Rare Occurrence of Classical Hodgkin's Disease as a T Cell Lymphoma (original) (raw)
Abstract
Recent work identified Hodgkin and Reed-Sternberg (H/RS) cells in classical Hodgkin's disease (cHD) as clonal progeny of mature B cells. Therefore, it is generally assumed that cHD homogenously represents a B cell lymphoma. In a subset of cHD, however, H/RS cells expressing T cell–associated proteins may be candidates for alternative lineage derivation. Single H/RS cells with cytotoxic T cell phenotype were micromanipulated from three cases of cHD and analyzed by single cell polymerase chain reaction for immunoglobulin heavy (IgH) and light chain (IgL) gene rearrangements, T cell receptor (TCR)-β gene rearrangements, and germline configuration of the IgH and TCR-β loci. H/RS cells from two cases of cHD harbored clonal, somatically mutated Ig gene rearrangements, whereas TCR-β loci were in germline configuration. In contrast, H/RS cells from an additional case harbored clonal TCR-β variable/diversity/joining (VDJ) and DJ gene rearrangements, whereas the IgH locus was in germline configuration on both alleles. Thus, in two cases of cHD with H/RS cells expressing cytotoxic T cell molecules, the tumor cells are derived from mature B cells that aberrantly express T cell markers. In a third case, however, H/RS cells were derived from a T cell, demonstrating that cHD can also occur as a T cell lymphoma.
Keywords: Hodgkin's disease, T cell receptor genes, immunoglobulin genes, somatic hypermutation, Epstein-Barr virus
Introduction
In classical Hodgkin's disease (cHD), the malignant Hodgkin and Reed-Sternberg (H/RS) cells typically account for <1% of cells within a complex admixture of lymphocytes, plasma cells, histiocytes, and eosinophils 1. The origin of H/RS cells in cHD was enigmatic and a matter of debate for more than a decade. Although H/RS cells usually lack expression of B lineage markers, there is now strong evidence that H/RS cells represent the outgrowth of a dominant tumor clone derived from mature B cells 2 3. This conclusion is based on the amplification of clonally related Ig gene rearrangements from single micromanipulated H/RS cells 4 5. The presence and pattern of somatic mutations in the rearranged V genes identified germinal center B cells as the precursors of the tumor cells 5.
In a minority of cHD cases (∼5–15%), however, H/RS cells express cytotoxic T cell markers (granzyme B, perforin, and T cell intracellular antigen 1 [TIA-1]), raising the possibility that the tumor cells in these cases might originate from T lymphocytes 6 7 8 9. To clarify whether H/RS cells in such cases are indeed derived from T lymphocytes, H/RS cells were micromanipulated and subjected to single cell PCR analysis for rearranged Ig heavy chain (IgH), Igκ, and Igλ light chain genes, TCR-β VDJ and DJ gene rearrangements, as well as IgH and TCR-β germline configuration (i.e., absence of rearrangements). Whereas IgH VDJ gene rearrangements are specific for and restricted to B lineage cells, the presence of TCR-β VDJ gene rearrangements identifies a T cell. Studying H/RS cells from three such cases, a B cell genotype was found in two, a genotype revealing T cell origin in one case.
Materials and Methods
Clinical data on the three cases of cHD are summarized in Table .
Table 1.
Case Description of Patients with cHD
| Case | I | II | III |
|---|---|---|---|
| Age | 31 | 26 | 51 |
| Sex | Male | Male | Male |
| Presentation | First | First | First |
| LN biopsy site | Abdominal | Cervical | Inguinal |
| Stage | IIB | I | II |
| Hodgkin subtype | Mixed cellularity | Mixed cellularity | Nodular sclerosis |
| Phenotype of H/RS cells | |||
| CD30 | + | + | + |
| CD15 | + | + | + |
| CD20 | − | − | − |
| CD3 | − | − | − |
| TCR-α/β | − | ND | − |
| Granzyme B | + (90%) | + (60%) | + (40%) |
| Perforin | − | − | + (100%) |
| TIA-1 | + (60%) | + (>90%) | + (40%) |
| EBV | + | + | − |
| ALK | − | ND | − |
Immunostaining and Micromanipulation.
For immunostaining, 6–7-μm frozen tissue sections were stained using antibodies against CD30 (Fig. 1 A), CD20, LMP1, anaplastic lymphoma kinase (ALK)-1 (Dako), CD15 (Fig. 1 C; Becton Dickinson), CD3 (Ortho), TCR-α/β (T Cell Diagnostics), perforin (Neo Markers), granzyme B (Monosan), and TIA-1 (Immunotech). Stained cells were mobilized and aspirated with the help of a micropipette fixed to a hydraulic micromanipulator. Multiple cells were picked from each section. Buffer covering the sections was aspirated as negative controls for PCR analysis. For positive control of PCR, single B and T cells were either micromanipulated or sorted by flow cytometry.
Figure 1.
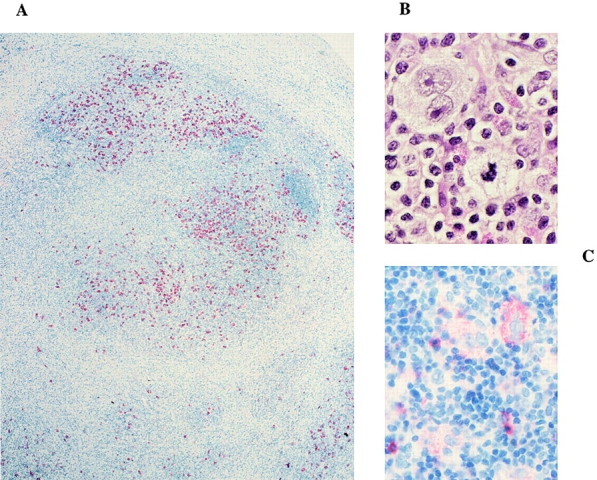
Immunostaining of cHD, case III. Histological stainings were as follows: (A) the tissue is stained for CD30 with the use of alkaline phosphatase (4-fold magnification); (B) hemalaun-eosin staining of some multinucleated Reed-Sternberg cells at 60-fold magnification; (C) staining for CD15 at 40-fold magnification. Staining of this case for expression of perforin is shown on the cover illustration of this issue.
Single Cell PCR.
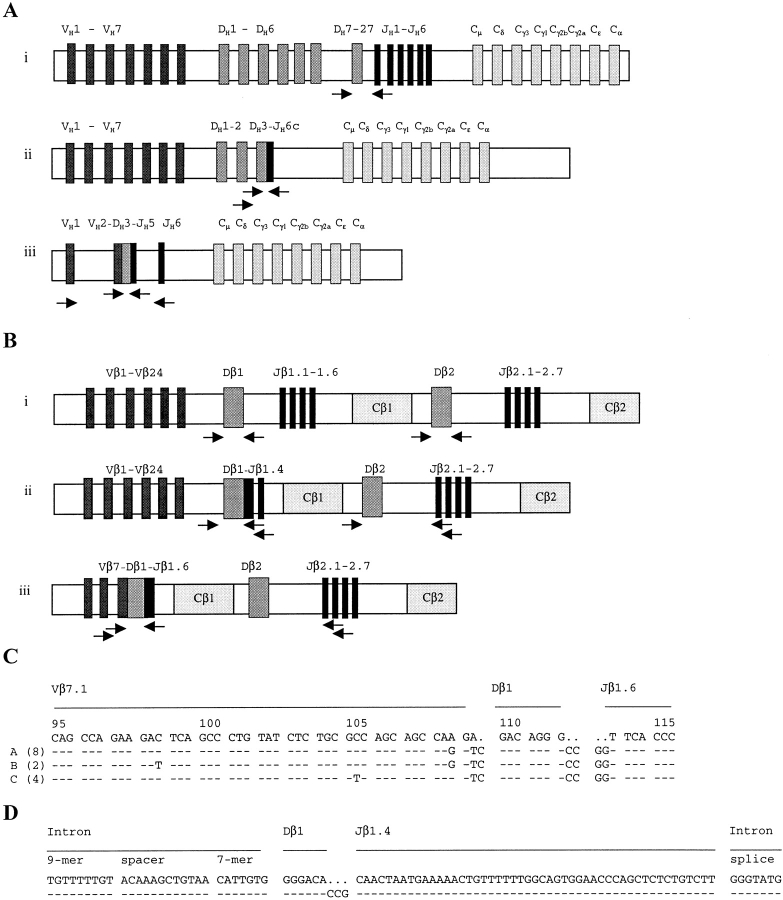
To analyze individual micromanipulated cells for IgH, Igκ, Igλ, as well as TCR-β VDJ and DJ gene rearrangements or germline configuration of the IgH and TCR-β loci, whole genome preamplification 10 was performed. Aliquots from these reactions were then subjected to two rounds of seminested PCR amplification as described previously. For analysis of the IgH and TCR-β loci, three PCR strategies were applied (Fig. 2A and Fig. B), one of which targets IgH (Fig. 2 A, iii) or TCR-β (Fig. 2 B, iii) VDJ rearrangements, a second IgH (Fig. 2 A, ii) or TCR-β (Fig. 2 B, ii) DJ rearrangements, and a third detects germline configuration of either the IgH (Fig. 2 A, i) or the TCR-β (Fig. 2 B, i) locus. Rearranged VH, Vκ, and Vλ genes were amplified using family-specific leader or framework region V gene primers and two sets of JH, Jκ, and Jλ primers in a seminested approach 5 11 12. DHJH rearrangements and germline configuration within the IgH locus were detected using seven DH family–specific primers and two sets of JH gene–specific primers in a seminested approach 5. In the case of germline configuration of the IgH locus, a 340-bp fragment was obtained with the DH7 primer, due to the close vicinity of the DH7-27 gene segment and JH1 (Fig. 2 A, i). DH family–specific primers were as follows: 5′-GTGTGCAGGCCTCRGTCTCTGTG-3′ for the DH1 gene family; 5′-GCACTGGGCTCAGAGTCCTCTC-3′ for the DH2 family; 5′-CCTCAGGTCAGCCCTGGACATC-3′ for the DH3 family; 5′-TGAGATCCCCAGGACGCAGCAC-3′ for the DH4 family; 5′-TCCCTGGGAAGCTCCTCCTGAC-3′ for the DH5 family; 5′-GACACCAGACAGAGGGGCAGGC-3′ for the DH6 family; and 5′-AGAGTGACTGGCAGGGTTGAGG-3′ for the DH7–27 gene. Amplification of TCR-β VDJ gene rearrangements was carried out as described previously using a panel of 24 Vβ family–specific primers and two sets of Jβ gene–specific primers in a seminested approach 13. Germline configuration was detected separately for both Cβ loci (Fig. 2 B, i) using primers binding to intronic sequences flanking the Dβ1 (5′-CCCCTTCGCCAAACAGCCTTA-3′ as forward, 5′-GAGTGAGGCAGAGGCATTCTGAAC-3′ as external reverse, and 5′-GCAGAGGCATTCTGAACCAAATTG-3′ as internal reverse primer) or the Dβ2 gene (5′-TCAGGGTGATGCATGTTCCAAGGA-3′ as forward, 5′-GGGACCCTGCAAGACCACAGCT-3′ as external reverse, and 5′-ACTCTTCCCACCTGGTAGCTGCAT-3′ as internal reverse primer). DβJβ rearrangements were amplified using the primers specific for intronic sequences in the upstream regions of the Dβ1 and the Dβ2 genes, together with primers specific for the Jβ1 or Jβ2 gene clusters, respectively (Fig. 2 B, ii).
Figure 2.
Amplification of IgH gene and TCR-β gene rearrangements from H/RS cells. PCR strategies for amplification of PCR products which are specific for IgH (A) or TCR-β (B) germline configuration (i), DJ gene rearrangements (ii), and VDJ gene rearrangements (iii) are depicted. VH1-VH7 and Vβ1-Vβ24 represent the seven Ig VH gene families and the 24 TCR Vβ gene families; and JH1-JH6 and Jβ1.1-1.6 and Jβ2.1-2.7 indicate the Ig JH and the TCR Jβ genes, respectively. The IgH DJ (A, ii) and IgH VDJ (A, iii) rearrangements and TCR-β DJ (B, ii) and TCR-β VDJ (B, iii) rearrangements depict those amplified from cases I and III, respectively. Arrows indicate the PCR primers used (not to scale). (C) A fragment (codons 95–115) of the sequence alignment of the clonal TCR Vβ7.1–Dβ1–Jβ1.6 rearrangement amplified from single H/RS cells of case III is given. The germline sequence of Vβ7.1, Dβ1, and Jβ1.6 genes (top) is compared with the clonal sequence variants (A, B, and C) obtained from eight, two, and four H/RS cells of case III, respectively. The three sequence variants (A, B, and C) differ by single nucleotide substitutions in codons 98, 105, and 108 (complete sequence data are available from GenBank/EMBL/DDBJ under accession nos. AJ243645–AJ243647). (D) Sequence alignment of the clonal TCR Dβ1–Jβ1.4 rearrangement amplified from 14 H/RS cells of case III is given. In contrast to the clonal Vβ7.1 gene rearrangement (C), the Dβ1–Jβ1.4 gene rearrangement does not exhibit intraclonal diversity (complete sequence data is available from GenBank/EMBL/DDBJ under accession no. AJ243648).
EBV infection of single H/RS cells was examined by amplification of a fragment of the EBV nuclear antigen 1 (EBNA1) gene by seminested PCR (5′-GGTCGCCGGTGTGTTCGTATATGG-3′ as forward, 5′-GCGGCAGCCCCTTCCACCATAG-3′ as external reverse, and 5′-AGGGAGGCAAATCTACTCCATCGTC-3′ as internal reverse primer).
PCR products were gel-purified and directly sequenced.
Results
H/RS cells from all three cases studied exhibit the typical immunophenotype of the tumor cells in cHD, as they coexpress CD30 and CD15 (for case III, see Fig. 1) but lack expression of the B cell antigen CD20 (Table ). H/RS cells from cases I and II express in part granzyme B (see cover of this issue), and TIA-1 and are EBV-positive, whereas H/RS cells from case III express perforin in addition to granzyme B and TIA-1 and are EBV-negative (Table ). Since the majority of anaplastic large cell lymphomas express one or more of the cytotoxic T cell markers studied here 7 8 9, diagnosis of cHD was further corroborated by the lack of nucleophosmin(NPM)-ALK gene expression in the H/RS cells. The NPM-ALK fusion protein arises from the translocation t(2; 5) (p23; q35), which is typically seen in anaplastic large cell lymphoma but not in cHD 14.
Efficiency of PCR amplification from single micromanipulated cells for all three cases was similar to that usually encountered (i.e., <40%, likely due to technical matters such as partial degradation or inaccessibility of DNA [3–5, 11–13]; Table ).
Table 2.
Summary of Single Cell PCR Analysis of Three Cases of cHD
| Case | Gene locus | Staining for micromanipulation | H/RS cells positive | PCR products | Rearrangements or germline configuration | Buffer controls positive | |
|---|---|---|---|---|---|---|---|
| Total | Sequenced | Repeated | Unique | ||||
| I | IgH | CD30 | 21/50 | 19 VH2 | 17 | 17 VH2 | 0/15 |
| 1 VH3 | 1 | 1 VH3 | |||||
| 1 VH4 | 1 | 1 VH4 | |||||
| 9/14 | 9 DH3–JH | 9 | 9 DH3-9–JH6c | 0/4 | |||
| Granzyme B | 3/15 | 3 VH2 | 3 | 3 VH2 | |||
| Igκ | CD30 | 0/16 | 0/4 | ||||
| Igλ | CD30 | 0/8 | 0/4 | ||||
| TCR Cβ1 | CD30 | 9/20 | 9 germline | 9 | 9 germline | 0/14 | |
| Granzyme B | 3/15 | 3 germline | 3 | 3 germline | |||
| TCR Cβ2 | CD30 | 9/20 | 9 germline | 9 | 9 germline | 0/14 | |
| Granzyme B | 3/15 | 3 germline | 2 | 2 germline | |||
| EBNA1 | CD30 | 7/10 | 7 EBNA1 | 0/4 | |||
| II | IgH | CD30 | 8/20 | 6 VH1 | 6 | 6 VH1 | 0/6 |
| 7 VH3 | 6 | 6 VH3 | |||||
| TIA-1 | 3/10 | 2 VH1 | 2 | 2 VH1 | 0/6 | ||
| 3 VH3 | 2 | 2 VH3 | |||||
| Igκ | CD30 | 9/20 | 9 Vκ1 | 7 | 7 Vκ1 | 0/6 | |
| TIA-1 | 2/10 | 3 Vκ1 | 2 | 2 Vκ1 | |||
| TCR Cβ1 | CD30 | 4/20 | 4 germline | 0/4 | |||
| TIA-1 | 6/10 | 6 germline | |||||
| TCR Cβ2 | CD30 | 9/20 | 9 germline | 0/4 | |||
| TIA-1 | 4/10 | 4 germline | |||||
| EBNA1 | CD30 | 8/20 | 8 EBNA1 | 0/6 | |||
| TIA-1 | 5/10 | 5 EBNA1 | |||||
| III | IgH | CD30 | 1/30 | 1 VH3 | 1 | 1 VH3 | 0/10 |
| 6/15 | 6 germline | 6 | 6 germline | 0/4 | |||
| Igκ | CD30 | 0/30 | 0/10 | ||||
| Igλ | CD30 | 0/30 | 0/10 | ||||
| TCR Cβ1 | CD30 | 20/30 | 14 Vβ7 | 14 | 14 Vβ7 | 1/10 | |
| 14 Dβ1–Jβ1 | 14 | 14 Dβ1–Jβ1.4 | |||||
| 1 germline | 1 | 1 germline | |||||
| TCR Cβ2 | CD30 | 17/30 | 17 germline | 7 | 7 germline | 0/10 | |
| EBNA1 | CD30 | 0/10 | 0/4 | ||||
| Controls: | Micromanipulated | Sorted by flow cytometry | |||||
| Gene locus | B cells | T cells | B cells | T cells | |||
| IgH | VDJ | 5/22 | 0/12 | 8/15 | |||
| DJ/germline | 9/10 | ||||||
| Igκ | 3/14 | 4/7 | |||||
| Igλ | 3/14 | 3/7 | |||||
| TCR-β | VDJ | 6/22 | 7/9 | ||||
| DJ/germline | 16/22 | 16/18 |
From case I, 50 CD30+ and 15 granzyme B+ H/RS cells were micromanipulated and analyzed by single cell PCR. From multiple H/RS cells of both subsets, a clonal Ig VH2-5–DH3-10–JH5b and a clonal DH3-9–JH6c gene rearrangement were amplified. The VH2 gene was rearranged in-frame, all sequences were identical, and the rearrangement was rendered nonfunctional by a somatic mutation generating a translation stop in codon 91 of framework region III (Table ). Thus, case I represents another example in which the H/RS cells have lost their capacity to express antigen receptor due to deleterious somatic mutations 5. No IgL, TCR-β VDJ, or DJ gene rearrangements were obtained analyzing multiple cells (Table ). Fragments corresponding to germline configuration of the IgH locus were not obtained, but fragments specific for germline configuration of the TCR Cβ1 and Cβ2 loci were repeatedly amplified (Table ). For the TCR Cβ2 locus, germline configuration could be assigned to both alleles because of the detection of two polymorphic forms of the Dβ2 gene (G and/or A at position 13 of the Dβ2 gene; see reference 15).
Table 3.
Sequence Analysis of Clonal Ig and TCR-β Gene Rearrangements
| Case | Gene locus | Rearrangement | Potentially functional | Percent mutation | Remarks |
|---|---|---|---|---|---|
| I | IgH | VH2-5–DH3-10–JH5b | No | 4.3 | Stop (codon 91) |
| DH3-9–JH6c | n.a. | 0.9 | |||
| TCR-β loci in germline | |||||
| II | IgH | VH1-8–DH3-22–JH6b | No | 3.5 | 47-bp deletion; |
| IgH | VH3-53–DH2-2–JH6b | No | 1.9 | Stop in CDR3 | |
| Igκ | Vκ1 (L12)–Jκ1 | No | 1.3 | Stop in CDR3 | |
| TCR-β loci in germline | |||||
| III | TCR-β | Vβ7.1–Dβ1–Jβ1.6 | Yes | 0.3–0.7 | Sequence variations involving codons 98, 105, and 108; three variants |
| TCR-β | Dβ1–Jβ1.4 | n.a. | 0 | ||
| IgH locus in germline |
Case II, which we studied previously for Ig gene rearrangements 4, was retrospectively found to express granzyme B and TIA-1 in a fraction of H/RS cells (Table ). 20 CD30+ H/RS cells and 10 TIA-1+ H/RS cells were micromanipulated and analyzed for IgH and Igκ gene rearrangements and configuration of the TCR-β loci. Two clonal IgH and one clonal Igκ gene rearrangement were amplified from H/RS cells regardless of their phenotype (Table and Table ). VH and Vκ genes were somatically mutated. The Vκ region gene and one of the VH gene rearrangements, which had both likely been originally productive, were rendered nonfunctional by somatic mutations (Table ). For unknown reasons, a second clonal Vκ gene rearrangement amplified in the first analysis was not obtained in the present study. No TCR-β gene rearrangements, but instead germline configuration for the TCR Cβ1 and Cβ2 loci, were detected (Table ).
Taking cases I and II together, amplification of clonal VH gene rearrangements rendered non-functional by deleterious somatic mutations together with the detection of germline configuration in both TCR-β loci identify the H/RS cells in these cases as the progeny of germinal center B cells that have lost their capacity to express antigen receptor due to “crippling” somatic mutations.
From case III, in which virtually all H/RS cell express perforin (Table ), 30 CD30+ H/RS cells were analyzed for IgH, Igκ, and Igλ gene rearrangements (half of the cells were in addition analyzed with VH leader, IgH DJ, and IgH germline primer collections). Only one IgH VDJ gene amplificate was obtained, likely representing cellular or other contamination (Table ). No IgH DJ gene rearrangement was obtained. However, a fragment specific for germline configuration was repeatedly amplified from H/RS cells. Within these IgH germline fragments, two distinct sequences of the JHψ1 pseudogene (G and/or A at position 45 of the JHψ1 pseudogene; see reference 16) were detected, suggesting that both alleles of the IgH locus are in germline configuration. In contrast, analysis of the TCR-β loci yielded two clonal rearrangements involving the two alleles of the TCR Cβ1 locus in about half of the H/RS cells. One allele harbors a clonal Vβ7.1–Dβ1–Jβ1.6 rearrangement (Fig. 2 C), whereas the other carries a clonal Dβ1–Jβ1.4 gene rearrangement (Fig. 2 D). The Vβ7.1 gene rearrangement is potentially functional. Unexpectedly, it exhibits intraclonal diversity (Fig. 2 C). Three different sequences were obtained (Fig. 2 C, sequences A–C). The three sequence variants were confirmed by repeated reamplification and sequencing from distinct aliquots of the whole genome amplification. Taken together, in case III the single cell PCR results identify a T cell as the progenitor of the tumor clone, thus classifying this case of cHD as a T cell lymphoma.
Discussion
Derivation of H/RS cells from mature B cells was previously demonstrated in 18 out of 18 informative unselected cases of cHD analyzed in Cologne and Frankfurt 3 4 5 17 18 19. In addition, recent results obtained by others indicate B cell derivation of H/RS cells in 24 out of 25 cases of cHD 20. On the other hand, in a nonselected collection of 13 cases of primary cHD, Daus and colleagues did not detect clonal TCR-γ gene rearrangements in micromanipulated H/RS cells from any of these cases 21. Therefore, these data collectively indicate that cHD represents a homogenous entity as a B cell lymphoma.
However, there are some observations raising the possibility that—in a subset of cHD—the tumor cells might stem from T lymphocytes. Some putative H/RS cell lines, for example, are derived from T cells 2. For these cell lines, proof of derivation from H/RS cells in the patients is missing. Furthermore, a TCR-α gene rearrangement was amplified from whole tissue DNA of lymphomatoid papulosis, cHD, and anaplastic large cell lymphoma occurring sequentially in one individual patient, suggesting a common T cell derivation of the three diseases 22. However, assignment of T cell genotype to H/RS cells of the Hodgkin's lymphoma was not conclusive: anaplastic large cell lymphoma versus classic cHD is often a difficult differential diagnosis 1 14. In this case, a clear discrimination between the two entities was particularly complicated because H/RS cells coexpressing CD30 and CD15 were found in both lymphomas. Furthermore, the TCR-α gene rearrangement identified in the Hodgkin's disease biopsy with the help of clone-specific primers represented a faint band, which might have arisen from a few contaminating cells originating from either the lymphomatoid papulosis or the anaplastic large cell lymphoma instead of the cHD. Finally, H/RS cells in a minority of cHD cases express cytotoxic granular molecules, in particular granzyme B, perforin, and TIA-1, which are otherwise typically found in cytotoxic T lymphocytes 6 7 8 9. Three such cases were analyzed here.
In cases I and II, molecular analysis of Ig and TCR-β loci revealed that the H/RS cells, despite expression of granzyme B and TIA-1 (Table ), were derived from germinal center B cells. Thus, expression of granzyme B and TIA-1 does not necessarily reflect a T cell origin of H/RS cells and shows that granzyme B and TIA-1 are aberrantly expressed by B lineage–derived H/RS cells. Similarly, expression of molecules thought to be specific for cells of the dendritic/myeloid lineage 23 by H/RS cells does not apparently reflect a derivation of these cells from dendritic or myeloid cells. Thus, H/RS cells can mimic cells of various hematopoietic lineages in terms of cell surface marker expression.
In case III, the H/RS cells harbor a clonal TCR-β VDJ and a clonal DJ gene rearrangement but no clonal Ig gene rearrangement. Furthermore, the IgH locus was found in germline configuration biallelically, directly demonstrating the absence of clonal IgH gene rearrangements. Given that the presence of a TCR-β VDJ gene rearrangement defines a T cell, the H/RS cells in this case are derived from a T lymphocyte. The Vβ7.1 gene rearrangement amplified from the T cell tumor clone exhibits significant intraclonal diversity. This was not expected, since TCR genes in T cells are usually not subject to somatic hypermutation, although there are some reports claiming the rare occurrence of somatically mutated TCR genes 24 25 26. Notably, somatic mutations were not observed in the Dβ1–Jβ1.4 rearrangement or in intronic sequences flanking the germline Dβ2 gene (830-bp sequences; see Table ). The distribution of somatic mutations among the distinct gene fragments of the TCR-β loci argues in favor of somatic hypermutation rather than some other type of somatic mutation (e.g., “genomic instability”) as the cause for the mutations in the TCR-β VDJ genes: in analogy to the Ig loci, somatic hypermutation would be expected to preferentially target VDJ joints rather than DJ rearrangements or germline genes, whereas genomic instability should not specifically target rearranged VDJ genes. Whether these mutations indeed reflect somatic hypermutation outside Ig loci, however, remains unclear.
In ∼5–15% of all cases of cHD, the H/RS cells exhibit cytotoxic T cell phenotype. Studying three of these cases, we found one to be T cell derived. Apart from this, 18 out of 18 nonselected cases of cHD in our collection are B lineage derived. On this basis, a rough estimate would be that cHD occurs as a T cell lymphoma at a low frequency (i.e., <5%).
This study establishes that rare cases of cHD derived from T cells indeed exist, indicating that cHD as defined by histopathology is not a uniform disease. It is remarkable that the transformation of both T and B cells can lead to the H/RS cell phenotype. Whether this reflects an initial transforming event inside the germinal center microenvironment in both cases and whether T and B cell derived H/RS cells can be distinguished in terms of gene expression patterns remain to be established.
Acknowledgments
We thank Michaela Fahrig, Christiane Gerhard, Julia Jesdinsky, and Tanja Schaffer for expert technical assistance; Manuel Montesinos-Rongen, Dr. Berit Jungnickel, and Dr. Ulf Klein for encouraging discussions; and Dr. Tilmann Spieker for performing EBV-encoded small RNA (EBER) in situ hybridization.
This work was supported by the Deutsche Forschungsgemeinschaft through SFB 502 and the Deutsche Krebshilfe, Dr. Mildred Scheel Stiftung.
References
- Burke J.S. Hodgkin's diseasehistopathology and differential diagnosis. In: Knowles D.M., editor. Neoplastic Hematopathology. Lippincott, Williams and Wilkins; Baltimore: 1992. pp. 497–533. [Google Scholar]
- Küppers R., Rajewsky K. The origin of Hodgkin and H/RS cells in Hodgkin's disease. Annu. Rev. Immunol. 1998;6:471–493. doi: 10.1146/annurev.immunol.16.1.471. [DOI] [PubMed] [Google Scholar]
- Bräuninger A., Hansmann M.-L., Strickler J.G., Dummer R., Burg G., Rajewsky K., Küppers R. Identification of common germinal-center B-cell precursors in two patients with both Hodgkin's disease and non-Hodgkin's lymphoma. N. Engl. J. Med. 1999;340:1239–1247. doi: 10.1056/NEJM199904223401604. [DOI] [PubMed] [Google Scholar]
- Küppers R., Rajewsky K., Zhao M., Simons G., Laumann R., Fischer R., Hansmann M.-L. Hodgkin diseaseHodgkin and H/RS cells picked from histological sections show immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc. Natl. Acad. Sci. USA. 1994;91:10962–10966. doi: 10.1073/pnas.91.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzler H., Küppers R., Hansmann M.-L., Rajewsky K. Hodgkin and H/RS cells in Hodgkin's disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J. Exp. Med. 1996;184:495–505. doi: 10.1084/jem.184.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudejans J.J., Kummer J.A., Jiwa M., van der Valk P., Ossenkoppele G.J., Kluin P.M., Kluin-Nelemans J.C., Meijer C.J. Granzyme B expression in H/RS cells of Hodgkin's disease. Am. J. Pathol. 1996;148:233–240. [PMC free article] [PubMed] [Google Scholar]
- Foss H.D., Anagnostopoulos I., Araujo I., Assaf C., Demel G., Kummer J.A., Hummel M., Stein H. Anaplastic large-cell lymphomas of T-cell and null-cell phenotype express cytotoxic molecules. Blood. 1996;88:4005–4011. [PubMed] [Google Scholar]
- Felgar R., Macon W.R., Kinney M.C., Roberts S., Pasha T., Salhany K.E. TIA-1 expression in lymphoid neoplasms. Identification of subsets with cytotoxic T lymphocyte or natural killer cell differentiation. Am. J. Pathol. 1997;150:1893–1899. [PMC free article] [PubMed] [Google Scholar]
- Krenacs L., Wellmann A., Sobara L., Himmelmann A.W., Bagdi E., Jaffé E.S., Raffeld M. Cytotoxic cell antigen expression in anaplastic large-cell lymphomas of T- and null-cell type and Hodgkin's diseaseevidence for distinct cellular origin. Blood. 1997;89:980–989. [PubMed] [Google Scholar]
- Zhang L., Cui X., Schmitt K., Hubert R., Navidi W., Arnheim N. Whole genome amplification from a single cellimplication for genetic analysis. Proc. Natl. Acad. Sci. USA. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräuninger A., Küppers R., Spieker T., Siebert R., Strickler J.G., Schlegelberger B., Rajewsky K., Hansmann M.-L. Molecular analysis of single B cells from T-cell-rich B-cell lymphoma reveals the derivation of the tumor cells from mutating germinal center B cells and exemplifies means by which immunoglobulin genes are modified in the germinal center B cells. Blood. 1999;93:2679–2687. [PubMed] [Google Scholar]
- Braeuninger A., Küppers R., Strickler J.G., Wacker H.W., Rajewsky K., Hansmann M.-L. Hodgkin and Reed Sternberg cells in lymphocyte predominant Hodgkin disease represent clonal populations of germinal center-derived tumor B cells Proc. Natl. Acad. Sci. USA. 94 1997. 9337 9342[published erratum at 94:14211] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roers A., Montesinos-Rongen M., Hansmann M.-L., Rajewsky K., Küppers R. Amplification of TCRβ gene rearrangements from micromanipulated single cellsT cells rosetting around Hodgkin and H/RS cells in Hodgkin's disease are polyclonal. Eur. J. Immunol. 1998;28:2424–2431. doi: 10.1002/(SICI)1521-4141(199808)28:08<2424::AID-IMMU2424>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Weiss L.M., Lopategui J.R., Sun L.H., Kamel O.W., Koo C.H., Glackin C. Absence of the t(2; 5) in Hodgkin's disease. Blood. 1995;85:2845–2847. [PubMed] [Google Scholar]
- Tunnacliffe A., Rabbitts T.H. Sequence of the Dβ2-Jβ2 region of the human T-cell receptor β-chain locus. Nucleic Acids Res. 1985;13:6651–6661. doi: 10.1093/nar/13.18.6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravetch J.V., Siebenlist U., Korsmeyer S., Waldmann T., Leder P. Structure of the human immunoglobulin μ locuscharacterization of embryonic and rearranged J and D genes. Cell. 1981;27:583–591. doi: 10.1016/0092-8674(81)90400-1. [DOI] [PubMed] [Google Scholar]
- Kanzler H., Hansmann M.-L., Kapp U., Wolf J., Diehl V., Rajewsky K., Küppers R. Molecular single cell analysis demonstrates the derivation of a peripheral blood-derived cell line (L1236) from the Hodgkin/H/RS cells of a Hodgkin's lymphoma patient. Blood. 1996;87:3429–3436. [PubMed] [Google Scholar]
- Irsch J., Nitsch S., Hansmann M.-L., Rajewsky K., Tesch H., Diehl V., Jox A., Küppers R., Radbruch A. Isolation of viable Hodgkin and H/RS cells from Hodgkin disease tissues. Proc. Natl. Acad. Sci. USA. 1998;95:10117–10122. doi: 10.1073/pnas.95.17.10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vockerodt M., Soares M., Kanzler H., Küppers R., Kube D., Hansmann M.-L., Diehl V., Tesch H. Detection of clonal Hodgkin and H/RS cells with identical somatically mutated and rearranged VH genes in different biopsies in relapsed Hodgkin's disease. Blood. 1998;92:2899–2907. [PubMed] [Google Scholar]
- Hummel M., Marafioti T., Stein H. Clonality of Reed-Sternberg cells in Hodgkin's disease. N. Engl. J. Med. 1999;340:394–395. doi: 10.1056/NEJM199902043400518. [DOI] [PubMed] [Google Scholar]
- Daus H., Trümper L., Roth J., von Bonin F., Möller P., Gause A., Pfreundschuh M. Hodgkin and Reed-Sternberg (H/RS) cells do not carry TCRγ rearrangementsevidence from a single cell polymerase chain reaction examination. Blood. 1995;85:1590–1595. [PubMed] [Google Scholar]
- Davis T., Morton C.C., Miller-Cassman R., Balk S.P., Kadin M.E. Hodgkin's disease, lymphomatoid papulosis, and cutaneous T cell lymphoma derived from a common T-cell clone. N. Engl. J. Med. 1992;326:1115–1122. doi: 10.1056/NEJM199204233261704. [DOI] [PubMed] [Google Scholar]
- Van den Berg A., Visser L., Poppema S. High expression of the CC chemokine TARC in H/RS cells. Am. J. Pathol. 1999;154:1685–1691. doi: 10.1016/S0002-9440(10)65424-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Xue W., Kelsoe G. Locus-specific somatic hypermutation in germinal center T cells. Nature. 1994;372:556–558. doi: 10.1038/372556a0. [DOI] [PubMed] [Google Scholar]
- Cheynier R., Henrichwark S., Wain-Hobson S. Somatic hypermutation of the T cell receptor Vβ gene in microdissected splenic white pulps from HIV-1-positive patients. Eur. J. Immunol. 1998;28:1604–1610. doi: 10.1002/(SICI)1521-4141(199805)28:05<1604::AID-IMMU1604>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Marshall B., Schulz R., Zhou M., Mellor A. Alternative splicing and hypermutation of a nonproductively rearranged TCR α-chain in a T cell hybridoma. J. Immunol. 1999;162:871–877. [PubMed] [Google Scholar]