Rapid Identification of Bacteria in Blood Cultures by Using Fluorescently Labeled Oligonucleotide Probes (original) (raw)
Abstract
The applicability of whole-cell hybridization for the identification of pathogenic bacteria in blood from septic patients was examined. Oligonucleotide probes, fluorescently labeled with fluorescein isothiocyanate, directed against the variable regions of the 16S rRNAs of the following bacterial species and/or genera were used: Streptococcus spp., Enterococcus faecalis, Staphylococcus aureus, coagulase-negative staphylococci (CoNS), Escherichia coli, Pseudomonas aeruginosa, and the Enterobacteriaceae family. A probe specific for the rRNAs of almost all bacteria and its complementary, reversed counterpart was used as positive and negative control, respectively. The probes were used in conjunction with a fast and simple-to-use protocol for whole-cell hybridization. This protocol yields an identification after 25 to 45 min, depending on whether the bacterium is gram positive or gram negative. A total of 182 blood samples which tested positive in a blood culture machine were investigated. All probes except for the ones for S. aureus and the CoNS showed sensitivities and specificities of 1.000. It was concluded that whole-cell hybridization is well suited for the fast screening of septic blood containing streptococci and/or enterococci or gram-negative rods.
Septicemia is a pathological condition in which viable and multiplying bacteria may be present in the bloodstream. This condition may occur after trauma or surgery (especially of the visceral organs), immunosuppression, and obstetrical complications (5). It is a potentially life-threatening condition, and appropriate information on the causative agent should be available to the clinician as soon as possible. Bacteriological analysis of blood samples is routinely carried out by selective culturing of blood which has previously been incubated on a general medium in a blood culture system. Currently, three continuously reading, automated blood culture systems are available in clinical microbiology: the BACTEC 9240 (Becton Dickinson Instruments), the BacT/Alert (Organon Teknika), and the Extra Sensing Power (Difco Laboratories) systems. All of these machines measure the rate of production of a bacterium-specific metabolite in the culture bottle containing the patient's blood supplemented with general nutrient broth. Subsequently, if bacterial metabolic activity is detected, the positive blood culture sample is plated on an appropriate selective medium for further analysis. When the positive blood culture is taken as a starting point, microbial identification takes 24 to 72 h to complete. Reduction of the analysis time may result in reduction of the use of broad-spectrum antibiotics, as the genus or species of a pathogen gives an indirect indication of the most appropriate antibiotic to be used. This may result in a lower frequency of emergence of resistance against broad-spectrum antibiotics. Furthermore, it may result in lower cost because suppressive empiric therapy may be replaced by tailored and less expensive antibiotics with smaller spectra of activity. Several methods for rapid detection of pathogens in human blood have been described previously, and most of them have used PCR (8) or fluorescently labeled probes (4). Although these methods are fast and accurate, routine bacteriological analysis still relies heavily on classical culturing techniques. It was therefore decided that for a molecular biological method to be successfully implemented in routine bacteriology it should be fast (maximally, 1 h) and very easy to use (e.g., as complex as the preparation of a Gram-stained slide). In this study a simple and direct whole-cell hybridization assay with species- and genus-specific, fluorescently labeled oligonucleotide probes was developed and validated. The probes described in this paper comprise fluorescein-labeled, single-stranded oligonucleotides complementary to a genus- or a species-specific sequence on the 16S rRNA of the target organism. This method was subsequently applied to a set of positive blood cultures to test whether fast identification of the microbial agent present in these blood cultures was feasible. The principal results of this study show that identification of the microbial agent in a positive blood culture is possible within 25 min (gram-positive streptococci) and 45 min (gram-negative rods). Furthermore, except for the genus Staphylococcus, a 100% correlation between the results obtained by traditional culturing on selective media and the method presented here was achieved.
MATERIALS AND METHODS
Blood samples.
During the time of this study (from 1 September 1998 to 31 January 1999) a total of 182 blood samples which tested positive in a BacT/Alert blood culture system were processed simultaneously by both whole-cell hybridization and accepted culturing methods.
Culturing.
Microorganisms cultivated from positive blood cultures were identified with the API test system (BioMerieux, Marcy l'Etoile, France) or by using standard microbiological methods.
Probes.
The characteristics of the probes used in this study are listed in Table 1. All probes consist of a single-stranded oligonucleotide sequence covalently linked to fluorescein isothiocyanate at the 5′ end. Probes were synthesized by EuroGentec BV (Maastricht, The Netherlands). The _Enterococcus faecalis_-specific probe sequences is also present in Enterococcus sulfuricus. However, because of the very low incidence of Enterococcus sulfuricus as a causative agent of sepsis, this cross-reactivity was not considered diagnostically significant.
TABLE 1.
Oligonucleotide probes used for hybridization of some pathogens normally detected in blood from septic patients
| Probea | Sequence (5′→3′) | Target(s) | Referenceb |
|---|---|---|---|
| EUB | GCTGCCTCCCGTAGGAGT | Bacterial kingdom | 2 |
| Non-EUB | ACTCCTACGGGAGGCAGC | Negative control | 2 |
| STREP | GTTAGCCGTCCCTTTCTGG | Streptococcus spp. | 6 |
| EFAEC | TTATCCCCCTCTGATGGG | Enterococcus faecalis, Enterococcus sulfuricus | |
| STAUR | AGAGAAGCAAGCTTCTCGTCCG | Staphylococcus aureus | 3 |
| CoNS (RDR512)c | CGACGGCTAGCTCCAAATGGTTACT | CoNS | 7 |
| ECOLI | GCAAAGGTATTAACTTTACTCCC | Escherichia coli | 9 |
| PSEUDAER | GGACGTTATCCCCCACTAT | Pseudomonas aeruginosa | |
| ENTBAC | CATGAATCACAAAGTGGTAAGCGCC | Enterobacteriaceae | 10 |
The ENTBAC probe was designed to hybridize to all members of the family Enterobacteriaceae including Escherichia coli. A search of the current list of sequences in the BLAStn database (1) revealed that the sequence complementary to the ENTBAC probe is highly conserved among members of the Enterobacteriaceae family, including the genera Escherichia, Proteus, Yersinia, and Klebsiella. A few non-Enterobacteriaceae (Plesiomonas shigelloides) and an insect symbiont also showed 100% homology with this probe. However, these organisms have not previously been associated with sepsis.
Whole-cell hybridization.
After Gram staining of an initial streak-out preparation of a positive blood culture, the subsequent permeabilization protocol and a set of appropriate probes were chosen (see Table 2). A total of 15 μl from a positive blood culture was pipetted onto a glass slide and was subsequently streaked out. After air drying of the slide, the cells on the slide were fixed in a 4% formaldehyde solution in 96% ethanol. Gram-positive streptococci were permeabilized by incubating the fixed slide in a permeabilization buffer (1 mg of lysozyme per ml) for 5 min. Gram-positive staphylococci were permeabilized by incubating the fixed slide in a second permeabilization buffer (10 U of lysostaphin per ml) for 20 min. Gram-negative rods were not permeabilized. Isolates with other (Gram staining) morphologies were not considered in this study because of the low incidence of these groups of bacteria as a cause of septicemia. After permeabilization, the cells on the slide were hybridized at 50°C. Gram-negative rods were hybridized for 45 min, gram-positive staphylococci were hybridized for 2 h, and gram-positive streptococci were hybridized for 5 min. A different set of probes specific for each type of gram-positive or gram-negative bacteria was chosen (see Table 2). However, a probe (i.e., the EUB probe) specific for almost all bacteria and the reverse complementary probe (i.e., the non-EUB probe) were included as positive and negative controls, respectively, irrespective of the type identified by Gram staining. Prior to use the probes were diluted to a concentration of 10 ng/ml in hybridization buffer (20 mM Tris-HCl, 0.9 M NaCl, 0.1% sodium dodecyl sulfate [pH 7.2]). After hybridization, the slides were washed for 10 min at 50°C in washing buffer (20 mM Tris-HCl, 0.9 M NaCl [pH 7.2]) and mounted with VectaShield (Vector Laboratories, Burlingame, Calif.). Immediately thereafter, the slides were evaluated with an epifluorescence microscope.
TABLE 2.
Performance of the method
| Application criteriona | Probe | Isolated microorganism | nb | Sensitivityc | Specificityd |
|---|---|---|---|---|---|
| Each assay | EUB | All bacteria | 182 | 0.948 | 1.000 |
| Non-EUB | Negative control | 182 | NCe | NC | |
| Gram-positive chains | STREP | Streptococcus mitis | 10 | 1.000 | 1.000 |
| STREP | Streptococcus pyogenes | 3 | |||
| STREP | Streptococcus oralis | 1 | |||
| STREP | Streptococcus dysgalactiae | 1 | |||
| STREP | Streptococcus sanguis | 1 | |||
| STREP | Streptococcus constellatus | 1 | |||
| STREP | Streptococcus agalactiae | 1 | |||
| STREP | Streptococcus milleri | 2 | |||
| EFAEC | Enterococcus faecalis | 10 | 1.000 | 1.000 | |
| Gram-positive clumps | STAUR | Staphylococcus aureus | 13 | 0.667 | 1.000 |
| CoNS | CoNS | 73 | 1.000 | 0.111 | |
| Gram-negative rods | ECOLI | Escherichia coli | 23 | 1.000 | 1.000 |
| PSEUDAER | Pseudomonas aeruginosa | 4 | 1.000 | 1.000 | |
| ENTBAC | Klebsiella pneumoniaef | 12 | 1.000 | 1.000 | |
| ENTBAC | Serratia marcescensf | 4 | |||
| ENTBAC | Proteus mirabilisf | 3 | |||
| ENTBAC | Serratia liquefaciensf | 2 | |||
| ENTBAC | Enterobacter cloaceaef | 2 |
Data analysis.
For each probe, the sensitivity (defined as the proportion of blood samples that contain the target organism and that give a positive result by fluorescent in situ hybridization [FISH] analysis) and the specificity (defined as the proportion of blood samples that do not contain the target organism and that give a negative result by FISH analysis) were calculated. The results obtained during the simultaneous culturing of the positive blood samples were used as the “gold standard.”
RESULTS AND DISCUSSION
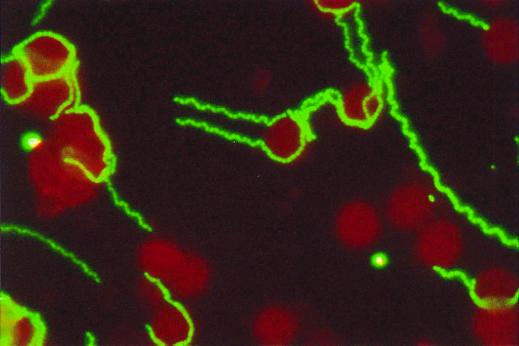
The results of this study show that bacterial identification by whole-cell hybridization dramatically increases the speed of the diagnosis. Figure 1 shows a typical example of the microscopic image obtained after hybridization of a blood sample obtained from a patient with Streptococcus pneumoniae sepsis with the STREP probe. A clear-cut positive signal was obtained by the protocol described above. Repeated microscopic evaluation by different observers confirmed the unambiguity of the interpretation of the images obtained by this method. The results of the study are listed in Table 2. The observation that all strains hybridize positively to the EUB probe indicates that the hybridization protocol is applicable for whole-cell hybridization of the bacterial species and genera tested in this study. The sensitivity of the EUB probe does not equal 1.000. This observation is explained by the fact that for 9 of the 182 blood samples, the bacterial density was too low to yield a microscopic image suitable for examination, although a pathogen was identified by subculturing of the sample.
FIG. 1.
Photomicrograph of a typical result. Cells of S. pneumoniae show intense fluorescence after 5 min of incubation with the STREP probe at 50°C. The fluorochrome was fluorescein isothiocyanate. Magnification, = 10 × 100.
The results obtained with the non-EUB probe indicate the absence of nonspecific interaction between the probe and constituents of the cellular matrix.
The choice of the appropriate set of probes on the basis of the microscopic aspect of the bacteria by Gram staining was performed without error with respect to the gram-negative rods. When gram-positive coccoid bacteria were observed, the STREP and the EFAEC probes were first applied because of the short analysis time (i.e., maximally, 25 min) required for this probe set. In case no signal was emitted by these probes, the STAUR and CoNS probes were applied. This procedure may increase the total analysis time for staphylococci by 25 min. However, this extra time was accepted since it comprises only a minor proportion of the time needed for the identification of the staphylococci solely (i.e., 2 h and 20 min).
All probes except for the _Staphylococcus aureus_-specific probe scored a sensitivity of 1.000. This implies that this method is especially applicable for the identification of gram-negative rods and gram-positive streptococci.
Furthermore, the short hybridization time needed for positively hybridizing streptococci and/or enterococci renders this method particularly useful as an initial screening method for positive blood cultures. Both the specificity and the sensitivity of this method for the identification of gram-negative rods amount to 1.000. However, it must be realized that the low incidence of P. aeruginosa in septic patients may hamper the significance of the sensitivities and specificities obtained with this probe. Previous tests (data not shown) with 20 sterile blood samples, each spiked with Pseudomonas aeruginosa, show that the PSEUDAER probe performs as well as the other probes for the identification of gram-negative rods. The low specificity score for the probe specific for S. aureus may be explained by differences in the permeability of the cell wall. The permeabilization protocol is optimized to yield positive hybridization signals for both S. aureus and coagulase-negative staphylococci (CoNS). A small proportion of the S. aureus strains emit only moderate fluorescence signals with the positive control probe (which normally emits intense fluorescence signals). This is probably due to the strain-specific characteristics of the cell wall composition. However, intensification of the permeabilization by increasing the concentration of lytic enzymes or elongation of the permeabilization time resulted in complete lysis of the majority of the CoNS strains and was therefore not useful. Furthermore, from Table 2 it can be concluded that the specificity of FISH analysis is only as good as the type and number of probes used. Since FISH analysis is a “fixed-end” methodology, it detects only the target organisms at which it is aimed. Classical subculturing is an “open-end” methodology and will therefore detect a broader range of possible pathogens. Both this observation and the short analysis times render FISH analysis of positive blood especially suited as a screening tool for reducing the number of blood samples for subsequent subculturing.
From Table 2 it can also be observed that the specificities of all probes except the CoNS-specific probe are 1.000. This implies that the FISH method did not yield false-negative results for any of the probes used except the CoNS-specific probe. This probe showed a tendency to cross-react with S. aureus. This property could not be eliminated by raising the hybridization temperature to a maximum of 65°C (at temperature intervals of 2°C). Also, addition of formamide to a maximal concentration of 20% (vol/vol) (at concentration intervals of 5%) did not result in an enhancement of the specificity of the FISH method.
We therefore conclude that for the detection of Streptococcus spp., Enterococcus spp., and rod-shaped, gram-negative bacteria, fluorescent whole-cell hybridization is a fast and very easy-to-apply method for the screening of positive blood cultures. Its simplicity (it requires the same number of actions and takes the same hands-on time as the preparation of a sample for Gram staining) renders it especially useful for implementation in routine bacteriology. In addition, this method does not require special equipment like thermal cyclers or UV spectrophotometers. Furthermore, a generally applicable protocol for whole-cell hybridization of staphylococci is probably not possible because of the large interstrain variability in cell wall structure, which renders this genus difficult to hybridize in situ. Other methodologies (e.g., PCR) are to be preferred as alternative strategies for determination of the identities of staphylococcus-shaped bacteria in blood cultures.
Although the species or genus name of a pathogen yields only indirect information on the expected antibiotic sensitivity, this information may be applied by the clinician in order to narrow the scope of applicable antibiotics. Research on rapid in situ detection of antibiotic resistance genes is in progress. Also, research on further shortening of the hybridization time and the use of combinations of several probes labeled with different fluorochromes in one assay is being carried out. Finally, application of this technique for identification of pathogens in material other than blood is being developed.
ACKNOWLEDGMENTS
We kindly acknowledge the advice of H. G. de Vries-Hospers and the Laboratory of Bacteriology of the Academic Hospital of Groningen for providing the positive blood samples.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Amman R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentley R W, Harland N M, Leigh J A, Collins M D. A Staphylococcus aureus specific oligonucleotide probe derived from 16S rRNA gene sequences. Lett Appl Microbiol. 1993;16:203–206. doi: 10.1111/j.1472-765x.1993.tb01397.x. [DOI] [PubMed] [Google Scholar]
- 4.Davis T E, Fuller D D. Direct identification of bacterial isolates in blood cultures by using a DNA probe. J Clin Microbiol. 1991;29:2193–2196. doi: 10.1128/jcm.29.10.2193-2196.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards J D. Management of septic shock. Br Med J. 1993;306:1661–1664. doi: 10.1136/bmj.306.6893.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franks A, Harmsen H J M, Raangs G C, Jansen G J, Schut F, Welling G W. Variations of bacterial populations in human faeces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336–3345. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laforgia N, Coppola B, Carbone R, Grassi A, Mautone A, Lolascon A. Rapid detection of neonatal sepsis using polymerase chain reaction. Acta Paediatr. 1997;86:1097–1099. doi: 10.1111/j.1651-2227.1997.tb14815.x. [DOI] [PubMed] [Google Scholar]
- 9.McGregor D P, Forster S, Steven J, Adair J, Leary S E C, Leslie D L, Harris W J, Titball R W. Simultaneous detection of microorganisms in soil suspension based on PCR amplification of bacterial 16S rRNA fragments. BioTechniques. 1996;21:463–471. doi: 10.2144/96213st04. [DOI] [PubMed] [Google Scholar]
- 10.Mittelman M W, Habash M, Lacroix J M, Khoury A E, Krajden M. Rapid detection of Enterobacteriaceae in urine by fluorescent 16S rRNA in situ hybridization on membrane filters. J Microbiol Methods. 1997;30:153–160. [Google Scholar]