Aczonin, a 550-Kd Putative Scaffolding Protein of Presynaptic Active Zones, Shares Homology Regions with Rim and Bassoon and Binds Profilin (original) (raw)
Abstract
Neurotransmitter exocytosis is restricted to the active zone, a specialized area of the presynaptic plasma membrane. We report the identification and initial characterization of aczonin, a neuron-specific 550-kD protein concentrated at the presynaptic active zone and associated with a detergent-resistant cytoskeletal subcellular fraction. Analysis of the amino acid sequences of chicken and mouse aczonin indicates an organization into multiple domains, including two pairs of Cys4 zinc fingers, a polyproline tract, and a PDZ domain and two C2 domains near the COOH terminus. The second C2 domain is subject to differential splicing. Aczonin binds profilin, an actin-binding protein implicated in actin cytoskeletal dynamics. Large parts of aczonin, including the zinc finger, PDZ, and C2 domains, are homologous to Rim or to Bassoon, two other proteins concentrated in presynaptic active zones. We propose that aczonin is a scaffolding protein involved in the organization of the molecular architecture of synaptic active zones and in the orchestration of neurotransmitter vesicle trafficking.
Keywords: synapse, neurotransmitter exocytosis, membrane traffic, PDZ domain, zinc finger
Neurotransmitter vesicle exocytosis is confined to synaptic specializations of distinctive architecture. Cell adhesion and extracellular matrix proteins keep the pre- and postsynaptic components in register and at a defined distance. At the postsynaptic side, a characteristic submembranous cytoskeletal structure, the postsynaptic density, is thought to mediate the anchoring and clustering of neurotransmitter receptors. The presynaptic specialization contains a machinery that mediates the rapid yet highly controlled exocytotic and reendocytotic trafficking of neurotransmitter vesicles. Exocytosis is restricted to the active zone, an area of the presynaptic plasma membrane where neurotransmitter vesicles are lined up in close vicinity to the cytoplasmic face of the membrane. Neurotransmitter release is triggered by Ca2+ influx through voltage-gated ion channels, which are also concentrated in the active zone membrane (Burns and Augustine 1995; Südhof 1995; Ziff 1997). Quick-freeze deep-etch electron microscopic studies have revealed a characteristic morphology of the presynaptic cortical cytoplasm. Besides other features, neurotransmitter vesicles were seen to be linked to each other and to cytoskeletal filaments by short cross-bridges, probably representing synapsin I, whereas vesicles directly at the active zone were often connected with the plasma membrane by longer strands (∼100 nm) of unknown identity (Landis et al. 1988; Hirokawa et al. 1989).
Recently, much has been learned about elementary molecular interactions that immediately contributes to the fusion of vesicles with and their reendocytosis from the presynaptic plasma membrane, such as the formation and structure of the fusion core complex or the involvement of dynamin in membrane scission. However, we need to better understand how the many interactions between individual vesicle, plasmalemmal, cytoskeletal, and cytosolic proteins and lipids are integrated into the high degree of spacial and temporal organization that must underlie neurotransmitter vesicle dynamics. Scaffolding proteins specific for active zones may play a role in this. One active zone–specific protein is Rim, a 170-kD protein that carries a PDZ domain and two C2 domains in its COOH-terminal part, and a pair of Cys4 zinc fingers at its NH2 terminus (Wang et al. 1997). Rim is associated with the plasma membrane, presumably through its PDZ and/or C2 domains, whereas its NH2-terminal region interacts with Rab3A, a small G protein involved in synaptic vesicle trafficking (Gonzalez and Scheller 1999). Another protein concentrated at the active zone, Bassoon, is 420 kD in size and associated with a cytoskeletal-like subcellular fraction (tom Dieck et al. 1998). Bassoon contains two pairs of Cys4 zinc fingers with sequence homology to those of Rim and rabphilin-3A (another Rab3A-binding protein), but no functional properties of Bassoon have yet been reported. Only immunomorphological but no molecular information is available for Piccolo, a third large protein concentrated at active zones (Cases-Langhoff et al. 1996).
Here, we describe the identification and initial characterization of a new active zone–specific protein, aczonin. Aczonin shares extensive regions of homology either with Bassoon or Rim, but also possesses unique sequence regions, including a polyproline stretch. Probably through this polyproline stretch, aczonin binds profilin, a protein involved in actin cytoskeletal dynamics. Aczonin is mainly associated with a detergent-resistant cytoskeletal-like subcellular fraction. We propose that aczonin is a scaffolding protein that interacts with multiple partner molecules and is involved in organizing the interplay between neurotransmitter vesicles, the cytoskeleton, and the plasma membrane at synaptic active zones.
Materials and Methods
cDNA Cloning and Northern Blot Analysis
Immunoscreening of a chicken brain cDNA expression library in λgt11 with a rabbit serum against the aqueous Triton X-114 fraction of chicken brain synaptic plasma membranes (Lichte et al. 1992) identified a strongly immunopositive clone designated 5.8. Starting from clone 5.8 (later determined to be colinear to codons 1505–1805 of mouse aczonin), full-length mouse cDNA and nearly full-length chicken (lacking ∼80 NH2-terminal codons) cDNA contigs were built by several rounds of hybridization and PCR screening of cDNA libraries from chicken (Lichte et al. 1992) and mouse brain (newborn whole brain, Stratagene; adult whole brain, Clontech). Database searches identified a human expressed sequence tag (EST) (IMAGE clone 192540; identical to STS WI-15215) that was obtained through Genome Systems, Inc., and completely sequenced. Total and poly(A)+ RNA preparation from chicken tissues and Northern blot analysis with 32P-labeled hybridization probes were performed according to conventional procedures. Human Northern blots were purchased from Clontech and hybridized according to the manufacturer's instructions. Chicken blots were hybridized with clone 5.8 and human blots with IMAGE clone 192540.
Antibodies
Two sequence regions that have little or no similarity with Bassoon and Rim (codons 485–754 [serum 1] and 1808–2150 [serum 2]) were amplified from mouse brain RNA and cloned into the His-tag vectors pQE31 and pQE30 (Qiagen). Subclone inserts were sequenced to confirm the absence of mutations. His-tag fusion proteins were expressed in bacteria, purified on nickel agarose, and used to immunize rabbits. Sera were affinity-purified with the same fusion proteins coupled to tresyl chloride–activated Sepharose (Sigma Chemical Co.). Commercial mAbs for mannosidase II (clone 53FC3; BAbCO), Na/K-ATPase β subunit (Upstate), Rab3 (clones 42.1 and 42.2; Transduction Laboratories, Inc., and Synaptic Systems), Rab5 (Transduction Laboratories, Inc.), transferrin receptor (clone H68.4; Zymed), and α-tubulin (Amersham Pharmacia Biotech), and sera for synaptophysin (Biometra) and rabphilin-3A (Synaptic Systems) were purchased from the sources indicated. An affinity-purified Mena antibody (LKE) was donated by Frank Gertler (Massachusetts Institute of Technology, Cambridge, MA), isoform-specific antisera against profilins I and II were gifts of Walter Witke (European Molecular Biology Laboratory, Heidelberg, Germany), and a KDEL receptor mAb was the gift of Wanjin Hong (University of Singapore, Singapore).
Immunoblotting and Subcellular Fractionation
To determine the tissue distribution of aczonin, tissues were homogenized in 0.32 M sucrose, 1 mM EDTA, 10 mM Tris, pH 7.4, 0.5 mM PMSF, 2 μg/ml pepstatin A, 2 μg/ml leupeptin, with a glass-Teflon homogenizer, or for muscle and heart, a turning-knife homogenizer. After spinning for 3 min at 900 g, 80 μg protein of each supernatant was resolved by SDS-PAGE (5% polyacrylamide), transferred to nitrocellulose, and the blot developed with affinity-purified rabbit antiaczonin and the ECL kit (Amersham Pharmacia Biotech).
For 120,000 g fractionation and reextraction experiments, 900 g supernatants of brain homogenates (in 150 mM NaCl, 1 mM EDTA, 10 mM Tris, pH 7.4, 0.5 mM PMSF, 2 μg/ml pepstatin A, 2 μg/ml leupeptin) were subjected to a 120,000 g centrifugation for 30 min at 4°C. Pellets were washed by resuspending in homogenization buffer followed by a second 120,000 g spin, and then resuspended either in homogenization buffer or in various solubilization buffers (see legend to Fig. 5), either for 20 min at 4°C or for 30 min at room temperature. The 120,000 g centrifugation was then repeated. Equal aliquots of pellets and supernatants were analyzed by immunoblotting as described above.
Figure 5.
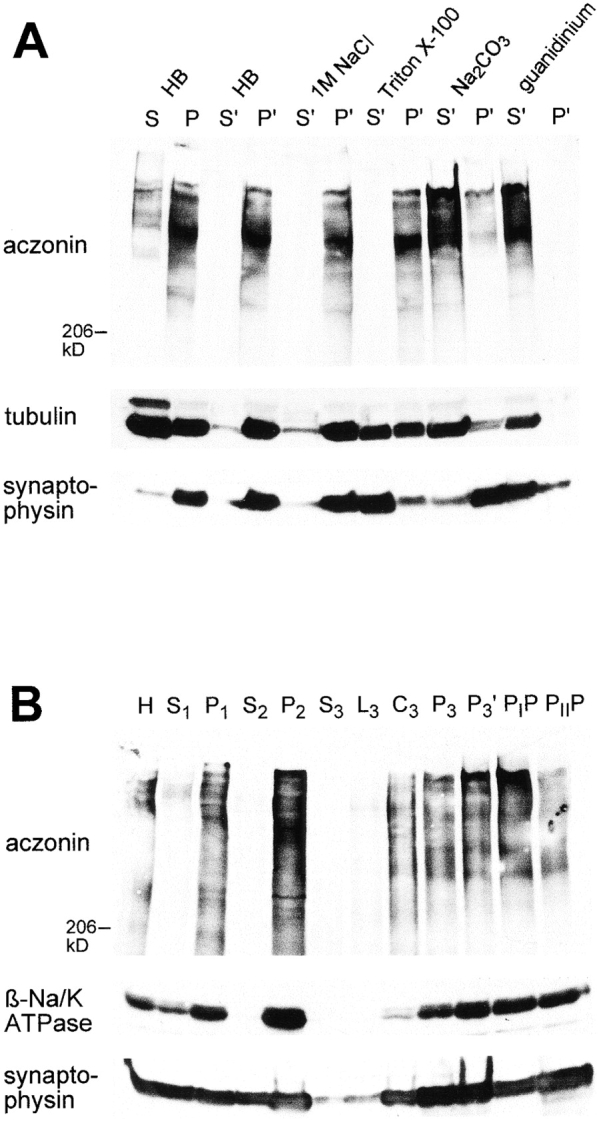
Distribution of aczonin in subcellular fractionation. (A) Mouse brain homogenate was subjected to 120,000 g fractionation (S, supernatant; P, pellet) in a detergent-free homogenization buffer (HB) containing 150 mM NaCl as described in Materials and Methods. The pellet fraction P was resuspended in the homogenization buffer (HB) or in various extraction buffers (1 M NaCl in homogenization buffer; 1% Triton X-100 in homogenization buffer without NaCl; 100 mM Na2CO3, pH 11.5; 6 M guanidinium chloride) and recentrifuged at 120,000 g. Supernatant and pellet fractions after recentrifugation are termed S′ and P′. Equal aliquots of all fractions were analyzed by SDS-PAGE and immunoblotting with aczonin, tubulin, and synaptophysin antibodies. In the experiment shown, extraction was carried out at 4°C for 20 min. The same distribution was obtained when extraction was performed at room temperature for 30 min. In additional experiments not shown, aczonin could be partially extracted from the pellet by 8 M urea, but not by 3% NP-40. (B) Synaptic vesicles were purified from rat brain according to Hell et al. 1988: H, homogenate; S1 and P1, 47,000 g supernatant and pellet derived from H; S2 and P2, 120,000 g supernatant and pellet derived from S1; supernatant S3, fluffy layer L3, cushion C3, and pellet P3 from the 260,000 g spin of S2; P3′, resuspended and cleared P3 before controlled-pore glass chromatography; PIP and PIIP, pools from breakthrough peak and vesicle peak of the controlled-pore glass chromatography. 30 μg protein was applied per lane and analyzed by immunoblotting as indicated.
Preparative subcellular fractionation procedures for the purification of synaptic vesicles or synaptic plasma membranes were performed according to Hell et al. 1988 and Babitch et al. 1976, respectively (see also Lichte et al. 1992, and Kutzleb et al. 1998).
Immunomorphological Analysis
Immunohistochemical procedures for light and electron microscopical analysis of rat brain were as described previously (Kutzleb et al. 1998). Identical results were obtained with affinity-purified antibodies against two different aczonin sequence regions (see above). Images shown in Fig. 4 were obtained with serum 2. Preimmune antibodies and preincubation of the immune antibodies with an excess of the recombinant antigen were employed as controls.
Figure 4.
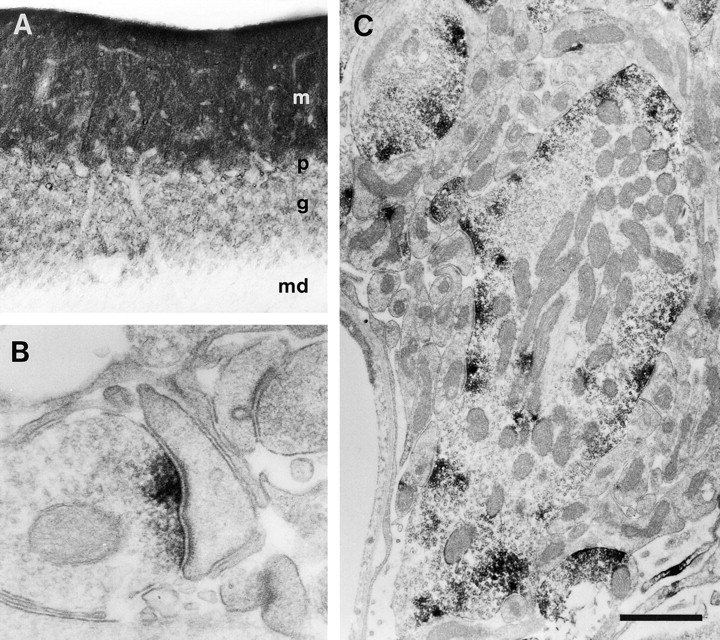
Immunohisto-chemical localization of aczonin in rat brain. Light microscopic inspection of the cerebellar cortex (A) shows finely punctate staining of the molecular layer (m), and ring-shaped or patchy immunopositive structures in the granule cell layer (g), whereas the medulla (md) is immunonegative. p indicates Purkinje cell layer. Electron microscopy shows that immunoperoxidase reaction product is restricted to the presynaptic active zones (B) of an asymmetric synapse with a dendritic spine in the molecular layer of the dentate gyrus or (C) of a mossy fiber terminal in a cerebellar glomerulus. In B, note that aczonin immunoreactivity is focused to the two junctional zones of the perforated synaptic specialization. Bar: 115 μm (A); 0.33 μm (B); or 1 μm (C).
Cell culture, immunofluorescence analysis, and brefeldin A treatment were performed according to conventional procedures. PC12 and NS20Y cells were fixed in 4% paraformaldehyde in PBS, and permeabilized with either 0.04% saponin or 0.2% Triton X-100. For double-labeling experiments, cells were incubated simultaneously with both primary antibodies. Antiaczonin was visualized with a biotinylated goat anti–rabbit secondary antibody (Vector Labs) followed by streptavidin-FITC. Antimannosidase II, anti-KDEL receptor, or antitransferrin receptor marker antibodies were visualized with a Cy3-conjugated goat anti–mouse antibody (Dianova).
Protein Binding Experiments
Recombinant Protein Constructs.
The mouse Rab3A sequence was taken from Baumert et al. 1993 and the mouse profilin I sequence from Sri Widada et al. 1989. The mouse profilin II sequence was identified by expressed sequence tag (EST) database screening (sequence data available from EMBL/GenBank/DDBJ under accession no. AA032658; 93% predicted amino acid sequence identity to human profilin II). Full-length coding sequences of these three proteins were amplified from mouse brain RNA and subcloned, with NH2-terminal His-tags, into pQE-31 (Qiagen). Sequences encompassing codons 374–654 and codons 863–1115 of mouse aczonin, and codons 11–399 of rat Rim (Wang et al. 1997) were amplified from mouse brain and subcloned into pGEX-4T (Amersham Pharmacia Biotech). All subclones employed for expression were verified by sequencing to be free of mutations.
Profilin Binding Experiments.
Mouse brain was homogenized in lysis buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 4 mM MgCl2, 2 mM EDTA, 10 mM NaF, 1 mM Na3VO4, 2 mM PMSF, 2 μg/ml pepstatin A, 2 μg/ml leupeptin, 2 μg/ml aprotinin, 2 mM benzamidine, and 0.2% Triton X-100). The 120,000 g supernatant was adjusted to 4 mg/ml total protein. The pellet was resuspended in 0.1 M Na2CO3 (pH 11.5) for 30 min at room temperature, dialyzed against several changes of lysis buffer with stepwise decreasing pH until pH 7.4, and adjusted to 6 mg/ml. Recombinant profilins or BSA (MBI Fermentas) were coupled to _N_-hydroxysuccinimide–activated Hi-Trap columns (Amersham Pharmacia Biotech) following the manufacturer's instructions. 20-μl aliquots of protein-coupled resins were preblocked with 3% BSA in PBS, washed with lysis buffer, and incubated with brain lysate for 4 h at 4°C under constant agitation. After spin, pellets were washed six times in lysis buffer and resuspended in SDS sample buffer. 1/20 vol of supernatants and 1/2 vol of pellets were analyzed by SDS-PAGE and immunoblotting. In poly-amino acid blocking experiments, resins were preincubated with 100 μl of 5 mg/ml polyproline (1–10 kD; Sigma Chemical Co.) or polyalanine (1–5 kD; Sigma Chemical Co.) in lysis buffer overnight at 4°C and washed once with 1 ml lysis buffer before incubation with lysates. Alternatively, profilins and other His-tagged protein constructs (see Results) were immobilized on nickel agarose (20 μl of resin per sample), incubated with lysates additionally containing 5 mM imidazole, and washed with lysis buffer additionally containing 50 mM imidazole.
Rab3A Binding Experiments.
For Rab3A overlay assay, equal quantities (2 μg) of glutathione _S_-transferase (GST)1 fusion proteins with similar-sized inserts from aczonin (amino acids 374–654 and 863–1115), Rim (11–399) as positive control, and paralemmin and HSB (Kutzleb et al. 1998) as negative controls, were resolved by SDS-PAGE and transferred to nitrocellulose. Blots were renatured overnight at 4°C in PBS with 50 μM ZnCl2 and 0.5 mM MgCl2, blocked for 2 h in overlay buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 2 mM MgCl2, 0.1% Tween 20, and 1% BSA) with 5% nonfat dry milk, rinsed in water, and incubated overnight at 4°C with 12 μg/ml of His-tagged Rab3A in the overlay buffer containing 2 μg/ml each of pepstatin A, leupeptin, and aprotinin, in the presence of 0.5 mM GTPγS, 1 mM GDP, or without nucleotides. Blots were washed with overlay buffer and processed for immunodetection using His-tag antibody (Qiagen) or Rab3A antibody (Synaptic Systems) and the ECL kit (Amersham Pharmacia Biotech). Recombinant Rab3A was also immobilized on nickel agarose and employed in precipitation experiments with brain lysates as described above for profilin, in the presence of either 0.5 mM GTPγS or 1 mM GDP. Immunoprecipitations with affinity-purified antiaczonin were performed with Pansorbin (Calbiochem) according to conventional procedures using mouse brain lysate prepared as described above, with the addition of 0.5% Triton X-100 and 0.5% BSA, in the presence of either 0.5 mM GTPγS or 1 mM GDP.
Results
Primary Structure of Mouse and Chicken Aczonin: Sequence Organization, Partial Homology with Bassoon and Rim, and Differential Splicing
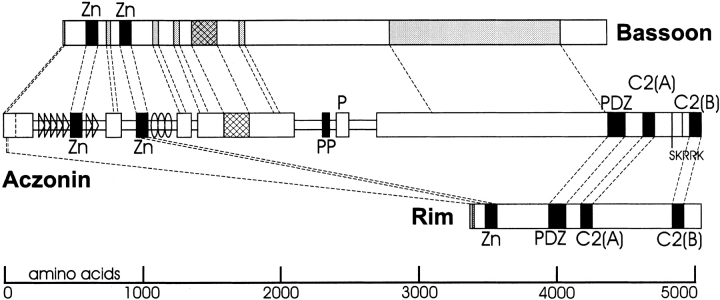
Aczonin was identified as a new molecular constituent of neuronal synapses by immunoscreening a brain cDNA expression library with antisera raised against synaptic plasma membranes (Lichte et al. 1992). An alignment of the cDNA-deduced mouse and chicken aczonin sequences with Bassoon is presented in Fig. 1, and an overview of the regional organization of the aczonin sequence and its similarities with Bassoon and Rim is given in Fig. 2. The longest splicing variant of mouse aczonin is predicted to be a 550-kD hydrophilic polypeptide of balanced charge (pI 6.4) featuring two mutually homologous pairs of Cys4 zinc fingers in the NH2-terminal region, a polyproline stretch in the middle (22 uninterrupted proline residues in the mouse and 11 in chicken), and a PDZ domain and two C2 motifs in the COOH-terminal region.
Figure 1.

Sequence alignment of mouse aczonin (mACZ), chicken aczonin (cACZ), and rat Bassoon (Bsn). The long splicing variants are shown (mouse, L; chicken, XL), and the position at mouse codon 4829 is indicated by an asterisk where the QQLRIQP sequence can instead be followed by the short SKRRK COOH terminus. Overlining beginning at mouse codon 430 marks three 10-mer repeat units deleted in some mouse cDNAs. The chicken sequence is incomplete for the ∼80 NH2-terminal codons. Upstream of the putative start codon, the mouse cDNA contig continues for 304 nucleotides of GC-rich sequence with no in-frame stop codon. Additional rescreenings did not yield sequences reaching further upstream. Between aczonin and Bassoon, the first nine codons are synonymous, whereas the upstream cDNA sequences are completely dissimilar, also suggesting that the codon assumed here as methionine 1 is the true start codon. Specific sequence motifs (see Fig. 2) are framed by arrowheads above the mouse sequence (except the zinc finger and polyproline motifs that are self-evident) and designated at the right margin. The rat Bassoon sequence is taken from tom Dieck et al. 1998. EMBL/GenBank/DDBJ sequence database accession numbers are Y19185-6 (mouse aczonin-L and S), Y19187 (chicken aczonin-XL), and Y19188 (partial human aczonin; data not shown).
Figure 2.
Regional organization of aczonin and partial homology to Bassoon and Rim. For aczonin, wide bars indicate sequence regions with high similarity between chicken and mouse, and narrow bars indicate sequences with low interspecies conservation. Triangles mark regions with 10-mer repeats, and ovoids mark the region with 22-mer repeats in the chicken sequence (actual repeat units are shorter and more numerous than these symbols). Black boxes represent zinc finger (Zn), polyproline (PP), PDZ, and C2 modules as indicated. Shaded boxes in Bassoon and Rim indicate additional sequence regions with similarity to aczonin. Cross-hatched boxes indicate a sequence region of particularly high conservation between mouse aczonin, chicken aczonin, and Bassoon. Regions of sequence similarity are connected by dashed lines. In aczonin, a vertical dashed line near the NH2 terminus indicates the end of the chicken sequence, and two vertical lines between the C2 modules indicate the sites of differential splicing.
Sequence comparison between mouse and chicken reveals an organization into regions of high or low interspecies conservation (Fig. 2). In particular, the sequences surrounding the two zinc finger pairs and the polyproline stretch are poorly conserved, suggesting that they mainly serve as spacers. This interpretation is further supported by the finding that the two zinc finger pairs are flanked by blocks of sequence repeats. Upstream of the first zinc finger pair is a series of degenerate proline- and glutamine-rich 10-mer repeats (consensus, KPxPQQPGPx, other residues often small or aliphatic amino acids). These repeats are found in different numbers (15–25) in chicken, mouse, and humans. We also isolated mouse cDNA sequence variants differing in the presence or absence of three 10-mer repeat units (Fig. 1), which may reflect differential splicing or a genomic polymorphism. Downstream from the second zinc finger pair, the chicken and mouse sequences diverge again for several hundred amino acids, and the mouse sequence is shorter, whereas the chicken sequence has expanded by eight lysine-rich ∼22-mer repeats (core motif, VQKED … SADKI). We suppose that both repeat series served to expand the spacer sequences during evolution, although it remains possible that the 10-mer repeats have an additional function.
Aczonin shares regions of homology with two other proteins concentrated at active zones, Rim and Bassoon (Fig. 2). Rim possesses a similar COOH-terminal array of one PDZ and two C2 domains, and both Cys4 zinc finger pairs of aczonin have sequence similarity with the single pair of Cys4 zinc fingers of Rim. Outside these circumscript molecular modules, we do not detect significant sequence similarity between aczonin and Rim, except a motif of 19 amino acids with 68% identity close to the NH2 termini of both proteins (DLSQLSEEERRQIAAVMSR; Fig. 1 and Fig. 2). Upstream of the PDZ domain, many but not all sequences highly conserved between mouse and chicken aczonin are homologous to Bassoon. This includes the two zinc finger pairs and the extreme NH2 terminus, but not the polyproline stretch and another proline-rich region downstream from it. A highly charged region between amino acids 1550–1700 of mouse aczonin is particularly conserved between mouse and chicken aczonin and Bassoon. In summary, the aczonin sequence can be structured into Rim-related, Bassoon-related, and aczonin-specific regions or motifs. The zinc finger motifs are similar in all three proteins.
Two sequence regions near the COOH terminus are subject to differential splicing. Splicing at codon 4829 of the mouse sequence can abort the downstream 210 amino acids, including the C2B domain, and replace them by a short SKRRK COOH terminus. Out of nine adult mouse brain cDNAs from this region that were sequenced, eight encode the shorter (aczonin-S) and one the longer COOH terminus (aczonin-L). From chicken, only one cDNA was isolated. It encodes the longer COOH terminus plus 61 additional codons (AHKS … PEGA) inserted at a position corresponding to mouse codon 4829 (aczonin-XL). Reverse transcription PCR with primers between positions corresponding to codons 4751 and 4946 of the mouse sequence indicated that mRNAs with and without this insert are expressed in similar quantities in chicken brain. In the human aczonin gene (see Discussion), the sequence encoding the short SKRRK COOH terminus is contiguous with the sequences immediately upstream in the cDNA, whereas the alternative COOH-terminal sequences, including the XL insert, are encoded by exons further downstream.
Aczonin Is a Brain-specific Protein
Northern blot analysis of RNAs from various chicken and human tissues showed that aczonin mRNA, which is very large and therefore partially degraded, is most highly expressed in the brain and detectable at low abundance in several endocrine glands but in none of the other tissues analyzed (Fig. 3A and Fig. B). In both species, testis mRNA gave a unique band pattern, with smaller molecular sizes than in the other tissues. Antisera were raised against recombinant aczonin partial sequences and employed for Western blot analysis of mouse tissue homogenates. A very large protein far above the 206-kD marker, apparently partially degraded, was detected in different brain regions and, after longer exposure, very weakly in stomach but in none of the other tissues analyzed, including adrenal gland, testis, and pancreas (Fig. 3 C). In endocrine cells, aczonin protein may be poorly translated from the mRNA or rapidly degraded. Aczonin mRNA and protein are found in similar abundances in forebrain, cerebellum, and brainstem (Fig. 3A and Fig. C), indicating expression throughout the brain.
Figure 3.
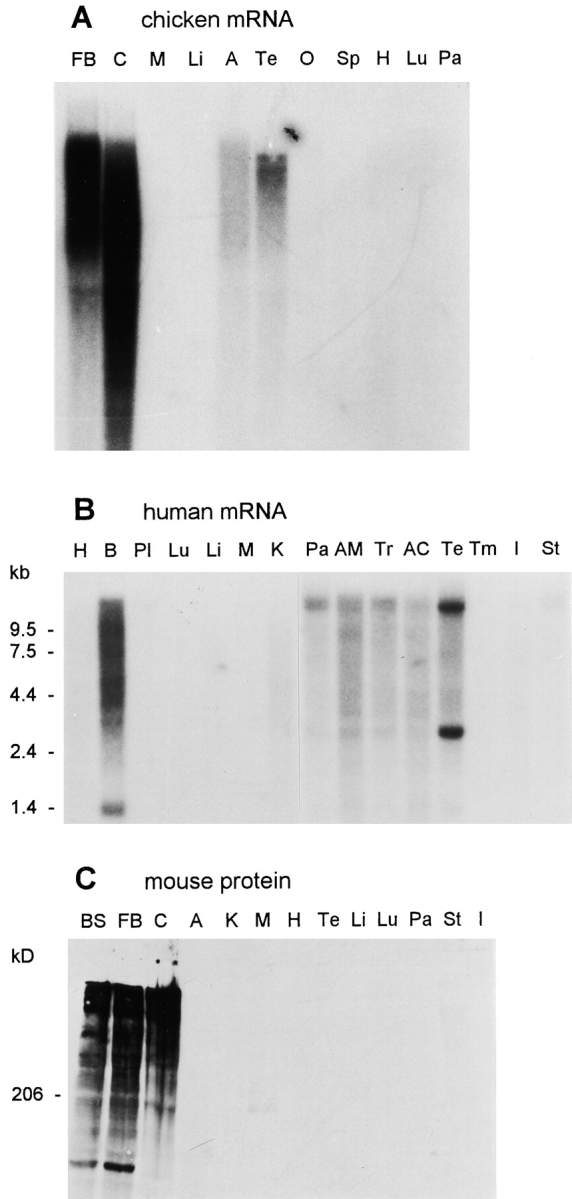
Tissue specificity of aczonin mRNA and protein expression. (A) Chicken aczonin mRNA (10 μg poly(A)+ RNA per lane). (B) Human aczonin mRNA (2 μg poly(A)+ RNA per lane). (C) Mouse aczonin protein (80 μg of tissue homogenate protein per lane). Tissue abbreviations are: A, adrenal gland; AC, adrenal cortex; AM, adrenal medulla; B, brain; BS, brain stem; C, cerebellum; FB, forebrain; H, heart; I, small intestine; K, kidney; Li, liver; Lu, lung; M, muscle; O, ovary; Pa, pancreas; Pl, placenta; Sp, spleen; St, stomach; Te, testis; Tm, thymus; and Tr, thyroid. Smears are attributed to partial degradation of these very long mRNA and protein molecules. Long exposures are shown to illustrate tissue specificity.
Aczonin Is Concentrated at the Active Zones of Synaptic Terminals
Antisera raised against aczonin-specific sequences labeled neuropil-rich areas throughout the rat brain. Cell bodies and myelin-rich areas were spared (Fig. 4 A). The examination of the cerebellum and the dentate gyrus by electron microscopy revealed that immunoreactivity concentrates at the presynaptic side of synaptic specializations. Immunoperoxidase reaction product is restricted to a space reaching from the plasma membrane of the active zone into the interior of the synapse by only a few neurotransmitter vesicle diameters (Fig. 4B and Fig. C). Aczonin immunoreactivity was found in many but not all synapses. In the glomeruli of the cerebellum, all mossy fiber terminals were decorated, whereas only a fraction of synapses between Golgi and granule cells showed reaction product (Fig. 4 C). Terminals of parallel fibers, which form contacts with Purkinje cell dendrites in the molecular layer of the cerebellum, were also labeled in many but not all cases. In the dentate gyrus only a subpopulation of granule cell mossy fiber terminals was immunopositive.
Aczonin Is Associated with a Detergent-resistant Cytoskeletal-like Subcellular Fraction in Brain, and with Intracellular Membranes in Neuronal Cell Lines
120,000 g fractionation of brain homogenate showed that ∼90% of total aczonin was recovered in the pellet and ∼10% in the supernatant (Fig. 5 A, fractions P and S). From the pellet, aczonin could not be extracted with 1 M NaCl or with 1% Triton X-100, but could be with 0.1 M sodium carbonate (pH 11.5). In this behavior, aczonin differed from the intrinsic membrane protein, synaptophysin, which was almost completely solubilized by the detergent but not by sodium carbonate, and was similar to the cytoskeletal control protein, tubulin (Fig. 5 A). This result suggests that aczonin is associated through polar interactions with a detergent-resistant cytoskeletal-like subcellular fraction.
The distribution of aczonin in a subcellular fractionation course leading to the purification of synaptic vesicles closely follows that of the plasma membrane marker, the Na/K-ATPase β subunit, and differs from the synaptic vesicle marker, synaptophysin, which partitions markedly also into the light fractions. In the final step of this procedure, controlled-pore glass gel filtration, synaptophysin is enriched in the small-vesicular fraction permeating the gel (fraction PIIP), whereas aczonin is enriched in the PIP exclusion peak, even more so than the plasma membrane marker. Codistribution with a plasma membrane marker was also observed in a fractionation procedure leading to synaptic plasma membranes (data not shown). These observations indicate that aczonin is not firmly associated with free synaptic vesicles, but rather with larger structures that sediment faster than vesicles in centrifugation and are excluded by the controlled-pore glass chromatography matrix, such as large cytoskeletal aggregates, the plasma membrane, or both.
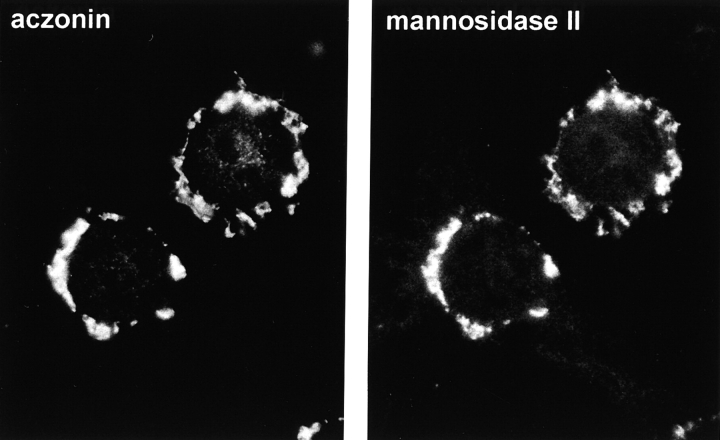
Neuronal cell lines also express aczonin. In these cells, it is associated with endomembrane structures within the cell body. In PC12 neuroendocrine cells, aczonin immunofluorescence is congruent with that of mannosidase II, a marker of the Golgi complex (Fig. 6). This aczonin and mannosidase II–positive membrane structure is fragmented within ∼5 min by brefeldin A, but is unaffected by wortmannin (not shown), further supporting its identification as the Golgi complex or a closely apposed structure such as the TGN. In NS20Y neuroblastoma cells, aczonin instead decorates more finely punctate structures that cluster around the nucleus or at the bases of processes but spare the Golgi region. This apparently vesicular compartment is not labeled by KDEL receptor (marker for ER–Golgi intermediate compartment and _cis_-Golgi) or transferrin receptor (recycling endosomes) immunofluorescence (data not shown). These observations suggest that aczonin, although not an intrinsic membrane protein, associates with membranes, membrane proteins, or the membrane-associated cytoskeleton during earlier stages of the secretory pathway, and in this way probably reaches its presynaptic destination in neurons.
Figure 6.
In neuronal cell lines, aczonin is associated with endomembranes. In PC12 cells as shown, double-immunofluorescence demonstrates colocalization with the Golgi complex marker, mannosidase II.
Aczonin Binds Profilin
A polyproline tract is conserved between chicken, mouse, and human aczonin, whereas extensive flanking regions are not or poorly conserved (Fig. 1 and Fig. 2). Synthetic polyproline (Petrella et al. 1996) and proline tracts in a number of proteins (Mahoney et al. 1997) are known to bind profilin, a small protein that is implicated in actin cytoskeletal dynamics, and which in neurons is concentrated in synaptic terminals (Faivre-Sarrailh et al. 1993). Efficient profilin binding is mediated by homoproline tracts of >10 residues or PPPPPG tandem repeats. Because it is well-established that isolated homopolymeric proline tracts like those in mouse and chicken aczonin bind profilin, we concentrated on probing for profilin binding to native, full-length aczonin.
Profilin exists in two isoforms, profilin I being expressed in many tissues including brain, and profilin II predominating in brain and skeletal muscle (Witke et al. 1998). We expressed both isoforms as His-tag fusion proteins, coupled them to Sepharose resin, and could specifically precipitate aczonin from brain lysates with both (Fig. 7). Aczonin precipitation is blocked by preincubation of the profilin resins with synthetic polyproline but not by polyalanine, indicating that polyproline and aczonin compete for the same binding site on profilin, aczonin probably through its polyproline tract (Fig. 7).
Figure 7.
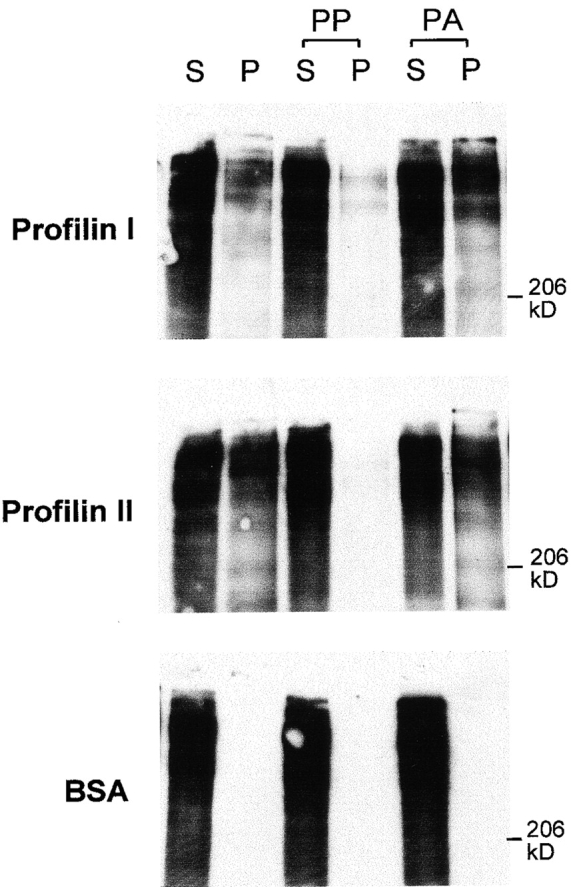
Aczonin binds profilin. Recombinant profilins I and II covalently coupled to Sepharose precipitate aczonin from mouse brain lysate (S, supernatant; P, pellet). Profilin binding is blocked by preincubation of the profilin resin with polyproline (PP), but not by polyalanine (PA). Immobilized BSA as a negative control does not precipitate aczonin. See Results for additional control experiments not shown.
In several independent experiments with equal quantities of covalently immobilized proteins as shown in Fig. 7, profilins I and II precipitated aczonin with similar efficiencies. In another set of experiments where equal quantities of recombinant profilins were immobilized on nickel agarose, profilin II consistently precipitated more aczonin than profilin I did. The established profilin-binding protein, Mena (Gertler et al. 1996), for which we probed as a positive control target protein in these experiments, was also more efficiently precipitated from brain lysate by profilin II than by profilin I (data not shown).
The specificity of profilin binding to aczonin was underpinned by additional controls (data not shown). Two additional His-tagged negative control constructs immobilized on nickel agarose, recombinant Rab3A (see below), and a 91 amino acid sequence from neurobeachin (construct C3; Wang, X., and M.W. Kilimann, unpublished data) did not precipitate either aczonin or Mena. A negative control target protein, neurobeachin, probed for by Western blotting, was not precipitated by the profilin constructs or by the negative control constructs. Profilins I and II bound both aczonin from the soluble fraction of a brain lysate (corresponding to fraction S of Fig. 5 A), and aczonin from the sedimentable fraction solubilized by 0.1 M Na2CO3 (fraction S′/Na2CO3 of Fig. 5 A) and then back-dialyzed against lysis buffer.
No Detectable Binding of Rab3A to Aczonin
The two zinc finger pairs of aczonin, but not their flanking sequences, have significant sequence similarity to the zinc finger structures of rabphilin-3A and Rim. Sequence regions from these two proteins encompassing the zinc fingers have been shown to bind Rab3A, a small G protein involved in synaptic vesicle trafficking. However, in three different types of experiments we were unable to detect binding of Rab3A and related Rab proteins to aczonin (data not shown). We expressed the two zinc finger pairs from mouse aczonin together with extensive flanking sequences (amino acids 374–654 and 863–1115, respectively) as GST fusion proteins, and probed for binding to recombinant Rab3A by blot overlay assay. No binding of recombinant Rab3A to these constructs was detected, in contrast to a corresponding Rim sequence (amino acids 11–399 fused to GST) employed as a positive control, which displayed pronounced GTP-dependent binding of Rab3A on the same blot. Recombinant His-tagged Rab3A immobilized on nickel agarose also did not precipitate detectable amounts of holo-aczonin from brain lysate; however, as positive controls, Rab3A precipitated rabphilin-3A, and resin-bound recombinant profilin (see above) precipitated aczonin in the same experiment. Finally, immunoprecipitation of aczonin from brain lysate did not bring down Rab3A, Rab3B, Rab3C, or Rab5 in quantities detectable by Western blot analysis.
Discussion
Aczonin is a very large multidomain protein of ∼5,000 amino acids which is specifically concentrated at presynaptic active zones and firmly associated with a detergent-resistant, cytoskeletal-like subcellular fraction. It may be a scaffolding protein that structures the presynaptic cortical cytoplasm, interacts with multiple partner molecules, and orchestrates the interplay of neurotransmitter vesicles with the cytoskeleton, the plasma membrane, and probably cytosolic proteins at the active zone. From the aczonin sequence, several motifs stand out that also suggest links to plasmalemmal, vesicular, and cytoskeletal proteins.
The overall pattern of one PDZ and two C2 domains in the COOH-terminal region is similar to Rim. Whereas many PDZ domain proteins have been identified that are involved in the organization of postsynaptic protein arrays (Craven and Bredt 1998), few presynaptic PDZ proteins are yet known. Rim and aczonin are the only two proteins known to contain both PDZ and C2 domains. C2 domains may bind proteins or membrane lipids. Alignment of the aczonin C2 domains with those of various other proteins shows that those of Rim are also the closest relatives in terms of sequence similarity (47% identity in the C2A and 42% identity in the C2B domains). Synaptotagmins are also close relatives with 35–42% identity, whereas Munc13 C2 sequences are less related, with 17–29% identity. All five aspartate residues critical for calcium chelation by C2 domains (Rizo and Südhof 1998) are conserved in the aczonin C2A domain. In the aczonin C2B domain, four of them are replaced by uncharged residues, as is the case in both C2 domains of Rim. Differential splicing of sequences between the two C2 modules is another feature that aczonin shares with Rim. In addition, the C2B domain of aczonin is subject to differential splicing, and the relative frequencies of short and long variants isolated from a mouse brain cDNA library suggest that aczonin-S, with a single COOH-terminal C2A domain, is more common. PDZ domains of other proteins are known to bind to the cytoplasmic COOH termini of transmembrane proteins or to signaling proteins (Oschkinat 1999). According to sequence alignment with a number of PDZ domains, that of Rim again seems to be the closest relative (36–41% identity with mouse and chicken aczonin, respectively). The PDZ and/or C2 domains are likely to anchor aczonin to the plasma membrane, and it will be of interest to identify their binding partners.
The two Cys4 zinc finger pairs of aczonin are homologous to each other, to two similar motifs in Bassoon, and to the single Cys4 zinc finger pairs of Rim, rabphilin-3A, and Noc2. According to cysteine residue spacing (CX2CX17CX2CX4CX2CX15CX2C in aczonin) and additional sequence similarity, the zinc finger pairs of these proteins constitute a distinct subfamily of zinc finger motifs, and the three-dimensional structure of its prototype, the rabphilin-3A zinc finger pair, has been solved recently (Ostermeier and Brünger 1999). A larger, related subfamily with the same cysteine residue spacing (four-residue interval between the two central cysteines) but a different sequence signature binds to the membrane lipid, phosphatidylinositol 3-phosphate, and is found, for example, in the synaptosomal-associated protein of molecular mass 25 kD (SNAP-25)–binding protein Hrs-2 and the Rab5-binding protein EEA1 (FYVE finger; Misra and Hurley 1999; see also sequence alignment in Wang et al. 1997). Rim, rabphilin-3A, and Noc2 have been shown to be functionally implicated in calcium-stimulated exocytosis, and sequence regions encompassing the zinc finger motifs of Rim and rabphilin-3A bind Rab3A, but Noc2 does not (McKiernan et al. 1996; Stahl et al. 1996; Kotake et al. 1997; Wang et al. 1997). We were unable to detect binding of Rab3A to aczonin, suggesting that the Bassoon/aczonin/rabphilin/Rim/Noc2-type zinc fingers (proposed designation: “BARRN fingers”) are not sufficient, and perhaps not necessary for Rab3A binding. They may be, at least in rabphilin-3A and Rim, effector domains that mediate an interaction between Rab3A binding and other molecular targets such as synaptic vesicle proteins or lipids. In agreement with this notion, the three-dimensional structure of the Rab3A-binding region of rabphilin-3A in complex with Rab3A shows that Rab3A is contacted by sequences upstream and downstream from the zinc finger, but not by the zinc finger motif itself (Ostermeier and Brünger 1999). The SGAWFF sequence motif that is part of the downstream Rab3A-binding interface of rabphilin-3A and is conserved in Rim but also in Noc2, is not present in either aczonin or Bassoon.
Aczonin contains several proline-rich regions that may include targets for the binding of SH3- or WW-domain–containing proteins, such as the NH2-terminal 1,100 amino acids around and upstream of the zinc fingers or a conserved sequence region at amino acids 2380–2500. Most strikingly, a polyproline stretch and short flanking sequences in the middle of the aczonin molecule are conserved between chicken, mouse, and humans, whereas several hundred amino acids around them are not or poorly conserved. This suggested an interaction with profilin, and we could indeed demonstrate that both profilin isoforms bind to aczonin and that this binding is blocked by homopolymeric proline but not by polyalanine. Profilin is an actin and phosphoinositide-binding protein expressed in many cell types, including neurons, where it is concentrated in synaptic terminals (Faivre-Sarrailh et al. 1993). It is believed to play an important role in the local remodeling of the actin cytoskeleton, although its exact mechanistic role(s) in interplay with actin, membrane lipids, and multiple profilin-binding proteins is insufficiently understood. Depending on specific conditions, profilin may either promote actin polymerization or depolymerization (for reviews see Mahoney et al. 1997, and Witke et al. 1998). Actin filaments are abundant in synaptic terminals but may be rarefied at the active zone (Landis et al. 1988). Control of the presynaptic microfilament architecture may be important for neurotransmitter vesicle dynamics, particularly in the vesicle reserve pool (Bernstein and Bamburg 1989; Wang et al. 1996; Lamaze et al. 1997; Bernstein et al. 1998; Job and Lagnado 1998; Kim and Lisman 1999). Binding of profilin to several presynaptic proteins, including the synapsins, clathrin, and dynamin has been described recently (Witke et al. 1998). In interplay with other presynaptic profilin-binding proteins, and possibly also with other proteins binding to its proline-rich sequences (Mahoney et al. 1999), aczonin may recruit profilin to or control its availability at the active zone.
At the gene level, a polyproline tract is encoded by a triplet repeat that has an intrinsic tendency to expand or contract. Therefore, it may be argued that the polyproline tract of aczonin has arisen by serendipity and may be physiologically meaningless at the protein level, although like any polyproline sequence, it binds profilin. However, it is clearly accessible for profilin binding in the context of the complete aczonin molecule, and it is highly conserved in evolution, even between birds and mammals (chicken, 11; mouse, 22; humans, 22 uninterrupted proline residues and additional prolines immediately upstream or downstream), whereas its flanking sequences are not. Leucine residues at or near the ends of proline runs, as in the aczonin sequences, are also found in other profilin-binding proteins (Mahoney et al. 1997). In contrast, for example, a polyglutamine tract in Bassoon is more heterogeneous in length, with 11 residues in the mouse, 24 in the rat, but only 5 in the human sequence (Hashida et al. 1998), whereas its flanking sequences are highly conserved. Therefore, the Bassoon polyglutamine is more likely to represent an autonomously fluctuating microsatellite-repeat DNA element with little physiological significance at the protein level.
The zinc finger region of aczonin is flanked by oligopeptide repeats. Dekapeptide repeats upstream of the first zinc finger motif differ in number between chicken, humans, and mouse and even among different mouse cDNAs, whereas 22-mer repeats downstream from the second zinc finger pair are found only in chicken but not in mouse. Therefore, it seems likely that these repeats primarily served as an evolutionary mechanism to rapidly expand spacer sequences around the zinc finger modules. The 10-mer repeat region is similar (45% predicted amino acid sequence identity) to an untranslated repetitive sequence region in the bovine herpesvirus type 1 BICP22 gene (Schwyzer et al. 1994). The Bassoon sequence contains a tract of heptapeptide repeats downstream from its second zinc finger pair (three copies in rat and five in mouse, unrelated in sequence to the aczonin repeats) that may also serve as a spacer. Long spacers could position binding partners of the zinc finger modules and of the polyproline tract at defined distances from the main body or along the backbone of aczonin, or they could facilitate the accommodation of bulky binding partners such as vesicles or large protein complexes.
We have also determined a partial cDNA sequence from the NH2-terminal region of human aczonin (codons 34–794). A partial human cDNA sequence representing the 1,213 COOH-terminal amino acids of the short splicing variant of human aczonin has been reported (KIAA0559; Nagase et al. 1998) (sequence data available from EMBL/GenBank/DDBJ under accession no. AB011131). Human aczonin genomic sequences connecting both partial cDNAs are also available (sequence data available from EMBL/GenBank/DDBJ under accession nos. AC004082 and AC004903), extending over several hundred kilobases and revealing a remarkable gene structure with some very large exons. The Bassoon gene also has unusually large exons (tom Dieck et al. 1998). Several sequence-tagged site markers (WI-15215 and UniGene Cluster Hs.12376) from both human cDNA sequence regions have been concordantly mapped to a region on human chromosome 7 corresponding to cytogenetic bands 7q11.23–q22.1 (NCBI GeneMap'98). However, no human neurological disease loci have yet been mapped to this region according to Online Mendelian Inheritance in Man (OMIM) (all databases accessible via http://www.ncbi.nlm.nih.gov).
Computer-assisted secondary structure prediction from the aczonin sequence indicates a high potential of flexibility, but also some sequence stretches with high coiled-coil potential that are shared with Bassoon. For example, anchored at the plasma membrane to ion channels or other transmembrane proteins through its PDZ domain, aczonin could potentially reach into the synaptic terminal across several neurotransmitter vesicle diameters. To give an upper-limit estimate, 5,000 amino acids could form an extended α-helix 750-nm long, i.e., 15 vesicle diameters. Thus, aczonin may play a role in organizing the supramolecular structure of the cortical cytomatrix at the active zone. It could constitute or be part of the longer strands seen in quick-freeze deep-etch electron microscopy to tether synaptic vesicles to the active zone (Landis et al. 1988; Hirokawa et al. 1989). In lamprey reticulospinal axons, Pieribone et al. 1995 observed a 300-nm-thick layer of synaptic vesicles adhering to the active zone independently of synapsin I. Aczonin, alone or together with proteins like Bassoon and Rim, may be responsible for the plasma membrane anchoring of this pool. Proteins tethering vesicles to target membranes at distances longer than the reach of soluble _N_-ethylmaleimide–sensitive factor attachment protein receptors (SNAREs), as observed recently at the Golgi complex (Orci et al. 1998; Sönnichsen et al. 1998), may be important for rapid and efficient vesicle trafficking and also for targeting specificity (Christoforidis et al. 1999; Yang et al. 1999). The synaptic target membrane SNAREs, syntaxin-1 and SNAP-25, are distributed all over the axonal plasmalemma (Garcia et al. 1995), so aczonin and related proteins may be essential to specifically target synaptic vesicles to the active zone.
According to immunoblot analysis and immunolight microscopy, aczonin is found throughout the brain. By immunoelectron microscopy of selected brain areas, we detect it very consistently in the mossy fiber terminals of cerebellar glomeruli, but only in a fraction of the terminals of other synapse populations. It remains to be clarified whether aczonin is present in all synapses, albeit at different concentrations, or only in specific populations, and with what functional features of synapses its level of expression and its different splicing variants correlate.
It will be of particular interest to understand the relationship between aczonin and its partial homologues, Bassoon and Rim. It will also be interesting to see whether Piccolo, which resembles both aczonin and Bassoon in molecular size, subcellular distribution, and immunomorphology, is identical to aczonin or whether it constitutes an additional member of this protein family. It is conceivable that these proteins can partially substitute for each other in different types of synapses, that their homologous domains interact with different isoforms or homologues of partner proteins, or that they work hand in hand within the same synapse. For example, aczonin, with a longer reach into the presynaptic cytoplasm than Rim, may usher vesicles towards the plasma membrane without interfering with Rab3 bound to them, and hand them over to Rim immediately before docking.
Acknowledgments
We thank Frank Gertler, Walter Witke, and Wanjin Hong for antibodies.
This work was supported by the Deutsche Forschungsgemeinschaft, the University of Bochum Medical School (FoRUM intramural research funds), and the Fonds der Chemischen Industrie.
Footnotes
1.used in this paper: GST, glutathione S-transferase
References
- Babitch J.A., Breithaupt T.B., Chiu T.-C., Garadi R., Helseth D.L. Preparation of chick brain synaptosomes and synaptosomal membranes. Biochim. Biophys. Acta. 1976;433:75–89. doi: 10.1016/0005-2736(76)90179-6. [DOI] [PubMed] [Google Scholar]
- Baumert M., Fischer von Mollard G., Jahn R., Südhof T.C. Structure of the murine rab3A genecorrelation of genomic organization with antibody epitopes. Biochem. J. 1993;293:157–163. doi: 10.1042/bj2930157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.W., Bamburg J.R. Cycling of actin assembly in synaptosomes and neurotransmitter release. Neuron. 1989;3:257–265. doi: 10.1016/0896-6273(89)90039-1. [DOI] [PubMed] [Google Scholar]
- Bernstein B.W., DeWit M., Bamburg J.R. Actin disassembles reversibly during electrically induced recycling of synaptic vesicles in cultured neurons. Mol. Brain Res. 1998;53:236–250. doi: 10.1016/s0169-328x(97)00319-7. [DOI] [PubMed] [Google Scholar]
- Burns M.E., Augustine G.J. Synaptic structure and functiondynamic organization yields architectural precision. Cell. 1995;83:187–194. doi: 10.1016/0092-8674(95)90160-4. [DOI] [PubMed] [Google Scholar]
- Cases-Langhoff C., Voss B., Garner A.M., Appeltauer U., Takei K., Kindler S., Veh R.W., De Camilli P., Gundelfinger E.D., Garner C.C. Piccolo, a novel 420 kDa protein associated with the presynaptic cytomatrix. Eur. J. Cell Biol. 1996;69:214–223. [PubMed] [Google Scholar]
- Christoforidis S., McBride H.M., Burgoyne R.D., Zerial M. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 1999;397:621–625. doi: 10.1038/17618. [DOI] [PubMed] [Google Scholar]
- Craven S.E., Bredt D.S. PDZ domains organize synaptic signaling pathways. Cell. 1998;93:495–498. doi: 10.1016/s0092-8674(00)81179-4. [DOI] [PubMed] [Google Scholar]
- Faivre-Sarrailh C., Lena J.Y., Had L., Vignes M., Lindberg U. Location of profilin at presynaptic sites in the cerebellar cortex; implication for the regulation of the actin-polymerization state during axonal elongation and synaptogenesis. J. Neurocytol. 1993;22:1060–1072. doi: 10.1007/BF01235749. [DOI] [PubMed] [Google Scholar]
- Garcia E.P., McPherson P.S., Chilcote T.J., Takei K., De Camilli P. rbSec1A and B colocalize with syntaxin 1 and SNAP-25 throughout the axon, but are not in a stable complex with syntaxin. J. Cell Biol. 1995;129:105–120. doi: 10.1083/jcb.129.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertler F.B., Niebuhr K., Reinhard M., Wehland J., Soriano P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell. 1996;87:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- Gonzalez L., Jr., Scheller R.H. Regulation of membrane traffickingstructural insights from a Rab/effector complex. Cell. 1999;96:755–758. doi: 10.1016/s0092-8674(00)80585-1. [DOI] [PubMed] [Google Scholar]
- Hashida H., Goto J., Zhao N., Takahashi N., Hirai M., Kanazawa I., Sakaki Y. Cloning and mapping of ZNF231, a novel brain-specific gene encoding neuronal double zinc finger protein whose expression is enhanced in a neurodegenerative disorder, multiple system atrophy (MSA) Genomics. 1998;54:50–58. doi: 10.1006/geno.1998.5516. [DOI] [PubMed] [Google Scholar]
- Hell J.W., Maycox P.R., Stadler H., Jahn R. Uptake of GABA by rat brain synaptic vesicles isolated by a new procedure. EMBO (Eur. Mol. Biol. Organ.) J. 1988;7:3023–3029. doi: 10.1002/j.1460-2075.1988.tb03166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N., Sobue K., Kanda K., Harada A., Yorifuji H. The cytoskeletal architecture of the presynaptic terminal and molecular structure of synapsin 1. J. Cell Biol. 1989;108:111–126. doi: 10.1083/jcb.108.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job C., Lagnado L. Calcium and protein kinase C regulate the actin cytoskeleton in the synaptic terminal of retinal bipolar cells. J. Cell Biol. 1998;143:1661–1672. doi: 10.1083/jcb.143.6.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.-H., Lisman J.E. A role of actin filament in synaptic transmission and long-term potentiation. J. Neurosci. 1999;19:4314–4324. doi: 10.1523/JNEUROSCI.19-11-04314.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake K., Ozaki N., Mizuta M., Sekiya S., Inagaki N., Seino S. Noc2, a putative zinc finger protein involved in exocytosis in endocrine cells. J. Biol. Chem. 1997;272:29407–29410. doi: 10.1074/jbc.272.47.29407. [DOI] [PubMed] [Google Scholar]
- Kutzleb C., Sanders G., Yamamoto R., Wang X., Lichte B., Petrasch-Parwez E., Kilimann M.W. Paralemmin, a prenyl-palmitoyl-anchored phosphoprotein abundant in neurons and implicated in plasma membrane dynamics and cell process formation. J. Cell Biol. 1998;143:795–813. doi: 10.1083/jcb.143.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamaze C., Fujimoto L.M., Yin H.L., Schmid S.L. The actin cytoskeleton is required for receptor-mediated endocytosis in mammalian cells. J. Biol. Chem. 1997;272:20332–20335. doi: 10.1074/jbc.272.33.20332. [DOI] [PubMed] [Google Scholar]
- Landis D.M.D., Hall A.K., Weinstein L.A., Reese T.S. The organization of cytoplasm at the presynaptic active zone of a central nervous system synapse. Neuron. 1988;1:201–209. doi: 10.1016/0896-6273(88)90140-7. [DOI] [PubMed] [Google Scholar]
- Lichte B., Veh R.W., Meyer H.E., Kilimann M.W. Amphiphysin, a novel protein associated with synaptic vesicles. EMBO (Eur. Mol. Biol. Organ.) J. 1992;11:2521–2530. doi: 10.1002/j.1460-2075.1992.tb05317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney N.M., Janmey P.A., Almo S.C. Structure of the profilin-poly-L-proline complex involved in morphogenesis and cytoskeletal regulation. Nat. Struct. Biol. 1997;4:953–960. doi: 10.1038/nsb1197-953. [DOI] [PubMed] [Google Scholar]
- Mahoney N.M., Rozwarski D.A., Fedorov E., Fedorov A.A., Almo S.C. Profilin binds proline-rich ligands in two distinct amide backbone orientations. Nat. Struct. Biol. 1999;6:666–671. doi: 10.1038/10722. [DOI] [PubMed] [Google Scholar]
- McKiernan C.J., Stabila P.F., Macara I.G. Role of Rab3A-binding domain in targeting of rabphilin-3A to vesicle membranes of PC12 cells. Mol. Cell. Biol. 1996;16:4985–4995. doi: 10.1128/mcb.16.9.4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S., Hurley J.H. Crystal structure of a phosphatidylinositol-3-phosphate-specific membrane targeting motif, the FYVE domain of Vps27p. Cell. 1999;97:657–666. doi: 10.1016/s0092-8674(00)80776-x. [DOI] [PubMed] [Google Scholar]
- Nagase T., Ishikawa K., Miyajima N., Tanaka A., Kotani H., Nomura N., Osamu O. Prediction of the coding sequences of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1998;5:31–39. doi: 10.1093/dnares/5.1.31. [DOI] [PubMed] [Google Scholar]
- Orci L., Perrelet A., Rothman J.E. Vesicles on stringsmorphological evidence for processive transport within the Golgi stack. Proc. Natl. Acad. Sci. USA. 1998;95:2279–2283. doi: 10.1073/pnas.95.5.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oschkinat H. A new type of PDZ domain recognition. Nat. Struct. Biol. 1999;6:408–410. doi: 10.1038/8203. [DOI] [PubMed] [Google Scholar]
- Ostermeier C., Brünger A.T. Structural basis of Rab effector specificitycrystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A. Cell. 1999;96:363–374. doi: 10.1016/s0092-8674(00)80549-8. [DOI] [PubMed] [Google Scholar]
- Petrella E.C., Machesky L.M., Kaiser D.A., Pollard T.D. Structural requirements and thermodynamics of the interaction of proline peptides with profilin. Biochemistry. 1996;35:16535–16543. doi: 10.1021/bi961498d. [DOI] [PubMed] [Google Scholar]
- Pieribone V.A., Shupliakov O., Brodin L., Hilfiker-Rothenfluh S., Czernik A.J., Greengard P. Distinct pools of synaptic vesicles in neurotransmitter release. Nature. 1995;375:493–497. doi: 10.1038/375493a0. [DOI] [PubMed] [Google Scholar]
- Rizo J., Südhof T.C. C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- Schwyzer M., Wirth U.V., Vogt B., Fraefel C. BICP22 of bovine herpesvirus 1 is encoded by a spliced 1.7 kb RNA which exhibits immediate early and late transcription kinetics. J. Gen. Virol. 1994;75:1703–1711. doi: 10.1099/0022-1317-75-7-1703. [DOI] [PubMed] [Google Scholar]
- Sönnichsen B., Lowe M., Levine T., Jämsä E., Dirac-Svejstrup B., Warren G. A role for giantin in docking COPI vesicles to Golgi membranes. J. Cell Biol. 1998;140:1013–1021. doi: 10.1083/jcb.140.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sri Widada J., Ferraz C., Liautard J.P. Total coding sequence of profilin cDNA from Mus musculus macrophage. Nucleic Acids Res. 1989;17:2855. doi: 10.1093/nar/17.7.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl B., Chou J.H., Li C., Südhof T.C., Jahn R. Rab3 reversibly recruits rabphilin to synaptic vesicles by a mechanism analogous to raf recruitment by ras. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:1799–1809. [PMC free article] [PubMed] [Google Scholar]
- Südhof T.C. The synaptic vesicle cyclea cascade of protein–protein interactions. Nature. 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- tom Dieck S., Sanmarti-Vila L., Langnaese K., Richter K., Kindler S., Soyke A., Wex H., Smalla K.H., Kämpf U., Fränzer J.-T. Bassoon, a novel zinc-finger CAG/glutamine repeat protein selectively localized at the active zone of presynaptic nerve terminals. J. Cell Biol. 1998;142:499–509. doi: 10.1083/jcb.142.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-H., Zheng J.Q., Poo M.-M. Effects of cytochalasin treatment on short-term synaptic plasticity at developing neuromuscular junctions in frogs. J. Physiol. 1996;491:187–195. doi: 10.1113/jphysiol.1996.sp021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Okamoto M., Schmitz F., Hofmann K., Südhof T.C. Rim is a putative Rab3 effector in regulating synaptic-vesicle fusion. Nature. 1997;388:593–598. doi: 10.1038/41580. [DOI] [PubMed] [Google Scholar]
- Witke W., Podtelejnikov A.V., Di Nardo A., Sutherland J.D., Gurniak C.B., Dotti C., Mann M. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:967–976. doi: 10.1093/emboj/17.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Gonzalez L., Jr., Prekeris R., Steegmaier M., Advani R.J., Scheller R.H. SNARE interactions are not selectiveimplications for membrane fusion specificity. J. Biol. Chem. 1999;274:5649–5653. doi: 10.1074/jbc.274.9.5649. [DOI] [PubMed] [Google Scholar]
- Ziff E.B. Enlightening the postsynaptic density. Neuron. 1997;19:1163–1174. doi: 10.1016/s0896-6273(00)80409-2. [DOI] [PubMed] [Google Scholar]