Distribution of Bacterial Growth Activity in Flow-Chamber Biofilms (original) (raw)
Abstract
In microbial communities such as those found in biofilms, individual organisms most often display heterogeneous behavior with respect to their metabolic activity, growth status, gene expression pattern, etc. In that context, a novel reporter system for monitoring of cellular growth activity has been designed. It comprises a transposon cassette carrying fusions between the growth rate-regulated _Escherichia coli rrnB_P1 promoter and different variant gfp genes. It is shown that the P1 promoter is regulated in the same way in E. coli and Pseudomonas putida, making it useful for monitoring of growth activity in organisms outside the group of enteric bacteria. Construction of fusions to genes encoding unstable Gfp proteins opened up the possibility of the monitoring of rates of rRNA synthesis and, in this way, allowing on-line determination of the distribution of growth activity in a complex community. With the use of these reporter tools, it is demonstrated that individual cells of a toluene-degrading P. putida strain growing in a benzyl alcohol-supplemented biofilm have different levels of growth activity which develop as the biofilm gets older. Cells that eventually grow very slowly or not at all may be stimulated to restart growth if provided with a more easily metabolizable carbon source. Thus, the dynamics of biofilm growth activity has been tracked to the level of individual cells, cell clusters, and microcolonies.
Biofilms play an important role in almost all aspects of microbiology and may appear as either beneficial or potentially harmful populations of microorganisms. The bacteria constituting the biofilms in our intestines, sewage treatment plants, bioremediation plants, etc., are mostly beneficial. Harmful biofilms are also abundant, ranging from relatively harmless dental plaque to Pseudomonas aeruginosa biofilms in the alveoli of cystic fibrosis patients, but biofouling of ships and offshore material is also a serious problem in the oil and shipping industries.
In order to improve the performance of some beneficial biofilms and to avoid or remove harmful biofilms, it is important to understand the mechanisms of biofilm formation, growth, and maintenance. Microbial biofilms consisting of either single or multiple species are structurally organized in patterns, which depend on several factors such as nutrient supply, flow rate, pH, temperature, etc. In such dynamic systems, the individual cells experience conditions determined by the outer environment, the already existing structures, and the local microbial activities. Furthermore, subpopulations may form locally which are completely different from the majority of the community. Microbial biofilms have been investigated either by visual inspection of biofilms cultivated in flow cells (6, 28, 37) or as disrupted samples withdrawn from either natural systems such as drinking water pipes (26, 32) or artificial model systems such as the Robbins device system (25).
In either case, the results of such analyses are primarily descriptive, yielding limited information about parameters like growth states of individual cells. Several methods of assessing different types of single-cell activity have been described, such as the use of (i) 5-cyano-2,3-ditolyl tetrazolium chloride staining (33) to identify actively respiring cells, (ii) ribosomal probing to monitor the number of ribosomes as a measure of growth rate (13, 30), or (iii) classical visual markers such as lacZ to monitor the expression of several genes in situ (31). Energy charge has also been used to measure cell activity by cryosectioning of biofilms (23). More recently, the green fluorescent protein Gfp (7, 10) has been used for in situ investigation of living biofilms (8, 9, 15, 28, 29, 35). While some of these methods are useful for actively growing cells, they may not prove effective for investigation of starving cells or cells exposed to changing environments. The intracellular marker compounds may accumulate and thus reflect the history of the cells rather than the present growth physiological state of the organisms.
We have recently developed new derivatives of the green fluorescent protein which, unlike the native protein, have short half-lives (3). The Gfp protein has been modified by the addition of a few amino acid residues to its C-terminal end, rendering it a target for indigenous tail-specific proteases. Through the construction of an expression cassette consisting of this novel gfp gene expressed from an Escherichia coli rRNA promoter, a reporter system was obtained which generates green fluorescence in fast-growing cells of both E. coli and Pseudomonas putida. By use of these monitor strains, we have been able to discriminate between fast- and slow-growing cells in structured surface communities of microbial biofilms and to determine the spatial and temporal distribution of growth activity.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study and their relevant characteristics are listed in Table 1. Strains were grown in FAB medium [containing 1 mM MgCl2, 0.1 mM CaCl2, 0.01 mM Fe-EDTA (Sigma E6760; Sigma, St. Louis, Mo.), 0.15 mM (NH4)SO4, 0.33 mM Na2HPO4, 0.2 mM KH2PO4, and 0.5 mM NaCl], and unless otherwise mentioned, 10 mM Na-citrate was added as a carbon source. When required, antibiotics were added at final concentrations of 50 μg/ml for nalidixic acid and 25 μg/ml for kanamycin.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli K-12 strains | ||
| HB101 | E. coli K-12/B hybrid; Smr_recA thi pro leu hsd_RM+ | 5 |
| CC118 λ_pir_ | Δ(ara-leu) araD ΔlacX74 galE galK phoA thi-1 rpsE rpoB argE(Am) recA, λ_pir_ lysogen | 14 |
| P. putida strains | ||
| R1 (JB156) | Natural isolate made Nalr | 9 |
| SM1639 | P. putida R1 (Nalr) × HB101(RK600) × CC118 λ_pir_(pSM1695)a; P. putida R1 with mini Tn_5_-Km-_rrnB_P1-gfp(AAV)-T0-T1 cassette randomly inserted into chromosome; Nalr Kmr | This study |
| SM1699 | P. putida R1 (Nalr) × HB101(RK600) × CC118 λ_pir_(pSM1621)a; P. putida R1 with mini Tn_5_-Km-_rrnB_P1-_gfp_mut3b*-T0-T1 cassette randomly inserted into chromosome; Nalr Kmr | This study |
| Plasmids | ||
| RK600 | Cmr ColE1 oriV RP4 oriT; helper plasmid in triparental matings | 22 |
| pUT-mini Tn5-Km | Apr Kmr; transposon delivery vector for mini Tn_5_-Km | 20 |
| pL1OW2Not | Kmr; low-copy cloning vector with pUC18-_Not_I polylinker | 18 |
| pJBA25 | Apr; pUC18Not-RBSII-_gfp_mut3b*-T0-T1 | J. B. Andersen |
| pJBA44 | Apr; pUC18Not-P_lac_UV5-RBSII-_gfp_mut3b*-T0-T1 | J. B. Andersen |
| pSM1606 | Kmr; pLOW2Not-_rrnB_P1-RBSII-gfp(AAV)-T0-T1 | This study |
| pSM1621 | Apr Kmr; delivery plasmid for mini Tn_5_-Km-_rrnB_P1-RBSII-gfp(AAV)-T0-T1 | This study |
| pSM1690 | Kmr; pLOW2Not-_rrnB_P1-RBSII-_gfp_mut3b*-T0-T1 | This study |
| pSM1695 | Apr Kmr; delivery plasmid for mini Tn_5_-Km-_rrnB_P1-RBSII-_gfp_mut3b*-T0-T1 | This study |
Constructions.
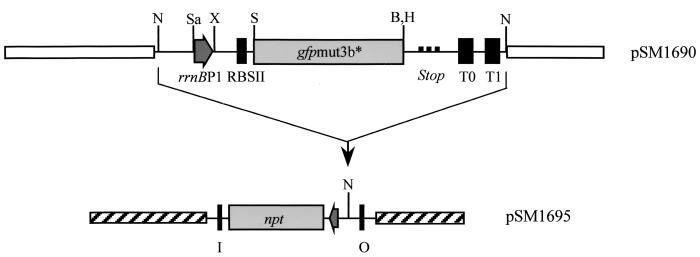
A linker fragment of 72 nucleotides, containing the _E. coli rrnB_P1 promoter region, corresponding to nucleotides 1156 to 1228 in the published GenBank sequence (accession no. J01695), was synthesized with a DNA synthesizer (Applied Biosystems 373A). The linker was designed with _Sac_I- and _Xba_I-compatible protruding ends. The promoter fragment contains nucleotides −70 to +3 relative to the transcription initiation site (4). Due to the high level of expression from the _rrnB_P1 promoter, the linker could not be inserted into our general high-copy-number gfp cloning vectors (pUC18 derivatives [3]). Therefore, a _Not_I fragment carrying the synthetic ribosomal binding site (RBSII), the _gfp_mut3b* gene, and the transcriptional terminators T0 and T1 was moved from pJBA25 (3) into the _Not_I site of the low-copy-number plasmid pLOW2 (18). The pLOW2 derivative carrying the RBSII-_gfp_mut3b*-T0-T1 cassette was subsequently digested with _Sac_I and _Xba_I, and the 72-base _rrnB_P1 promoter fragment was inserted to produce plasmid pSM1690 (see Fig. 1 for details). Plasmid pSM1690 was digested with _Not_I, and the fragment carrying _rrnB_P1-RBSII-_gfp_mut3b*-T0-T1 was inserted into the unique _Not_I site of pUT-mini Tn_5_-Km, resulting in transposon delivery vector pSM1695.
FIG. 1.
Schematic representation of plasmid SM1690 and the resulting transposon delivery plasmid pSM1695. The organization of relevant regions in the _rrnB_P1-_gfp_mut3b* cassette, as well as regions of the delivery plasmid, is shown. _rrnB_P1, 72-bp ribosomal promoter; _gfp_mut3b*, Gfp marker gene; RBSII, synthetic ribosomal binding site; Stop, stop codon in all three reading frames; T0 and T1, termination sites; I and O, 19-bp transposon inverted repeats; npt, gene encoding kanamycin resistance. Relevant restriction sites: N, _Not_I; Sa, _Sac_I; X, _Xba_I; S, _Sph_I; B, _Bam_HI; H, _Hin_dIII. Note that the _rrnB_P1-_gfp_mut3b* cassette is inserted into the _Not_I site of the delivery vector in an orientation that prevents readthrough from chromosomal promoters located adjacent to the transposon insertion in the host chromosome.
Finally, by triparental mating of CC118 λ_pir_(pSM1695), HB101(RK600), and P. putida R1 (strain JB156), the _rrnB_P1-RBSII-_gfp_mut3b*-T0-T1 cassette was inserted at random positions on the chromosome of P. putida R1 (JB156). One normally growing transconjugant colony was picked and designated SM1699. For construction of a P. putida R1 derivative carrying the _rrnB_P1-RBSII-gfp(AAV)-T0-T1 cassette, we used a procedure similar to the one described above, resulting in SM1639.
The chosen transposant clones showed no sign of phenotypic changes relative to the parent strain when tested in both liquid medium and flow chamber biofilms. The orientation of the gfp reporter cassette within the mini Tn_5_ transposon was such that no transcription from flanking chromosomal sequences could interfere with gfp expression.
Runout experiment.
Exponential growth was allowed for at least 10 generations at 30°C before the cultures were diluted (to an optical density at 450 nm [OD450] of 0.05) into prewarmed FAB medium containing 8 mM sodium citrate. After entering the stationary phase at an OD450 of ∼2.0, the cultures were monitored for more than 5 h. During the entire experiment, samples were taken for measurements of OD450 and green fluorescence emitted by the cells. To avoid autofluorescence excreted by the cells into the medium, 2-ml cell suspensions were harvested by centrifugation and resuspended in 0.9% NaCl. Both resuspended cells and the supernatants were measured in a fluorometer (RF-1501; Shimadzu, Kyoto, Japan). To determine the relative fluorescence activities of the cells, the OD450 values of the resuspended cells were also determined.
Chemostat experiments.
To determine specific activities of the different _rrnB_P1-gfp gene fusions at different growth rates, cells were grown in chemostats. A chemostat was made from a 50-ml tube (syringe) with a rubber stopper containing a glass tube for intake of air, which passed through a 0.2-μm-pore-size filter before entering the chemostat, and another tube for outlet of the effluent culture. Furthermore, we introduced a thin hypodermic needle for injection of medium and another for withdrawal of samples from the chemostat.
Five parallel chemostats were run at the same time. Tubings with different bore diameters were used in the same peristaltic pump (Watson Marlow 205S; Watson-Marlow Inc., Wilmington, Mass.) to obtain different dilution rates (resulting in different growth rates). The chemostats were each inoculated with approximately 15 ml of an overnight culture, and then medium flow was started. The volume of each operating chemostat varied between 40 and 45 ml. For each chemostat, the exact flow rate and chemostat volume were determined. The growth rates were calculated by dividing the flow rate by the chemostat volume. The chemostat cultures were run for at least 60 h before the first sample was taken from each chemostat. This corresponds to approximately 5 or 6 generations for the slowest-growing cultures and approximately 50 generations for the chemostat cultures with the highest growth rates. Samples were taken at least three times from each chemostat with intervals of approximately 12 h. All samples were harvested as described above, and OD450 and fluorescence were determined from the resuspended cells, as well as from the supernatant.
Flow chamber experiments.
Biofilms were cultivated in four-channel flow cells (37) with individual channel dimensions of 1 by 4 by 40 mm supplied with a flow of FAB medium supplemented with benzyl alcohol (Merck KGaA, Darmstadt, Germany) as a carbon source to a final concentration of 0.5 mM.
The flow system was assembled and prepared as described by Christensen et al. (9). The substratum consisted of a microscope glass coverslip (Knittel 24x50 st1; Knittel Gläser, Braunschweig, Germany). Flow cells were inoculated with an overnight culture of P. putida JB156, SM1639, or SM1699 diluted to an OD450 of 0.1 in 0.9% NaCl. After inoculation, the medium flow was arrested for 1 h. Medium flow was then started, and the substrate was pumped through the flow cells at a constant rate of 0.2 mm/s using a Watson Marlow 205S peristaltic pump.
Embedding and 16S rRNA hybridization of hydrated biofilm samples.
For 16S rRNA hybridization, probe PP986 (specific for P. putida subgroup A [16]) labeled with the indocarbocyanine fluorescent dye CY3 was used. In situ detection of biofilm cells expressing Gfp in combination with 16S rRNA hybridization was performed by fixing and embedding of the biofilm, followed by hybridization as previously described (9).
Microscopy and image analysis.
All microscopic observation and image acquisition were performed on a scanning confocal laser microscope (SCLM) (TCS4D; Leica Lasertechnik, GmbH, Heidelberg, Germany) equipped with detectors and filter sets for simultaneous monitoring of fluorescein isothiocyanate/Gfp and CY3.
Simulated fluorescence projections (shadow projections) and vertical cross sections through the biofilms were generated by using the IMARIS software package (Bitplane AG, Zürich, Switzerland) running on a Silicon Graphics Indigo2 workstation (Silicon Graphics, Mountain View, Calif.). Images were further processed for display by using Photoshop software (Adobe, Mountain View, Calif.).
RESULTS
Construction of monitor strains.
In order to monitor the growth activity of bacteria in a complex environment, it is important that the monitoring system respond rapidly to local environmental changes and, at the same time, be suitable for detection at the level of single cells.
We therefore employed the growth rate-regulated E. coli ribosomal P1 promoter (4, 17) to drive expression of the marker genes. An oligonucleotide linker containing the growth rate-regulated 72-bp _rrnB_P1 promoter sequence (4) was synthesized and inserted upstream of the various monitor genes to enable monitoring of ribosomal synthesis, i.e., bacterial growth activity.
The gene encoding the green fluorescent protein is an excellent marker gene, since no external substrates or inducers are needed for its expression and detection. However, Gfp is a very stable protein (3, 36) which is only lost from cells by dilution during cell growth. Recently, we described new variant Gfp proteins (3) which have considerably shorter half-lives than wild-type Gfp. We chose one of these, Gfp(AAV), as a rapidly degradable marker protein.
The rrnB_P1::gfp monitor cassettes were introduced into the pUT-mini Tn_5 transposon delivery system (14, 20) and subsequently inserted into the chromosome of our model biofilm organism, P. putida R1. Materials and Methods and Fig. 1 contain the details of the various constructions and the mating procedure. The resulting Pseudomonas transconjugants (monitor strains) were tested prior to biofilm growth to determine their behavior in nutrient-limited chemostats and in energy exhaustion batch experiments to verify that growth rate control of the E. coli P1 promoter took place in P. putida R1 as well.
Gfp expression in liquid cultures.
In chemostats, cells growing with fixed rates determined by the substrate dilution rate expressed fluorescence signals which could be determined as a function of the growth rate. Each strain was grown in five parallel chemostats with different dilution rates. After initial stabilization of the chemostats (60 h or more), samples were taken for determination of the fluorescence signal emitted by the culture. As the wild-type strain, even in the absence of the gfp gene, exhibits considerable fluorescence, we also did a chemostat analysis of wild-type P. putida R1 (Nalr) (JBA156). For determination of a maximum reference activity, we included measurements of bacteria grown exponentially in batch cultures in minimal medium supplemented with surplus (10 mM) citrate.
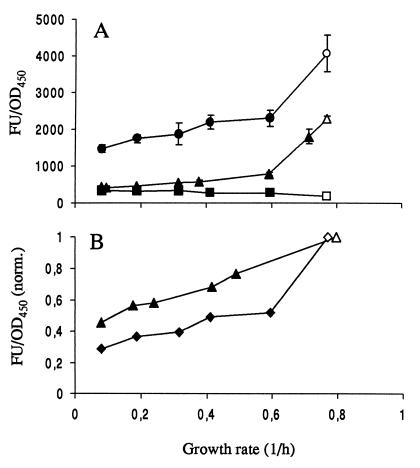
The fluorescence data for P. putida JB156 (wild type, Nalr), SM1699, and SM1639, harboring the _rrnB_P1-_gfp_mut3b* or _rrnB_P1-gfp(AAV) cassette, are plotted in Fig. 2A. The strains expressing stable _gfp_mut3b* fused to the _rrnB_P1 promoter showed reduced signal intensity as the growth rate was reduced. However, even at the slowest growth rates (in the present experiment, approximately 0.1 h−1), the signal intensity was more than three times stronger than the background autofluorescence signal emitted by wild-type strain JB156 itself. In contrast, the strain carrying the _rrnB_P1-gfp(AAV) cassette showed a detectable fluorescence signal only at growth rates above 0.4 h−1.
FIG. 2.
(A) Variation in fluorescence intensity [wt] as a function of growth rate of strains JB156 (wild-type [wt] P. putida R1 [Nalr]; ■), SM1699 (P. putida R1::P_rrnB_P1-_gfp_mut3b* wt; ●), and SM1639 [P. putida R1::P_rrnB_P1-gfp(AAV); ▴]. The growth rate was varied by growing cells in chemostats at different dilution rates (closed symbols) and exponentially in batch cultures (open symbols). Each point is the average of at least three independent measurements. Error bars indicate standard deviations. (B) Fluorescence intensity of strain SM1699 with the background from JB156 subtracted (⧫) compared with the fluorescence intensities of 16S rRNA hybridizations of wt P. putida R1 (JB156) cells (▴). Fluorescence intensity is normalized to 1 for cells grown in batch cultures. The ordinate axes are the same in panels A and B. FU, fluorescence units.
Since Gfpmut3b and ribosomes are rather stable compounds and are both under the control of the rRNA promoter, we expected comparable correlations between the ribosome and Gfpmut3b concentrations and the growth rate. We therefore compared the ribosome concentration that was estimated from hybridization data by using oligonucleotide probes targeting 16S rRNA (for a description of the technique, see, e.g., reference 27) with the fluorescence signal emitted by strain SM1699 carrying the cassette with _rrnB_P1 fused to stable _gfp_mut3b*. For comparison of the two different experiments, fluorescence intensity was normalized to that of cells exponentially growing in citrate medium. The results obtained for SM1699 (P. putida RI; _gfp_mut3b*) corresponded well to data obtained from rRNA probing experiments conducted under similar conditions (Fig. 2B). However, the fluorescence signal emitted by strain SM1699 decreased more with decreasing growth rate than the corresponding signals obtained from the rRNA hybridizations.
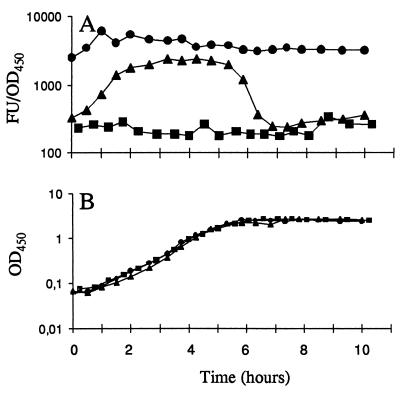
Energy exhaustion experiments in batch cultures, in which the cells experience the changing growth conditions from exponential growth to stationary phase, were employed to test the regulation of the P1 promoter and the stability of the Gfp proteins. Cultures were grown exponentially in the presence of a carbon source (citrate in these experiments) excess for at least 10 generations with repeated dilutions. In the final dilution, the cells were inoculated in medium with a carbon source limitation calculated to limit carbon source availability at an OD450 of approximately 2.0. Throughout the experiment, samples were withdrawn for measurement of OD450 and the Gfp fluorescence signal (Fig. 3A and B). When expressed from the _rrnB_P1 promoter, the stable Gfpmut3b protein remained relatively stable during the time of observation after the cells had exhausted the citrate supply. In striking contrast, the strain encoding the ribosomal promoter fused to gfp(AAV) showed a marked drop in fluorescence just prior to entering stationary phase, in accordance with published data for the phenotype of the _rrnB_P1 promoter (4). After less than 1 h in stationary phase, this strain had Gfp fluorescence below the detection limit. Consequently, the strain carrying the _rrnB_P1-gfp(AAV) fusion seems to be a useful marker for kinetic studies of changes in microbial growth behavior as changes in environmental conditions occur.
FIG. 3.
Energy exhaustion experiments demonstrating growth phase-dependent expression of the _rrnB_P1 promoter. Cells were grown in AB minimal medium with sodium citrate as the limiting energy source. Symbols: ■, JM156 (wild-type P. putida R1); ●, SM1699 (P. putida R1::P_rrnB_P1-_gfp_mut3b*); ▴, SM1639 [P. putida R1::P_rrnB_P1-gfp(AAV)]. Panels: A, fluorescence units (FU) per OD450 unit of the three strains; B, growth curves. The ordinate axes are the same in panels A and B.
Assessment of single-cell activity in biofilms.
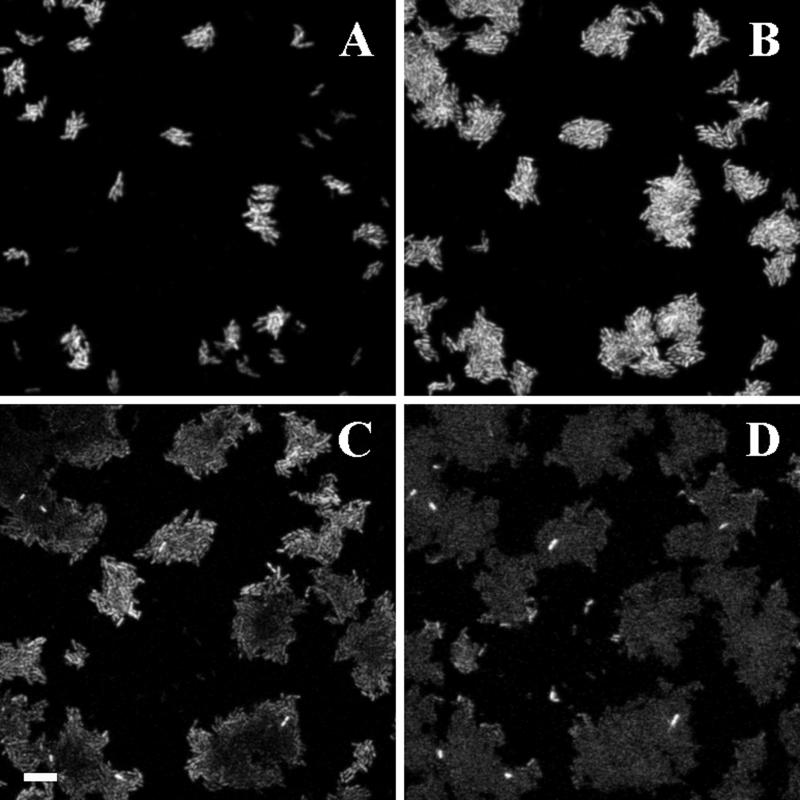
For determinations of bacterial activity at the single-cell level in complex biofilms, bacteria containing _rrnB_P1 fused to either the wild-type or the unstable variant gfp gene were introduced into flow chambers. To ensure that the signal emitted was caused by the Gfp protein and not by autofluorescence, we included the wild-type strain JB156 as a control. The autofluorescence of the wild-type strain was barely detectable in flow chamber experiments (Fig. 4A and B).
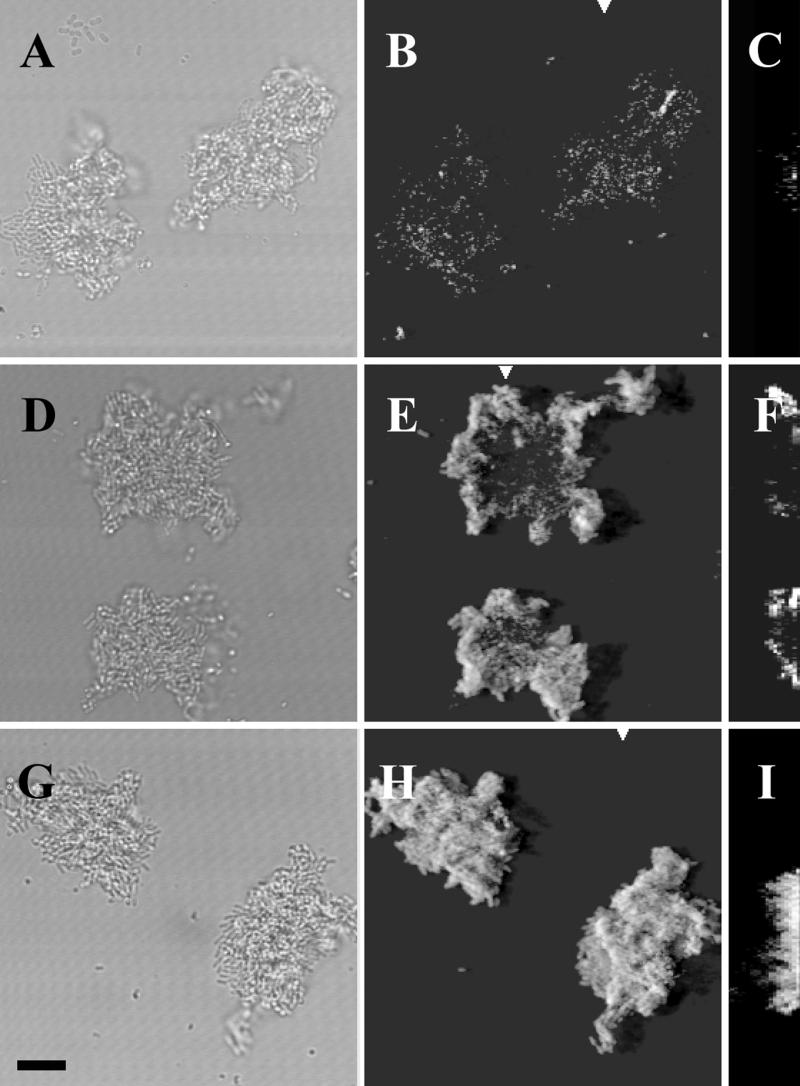
FIG. 4.
Demonstration of microbial growth activity in flow chamber monoculture biofilms. Comparison of different monitor strains. The panels represent flow chamber biofilms with the following strains: A, B, and C, JM156 (P. putida R1 [Nalr]); D, E, and F, SM1639 [P. putida R1::P_rrnB_P1-gfp(AAV)]; G, H, and I, SM1699 (P. putida R1::P_rrnB_P1-_gfp_mut3b*). Panels A, D, and G are bright-field images. Panels B, E, and H are SCLM images of the green fluorescent cells presented as simulated fluorescent projections where a long shadow indicates a large, high microcolony. Panels C, F, and I are vertical sections through the biofilm collected at positions indicated by the white triangles. The substratum is located at the left edge of panels C, F, and I. Vertical sections are included to provide information on vertical microcolony size. All images were recorded with the same SCLM settings, i.e., the same laser power and PMT setting. The bar indicates 10 μm, and the scale applies to all of the panels.
When the bacterial colonies in the flow channels reached a certain critical size (Fig. 4D and E), the light emitted by the cells carrying the _rrnB_P1-gfp(AAV) cassette decreased in the center of the colony, indicating that the growth activity of these cells was reduced compared to that of cells in the periphery of the colony. In fact, the signal intensity of the cells in the internal part of the microcolony was similar to the fluorescence activity of wild-type cells without Gfp (JB156) (compare Fig. 4E with 4B). When the cells carried _rrnB_P1 fused to the stable _gfp_mut3b* gene, a reduction in activity (or signal intensity) could not be detected in colonies comparable in size (Fig. 4G and H). The cells at the centers of these colonies continued to fluoresce long after the critical colony size (determined for the unstable gfp fusion) had been reached (data not shown).
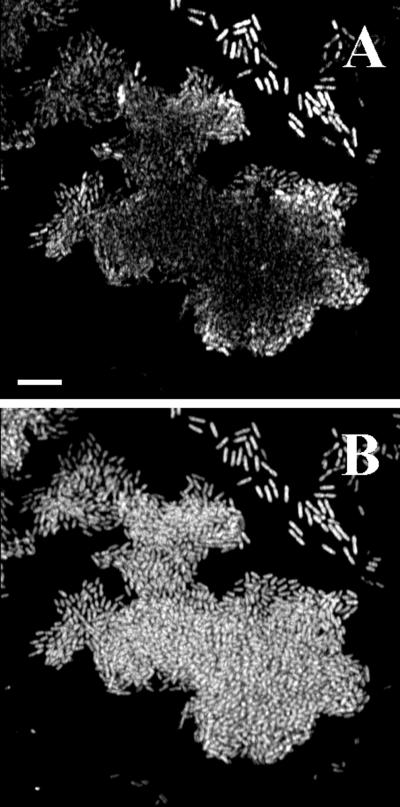
The sensitivity of our monitor system with strain SM1639 [_rrnB_P1-gfp(AAV)] was also compared to 16S rRNA hybridizations. A biofilm in a flow channel with microcolonies having reached the critical size (i.e., only cells at the periphery of each colony were green fluorescent) was fixed and embedded in acrylamide, leaving the biofilm in its native hydrated structure. Subsequently, the embedded biofilm was subjected to 16S rRNA hybridization using a _P. putida_-specific probe (PP986) labeled with the indocyanine dye CY3. This permits monitoring of the hybridization signal (proportional to the ribosome number) in combination with the Gfp signal emitted by the cells. Figure 5 shows a horizontal cross section of a representative microcolony from such a hybridization experiment. The hybridization signal from the center of the colony is only slightly weaker than that from the edges, whereas the green fluorescence signal varies from bright fluorescence at the border of the colony to very faint fluorescence in the middle. Thus, the strain carrying the new _rrnB_P1-gfp(AAV) cassette seems to provide online monitoring of changes from high to low bacterial activity. Such changes cannot be monitored by rRNA hybridization, because the ribosomes remain intact for a considerable time after de novo synthesis of rRNA has ceased.
FIG. 5.
Comparison of microbial activity in flow chamber biofilms monitored either as green fluorescence [Gfp(AAV) expression] emitted from strain SM1639 (P. putida R1::_rrnB_P1-gfp(AAV)] or by fluorescent rRNA hybridization of the same biofilm cells. Approximately 20 h after initial colonization of a flow chamber, the biofilm was fixed and embedded. The oligonucleotide probe PP986 labeled with CY3 was used for the hybridizations. Panel A represents Gfp(AAV) expression and panel B represents the hybridization signal in a horizontal SCLM cross section of the glass surface-attached cells in such an embedded biofilm. Bar, 10 μm.
Time course analysis of microbial activity during microcolony development.
Using the monitor system described above, the time course of a developing biofilm was monitored. With a confocal microscope equipped with a programmable XY specimen holder, we monitored several randomly selected positions in the biofilms. One such spot is shown in Fig. 6. During the early phases, single cells and small colonies consisting of only a few cells are seen. Virtually all of the bacteria in these phases are brightly green fluorescent as an indication of relatively high growth rates. After approximately 20 h (Fig. 6C), the colonies have become larger and the fluorescence from the central parts of these colonies is reduced, probably due to a lowered rate of Gfp synthesis. At the surface of the colonies, most cells are still fluorescent, indicating locally higher activity closer to the void space. The smaller microcolonies, however, are still fluorescent throughout and remain so for some time. Eventually (Fig. 6D), all of the cells in the microcolonies turn pale and only a few bright green cells are left. At this stage, most of the cells seem to have reduced their growth activity to a low level. This condition will persist if the conditions otherwise are kept constant.
FIG. 6.
On-line monitoring of establishment and changes in microbial activity in a flow chamber biofilm consisting of SM1639 [P. putida R1::P_rrnB_P1-gfp(AAV)]. The fluorescent signals emitted by the cells were visualized with SCLM. The images in panels A to D were recorded at the same flow chamber position 12, 17, 20, and 24.5 h after inoculation, respectively. Each image is presented as a horizontal cross section through a microcolony of cells located at the glass surface in the flow chamber. All images were recorded with the same SCLM setting. Bar, 10 μm.
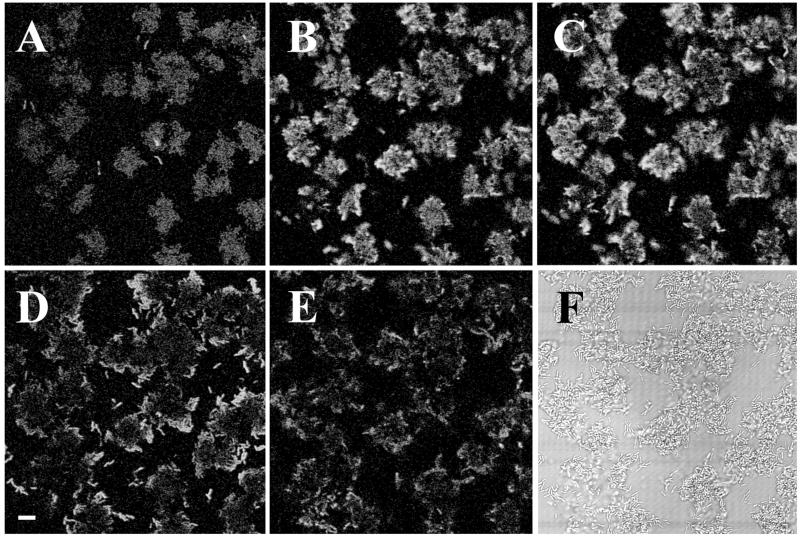
The cause of reduced growth activity in the biofilm colonies may be decreased penetration of nutrients through the biomass as the biofilm gets older and thicker. It is also possible that the biofilm cells gradually lose their capacity to become reactivated by nutrient supply. To investigate these possibilities, a developing biofilm consisting of P. putida R1 SM1639 [_rrnB_P1 gfp(AAV)] growing on a poor carbon source (benzyl alcohol) was monitored by confocal microscopy to the point where green fluorescence had faded (Fig. 7A). At this time, the carbon source in the medium was changed to citrate while the flow rate was kept constant. After a short period (approximately 2.5 h), the cells started to become green fluorescent again and remained so for approximately 2 h (Fig. 7B to D). Several hours after the upshift, fluorescence faded again and remained at a low level thereafter.
FIG. 7.
Effect of upshift in a P. putida R1::P_rrnB_P1-gfp(AAV) biofilm in a flow cell. Microcolonies were observed by SCLM. Biofilms were established and grown in AB minimal medium supplemented with 0.5 mM benzyl alcohol. A to E, 19, 23, 25, 26, and 28 h after inoculation, respectively; F, bright-field image of the same spot as shown in panel E. At 20 h, the medium was changed to FAB supplemented with 4 mM sodium citrate. Images were recorded as described in the legend to Fig. 6. Bar, 10 μm.
DISCUSSION
The in situ rRNA hybridization technique has created a revolution in the field of microbial ecology with respect to identification of individual bacteria in the most complex environments. The ribosome target for this method has turned out to be nearly ideal, since the cellular content of this organelle most often is sufficiently high to allow unambiguous identification at the single-cell level (1, 2). It was also realized very early in the development of the technology that in addition to species identification, quantitative rRNA hybridization might provide information about the growth state of the cells due to the direct correlation between ribosome concentration and cellular growth rate, such as it was known for several bacteria (27, 30).
The ribosome counting method should, however, be used and interpreted with caution. First, when using it in the context of new or even unknown organisms, it is essential to document that the correlation between growth rate and ribosome number is valid. This requires isolation of pure cultures and tests in chemostats or other types of conditions creating different cellular growth rates. Second, the stability of ribosomes under a variety of conditions may be different in different bacteria, and consequently, single determinations of cellular ribosome concentrations are not directly comparable. In fact, it is only in cells growing under steady-state conditions that the concentration of ribosomes truly reflects the growth state of the cells (27, 34). Third, the correlation between growth rate and ribosome concentration may be relevant only for bacteria able to grow fast under optimal conditions (carrying several copies of the rRNA operon). Thus, environmental monitoring of growth activity using this approach may, at best, only give indications of overall tendencies for fast-growing bacteria and only under conditions in which bacteria have a significant nutrient supply.
In order to expand the use of the well-known stringent control of ribosome synthesis, a method allowing determinations of rates of rRNA synthesis rather than the accumulated concentration was developed to better monitor the actual growth state of the cells, in particular if the conditions are heterogeneous with respect to nutrient status or if the outer environmental conditions change over time.
In our design of such a system for monitoring of bacterial growth activity, an rRNA promoter system, the rrnB operon from E. coli, was chosen because it is very well characterized and the promoter sequences are easily manipulated. The rrnB ribosomal genes are expressed from the _rrnB_P2P1 promoters. The ribosomal promoters are among the strongest described in E. coli, and the background expression is rather low when the cells are in late stationary phase. However, only the P1 promoter is growth rate regulated (17), for which reason we used it for expression of the reporter gene, gfp. This growth rate control on transcription from the E. coli P1 promoter is also exerted in P. putida, as demonstrated in our chemostat experiments, which also showed that the fluorescent signal emitted by strain SM1699 seemed to decrease more with decreasing growth rate than the corresponding ribosome number estimated from data obtained from the rRNA hybridizations. The cause of this difference could be that whereas general ribosome synthesis is normally controlled by tandem promoters like P1 and P2, the Gfpmut3b expression in our test system with SM1699 is under the control of only the _rrnB_P1 promoter. In this way, a steeper gradient of fluorescence signal intensity between slow- and fast-growing cells was obtained.
In test tube cultures of bacteria, rates of rRNA synthesis can be determined in hybridization experiments in which the growing RNA chains are labelled over a short period with radioactive precursors. The higher the frequency of transcription of rRNA, the more radioactivity is incorporated. This scheme, however, is not relevant in a complex and heterogeneous microbial community. Instead, we employed an unstable monitor system which, on one hand, reflects the rate of synthesis and, on the other, will indicate cessation of rRNA synthesis through disappearance of the signal. The monitor system was based on the Gfpmut3b protein. In its natural form, the protein is very stable, and consequently, when it was expressed from the stringently regulated P1 promoter, the energy exhaustion experiments showed that no significant decrease in fluorescence intensity occurred after entry into the stationary phase. However, by fusing the ribosomal promoter to the gfp(AAV) gene, encoding an unstable variant of Gfp, entry into stationary phase resulted in an immediate reduction in the fluorescence signal intensity.
In chemostats, the cells are continuously growing and the amount of the stable Gfp in the cells will be determined by the rate of transcription from the P1 rRNA promoter relative to the growth rate. In cells carrying a fusion to the unstable variant Gfp, the overall amount of fluorescing Gfp was lower under similar conditions, reflecting the fact that in these cells the Gfp level is determined by the rate of synthesis as well as the rate of degradation. Cells harboring fusions between the ribosomal promoter and the stable _gfp_mut3b* gene are good candidates as tools for estimating bacterial growth rates when the culture is in exponential growth, as in chemostats. In nature, however, bacteria rarely experience such well-defined and stable environments, and in such scenarios, the unstable Gfp reporter system has several advantages.
Microbial surface communities exhibit, in many ways, different physiological traits compared to suspended cultures (11), and it is therefore of interest to investigate the cellular response patterns to changing environmental conditions. If the key regulatory pathways in surface-bound cells differ from those known to control macromolecular synthesis in planktonic cells, there is a possibility of misinterpreting measurements of specific macromolecular synthesis rates. Recently, we documented, however, that the general correlations between cellular growth and parameters such as cell volume, ribosome concentration, division frequency, and DNA content seem to be unaltered in cells of P. putida when they are transferred from liquid- to solid-phase growth (27). It is therefore assumed here that the connection between rRNA synthesis rates—monitored by our gene fusions—and cellular growth activity in biofilms does indeed mimic what was demonstrated in the chemostats.
Thus, with the Gfp(AAV) physiological probing system, the cellular growth activity levels in biofilm samples were monitored on a temporal basis. One notable observation in biofilm microcolonies at stages later than the initial colonization phase was the appearance of dark centers in the colonies. In order to rule out the possibility that these nonfluorescent centers were caused by low oxygen concentrations (the maturation of Gfp has been reported to be dependent on molecular oxygen [19]), we measured the oxygen levels needed for Gfp maturation. Even at concentrations lower than 1% of the atmospheric oxygen tension, the maturation of Gfp was not inhibited (data not shown). DeBeer et al. (12) and others have measured the oxygen concentrations at various depths of biofilms, and within the distances which apply to our biofilms (i.e., less than 100 μm), the oxygen levels have been reported to drop less than 30-fold. Taken together, it seems unlikely that oxygen limitation inhibits Gfp maturation, even in the central parts of the microcolonies. We therefore suggest that the dark centers resulted from cells with reduced growth rates and, consequently, lower de novo Gfp synthesis. Strains tagged with stable Gfp versions were quite unaffected for the stage of colony development, and fluorescence persisted for several days in such microcolonies, in analogy with the persistence of high concentrations of ribosomes as monitored by in situ rRNA hybridization.
The down-regulation of microbial activity in the biofilm cells seemed to occur in at least two steps. First, the cells in the centers of the largest microcolonies reduced their growth activity whereas cells in smaller colonies remained highly active. Second, after a while, the cells in the smaller colonies also showed reduced growth activity. Some of the microcolonies in Fig. 6D (24.5 h after inoculation) were smaller than many of the colonies observed 7.5 h before (Fig. 6B), but nevertheless, their activity was also down-regulated. This indicates that it was not just the size of each individual microcolony that determined the reduced growth activity. It is possible that the major cause was a general exhaustion of the nutrient supply as a consequence of the overall higher density of bacteria. Thus, even in a rather thin biofilm, such as those analyzed in the present study, the microcolonies cannot be considered independent units since the activity of the single organisms inside one microcolony was dependent on factors caused by the surrounding microcolonies, e.g., depletion of nutrients.
In many studies of bacterial growth physiology, shifts or perturbations of the growth conditions have proven very informative with respect to clarification of the cellular response repertoire. A shift-up condition obtained by addition of an easily metabolizable carbon source to a slow-growing or stationary culture may thus document the level of responsiveness and the effects on different parts of the macromolecular synthesis. In bacteria with stringent control of ribosome synthesis, it is expected that ribosome synthesis is rapidly increased after a nutritional shift up. In addition to such cellular responses, the distribution of reactions to a shift up in a heterogeneous surface community may contribute to the understanding of how local environments differ with respect to penetration and diffusion of nutrients. Finally, growth inhibition of cells in structured communities could be determined by signals different from the nutrients. If the structure or size of the microcolonies determines the biological activity of the cells, a shift-up response to increased nutrient availability would not be expected.
The shift to a better carbon source in the present experiments clearly activated the cells in the microcolonies, as indicated by the significant increase in fluorescence. Moreover, the activation appeared to penetrate all the way through the microcolonies, showing that even the cells in the centers, which were the first to reduce their growth activity, were ready to respond to the improved nutrient supply. Overall, these results indicate that all of the cells in developing biofilm microcolonies are ready to respond to renewed supplies of nutrients. In addition, these findings support our conclusion that the oxygen levels in the microcolonies are sufficient for Gfp maturation.
The experiments performed in the present study have revealed a modulated growth activity of bacteria involved in the establishment of a new biofilm. In the initial phases, all cells were highly active, with ribosomal promoter activity corresponding to that of rapidly growing cells. After primary colonization and formation of small microcolonies, activity gradually decreased, first in the central parts of the microcolonies and eventually also at the surface. By choosing other variants of the Gfp protein, it is possible to construct reporter proteins with longer half-lives than those of the proteins employed here (3, 21). When fused with the rRNA promoter, such reporter systems may display alternative levels of cellular activity, which may be useful in monitoring the fine-tuning of microcolony development.
ACKNOWLEDGMENTS
This work was supported by grants from the Danish Biotechnology Program.
We thank Anne Nielsen for technical assistance.
REFERENCES
- 1.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen J B, Sternberg C, Poulsen L K, Bjørn S P, Givskov M, Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartlett M S, Gourse R L. Growth rate-dependent control of the rrnB P1 core promoter in Escherichia coli. J Bacteriol. 1994;176:5560–5564. doi: 10.1128/jb.176.17.5560-5564.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 6.Caldwell D E, Lawrence J R. Study of attached cells in continuous-flow slide culture. In: Wimpenny J W T, editor. CRC handbook of laboratory systems for microbial ecology research. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 117–138. [Google Scholar]
- 7.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 8.Christensen B B, Sternberg C, Andersen J B, Eberl L, Møller S, Givskov M, Molin S. Establishment of new genetic traits in a microbial biofilm community. Appl Environ Microbiol. 1998;64:2247–2255. doi: 10.1128/aem.64.6.2247-2255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, A. T. Nielsen, M. Givskov, and S. Molin. Molecular tools for study of biofilm physiology. Methods Enzymol., in press. [DOI] [PubMed]
- 10.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 11.Dagostino L, Goodman A E, Marchall K C. Physiological responses induced in bacteria adhering to surfaces. Biofouling. 1991;4:113–119. [Google Scholar]
- 12.DeBeer D, Stoodley P, Roe F, Lewandowski Z. Effects of biofilm structure on oxygen distribution and mass transport. Biotech Bioeng. 1994;43:1131–1138. doi: 10.1002/bit.260431118. [DOI] [PubMed] [Google Scholar]
- 13.DeLong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for identification of single cells. Science. 1989;243:1360–1362. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 14.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eberl L, Schulze R, Ammendola A, Geisenberger O, Erhart R, Sternberg C, Molin S, Amann E. Use of green fluorescent protein as a marker for ecological studies of activated sludge communities. FEMS Microbiol Ecol. 1997;149:77–83. [Google Scholar]
- 16.Franklin F C H, Bagdasarian M M, Timmis K T. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci USA. 1981;78:7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gourse R L, de Boer H A, Nomura M. DNA determinants of rRNA synthesis in E. coli: growth rate dependent regulation, feedback inhibition, upstream activation, antitermination. Cell. 1986;44:197–205. doi: 10.1016/0092-8674(86)90498-8. [DOI] [PubMed] [Google Scholar]
- 18.Hansen L H, Sørensen S J, Jensen L B. Chromosomal insertion of the entire Escherichia coli lactose operon, into two strains of Pseudomonas, using a modified mini-Tn5 delivery system. Gene. 1997;186:167–173. doi: 10.1016/s0378-1119(96)00688-9. [DOI] [PubMed] [Google Scholar]
- 19.Heim R, Prasher D C, Tsien R Y. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing nonantibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keiler K C, Waller P R H, Sauer R T. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 22.Kessler B, de Lorenzo V, Timmis K N. A general system to integrate lacZ fusions into the chromosomes of Gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol Gen Genet. 1992;233:293–301. doi: 10.1007/BF00587591. [DOI] [PubMed] [Google Scholar]
- 23.Kinniment S L, Wimpenny J W T. Measurements of the distribution of adenylate concentrations and adenylate energy charge across Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 1992;58:1629–1635. doi: 10.1128/aem.58.5.1629-1635.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kristensen C S, Eberl L, Sanchez-Romero J M, Givskov M, Molin S, de Lorenzo V. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J Bacteriol. 1995;177:52–58. doi: 10.1128/jb.177.1.52-58.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lappin-Scott H M, Costerton J W. Bacterial biofilms and surface fouling. Biofouling. 1989;1:323–342. [Google Scholar]
- 26.LeChevallier M W, Babcock T M, Lee R G. Examination and characterization of distribution system biofilms. Appl Environ Microbiol. 1987;53:2714–2724. doi: 10.1128/aem.53.12.2714-2724.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Møller S, Kristensen C S, Poulsen L K, Carstensen J M, Molin S. Bacterial growth on surfaces: automated image analysis for quantification of growth rate-related parameters. Appl Environ Microbiol. 1995;61:741–748. doi: 10.1128/aem.61.2.741-748.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Møller S, Sternberg C, Andersen J B, Christensen B B, Molin S. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl Environ Microbiol. 1998;64:721–732. doi: 10.1128/aem.64.2.721-732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer R J, Jr, Phiefer C, Burlage R S, Sayler G S, White D C. Single-cell bioluminescence and GFP in biofilm research. In: Hastings J W, Kricka L J, Stanley P E, editors. Bioluminescence and chemiluminescence: molecular reporting with photons. Chichester, England: John Wiley & Sons; 1996. pp. 445–450. [Google Scholar]
- 30.Poulsen L K, Ballard G, Stahl D A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poulsen L K, Dalton H M, Angles M L, Marshall K C, Molin S, Goodman A E. Simultaneous determination of gene expression and bacterial identity in single cells in defined mixtures of pure cultures. Appl Environ Microbiol. 1997;63:3698–3702. doi: 10.1128/aem.63.9.3698-3702.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ridgway H F, Olson B H. Scanning electron microscope evidence for bacterial colonization of a drinking-water distribution system. Appl Environ Microbiol. 1981;41:274–287. doi: 10.1128/aem.41.1.274-287.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez G G, Phipps D, Ishiguro K, Ridgway F. Use of a fluorescent redox probe for direct visualization of actively respiring bacteria. Appl Environ Microbiol. 1992;58:1801–1808. doi: 10.1128/aem.58.6.1801-1808.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaechter M, Maaløe O, Kjeldgaard N O. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 35.Stretton S, Techkarnjanruk S, McLennan A M, Goodman A E. Use of green fluorescent protein to tag and investigate gene expression in marine bacteria. Appl Environ Microbiol. 1999;64:2554–2559. doi: 10.1128/aem.64.7.2554-2559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tombolini R, Unge A, Davey M E, de Bruijn F J, Jansson J K. Flow cytometric and microscopic analysis of GFP-tagged Pseudomonas fluorescens bacteria. FEMS Microbiol Ecol. 1997;22:17–28. [Google Scholar]
- 37.Wolfaardt G M, Lawrence J R, Robarts R D, Caldwell S J, Caldwell D E. Multicellular organization in a degradative biofilm community. Appl Environ Microbiol. 1994;60:434–446. doi: 10.1128/aem.60.2.434-446.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]