oligonucleotides wsc1Δup and wsc1Δdown. Integration was confirmed by using the KANMX2 internal test oligonucleotide and the wsc1Δtest oligonucleotide. Deletion of RHO1 was accomplished by using a GFP-HIS3 cassette (34) and the oligonucleotides rho1Δup and rho1Δdown to create rho1Δ::GFP-HIS3. Correct integration of the deletion cassette into the SEY62102 (diploid) strain was confirmed by using the oligonucleotides rho1Δtest and GFP-HIS3 internal; 2:2 segregation of viable and inviable meiotic products was observed after dissection of tetrads derived from heterozygous rho1::GFP-HIS3 RHO1 diploids. Deletion of PKC1 was achieved by using the oligonucleotides pkc1Δup and pkc1Δdown to generate the pkc1Δ::GFP-HIS3 cassette. Integration of the PCR product was confirmed by using the oligonucleotides pkc1Δtest and GFP-HIS internal. Deletion of MTL1 was accomplished by replacement of the MTL1 reading frame with a mtl1Δ::GFP-HIS3 cassette generated by the nucleotides mtl1Δup and mtl1Δdown. Correct integration was confirmed by using the GFP-HIS3 internal and mtl1Δtest nucleotides.
TABLE 1.
Oligonucleotide sequences
| Name | Sequence 5′-3′a |
|---|---|
| mid2Δup | GCAGTATCTACTGCACGTTCTTCCGTAAGTAGAGTTAGTTCTGATATCAAGCTTGCCTCG |
| mid2Δdown | TGTTTGGCGTTTGGTAATACGCTATCGCTATCTCTTATTCTGGTCGACACTGGATGGCGG |
| mid2Δtest | GCCTTCAATGAGTTCCAC |
| KANMX2 internal | CAACAGGCCAGCCATTAC |
| wsc1Δup | ATGAGACCGAACAAAACAAGTCTGCTTCTGGCGTTATTATCCGATATCAAGCTTGCCTCG |
| wsc1Δdown | TGGATTGACCACTGTTAAAACGTTGTTTTTCCCTCCTGGTCCGTCGACACTGGATGGCGG |
| wsc1Δtest | CGATACAGTAAACTCGAC |
| HA-insert | GAATTATCACCACGAAATTATTACCCATACGACGTCCCAGACTACGCTTAATCATATCCATTCATATC |
| EcoRI Insert | GCAATAAATCCAAGAATTCGGGTCTTTC |
| KpnI Insert | GGTAACGAATTATCACCACGAAAGGTACCATCATATCCATTCATATCATTTAG |
| rho1Δup | ATGTCACAACAAGTTGGTAACAGTATCAGAAGAAAGCTGGTAATCATGAGTAAAGGAGAAGAAC |
| rho1Δdown | CTATAACAAGACACACTTCTTCTTCTTCTTTTCAGTAGTGTTCTTGCGCGCCTCGTTCAGAATG |
| rho1Δtest | CGACCATCGATCATTCCT |
| rho1 clone for | TAATGCGGTAGCATTGGACA |
| rho1 clone rev | AACCTTCCAACAAAACTGAGG |
| pkc1Δup | ATGAGTTTTTCACAATTGGAGCAGAACATTAAAAAAAAGATAGCCATGAGTAAAGGAGAAGAAC |
| pkc1Δdown | TCATAAATCCAAATCATCTGGCATAAAGGAAAATCCTCTAAACTCGCGCGCCTCGTTCAGAATG |
| pkc1Δtest | CCTGCCAGTGTAATAAGT |
| GFP-HIS3 internal | GTATAGTTCATCCATGCC |
| mtl1 clone rev | CTTGCCTTCTCCAGAGG |
| mtl1 clone for prom. | CCACATCAGAGACTTGGG |
| mtl1 clone for start | GGTTGGTTCGATCTTCGT |
| mtl1Δup | CGGGTTGGTTCGATCTTCGTTTCCACTTTTAACTTACTCCCATGAGTAAAGGAGAAGAAC |
| mtl1Δdown | GGGTGATTTTAAGAAGAAAAGTTATGGCAAAGCTGCTTTCGGCGCGCCTCGTTCAGAATG |
| mtl1Δtest | GGTAGAAAAGTGTAGATG |
TABLE 2.
Strains used
| Strain | Genotype |
|---|---|
| L40 | MATa his3 trp1 leu2 ade2 LYS2::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ GAL4) |
| TK60 | L40/pBTM116-LEXA-SKN7/pRS425 |
| TK61 | L40/pBTM116-LEXA-SKN7/pRS425-MID2 |
| SEY62102 | MATα/MATa leu2/leu2 ura3/ura3 his3/his3 lys2/lys2 trp1/trp1 suc2/suc2 |
| SEY6210 | MATα leu2 ura3 his3 lys2 trp1 suc2 |
| SEY6210a | MATa leu2 ura3 his3 lys2 trp1 suc2 |
| TK82 | SEY6210a/pRS426 |
| TK83 | SEY6210a/pRS426-MID2 |
| TK84 | SEY6210a/pRS426-MID2-HA |
| TK85 | SEY6210a/pRS426-Δ_S/T-MID2-HA_ |
| TK86 | SEY6210a/pVT101U |
| TK87 | SEY6210a/pVT101U-MID2 |
| TK88 | SEY6210a mid2Δ::KANMX2 |
| TK89 | SEY6210a mid2Δ::KANMX2/pRS425 |
| TK90 | SEY6210a mid2Δ::KANMX2/pRS425-WSC1 |
| TK91 | SEY6210a mid2Δ::KANMX2/pRS425-WSC2 |
| TK92 | SEY6210a mid2Δ::KANMX2/pRS425-RHO1 |
| TK93 | SEY6210a mid2Δ::KANMX2/pFL44 |
| TK94 | SEY6210a mid2Δ::KANMX2/pFL44-MPK1-HA |
| TK95 | SEY6210a/pRS425 |
| TK96 | SEY6210a/pFL44 |
| TK97 | SEY6210a/pFL44-MPK1-HA |
| TK98 | SEY6210a/pRS426-MID2-GFP |
| TK99 | SEY6210a chs3Δ::LEU2/pVT101U |
| TK100 | SEY6210a chs3Δ::LEU2/pVT101U-MID2 |
| TK101 | SEY62102_kre6Δ::HIS3/KRE6 mid2Δ::KANMX2/MID2_ |
| TK102 | SEY62102_kre9Δ::HIS3/KRE9 mid2Δ::KANMX2/MID2_ |
| TK103 | SEY62102_fks1Δ::HIS3/FKS1 mid2Δ::KANMX2/MID2_ |
| TK104 | SEY62102_wsc1Δ::KANMX2 mid2Δ::KANMX2/MID2_ |
| TK105 | SEY62102_mtl1Δ::GFP-HIS3 mid2Δ::KANMX2/MID2_ |
The RHO1 and MTL1 genes were amplified from SEY6210 genomic DNA by using Expand polymerase (Boehringer Mannheim) and the oligonucleotides rho1 clone for and rho1 clone rev to generate RHO1 and mtl1 clone rev and either mtl1 clone for prom or mtl1 clone for start to generate clones of MTL1 containing 916 nucleotides of promoter sequence or a promoterless clone with only 65 nucleotides 5′ of the ATG codon, which was used for fusion to the ADH1 promoter.
Mid2p-HA was created by the insertion of a single copy of the hemagglutinin (HA) epitope (YPYDVPDYA) between residues 375 and 376 via site-directed mutagenesis (Kunkel method [25]) on MID2 contained in pBSII KS−, using the oligonucleotide HA-insert. Fidelity of incorporation of the HA epitope was confirmed by DNA sequencing. A _Kpn_I-_Sst_I fragment containing MID2-HA was then subcloned into pRS316 and pRS426 at corresponding _Kpn_I and _Sst_I sites. Mid2p-HA was demonstrated to be functional by the observation that pRS316-MID2-HA was fully able to complement a mid2Δ mutant in a test for hypersensitivity to α-factor (Sigma).
Removal of the serine/threonine-rich region was accomplished by generating an _Eco_RI restriction site by modification of the sequence AAGTTC (nucleotides 641 to 646 in the MID2 reading frame) to GAATTC via site-directed mutagenesis using the oligonucleotide EcoRI Insert. A second _Eco_RI restriction site occurs naturally at positions 102 to 107 in the reading frame. This construct was then digested with Eco_RI, releasing a 540-bp fragment coding for the bulk of the serine/threonine-rich region. The remaining plasmid fragment was gel purified (Pharmacia) and religated, creating the continuous in-frame ORF Δ_S/T-MID2.
Western blots and membrane association tests.
For membrane association tests, total cell extracts were prepared from mid-log-phase cultures grown in selective medium by vigorous vortexing in lysis buffer (50 mM Tris [pH 7.5], 1 mM EDTA, 5% glycerol) in the presence of glass beads and protease inhibitors (Complete protease inhibitor cocktail; Boehringer Mannheim). The resulting slurry was centrifuged at 2,500 × g at 4°C for 5 min to remove cell walls and unbroken cells. The resulting supernatant was then divided, and the individual aliquots were subjected to either centrifugation at 15,000 × g at 4°C for 30 min or treatment with sodium chloride, sodium carbonate, urea, or Triton X-100 and then subjected to centrifugation at 60,000 × g at 4°C for 30 min. Postcentrifugation, supernatants were withdrawn, and pellets were resuspended in a volume of lysis buffer equal to the supernatant. Samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then subjected to Western blotting. Immunodetection of Mid2p-HA was achieved by using anti-HA monoclonal antibody 12CA5 (Babco) at 1:1,000 dilution and horseradish peroxidase-conjugated anti-mouse secondary antibody (Amersham Life Sciences) at a 1:1,000. Bands were visualized using by enhanced chemiluminescence (Amersham Life Sciences). For other SDS-PAGE and Western blotting procedures, total cell lysates were prepared with lysis buffer (2% Triton-X100, 1% SDS, 100 mM NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA).
Localization of Mid2p.
A MID2-GFP fusion was generated by modifying the coding sequence of MID2 (contained in pBSII KS−) immediately upstream of the TAA stop codon (TTATTA) via site-directed mutagenesis to a _Kpn_I restriction site (GGTACC), using the oligonucleotide KpnI Insert. Clones positive for the incorporation of the _Kpn_I site were then confirmed by sequencing. Creation of an in-frame fusion of MID2 to GFP (F64L S65T; kindly provided by U. Stochaj) was accomplished by a three-way ligation involving pRS426 or pRS316 (with _Xho_I/_Eco_RI ends) MID2 (with _Xho_I/_Kpn_I ends) and GFP (with _Kpn_I/_Eco_RI ends). Correct orientation of the ligation products was confirmed by diagnostic restriction digests. Function of the MID2-GFP fusion was demonstrated by observation that pRS316-MID2-GFP fully complements _mid2_Δ mutants for α-factor hypersensitivity. Localization of Mid2p-GFP (green fluorescent protein) was accomplished by examination of live, mid-log-phase _mid2_Δ cells carrying pRS316-MID2-GFP and pRS426-MID2-GFP.
Calcofluor white tests.
To test strains for sensitivity to calcofluor white, mid-log-phase cells were diluted, then spotted either onto rich YEPD agar plates containing the indicated amount of calcofluor white or onto selective medium buffered with 10 g of morpholineethanesulfonic acid per liter and adjusted to pH 6.2. Plates were incubated for 48 to 72 h at 30°C in a dark environment.
Multicopy suppression of α-factor-induced death.
Cultures of cells containing either control plasmids or plasmids bearing genes of interest were diluted to 3 × 104 cells/ml in buffered (85 mM succinic acid, 19 mM sodium hydroxide) YNB liquid medium containing appropriate amino acids. Triplicate aliquots of each strain were removed and spread on selective solid medium. α-Factor was then added to liquid cultures to a final concentration of 1 μM, and these cultures were incubated at 30°C for 330 min before a second series of aliquots were removed and plated. Petri dishes were incubated for 48 h at 30°C, resulting in the formation of colonies derived from single cells. The number of colonies on each petri dish was counted, and survival of α-factor exposure was measured by the difference between the number of colonies on pre- and post-α-factor petri dishes for each strain.
Chitin assays.
Total cellular chitin was measured essentially as described by Bulawa et al. (9) and outlined here (8a). Washed cells (∼50 mg [wet weight]) were suspended in 500 μl of 6% KOH and incubated at 80°C for 90 min. After cooling to room temperature, 50 μl of glacial acetic acid was added. Insoluble material was washed twice with water and resuspended in 250 μl of 50 mM sodium phosphate (pH 6.3); 2 mg of Streptomyces griseus chitinase (Sigma) was added, and tubes were incubated at 25°C with gentle agitation for 2 h. Tubes were centrifuged at 15,000 × g for 5 min at room temperature, and 250 μl of supernatant was transferred to a fresh tube to which 1 mg of Helix pomatia β-glucuronidase (Sigma) was added. Tubes were incubated at 37°C for 2 h with gentle agitation and then assayed for _N_-acetylglucosamine content.
Measurement of Mpk1p-HA phosphotyrosine content.
Mid-log-phase cultures of cells expressing Mpk1p-HA or a vector-only control were exposed to calcofluor white (40 μg/ml for 30 min), α-factor (4 μM for 3 h), or high temperature (37°C for 3 h); 1 ml of culture was centrifuged (15,000 × g, 30 s, room temperature), the supernatant was aspirated, and the cell pellet was resuspended in 50 μl of 1× SDS loading buffer and boiled for 4 min. Cell debris was pelleted by centrifugation, and 10 μl of supernatant per sample was subjected to SDS-PAGE and Western blotting. Tyrosine phosphorylation of proteins was visualized by incubation of blots with the phosphotyrosine-specific antibody 4G10 (gift from J. Lee) at 1:3,300 dilution and anti-mouse horseradish peroxidase-conjugated secondary antibody at 1:2,000. Blots were then stripped and reprobed with the anti-HA monoclonal antibody 12CA5 to verify equal loading of Mpk1p-HA in each lane.
RESULTS
MID2 stimulates Skn7p transcriptional activity.
Skn7p, a transcription factor containing a region of homology to bacterial two-component response-regulator proteins, was isolated by Brown et al. (6) as a high-copy-number suppressor of growth defects in _kre9_Δ mutants. A screen was performed to identify genes which, when overexpressed, would stimulate the Skn7p-LexA-dependent transcription of a (lexAop)4-HIS3 reporter (38). In this procedure, the L40 reporter strain carrying Skn7p-LexA was transformed with a Yep13-based multicopy genomic bank. Plasmid clones were extracted from colonies which could grow on synthetic medium lacking histidine and including 10 mM 3-amino-1,2,4-triazole (3AT) to squelch His3p activity resulting from basal transcription of HIS3.
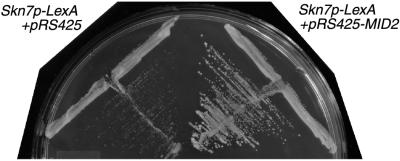
Ten groups of activator of SKN7 (ASK) clones, ASK1 to ASK10, were identified. Sequence analysis revealed that ASK5, ASK7, and ASK9 contained a common gene, YLR332W (MID2). We subcloned the MID2, gene including 627 nucleotides 5′ of the codon for the start methionine and 737 nucleotides 3′ of the stop codon, into pRS425 and directly tested the ability of MID2 to stimulate Skn7p-LexA transcriptional activity. Multicopy MID2 is able to strongly induce Skn7p-LexA activity compared to a vector-only control, permitting reporter strain growth on medium lacking histidine and containing 30 mM 3AT (Fig. 1). This effect is dependent on an interaction between MID2 and LEXA-SKN7 since overexpression of MID2 alone does not activate transcription of the reporter (not shown). We also measured the induction of β-galactosidase activity from the (lexAop)8-lacZ locus and found that multicopy MID2 was able to induce a 7- to 10-fold, Skn7-LexA-dependent increase in β-galactosidase activity (not shown).
FIG. 1.
Multicopy MID2 activates Skn7p-LexA-dependent transcription of HIS3, which allows growth on medium lacking histidine. Reporter strains containing Skn7p-LexA and either pRS425 (TK60) or pRS425-MID2 (TK61) were grown on selective medium lacking histidine and containing 30 mM 3AT.
Characterization and subcellular localization of Mid2p.
MID2 is predicted to encode a type I membrane-spanning protein containing an N-terminal secretion signal sequence followed by a domain of approximately 200 amino acids consisting of ∼62% serine and threonine residues. The C-terminal third of the protein contains a single predicted transmembrane domain and a short charged domain rich in aspartic acid residues, suggested to resemble a calcium binding domain (36). Although Mid2p has overall structural similarity to members of the WSC family, there are two important differences. Firstly, Mid2p does not contain an extracellular cysteine-rich motif that is characteristic of the Wsc proteins. Second, apart from the repetitive serine/threonine-rich region, there is no statistically significant amino acid residue sequence similarity between Mid2p and Wsc proteins.
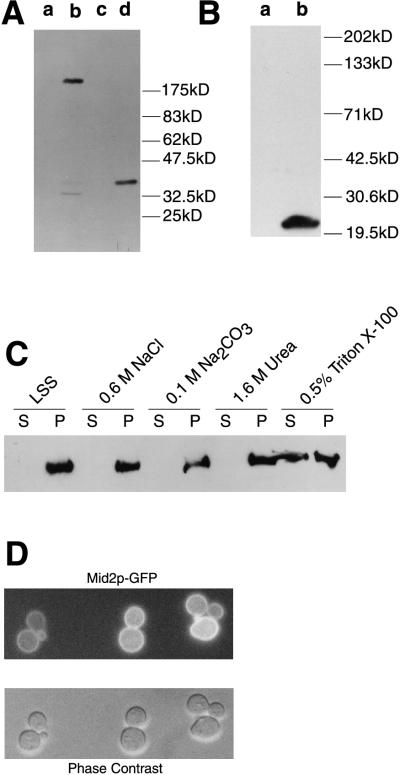
To examine physical characteristics of Mid2p, a functional HA-tagged protein (Mid2p-HA) was generated. After proteolytic processing of the signal peptide, Mid2p-HA is predicted to have a molecular mass of approximately 39 kDa. However, Mid2p-HA migrates with an apparent molecular mass of approximately 200 kDa on SDS-PAGE (Fig. 2A, lane b). The predicted type I orientation of Mid2p suggests that if Mid2p were plasma membrane localized, the serine/threonine-rich region would reside on the exterior face of the cell. Since serine/threonine-rich regions of extracellular protein domains can receive O-linked mannosylation as the protein travels through the secretory pathway (45), we examined Mid2p for evidence of this modification. When isolated from _pmt1_Δ _pmt2_Δ mutants (deficient in O-linked mannosylation [30]), Mid2p-HA migrates on SDS-PAGE close to the predicted size of 39 kDa (Fig. 2A, lane d), displaying a shift of roughly 160 kDa compared to Mid2p-HA expressed in wild-type cells. To verify that it was the serine/threonine domain that was the recipient of the O-mannosylation, we excised this region from Mid2p-HA, generating ΔS/T Mid2p-HA. On SDS-PAGE, ΔS/T Mid2p-HA also migrates near its predicted molecular mass of 23 kDa (Fig. 2B), strongly suggesting that the serine/threonine-rich region is the recipient site of O-linked mannosylation. ΔS/T Mid2p-HA localizes to the same subcellular location as Mid2p-HA (not shown; see below); however, pRS316-ΔS/T Mid2p-HA is unable to complement a _mid2_Δ mutant for sensitivity to α-factor, indicating that Mid2p requires the serine-threonine rich domain for activity. Together, these results suggest that extensive, functionally important modification of Mid2p occurs on the extracellular serine/threonine-rich region.
FIG. 2.
Cell biology of Mid2p. (A) Immunoblot analysis of cell extracts from TK82 (vector only) (lane a), TK84 (MID2-HA) (lane b), _pmt1Δ pmt2_Δ (vector only) (lane c), and _pmt1Δ pmt2_Δ (MID2-HA). (B) Immunoblot analysis of cell extracts from TK82 (vector only) (lane a) and TK85 (ΔS/T-Mid2p-HA) (lane b). (C) Immunoblot analysis of cell fractions from TK84 to demonstrate membrane association of Mid2p. LSS, low-speed-spin pellet fraction. (D) In cells expressing pRS426-MID2-GFP (TK98), Mid2p-GFP is localized to the cell periphery.
To verify the prediction that Mid2p is an integral membrane protein, partially purified extracts from cells expressing Mid2p-HA were fractionated into supernatant (soluble) and pellet (membrane-containing) portions by centrifugation. Mid2p-HA is found exclusively in the low-speed (15,000 × g)-spin pellet fraction, implying membrane association (Fig. 2C). To test whether this membrane association was peripheral or integral, partially purified, membrane-containing cell extracts were treated prior to ultracentrifugation with sodium chloride, sodium carbonate, or urea to disrupt peripheral interactions or with Triton X-100 to disrupt integral membrane association. Only Triton X-100 was capable of solubilizing a significant proportion of Mid2p-HA (Fig. 2C), strongly suggesting that Mid2p-HA is an integral membrane protein. There does not appear to be a fraction of Mid2p-HA covalently associated with the cell wall since treatment of purified cell wall fractions with β-1,3-glucanase (either laminarinase or Quantazyme) before solubilization of proteins by treatment with SDS does not release any detectable Mid2p-HA (not shown).
Direct immunofluorescence microscopy was performed to establish the subcellular localization of Mid2p. A functional Mid2p-GFP fusion protein was constructed by inserting GFP immediately upstream of the MID2 stop codon. Examination of fluorescing cells maintaining either centromeric (not shown) or multicopy (Fig. 2D) Mid2p-GFP reveals Mid2p-GFP distribution to be largely confined to the periphery of cells, consistent with a plasma membrane localization. Expression of native GFP alone produces a diffuse fluorescence pattern throughout the cell (not shown). Indirect immunofluorescence of cells expressing Mid2p-HA revealed an identical pattern of staining (not shown). No polarized localization of Mid2p-GFP to specific regions of the surface such as the bud tip or bud neck was detected; however, approximately 20% of α-factor-treated cells (n = 100) display faint preferential staining at the subapical region of the mating projection (not shown).
Interactions between MID2 and cell wall biosynthesis genes.
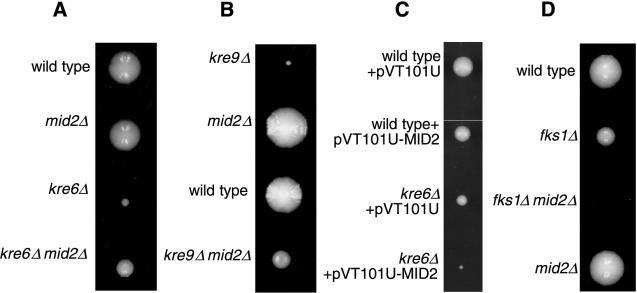
Since the O-mannosylated, extracellular domain of Mid2p is predicted to be oriented toward the cell wall, we explored a possible relationship between Mid2p and the cell wall by searching for genetic interactions between MID2 and genes known to be involved in cell wall construction. Deletion of MID2 in two viable but slow-growing β-1,6-glucan synthesis mutants, _kre6_Δ (42) and _kre9_Δ (5), partially restores growth rate (Fig. 3A, and B). This effect is not due to differential timing of spore germination since increased growth rate in the absence of MID2 (∼20% reduction in doubling time) is observed for _kre6_Δ cells cultured in liquid medium. Also, reintroduction of MID2 on a centromeric plasmid causes kre6Δ mid2Δ cells and kre9Δ mid2Δ cells to resume slower growth (not shown). Interestingly, high-copy-number expression of MID2 in the _kre6_Δ mutant has a strong negative effect on growth rate (Fig. 3C). Optical density measurement of cell growth in liquid culture revealed that high-copy-number expression of MID2(pRS426-MID2) in _kre6_Δ cells resulted in a ∼300% increase in doubling time compared to a vector-only control. This reduction in growth rate is much more pronounced in _kre6_Δ mutants than in wild-type cells, where doubling time increases by only 15% over a vector-only control for cells carrying pRS426-MID2. Interestingly, high-copy-number expression of MID2 in the growth-deficient pmt1Δ pmt2Δ mutants, where Mid2p is undermannosylated, also results in a reduction in growth rate. This finding suggests that although the serine/threonine-rich domain itself is indispensable for Mid2p activity, extensive O-linked mannosylation of this region may not be totally essential for Mid2p function.
FIG. 3.
Deletion of MID2 has effects on growth of different cell wall mutants. (A) Representative tetratype tetrad from TK101 (_kre6Δ mid2_Δ heterozygous diploid). (B) Representative tetratype tetrad from TK102 (_kre9Δ mid2_Δ heterozygous diploid). (C) Single cells containing the indicated plasmids were placed on selective agar and grown at 30°C for 4 days. Photo is representative of effect seen in three isolates each of three transformations. (D) Representative tetratype tetrad from TK103 (_fks1Δ mid2_Δ heterozygous diploid).
Examination of cell wall glucan content revealed that wild-type and _mid2_Δ cells have no differences in β-1,6- and β-1,3-glucan content. Similarly, _kre6_Δ and _kre6Δ mid2_Δ mutants have comparable levels of both glucan species (not shown), suggesting that at least for the _kre6_Δ mutant, increased growth rate induced by loss of MID2 is not caused by a restoration of β-1,6-glucan synthesis. To determine if MID2 genetically interacts with the β-1,3-glucan synthesis pathway, we examined the consequence of loss of MID2 in _fks1_Δ mutants (13, 17, 40). In contrast to the restorative effect that deletion of MID2 had in β-1,6-glucan mutants, loss of MID2 in _fks1_Δ cells causes inviability (Fig. 3D). This phenotype is not reversible by the addition of 1 M sorbitol or 30 mM calcium chloride (not shown). Overexpression of MID2 in an _fks1_Δ mutant does not result in an inhibition of growth like that seen in _kre6_Δ mutants with high-copy-number MID2 (not shown).
MID2 affects chitin synthesis under stress conditions.
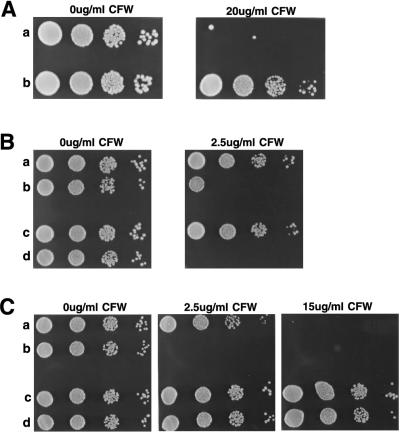
Although chitin comprises a small percentage of the cell wall weight, its contribution to wall integrity is vital. Calcofluor white is a fluorescent dye that intercalates into nascent chitin chains, preventing microfibril assembly (15). At sufficient concentrations, calcofluor white can kill cells through interference with cell wall assembly. _mid2_Δ cells display significant resistance to calcofluor white. At a calcofluor white concentration of 20 μg/ml on rich medium, wild-type cells are killed whereas _mid2_Δ cells can grow without apparent inhibition (Fig. 4A).
FIG. 4.
Dosage of MID2 affects sensitivity to calcofluor white. Mid-log-phase cells were diluted to a concentration of 3 × 106 cells/ml; 5 μl of this suspension and three subsequent 10-fold serial dilutions were each spotted onto the indicated medium. (A) SEY6210a (wild type) (row a) and TK88 (_mid2_Δ) (row b) cells were spotted onto YEPD containing 0 and 20 μg of calcofluor white (CFW) per ml. (B) TK82 [wild type (pRS426)] (row a), TK83 [wild type (pRS426-MID2)] (row b), TK86 [wild type (pVT101U)] (row c), and TK87 [wild type (pVT101U-MID2)] (row d) were spotted on uracil dropout medium containing 0 and 2.5 μg of calcofluor white per ml. (C) TK86 (row a), TK87 (row b), TK99 [_chs3_Δ (pVT101U)] (row c), and TK100 [_chs3_Δ (pVT101U-MID2)] (row d) were spotted onto uracil dropout medium containing 0, 2.5, and 15 μg of calcofluor white per ml.
Since resistance to calcofluor white is a phenotype often associated with defects in chitin synthesis, one possibility is that calcofluor white resistance of _mid2_Δ cells is a consequence of reduced chitin synthesis. Measurement of chitin levels in wild-type and _mid2_Δ cells revealed that they have identical chitin contents when grown in rich (YEPD) medium, suggesting that Mid2p is not required for basal chitin production under such optimum growth conditions (Table 3, condition A). Because exposure to calcofluor white increases chitin production in vivo (43), we tested whether MID2 is required for synthesis of supplemental chitin by measuring the chitin content of cells that had been challenged with sublethal concentrations of calcofluor white. We then calculated the amount of new chitin synthesized in response to calcofluor white challenge by subtracting the amount of chitin produced under nonstressed conditions from the total chitin measured after calcofluor white exposure. Interestingly, _mid2_Δ mutants contained 40% less new, stress-induced, total cell wall chitin than wild-type cells when cultures were grown in the presence of 10 μg of calcofluor white per ml for 2 h prior to harvesting (Table 3, condition B). This attenuation of calcofluor white-induced chitin synthesis is likely to be at least part of the cause of calcofluor white resistance in _mid2_Δ cells.
TABLE 3.
Measurement of total cellular chitin
| Condition | Strain | nmoles of GlcNAc/mg (wt) of cells (mean ± SD) | Increase from control (nmol of GlcNAc/mg [wet wt] of cells) | _MID2_-dependent change (%) |
|---|---|---|---|---|
| A (YEPD [control]) | Wild type | 3.54 ± 0.06 | ||
| _mid2_Δ | 3.60 ± 0.31 | |||
| B (YEPD + calcofluor white [10 μg/ml], 2 h) | Wild type | 8.96 ± 0.16 | 5.42 | |
| _mid2_Δ | 6.82 ± 0.14 | 3.22 | −40 | |
| C (YEPD) | _kre6_Δ | 9.50 ± 0.43 | 5.96 (from wild type) | |
| _kre6Δ mid2_Δ | 7.90 ± 0.08 | 4.30 (from _mid2_Δ) | −28 | |
| D (YEPD + 4 μM α-factor, 3 h) | Wild type | 5.05 ± 0.19 | 1.51 | |
| _mid2_Δ | 3.91 ± 0.05 | 0.31 | −79 | |
| E (YNB [uracil dropout]) | Wild type carrying pVT101U | 4.65 ± 0.65 | ||
| Wild type carrying pVT101U-MID2 | 11.87 ± 0.22 | +255 |
To determine if Mid2p is required for supplementary chitin synthesis under a broader range of cell wall stresses, we looked for Mid2p-dependent changes in cellular chitin content in two other circumstances. It has been observed that cell wall mutants typically have higher cell wall chitin levels than wild-type cells (39, 41). Analysis of chitin content revealed that _kre6_Δ cells have >2.5-fold more total chitin than wild-type cells. Similar to the effect seen in calcofluor white-challenged cells, loss of MID2 causes a small but significant decrease (∼28%) in extent of stress-induced chitin synthesis in _kre6_Δ mutants (Table 3, condition C). Another situation known to increase chitin production is shmoo formation in response to mating pheromone (44). After induction of projection formation by α-factor, MATa _mid2_Δ mutants had synthesized almost 80% less new chitin than wild-type cells (Table 3, condition D). These observations suggest that Mid2p is partially required for production of supplementary wall chitin under conditions of wall damage (by mutation or calcofluor white) or morphological change (shmoo formation).
To further address the relationship between MID2 and chitin synthesis, growth of cells overexpressing MID2 was examined on calcofluor white-containing medium. Cells harboring MID2 on a 2 μm plasmid have reduced viability at a calcofluor white concentration of 2.5 μg/ml, and cells expressing MID2 from the strong constitutive ADH1 promoter at 2 μm levels are completely inviable on calcofluor white at 2.5 μg/ml (Fig. 4B). Since calcofluor white resistance in _mid2_Δ cells is associated with reduced supplementary chitin synthesis, this finding suggests that overexpression of MID2 may cause an increase in wall chitin content, conferring hypersensitivity to this drug. Indeed, analysis of total chitin revealed that cells carrying ADH1 promoter-driven MID2 had approximately 250% more chitin than cells without multicopy MID2 (Table 1, condition E).
Because Chs3p is responsible for the bulk of lateral wall and bud scar chitin, and deletion of CHS3 leads to a 10-fold reduction in total cellular chitin content and strong resistance to calcofluor white, we next tested whether the hypersensitivity to calcofluor white caused by MID2 overexpression is mediated through Chs3p activity. Spot testing of _chs3_Δ cells containing multicopy MID2, with either its own or the ADH1 promoter, on calcofluor white-containing medium revealed that CHS3 is required for high-copy-number MID2 to confer calcofluor white hypersensitivity (Fig. 4C). This finding suggests that Mid2p may ultimately affect extra chitin synthesis through some form of regulation of Chs3p activity.
MID2 interacts with the cell integrity pathway.
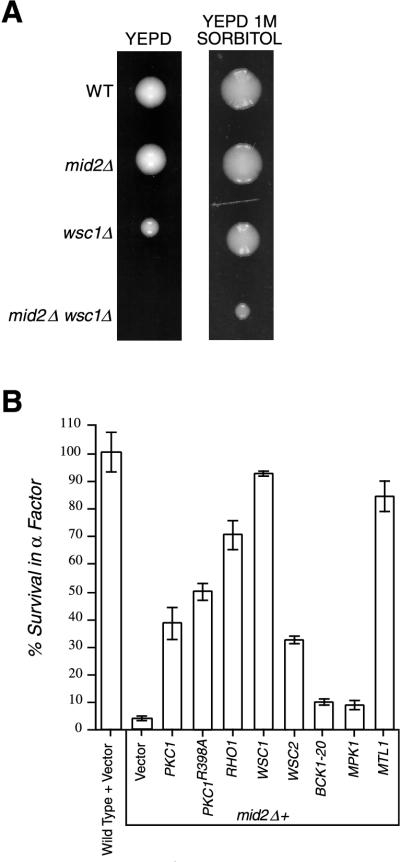
To identify additional MID2 interactors, we performed a screen for multicopy suppressors of the _mid2Δ fks1_Δ synthetic lethality [plasmids were selected for the ability to promote growth in _mid2Δ fks1_Δ (pRS316-FKS1) cells after loss of pRS316-FKS1 was promoted by replication onto medium containing 5-fluoro-orotic acid]. Two of the genes identified as _mid2Δ fks1_Δ suppressors were PKC1 and WSC1. Several genetic interactions between MID2 and WSC1 suggest that these genes possess overlapping activities. First, _mid2Δ wsc1_Δ cells are inviable at 22 or 30°C on YEPD medium. Inclusion of sorbitol (between 0.3 and 1 M) in medium permits partial restoration of growth, suggesting that _mid2Δ wsc1_Δ mutants, like _pkc1_Δ mutants, are prone to lysis without osmotic support (Fig. 5A). When transferred from osmotically supported medium to YEPD, _mid2Δ wsc1_Δ mutants arrest growth with a small bud. After 36 h on YEPD, approximately 85% of _mid2Δ wsc1_Δ cells have small buds, while between 6 and 8% of wild-type, _mid2_Δ and _wsc1_Δ cells display a small bud (more than 150 cells counted per genotype). Further suggesting a functional relationship between MID2 and WSC1, high-copy-number expression of MID2 partially suppresses the growth defect of _wsc1_Δ mutants (not shown). Finally, overexpression of WSC1 is able to relieve the sensitivity of _mid2_Δ cells to α-factor (Fig. 5B). Together, these findings indicate a functional overlap between Mid2p and Wsc1p.
FIG. 5.
Genetic interactions between MID2 and members of the cell integrity pathway. (A) Representative tetratype tetrads of TK104 (_mid2Δ wsc1_Δ heterozygous diploid) dissected onto either YEPD or YEPD plus 1 M sorbitol. YEPD plates were incubated for 60 h at 30°C, YEPD–1 M sorbitol plates were incubated for 80 h at 30°C. WT, wild type. (B) Members of the cell integrity pathway suppress α-factor-induced death in _mid2_Δ mutants. Wild-type cells containing pRS426 vector only and _mid2_Δ mutants carrying pRS426, YEP13-PKC1, pBM743-PKC1R398A (GAL-driven, hyperactive PKC1), pRS426-RHO1, pRS426-WSC1, pRS426-WSC2, pRS316-BCK1-20 (hyperactive BCK1), YEP352-MPK1, and pRS425-MTL1 in liquid medium were exposed to α-factor. Percentage of survival was measured by spreading liquid medium before and after α-factor exposure (330 mn) on petri dishes and counting colonies derived from single cells.
Since Wsc1p and its homologs have been implicated in Pkc1p activation, and the _mid2Δ wsc1_Δ small-budded terminal phenotype resembles that of cells lacking Pkc1p activity (28), we examined the possibility that Mid2p can also contribute to regulation of the PKC1-MPK1 pathway. Like _pkc1_Δ mutants, _mid2Δ pkc1_Δ cells are inviable on medium without osmotic support; however, these double mutants are able to grow at a rate similar to that of _pkc1_Δ mutants on medium containing 1 M sorbitol at both 22 and 30°C (not shown). Moreover, multicopy MID2 does not alter the growth rate of _pkc1_Δ cells (not shown), suggesting that genetically, PKC1 acts downstream of MID2.
To further explore the potential role of Mid2p in PKC1-MPK1 pathway regulation, we examined whether Mid2p influences Mpk1p activation. The Mpk1p MAP kinase is activated through tyrosine phosphorylation during mating projection formation (8, 49). We found that compared to wild-type cells, _mid2_Δ mutants have substantially less tyrosine phosphorylation on Mpk1p-HA after exposure of cells to α-factor (Fig. 6A). This observation suggests that underactivation of the cell integrity pathway might be, at least in part, responsible for pheromone-induced death in _mid2_Δ mutants. The finding that overexpression of PKC1, PKC1R398A (a hyperactive allele), RHO1, WSC1, and WSC2 can suppress α-factor-induced death in _mid2_Δ mutants (Fig. 5B) independently supports this conclusion. However, expression of either bck1-20 (a hyperactive allele of BCK1), or MPK1 does not suppress this phenotype, and at high concentrations of pheromone (>4 μM) only some of the most upstream elements of the cell integrity pathway, including RHO1, WSC1, and WSC2, can prevent mating pheromone-induced death (not shown).
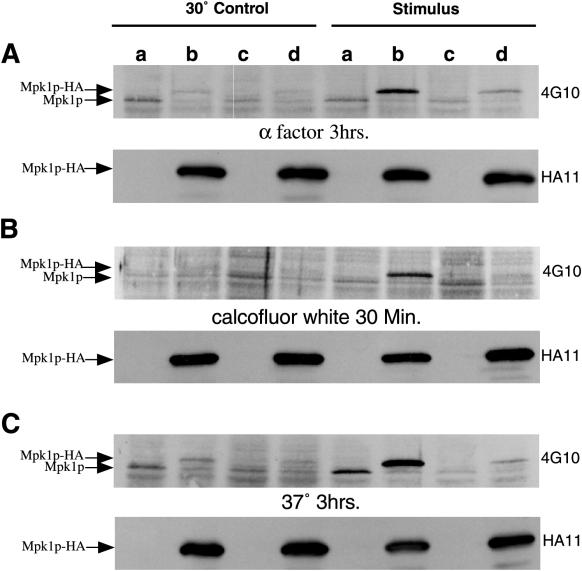
FIG. 6.
Immunoblot analysis of Mpk1p-HA tyrosine phosphorylation. Lanes are loaded with equal amounts of extracts from strains TK96 [wild type (pFL44)] (lanes a), TK97 [wild type (pFL44-MPK1-HA)] (lanes b), TK93 [_mid2_Δ (pFL44)] (lanes c), and TK94 [_mid2_Δ(pFL44-MPK1-HA)] (lanes d). Cultures exposed to α-factor (A), calcofluor white (B), or high-temperature growth (C) were harvested at the indicated times, and total cell proteins were subject to SDS-PAGE and Western blotting. In the top panel of each pair, tyrosine phosphorylation of Mpk1p-HA is detected by antiphosphotyrosine antibody 4G10. In the second panel of each pair, equal loading of Mpk1p-HA is demonstrated by anti-HA antibody HA11.
To explore the genetic relationship between MID2 and RHO1, we tested whether MID2 overexpression suppresses _rho1_Δ mutants. Because cells lacking Rho1p are inviable, even at low temperature on medium with osmotic support, _rho1_Δ heterozygous diploids were transformed with high-copy-number MID2 and then sporulated, and the resulting asci were dissected. No viable _rho1_Δ mutants were recovered from 36 tetrads analyzed, suggesting that high-copy-number MID2 is unable to suppress the lack of Rho1p. Since overexpression of RHO1 can suppress α-factor-induced death in _mid2_Δ mutants but high-copy-number MID2 cannot suppress inviability of _rho1_Δ mutants, it appears that Mid2p may act upstream of, or in parallel with, Rho1p.
Interestingly, Mpk1p-HA tyrosine phosphorylation is also increased in wild-type cells in response to exposure to calcofluor white. This effect is dependent on the presence of Mid2p, since there is a deficit of this phosphorylation on Mpk1-HA in _mid2_Δ cells (Fig. 6B). _MID2_-dependent increase in tyrosine phosphorylation of native Mpk1p in response to calcofluor white and mating pheromone is not readily apparent in Fig. 6A and B, perhaps due to lower relative abundance of native Mpk1p versus (2 μm-borne) Mpk1-HA. Also, for the chosen time points, a lower extent of tyrosine phosphorylation on Mpk1-HA is seen for calcofluor white and mating factor exposure than is observed for high-temperature growth (Fig. 6C), suggesting that differences in phosphorylation state for native Mpk1p in these panels may be below the detection threshold. In other assay conditions, we observe clear MID2 dependence for native Mpk1p tyrosine phosphorylation in response to calcofluor white and mating factor (not shown). Deletion of MPK1 results in calcofluor white hypersensitivity, likely due to gross disturbances in cell wall construction. Saliently, overexpression of MPK1, like overexpression of MID2, also results in a _CHS3_-dependent hypersensitivity to calcofluor white (not shown). This effect is _MID2_-dependent since overexpression of MPK1 does not bypass the resistance to calcofluor white displayed by _mid2_Δ cells (not shown).
Finally, we examined whether induction of tyrosine phosphorylation of Mpk1p-HA during periods of high-temperature stress requires MID2. Although _mid2_Δ cells do not have a growth defect at 37°C, induction of tyrosine phosphorylation of Mpk1p-HA was significantly impaired in _mid2_Δ mutants compared to wild-type cells (Fig. 6C). Together, these observations suggest a role for Mid2p in activation of the Mpk1p MAP kinase cascade under a variety of stress conditions.
Identification of a Mid2p functional homolog.
The S. cerevisiae genome contains a gene, YGR023W, which encodes a 551-amino-acid residue protein with both structural and amino acid sequence similarity to Mid2p [W. U. Blast V2.0 P(N) value of 1.2e−27]. We will refer to YGR023W as MTL1 (_MID2_-like 1). To initiate the characterization of MTL1, we disrupted its entire ORF. Unlike _mid2_Δ cells, _mtl1_Δ mutants in the SEY6210 strain background have no distinguishable phenotype when challenged with temperature extremes, oxidative and osmotic stresses, α-factor, calcofluor white, or mutation of the KRE6 or FKS1 cell wall synthesis gene (not shown). While _mtl1_Δ single mutants are not hypersensitive to caffeine, _mid2_Δ cells are mildly more susceptible than wild-type cells, and _mid2Δ mtl1_Δ double mutants show strong sensitivity to this drug. This phenotype appears to be the result of cell lysis since it is suppressible by the inclusion of 1 M sorbitol in the growth medium (not shown). Caffeine sensitivity of the _mid2Δ mtl1_Δ mutant is also suppressible by overexpression of WSC2; however, multicopy WSC1, RHO1, PKC1, BCK1, and MPK1 do not bypass this phenotype. Finally, although _mid2Δ mtl1_Δ double mutants are no more sensitive to α-factor than _mid2_Δ cells (not shown), high-copy-number expression of MTL1 from either its own promoter or the ADH1 promoter is able to suppress the caffeine (not shown) and α-factor sensitivity of _mid2_Δ cells (Fig. 5B), suggesting that MTL1 is a functional gene and that the activity of Mtl1p may be related to or overlap the activity of Mid2p.
DISCUSSION
Mid2p is an O-mannosylated, plasma membrane protein.
In this work, we offer evidence that Mid2p may potentially sense cell wall state and act to initiate a cellular response involving both chitin synthesis and the PKC1-MPK1 cell integrity pathway. Mid2p is a type I integral membrane protein which localizes to the plasma membrane and contains a large, extensively O-mannosylated extracellular serine/threonine-rich region. Removal of the serine/threonine-rich region does not affect the targeting of Mid2p; however, ΔS/T Mid2p is unable to complement _mid2_Δ cells, suggesting that this domain is required for Mid2p activity. This observation contrasts with the findings for Gas1p and Kre1p, two glycosyl phosphatidylinositol-anchored proteins involved in cell wall synthesis, where deletion of the serine/threonine-rich sequences does not greatly affect function of these proteins (4, 18). O-linked mannosylation could cause the extracellular region to adopt a stiff and extended conformation (23) that reaches from the plasma membrane toward the cell wall, perhaps interacting with it. In Mid2p, this region is unlikely to play a direct enzymatic role since it is largely composed of repetitive noncomplex amino acid sequence, and it lacks any similarity to known enzymatic motifs. An intriguing possibility is that the extracellular domain can act as a sensor of cell wall state.
MID2 interacts with genes required for cell wall construction and cell wall integrity signaling.
We isolated MID2 as an activator of the Skn7p transcription factor. Skn7p appears to affect a number of cellular processes, including cell wall biosynthesis. SKN7 was identified by Brown et al. (6) as a high-copy-number suppressor of the _kre9_Δ mutant, and it was later demonstrated that Skn7p may function in parallel with Pkc1p since overexpression of SKN7 can suppress the lysis defect of _pkc1_Δ mutants, and _skn7Δ pkc1_Δ cells are inviable, even on medium containing osmotic support (7). Recently, Alberts et al. (1) have shown that Rho1p and Skn7p physically interact and that the domain in Skn7p that mediates this interaction is important for activity. It is not clear how Mid2p might stimulate Skn7p transcriptional activity. Since RHO1 appears to have interactions with both SKN7 and MID2, one avenue of future research will be to examine whether Rho1p mediates a Mid2p-Skn7 interaction.
Deletion of MID2 causes significant changes in growth rate or viability for a variety of cell wall synthesis mutants. It is curious that for the β-1,6-glucan mutants examined, loss of MID2 increases growth rate, while for the β-1,3-glucan mutant _fks1_Δ, deletion of MID2 causes inviability. One possibility is that reduction of supplementary chitin levels caused by absence of Mid2p is a contributing factor. _fks1_Δ mutants seem to depend heavily on enhanced chitin synthesis to maintain viability since they are supersensitive to nikkomycin Z, a chitin synthase inhibitor (16). Conversely, there is some evidence that attenuation of the chitin synthesis stress response may actually be beneficial in cells lacking proper β-1,6-glucan synthesis, specifically in _kre9_Δ mutants (33). Consistent with this hypothesis, overexpression of MID2, which causes hyperaccumulation of chitin, is deleterious to β-1,6-glucan mutants but has little effect on the β-1,3-glucan-deficient mutant _fks1_Δ.
Screens to identify genes which interact with MID2 uncovered a relationship between MID2 and several components of a pathway known to promote cell integrity and polarized growth. Caffeine sensitivity of the _mid2_Δ and _mid2Δ mtl1_Δ mutants is suppressed by overexpression of WSC2, high-copy-number WSC1 and PKC1 suppress _mid2Δ fks1_Δ synthetic lethality, and multicopy PKC1, RHO1, WSC1, and WSC2 suppress lethality in mid2Δ MATa cells exposed to α-factor. Additionally, _mid2Δ wsc1_Δ mutants are inviable without osmotic support and exhibit defects similar to those observed in _pkc1_Δ mutants. Finally, we find that during shmoo formation, high-temperature growth, or exposure to calcofluor white, tyrosine phosphorylation of Mpk1p, a downstream target of Pkc1p, is reduced in _mid2_Δ cells. Interestingly, although _mid2_Δ mutants have reduced tyrosine phosphorylation of Mpk1p in response to high-temperature growth, these cells are not deficient for growth at high temperature. It is possible that there is sufficient remaining Mpk1p activity to allow cells to survive high-temperature stress or that some other mechanism is able to compensate for underactivation of this pathway. Strain differences may also play some role here, since we do not observe high-temperature sensitivity in _wsc1_Δ mutants as reported by Verna et al. (47).
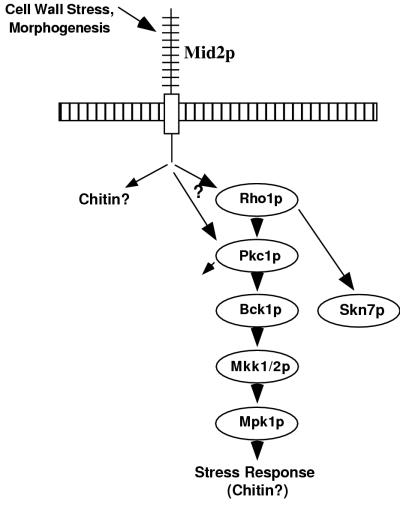
Although _mid2_Δ cells have no apparent vegetative growth defects, shmoo formation, mutation of cell wall synthesis genes, and exposure to calcofluor white cause _mid2_Δ mutants to manifest phenotypes. One possibility is that Mid2p and perhaps members of the WSC family act indirectly, through effects on cell wall structure, to activate the PKC1-MPK1 pathway. In an alternative model, Mid2p might sense cell wall stress and directly act to increase activity in the cell integrity pathway to counteract damage (Fig. 7). Under nonstressed conditions, Mid2p activity might be low, or not be required, and Wsc1p and its homologs may be largely responsible for Pkc1p activation. However, under circumstances of cell wall stress, in the absence of Mid2p, the cell integrity machinery would be unresponsive and would continue to function at a level more appropriate for low or nonstress situations.
FIG. 7.
Model of Mid2p activity. Mid2p responds to cell wall stress by activating the cell integrity pathway and increasing chitin synthesis.
This model explains why some of the phenotypes observed in _mid2_Δ mutants contrast with those displayed by other mutants in the cell integrity pathway. For example, strains carrying mutations such as _wsc1/2/3_Δ, _pkc1_Δ, _bck1_Δ, or _mpk1_Δ are prone to cell lysis in the absence of osmotic support. These genes are required for normal function of the cell integrity pathway under all growth conditions. In their absence, construction and maintenance of the cell wall is defective, making the wall highly susceptible to damage. _mid2_Δ mutants have apparently normal cell walls when grown in ordinary conditions. However, in stress situations, such as exposure to calcofluor white or α-factor, _mid2_Δ mutants have an attenuated response compared to wild-type cells, as indicated by the reduced amount of new chitin synthesized and by the reduced extent of tyrosine phosphorylation on Mpk1p.
Preliminary investigation of MTL1, the only S. cerevisiae gene encoding a protein with significant sequence similarity to Mid2p, revealed that it may have a function in common with MID2. Although _mtl1_Δ mutants do not display many of the phenotypes that _mid2_Δ cells do, sensitivity to caffeine is much greater in _mid2Δ mtl1_Δ double mutants than in either single mutant. Additionally, multicopy MTL1 can suppress α-factor sensitivity of _mid2_Δ cells. Further research may reveal whether Mtl1p is required for responding to different stresses than Mid2p, signals to a different pathway than Mid2p, or is important under physiological conditions different from those used in this study.
The effect of Mid2p on chitin synthesis depends ultimately on Chs3p, since overexpression of MID2 in _chs3_Δ mutants cannot confer hypersensitivity to calcofluor white. We suggest two ways in which Mid2p might affect chitin synthesis. Mid2p might directly interact with the chitin synthase complex, increasing its activity during stress periods. Alternatively, the activity of Chs3p might be regulated by a downstream target of Mid2p, such as Mpk1p.
Cells face a demanding variety of stresses in their natural environments and must respond accordingly. Our results provide new insights into the processes by which yeast cells sense and respond to threats against their physical integrity. Mid2p has been identified as a putative sensor of cell wall state that is required for stimulation of activity in the PKC1-MPK1 MAP kinase pathway under conditions of cell wall stress. Currently, the physiological roles of most serine/threonine-rich membrane proteins are poorly defined. An interesting possibility is that as a class, these proteins act as sensors for a range of environmental stresses and mediate a variety of cellular processes.
ACKNOWLEDGMENTS
We thank S. Nagahashi, M. Rajavel, T. Roemer, and P. A. Delley for helpful and stimulating discussion; R. C. Stewart and J. L. Brown for designing and performing the original ASK screen; S. Véronneau and T. Nguyen for sequencing; and S. Shahinian for critical input on the manuscript. We also thank C. Bulawa for assistance with the chitin assay protocol and for providing the chs3Δ::LEU2 plasmid, M. Snyder for providing multicopy MPK1-HA, BCK1, MKK1, and PKC1, and M. Rajavel for providing pRS316-BCK1-20, and pBM743-PKC1R398A.
This work was supported by an NSERC operating grant.
REFERENCES
- 1.Alberts A S, Bouquin N, Johnston L H, Treisman R. Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response regulator protein Skn7. J Biol Chem. 1998;273:8616–8622. doi: 10.1074/jbc.273.15.8616. [DOI] [PubMed] [Google Scholar]
- 2.Banuett F. Signalling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol Mol Biol Rev. 1998;62:249–274. doi: 10.1128/mmbr.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bickle M, Delley P A, Schidt A, Hall M N. Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 1998;17:2235–2245. doi: 10.1093/emboj/17.8.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boone C, Sdicu A, Laroche M, Bussey H. Isolation from Candida albicans of a functional homolog of the Saccharomyces cerevisiae KRE1 gene, which is involved in cell wall beta-glucan synthesis. J Bacteriol. 1991;173:6859–6864. doi: 10.1128/jb.173.21.6859-6864.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown J L, Bussey H. The yeast KRE9 gene encodes an O-glycoprotein involved in cell surface β-glucan assembly. Mol Cell Biol. 1993;13:6346–6356. doi: 10.1128/mcb.13.10.6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown J L, North S, Bussey H. SKN7, a yeast multicopy suppressor of a mutation affecting β-glucan assembly, encodes a product with domains homologous to prokaryotic two-component regulators and to heat shock transcription factors. J Bacteriol. 1993;175:6908–6915. doi: 10.1128/jb.175.21.6908-6915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown J L, Bussey H, Stewart R C. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 1994;13:5186–5194. doi: 10.1002/j.1460-2075.1994.tb06849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buehrer B M, Errede B. Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6517–6525. doi: 10.1128/mcb.17.11.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Bulawa, C. Personal communication.
- 9.Bulawa C E, Slater M, Cabib E, Au-Young J, Sburlati A, Adair W L, Robbins P. The S. cerevisiae structural gene for chitin synthase is not required for chitin synthesis in vivo. Cell. 1986;46:213–225. doi: 10.1016/0092-8674(86)90738-5. [DOI] [PubMed] [Google Scholar]
- 10.Cabib E, Drgon T, Dronova J, Ford R A, Kollar R. The yeast cell wall, a dynamic structure engaged in growth and morphogenesis. Biochem Soc Trans. 1997;25:200–204. doi: 10.1042/bst0250200. [DOI] [PubMed] [Google Scholar]
- 11.Costigan C, Gehrung S, Snyder M. A synthetic lethal screen identifies SLK1, a novel protein kinase homolog implicated in yeast cell morphogenesis and cell growth. Mol Cell Biol. 1992;12:1162–1178. doi: 10.1128/mcb.12.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniel J. Potentially rapid walking in cellular regulatory networks using the gene-gene interference method in yeast. Mol Gen Genet. 1993;240:245–257. doi: 10.1007/BF00277063. [DOI] [PubMed] [Google Scholar]
- 13.Douglas C M, Foor F, Marrinan J A, Morin N, Nielson J B, Dahl A M, Mazur P, Baginsky W, Li W, el-Sherbeini M, et al. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-beta-d-glucan synthase. Proc Natl Acad Sci USA. 1994;91:12907–12911. doi: 10.1073/pnas.91.26.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drgonova J, Drgon T, Tanaka K, Kollar R, Chen G C, Ford R A, Chan C S, Takai Y, Cabib E. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science. 1996;272:277–279. doi: 10.1126/science.272.5259.277. [DOI] [PubMed] [Google Scholar]
- 15.Elorza M V, Rico H, Sentandreu R. Calcofluor white alters the assembly of chitin fibrils in Saccharomyces cerevisiae and Candida albicans cells. J Gen Microbiol. 1983;129:1577–1582. doi: 10.1099/00221287-129-5-1577. [DOI] [PubMed] [Google Scholar]
- 16.el-Sherbeini M, Clemas J A. Nikkomycin Z supersensitivity of an echinocandin-resistant mutant of Saccharomyces cerevisiae. Antimicrob Agents Chemother. 1995;39:200–207. doi: 10.1128/aac.39.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eng W K, Faucette L, McLaughlin M M, Cafferkey R, Koltin Y, Morris R A, Young P R, Johnson R K, Livi G P. The yeast FKS1 gene encodes a novel membrane protein, mutations in which confer FK506 and cyclosporin A hypersensitivity and calcineurin-dependent growth. Gene. 1994;151:61–71. doi: 10.1016/0378-1119(94)90633-5. [DOI] [PubMed] [Google Scholar]
- 18.Gatti E, Popolo L, Vai M, Rota N, Alberghina L. O-linked oligosaccharides in yeast glycosyl phosphatidylinositol-anchored protein gp115 are clustered in a serine rich region not essential for its function. J Biol Chem. 1994;269:19695–19700. [PubMed] [Google Scholar]
- 19.Gray J V, Ogas J P, Kamada Y, Stone M, Levin D E, Herskowitz I. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 1997;16:4924–4937. doi: 10.1093/emboj/16.16.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Igual J C, Johnson A L, Johnston L H. Coordinated regulation of gene expression by the cell cycle transcription factor SWI4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 1996;15:5001–5013. [PMC free article] [PubMed] [Google Scholar]
- 21.Irie K, Takase M, Lee K S, Levin D E, Araki H, Matsumoto K, Oshima Y. MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C. Mol Cell Biol. 1993;13:3076–3083. doi: 10.1128/mcb.13.5.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacoby J J, Nilius S M, Heinisch J J. A screen for upstream components of the yeast protein kinase C signal transduction pathway identifies the product of the SLG1 gene. Mol Gen Genet. 1998;258:148–155. doi: 10.1007/s004380050717. [DOI] [PubMed] [Google Scholar]
- 23.Jentoft N. Why are proteins O-glycosylated? Trends Biochem Sci. 1990;15:291–294. doi: 10.1016/0968-0004(90)90014-3. [DOI] [PubMed] [Google Scholar]
- 24.Kamada Y, Qadota H, Python C P, Anraku Y, Ohya Y, Levin D E. Activation of yeast protein kinase C by Rho1 GTPase. J Biol Chem. 1996;271:9193–9196. doi: 10.1074/jbc.271.16.9193. [DOI] [PubMed] [Google Scholar]
- 25.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 26.Lee K S, Levin D E. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol Cell Biol. 1992;12:172–182. doi: 10.1128/mcb.12.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee K S, Irie K, Gotoh Y, Watanabe Y, Araki H, Nishida E, Matsumoto K, Levin D E. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol Cell Biol. 1993;13:3067–3075. doi: 10.1128/mcb.13.5.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levin D E, Bartlett-Heubusch E. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J Cell Biol. 1992;116:1221–1229. doi: 10.1083/jcb.116.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levin D E, Bowers B, Chen C Y, Kamada Y, Watanabe M. Dissecting the protein kinase C/MAP kinase signaling pathway of Saccharomyces cerevisiae. Cell Mol Biol Res. 1994;40:229–239. [PubMed] [Google Scholar]
- 30.Lussier M, Gentzsch M, Sdicu A M, Bussey H, Tanner W. Protein O-glycosylation in yeast. The PMT2 gene specifies a second protein O-mannosyltransferase that functions in addition to the PMT1-encoded activity. J Biol Chem. 1995;270:2770–2775. doi: 10.1074/jbc.270.6.2770. [DOI] [PubMed] [Google Scholar]
- 31.Manning B D, Padmanabha R, Snyder M. The Rho-GEF Rom2p localizes to sites of polarized cell growth and participates in cytoskeletal functions in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:1829–1844. doi: 10.1091/mbc.8.10.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcoux N, Bourbonnais Y, Charest P, Pallota D. Overexpression of MID2 suppresses the profilin deficient phenotype of yeast cells. Mol Microbiol. 1998;29:515–526. doi: 10.1046/j.1365-2958.1998.00944.x. [DOI] [PubMed] [Google Scholar]
- 33.Nagahashi S, Lussier M, Bussey H. Isolation of Candida glabrata homologs of the Saccharomyces cerevisiae KRE9 and KNH1 genes and their involvement in cell wall β-1,6-glucan synthesis. J Bacteriol. 1998;180:5020–5029. doi: 10.1128/jb.180.19.5020-5029.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niedenthal R K, Riles L, Johnston M, Hegemann J H. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 35.Nonaka H, Tanaka K, Hirano H, Fujiwara T, Kohno H, Umikawa M, Mino A, Takai Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 1995;14:5931–5938. doi: 10.1002/j.1460-2075.1995.tb00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ono T, Suzuki T, Anraku Y, Iida H. The MID2 gene encodes a putative integral membrane protein with a Ca(2+)-binding domain and shows mating pheromone-stimulated expression in Saccharomyces cerevisiae. Gene. 1994;151:203–208. doi: 10.1016/0378-1119(94)90657-2. [DOI] [PubMed] [Google Scholar]
- 37.Orlean P. Biogenesis of yeast wall and surface components. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces—cell cycle and cell biology. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 229–362. [Google Scholar]
- 38.Pagé N, Sheraton J, Brown J L, Stewart R C, Bussey H. Identification of ASK10 as a multicopy activator of Skn7p-dependent transcription of a HIS3 reporter gene. Yeast. 1996;12:267–272. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C267::AID-YEA897%3E3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 39.Popolo L, Gilardelli D, Bonfante P, Vai M. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1Δ mutant of Saccharomyces cerevisiae. J Bacteriol. 1997;179:463–469. doi: 10.1128/jb.179.2.463-469.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ram A F, Brekelmans S S, Oehlen L J, Klis F M. Identification of two cell cycle regulated genes affecting the beta 1,3-glucan content of cell walls in Saccharomyces cerevisiae. FEBS Lett. 1995;358:165–170. doi: 10.1016/0014-5793(94)01418-z. [DOI] [PubMed] [Google Scholar]
- 41.Ram A F, Kapteyn J C, Montijn R C, Caro L H, Douwes J E, Baginsky W, Mazur P, van den Ende H, Klis F M. Loss of the plasma membrane-bound protein Gas1p in Saccharomyces cerevisiae results in the release of β-1,3-glucan into the medium and induces a compensation mechanism to ensure cell wall integrity. J Bacteriol. 1998;180:1418–1424. doi: 10.1128/jb.180.6.1418-1424.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roemer T, Bussey H. Yeast β-glucan synthesis: KRE6 encodes a predicted type II membrane protein required for glucan synthesis in vivo and for glucan synthase activity in vitro. Proc Natl Acad Sci USA. 1991;88:11295–11299. doi: 10.1073/pnas.88.24.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roncero C, Duran A. Effect of calcofluor white and congo red on fungal wall morphogenesis: in vivo activation of chitin polymerization. J Bacteriol. 1985;170:1950–1954. doi: 10.1128/jb.163.3.1180-1185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schekman R, Brawley V. Localized deposition of chitin on the yeast cell surface in response to mating pheromone. Proc Natl Acad Sci USA. 1979;76:645–649. doi: 10.1073/pnas.76.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanner W, Lehle L. Protein glycosylation in yeast. Biochim Biophys Acta. 1987;906:81–99. doi: 10.1016/0304-4157(87)90006-2. [DOI] [PubMed] [Google Scholar]
- 46.Takeuchi J, Okada M, Toh-e A, Kikuchi Y. The SMS1 gene encoding a serine-rich transmembrane protein suppresses the temperature sensitivity of HTR1 disruptant in Saccharomyces cerevisiae. Biochim Biophys Acta. 1995;1260:94–96. doi: 10.1016/0167-4781(94)00188-9. [DOI] [PubMed] [Google Scholar]
- 47.Verna J, Lodder A, Lee K, Vagts A, Ballester R. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:13804–13809. doi: 10.1073/pnas.94.25.13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 49.Zarzov P, Mazzoni C, Mann C. The SLT2(MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO J. 1996;83:93–91. [PMC free article] [PubMed] [Google Scholar]