Infectivity of Moloney Murine Leukemia Virus Defective in Late Assembly Events Is Restored by Late Assembly Domains of Other Retroviruses (original) (raw)
Abstract
The p12 region of the Moloney murine leukemia virus (M-MuLV) Gag protein contains a PPPY motif important for efficient virion assembly and release. To probe the function of the PPPY motif, a series of insertions of homologous and heterologous motifs from other retroviruses were introduced at various positions in a mutant gag gene lacking the PPPY motif. The assembly defects of the PPPY deletion mutant could be rescued by insertion of a wild-type PPPY motif and flanking sequences at several ectopic positions in the Gag protein. The late assembly domain (L-domain) of Rous sarcoma virus (RSV) or human immunodeficiency virus type 1 (HIV-1) could also fully or partially restore M-MuLV assembly when introduced into matrix, p12, or nucleocapsid domains of the mutant M-MuLV Gag protein lacking the PPPY motif. Strikingly, mutant viruses carrying the RSV or the HIV-1 L-domain at the original location of the deleted PPPY motif were replication competent in rodent cells. These data suggest that the PPPY motif of M-MuLV acts in a partially position-independent manner and is functionally interchangeable with L-domains of other retroviruses. Electron microscopy studies revealed that deletion of the entire p12 region resulted in the formation of tube-like rather than spherical particles. Remarkably, the PPPY deletion mutant formed chain structures composed of multiple viral particles linked on the cell surface. Many of the mutants with heterologous L-domains released virions with wild-type morphology.
Virion particle assembly represents a crucial late stage of the virus life cycle (31). Many C-type retroviruses, such as human immunodeficiency virus type 1 (HIV-1), Rous sarcoma virus (RSV), and Moloney murine leukemia virus (M-MuLV), assemble at the plasma membrane, where budding occurs (31). During or soon after budding, the viral protease (PR) is activated and virion proteins are cleaved to form mature particles (2, 3, 33). The Gag protein plays an important role in the entire assembly process (31, 35). The M-MuLV Gag precursor protein comprises four separate domains, termed matrix (MA), p12, capsid (CA), and nucleocapsid (NC) (20), and all these regions are involved in different steps of viral assembly. The MA domain is thought to be responsible for transporting the Gag precursor and its associated proteins to the plasma membrane, CA directs the formation of the virion core, and NC binds the viral RNA genome and contains a major Gag-Gag interaction domain (31, 35). The p12 region contains a PPPY motif, which is required for the efficient assembly and release of M-MuLV virions (38).
Functionally, three assembly domains have been defined for the Gag precursor protein, each apparently responsible for a separate function in the assembly process. These domains are M (membrane association), L (late assembly function), and I (interaction between Gag proteins). L-domains, which are required for viral assembly and budding, have been identified by detailed mutational analysis in many retroviruses (19, 34, 36, 37). L-domains are found at quite different regions of Gag in different viruses. In RSV and Mason-Pfizer monkey virus (M-PMV), the L-domains contain a highly conserved PPPY motif as the core sequence and are located between the MA and CA domains of the Gag protein (34, 37). The L-domains of lentiviruses are located at the C terminus of the Gag precursor protein and have distinct core motifs, PTAPP in HIV-1 and YXXL in equine infectious anemia virus (EIAV) (19, 22). It has been shown that the L-domain sequences can be functionally interchanged among several different viruses (HIV-1, RSV and EIAV) despite the lack of sequence homology. Further, they can also function at unnatural sites in chimeric Gag proteins (21, 22, 36). Therefore, the various retroviral L-domains seem to act in a largely position-independent manner and through a common mechanism (31). However, the recovery of function by heterologous retrovirus L-domains was not complete, and none of the chimeric viruses has been reported to be replication competent (21, 22, 36). The molecular mechanisms by which the L-domains may function to facilitate virion release are unknown.
We have previously reported that mutations affecting the PPPY motif of the M-MuLV p12 protein cause defects in assembly or virion release (38). We have now examined the block to virus production in mutant M-MuLVs lacking the PPPY motif. The M-MuLV wild-type PPPY motif and its flanking region, the p2b region of RSV, or the first 18 amino acids of the HIV-1 p6 protein were inserted into several locations in the coding region of M-MuLV Gag. Analysis by electron microscopy (EM) showed that the PPPY motif functions at a very late stage in assembly, perhaps at the release step itself. We also found that the PPPY motif can be replaced by L-domains from other, unrelated retroviruses, despite the lack of sequence identity between the L-domains, and that these domains can function at several sites in M-MuLV Gag. Most strikingly, two of these chimeras were fully infectious; thus, all of the functions of PPPY in M-MuLV replication can be efficiently performed by L-domains from other retroviruses.
MATERIALS AND METHODS
Cells and viruses.
NIH 3T3 fibroblasts, 293T cells, and a subclone of the Rat2 cell line (Rat2-2 cells) (11) were maintained in Dulbecco's modified Eagle's medium containing penicillin-streptomycin and 10% fetal calf serum. Wild-type (WT) M-MuLV was harvested from 293T cells after transfection with the plasmid pNCS.
Plasmids and mutants.
pNCS contains an infectious copy of M-MuLV proviral DNA and carries a simian virus 40 replication origin to allow high-level expression in 293T cells (8). pNCSB was derived from pNCS by replacing the sequence CACTTTGAG (303 bp downstream from the 3′ end of the gag gene) with CACTTCGAA, creating a unique _Bst_BI restriction site. The resulting virus encoded by pNCSB had a wild-type phenotype. Plasmids DPY-pNCS and DPY-pNCSB were similar to the wild-type parents but contained a deletion that removed 5 amino acids (DPPPY) from p12 (Fig. 1) (38).
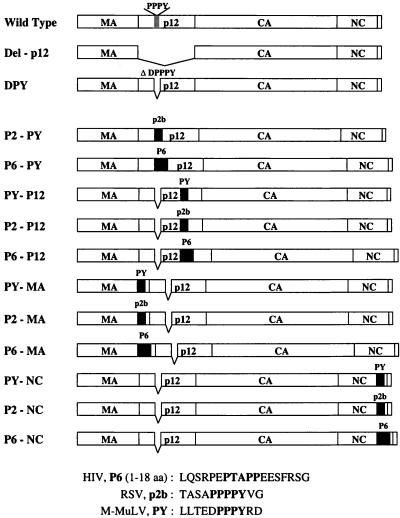
FIG. 1.
Mutations in the M-MuLV Gag polyprotein. The M-MuLV Gag protein is represented by rectangular boxes. The location of the PPPY motif in the p12 region of the wild-type Gag protein is indicated. The entire p12 region is deleted in the Del-p12 mutant. The DPY mutant contains a 5-amino acid deletion (DPPPY) which removes the PPPY motif; this mutant was used as the parent to generate the insertion mutants. Three different fragments were used for the insertion mutants: PY, an 11-amino-acid fragment containing the wild-type PPPY motif and flanking sequences from M-MuLV; p2b, the entire RSV p2b sequence (11 amino acids) including the PPPPY motif; and P6, the N-terminal 18 amino acids of the HIV-1 p6 protein including the PTAPP motif. The amino acid (aa) sequences of PY, p2b, and P6 are as shown. The insertions are represented by black boxes. The locations of insertions are as indicated, with the names of the mutants on the left.
Mutations in MA and p12 were generated by PCR using a subclone of the gag region between the unique _Bsr_GI and _Xho_I sites. The primers used to amplify the _Bsr_GI-_Xho_I fragment were BSRGI (forward) (5′-CCTTTGTACACCCTAAGCC-3′) and XHOIR (reverse) (5′-GTCTGGGCGCTCGAGGGGAAAAGCG-3′). Insertions in NC were also generated by PCR between the _Xho_I site and the _Bst_BI site of DPY-pNCSB. The primers used to amplify the _Xho_I-_Bst_BI region were: XHOI (forward) (5′-TTCCCCTCGAGCGCCCAGACTGG-3′) and BSTBI (reverse) (5′-TCCTGATCCTTCGAAGTGGATTTGGGCTTTTAG-3′). The Expand High-Fidelity PCR system (Boeheringer Mannheim) was used to limit possible random mutagenesis (4). The oligonucleotides used to introduce mutations are shown in Table 1. The forward oligonucleotides were labeled as XX-XX-F, while the reverse oligonucleotides were labeled as XX-XX-R. To generate an insertion between the _Bsr_GI and _Xho_I sites, two intermediate PCR fragments, XX-XX-F/XHOIR and BSRGI/XX-XX-R, were first synthesized using plasmid DPY-pNCS as a template. These two products, containing a large overlapping sequence, were used as PCR templates, and the entire fragment between _Bsr_GI and _Xho_I was amplified by the flanking oligonucleotide pair XHOIR and BSRGI. A similar strategy was applied to generate insertions in the NC region, using DPY-pNCSB plasmid as a template.
TABLE 1.
Mutagenic oligodeoxynucleotides used in this study
| Mutant | Mutagenic oligodeoxynucleotide | |
|---|---|---|
| Designation | Sequence | |
| P2-PY | P2-PY-F | 5′-ACAGCCTCGGCTCCTCCTCCTCCTTATGTGGGGAGGGACCCAAGACCACC-3′ |
| P2-PY-R | 5′-CACATAAGGAGGAGGAGGAGCCGAGGCTGTGTCTTCTGTAAGTAGGTC-3′ | |
| P6-PY | P6-PY-F | 5′-GAGCCAACAGCCCCACCAGAAGAGAGCTTCAGGTCTGGGAGGGACCCAAGACCACC-3′ |
| P6-PY-R | 5′-TTCTGGTGGGGCTGTTGGCTCTGGTCTGCTCTGAAGGTCTTCTGTAAGTAGGTC-3′ | |
| PY-P12 | PY-P12-F | 5′-CTACTTACAGAAGACCCCCCGCCTTATAGGGACGGAGAGGCACCGGACCC-3′ |
| PY-P12-R | 5′-CCTATAAGGCGGGGGGTCTTCTGTAAGTAGCGCAGGGGTCGCTTCTC-3′ | |
| P2-P12 | P2-P12-F | 5′-ACAGCCTCGGCTCCTCCTCCTCCTTATGTGGGGGGAGAGGCACCGGACCC-3′ |
| P2-P12-R | 5′-CACATAAGGAGGAGGAGGAGCCGAGGCTGTCGCAGGGGTCGCTTCTC-3′ | |
| P6-P12 | P6-P12-F | 5′-GAGCCAACAGCCCCACCAGAAGAGAGCTTCAGGTCTGGGGGAGAGGCACCGGACCC-3′ |
| P6-P12-R | 5′-TTCTGGTGGGGCTGTTGGCTCTGGTCTGCTCTGAAGCGCAGGGGTCGCTTCTC-3′ | |
| PY-MA | PY-MA-F | 5′-CTACTTACAGAAGACCCCCCGCCTTATAGGGACACCCCGCCTCGATCCTC-3′ |
| PY-MA-R | 5′-CCTATAAGGCGGGGGGTCTTCTGTAAGTAGCGAACGAGGAGGTTCAAG-3′ | |
| P2-MA | P2-MA-F | 5′-ACAGCCTCGGCTCCTCCTCCTCCTTATGTGGGGACCCCGCCTCGATCCTC-3′ |
| P2-MA-R | 5′-CACATAAGGAGGAGGAGGAGCCGAGGCTGTCGAACGAGGAGGTTCAAG-3′ | |
| P6-MA | P6-MA-F | 5′-GAGCCAACAGCCCCACCAGAAGAGAGCTTCAGGTCTGGGACCCCGCCTCGATCCTC-3′ |
| P6-MA-R | 5′-TTCTGGTGGGGCTGTTGGCTCTGGTCTGCTCTGAAGCGAACGAGGAGGTTCAAG-3′ | |
| PY-NC | PY-NC-F | 5′-CTACTTACAGAAGACCCCCCGCCTTATAGGGACACCTCCCTCCTGACCCTAG-3′ |
| PY-NC-R | 5′-CCTATAAGGCGGGGGGTCTTCTGTAAGTAGCTGGGGTCTTGGTCCCC-3′ | |
| P2-NC | P2-NC-F | 5′-ACAGCCTCGGCTCCTCCTCCTCCTTATGTGGGACCTCCCTCCTGACCCTAG-3′ |
| P2-NC-R | 5′-CACATAAGGAGGAGGAGGAGCCGAGGCTGTCTGGGGTCTTGGTCCCC-3′ | |
| P6-NC | P6-NC-F | 5′-GAGCCAACAGCCCCACCAGAAGAGAGCTTCAGGTCTGGGACCTCCCTCCTGACCCTAG-3′ |
| P6-NC-R | 5′-TTCTGGTGGGGCTGTTGGCTCTGGTCTGCTCTGAAGCTGGGGTCTTGGTCCCC-3′ |
The amino acid sequences inserted into the mutant Gag protein are shown in Fig. 1. The insertions were introduced into the following sites (where the slash indicates the precise location of the insertion; amino acid residues are numbered from the N terminus of the Pr65_gag_ protein): in MA, the site was between amino acids Ser123 and Thr124 in the C-terminal region (…ProProArgSer/ThrProProArg…); in p12, the site was between amino acids Ala185 and Gly186 in the central region (…AlaThrProAla/GlyGluAlaPro…); and in NC, the insertion site was between amino acids Gln530 and Thr531 in the C-terminal region (…ProArgProGln/ThrSerLeuLeu…). RSV P2 and HIV-1 P6 L-domains were also inserted into the original location of the PPPY motif in the p12 region of the DPY mutant, which lacks 5 amino acids (DPPPY). The insertion site was between amino acids Glu160 and Arg166 (…LeuLeuThrGlu/ArgAspProArg…). The presence of all mutations was confirmed by DNA sequencing.
Analysis of virus assembly and infection.
293T cells were transiently transfected by wild-type or mutant pNCS (or pNCSB) using the calcium phosphate method. The culture medium was harvested 48 h posttransfection, and a standard reverse transcriptase (RT) assay was performed to monitor virion production (32). The culture medium was also used to infect fresh NIH 3T3 cells or Rat2-2 cells to test the viability of mutant viruses. The infected cells were maintained for up to 17 days, and the virus yields in the culture supernatants were monitored by RT assay daily. Infections of NIH 3T3 cells and Rat2-2 cells were carried out in the presence of Polybrene (8 μg/ml).
To study viral DNA synthesis in the infected cells, fresh NIH 3T3 cells were acutely infected with equal amounts of virus normalized by RT activity in the culture medium from transfected 293T cells. Preintegrative viral DNAs were extracted from cells 24 h postinfection (16) and analyzed by Southern blotting.
Viral particle purification and analysis.
At 48 h after transfection of 293T cells with wild-type or mutant proviral DNAs, 6 ml of culture supernatant was collected from each 100-mm-diameter plate of transfected cells. Cell debris was removed by filtering the medium through a 0.45-μm-pore-size filter, and HEPES buffer was added to a final concentration of 20 mM. Virions were purified by ultracentrifugation through sucrose cushions as described previously (38).
The pelleted virus particles were lysed and subjected to Western blot analysis (38). The antisera used in the Western blot assays included goat anti-CA serum (NCI serum 79S-804), goat anti-p12 serum (NCI serum 78S-276), and goat anti-MA serum (NCI serum 78S-282). The monoclonal antiserum against the transmembrane (TM) envelope protein was collected from a rat hybridoma line (42-114) (14).
Quantitative study of virus infectivity.
293T cells at about 70% confluence in 100-mm diameter plates were transiently cotransfected with 10 μg of pSR-LLuc (a plasmid DNA encoding a retroviral vector transducing luciferase) (1) and 10 μg of a plasmid DNA encoding the indicated virus, using the calcium phosphate method. The cells were incubated in 10 ml of Dulbecco's modified Eagle's medium-fetal calf serum for 2 days, after which the RT activity in the culture supernatants was assayed. To test for virus helper function, the culture medium was filtered through 0.45-μm-pore-size filters after the addition of 20 mM HEPES (pH 7.4) and Polybrene (8 μg/ml) and 4 ml of the filtrate were used to infect 1.2 × 106 Rat2-2 cells in 100-mm plates for 2 h. Lysates of infected cells were prepared 2 days after infection and analyzed for specific luciferase activity using a luminometer (Lumat LB 9501; Berthold) and a luciferase assay system (Promega) as specified by the manufacturer. The total protein concentration in cell lysates was determined using a protein assay kit (Bio-Rad). The specific activity of luciferase was calculated as relative light units per milligram of total protein. The luciferase activity of the infected cells was corrected for variation in the RT levels of the supernatants of the transfected cells. Mock-transfected and infected cells (no plasmid DNA or virus added) were used to subtract background levels from the RT and luciferase activities. A 1:10 dilution of the supernatants gave a 10-fold reduction in the luciferase activity, suggesting that infections were done at a multiplicity in the linear range (data not shown).
EM analysis.
293T cells were transfected with wild-type or mutant pNCS plasmids, and 48 h after transfection the cells were fixed with 2% glutaraldehyde in 0.1 M sodium cacodylate. The cells were further fixed with 2% OsO4 for 2 h and then dehydrated with a graded series of ethanol dilutions ranging from 30 to 100% before being embedded in Epon resin. Thin sections were counterstained with 1% lead citrate and 2% uranyl acetate before being examined by transmission electron microscopy.
RESULTS
Construction of insertion mutants.
The M-MuLV p12 protein contains a PPPY motif that is important for virus assembly or release. Deletion or alanine substitution of this motif caused severe defects in virion production, Gag protein processing, and TM protein cleavage (38). To determine whether this domain acts in a position-independent manner, a series of insertion mutations were introduced into a mutant M-MuLV gag gene encoding a protein lacking the PPPY motif, specifically containing a deletion of the 5 amino acids DPPPY. A sequence encoding 11 amino acids, the PPPY motif, and flanking sequences from the wild-type M-MuLV was inserted into ectopic sites in the gag gene, resulting in a peptide inserted in the middle region of p12, the C terminus of MA, or the C terminus of NC (PY-P12, PY-MA, and PY-NC [Fig. 1]). The insertion sites were chosen insofar as possible to avoid effects on known protein functions.
To determine whether the M-MuLV L-domain is functionally interchangeable with the L-domains of other retroviruses, DNAs encoding the entire p2b protein of RSV (including the PPPPY motif) and the first 18 amino acids of the HIV-1 p6 protein (including the PTAPP motif) were inserted into the original location of the PPPY motif (P2-PY and P6-PY). The p2b and p6 L-domains were also inserted into the same locations of p12, MA, or NC as the wild-type M-MuLV PPPY motif in the mutants described above (P2-P12, P2-MA, P2-NC, P6-P12, P6-MA and P6-NC [Fig. 1]).
Virion production by chimeric mutants.
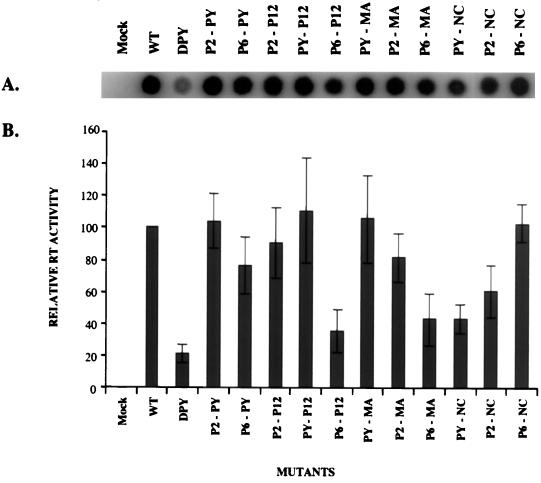
To test whether the newly inserted sequences could rescue the defects of the parental PPPY deletion mutant, 293T cells were transiently transfected with the mutant proviruses by the calcium phosphate precipitation method. Culture supernatants were collected 48 h after transfection, and the released particles were analyzed by the RT assay. All the insertion mutants showed increased RT activity compared to the PPPY deletion mutant DPY (Fig. 2A). Quantitation of the RT activity revealed that the PPPY deletion mutant produced less than 20% of the levels of virion-associated RT of the wild-type virus. The viruses carrying the inserted copy of the wild-type PPPY at the middle region of p12 (PY-P12) or the C-terminal region of MA (PY-MA) exhibited RT activity equal to that of the wild-type virus, while the virus carrying the same sequence in the C-terminal region of NC (PY-NC) produced RT activity approximately 50% of that of the wild type. Viruses carrying the RSV p2b at p12, MA, or NC yielded levels of RT comparable to those of viruses carrying the wild-type PPPY motif in the corresponding Gag regions (P2-P12, P2-MA, and P2-NC). Furthermore, insertion of p2b into the location of the original PPPY motif also restored 100% of the RT activity of the wild type (P2-PY). In contrast, viruses carrying the HIV-1 p6 L-domain at p12 or MA showed only 40 to 50% of the wild-type RT activity (P6-P12 and P6-MA). However, the viruses showed nearly normal RT activity when the p6 L-domain was inserted into the original PPPY location (80% of wild-type RT activity [P6-PY]) or the NC region (100% of wild-type RT activity [P6-NC]) (Fig. 2B). These results suggest that the M-MuLV L-domain can act in other locations and that its function can be efficiently provided by other retroviral L-domains. However, different L-domains displayed distinct preferences for their original location in the Gag protein in providing optimal function.
FIG. 2.
Viral assembly and particle release analyzed by the RT assay. (A) RT activity in the culture supernatant. The proviruses of the insertion mutants were transiently transfected into 293T cells by calcium phosphate precipitation, and supernatants were analyzed by the RT assay 48 h after transfection. (B) PhosphorImager quantitation of the relative RT activity in produced virions. The mean of three independent experiments is shown. Error bars indicate standard deviation of the three results. The amount of RT activity in the wild-type (WT) virus is set as 100.
Gag gene products in virions and transfected 293T cells.
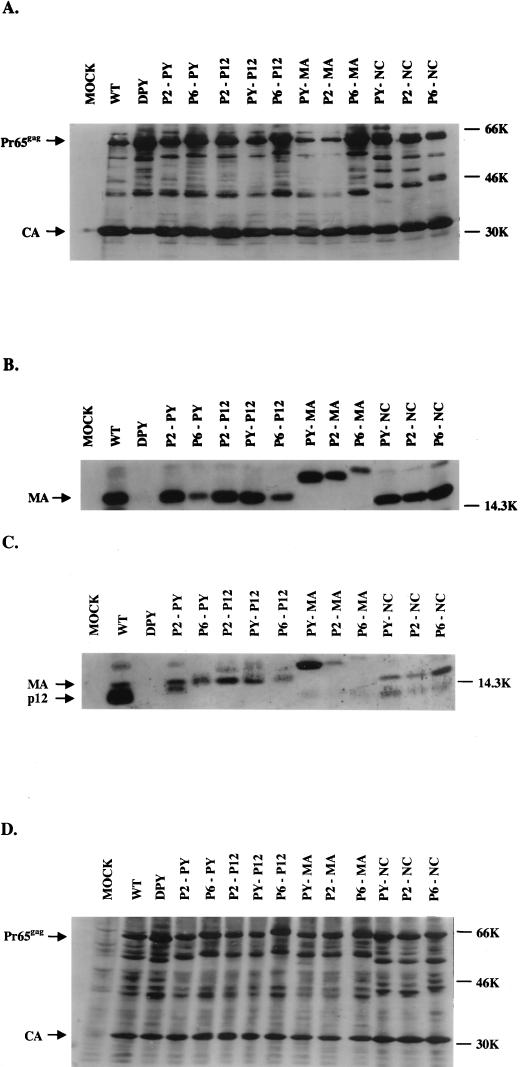
The parental PPPY mutant showed major defects in viral protein processing normally associated with virion maturation. To test whether the new insertions could rescue the defects of viral Gag protein processing, culture supernatants from transfected 293T cells were collected and the virions were purified by ultracentrifugation through sucrose cushions. The virion lysates were subjected to RT assays to monitor the recovery of virions through the purification process (data not shown), and the virion-associated proteins were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting. Anti-CA, anti-MA, or anti-p12 polyclonal antisera were used to detect the corresponding gag gene products. As previously reported (38), the PPPY deletion mutant (DPY) showed defects in Gag protein processing: an increased level of the unprocessed Pr65_gag_ precursor protein was observed, and nearly no mature p12 or MA proteins could be detected in the virions (Fig. 3A to C). Gag processing was almost fully restored in several mutants carrying the homologous sequence (PY-P12 and PY-MA), several carrying the RSV sequence (P2-PY, P2-P12, and P2-MA), and one carrying the HIV-1 sequence (P6-NC). However, other mutants, PY-NC, P2-NC, P6-PY, P6-P12 and P6-MA, still showed partial defects in Gag protein processing (Fig. 3A to C), and smaller amounts of MA proteins were detected in these virions (Fig. 3B). Insertions into MA and p12 caused the corresponding MA and p12 proteins to migrate differently in the SDS-PAGE (Fig. 3B and C). Only very weak p12 bands could be detected in those mutants with the PPPY motif deletion (Fig. 3C), suggesting that the PPPY motif and flanking regions may be the major recognition site for the anti-p12 antibody used in these blots. Insertion mutagenesis caused the p12 proteins of several mutants (PY-P12, P2-PY, P2-P12, P6-PY, and P6-P12) to migrate at similar positions to the MA protein in the SDS-PAGE gel and were thus hard to detect (Fig. 3C). However, the data clearly indicate that Gag protein processing could often be restored by inserting a new copy of this domain into other locations of the Gag protein or by inserting the L-domains of other retroviruses.
FIG. 3.
Western blot analysis of gag gene products in virions and in lysates of transfected cells. The virus particles were collected by ultracentrifugation from the culture supernatants of the transfected 293T cells. The virions were lysed, and proteins were subjected to Western blot analysis. Blots were probed with polyclonal antisera against CA (A), MA (B), and p12 (C). The anti-p12 antiserum also showed contaminating reactivity to MA. (D) Lysates of transfected 293T cells were also analyzed by Western blotting using anti-CA polyclonal serum. The positions of Pr65_gag_, CA, MA, and p12 are indicated. WT, wild type.
To ensure that the viral proteins were expressed normally in the transfected 293T cells, cell lysates were also prepared 48 h after transfection and the intracellular viral proteins were analyzed by Western blotting. The levels of CA and Pr65_gag_ proteins found in the cells expressing all of the mutants were similar to those seen in cells expressing the wild type. For mutants DPY, P6-P12, and P6-MA, slightly more unprocessed Pr65_gag_ precursor protein was detected in the lysates than in the controls, suggesting that in these mutants the defect in virion budding was more severe and that a larger amount of Gag protein was retained in the transfected cells (Fig. 3D).
TM protein processing in the new insertion mutants.
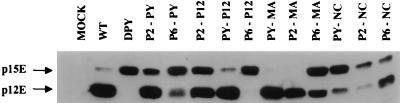
Mutants lacking the PPPY motif mutants exhibited a complete block in the cleavage of the C-terminal tail of the TM envelope protein from p15E to p12E (38) (Fig. 4). It has been shown that mutations blocking the proteolytic processing of the TM cytoplasmic tail result in noninfectious virus (24, 25). To test for restoration of this cleavage, virions were purified from the culture supernatants of transfected 293T cells by ultracentrifugation, and cleavage of the TM protein was analyzed by SDS-PAGE and Western blotting. Several of the insertion mutants, including PY-P12, PY-MA, P2-PY, and P2-MA, processed the TM protein almost as efficiently as the wild-type virus did (Fig. 4). For other mutants, the extent of recovery of TM cleavage was less than complete. P6-P12, P6-MA, PY-NC, and P2-NC showed only poor cleavage. These data correlated well with the Gag protein-processing results (Fig. 3). Therefore, several different L-domains inserted in different locations of the M-MuLV Gag protein can complement the defects of the TM envelope protein processing of the PPPY mutant.
FIG. 4.
Analysis of TM envelope protein in mutant virions. Virions were harvested, and proteins were analyzed by Western blotting. A TM-specific monoclonal antiserum was used to detect p15E and p12E forms of the TM protein. The positions of p15E and p12E are indicated. WT, wild type.
Viral DNA synthesis in infected NIH 3T3 cells.
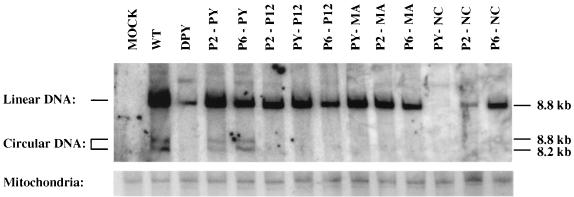
The DPY mutant carrying a deletion of the PPPY motif is replication defective. The deletion not only severely reduced virion assembly but also blocked the ability of the virions to carry out synthesis of most of the linear and all the circular viral DNAs in infected NIH 3T3 cells. To determine whether the insertions could rescue the defects of the DPY mutant in the early events of the virus life cycle, virus supernatants from transfected 293T cells were collected and used to acutely infect fresh NIH 3T3 cells. The virus levels in these infections were normalized by RT activity. The low-molecular-weight DNAs were extracted 24 h after infection, and the viral DNAs were analyzed by Southern blotting with a virus-specific DNA probe. The same Southern blot membrane was then stripped and hybridized with a mitochondrial DNA probe to monitor recovery of the DNA. For the DPY mutant, the levels of linear viral DNA were less than 10% of those seen for the wild-type virus and no circular DNAs were detected, consistent with the results reported previously (38). Mutants P2-PY and P6-PY, in which the RSV p2b and HIV-1 p6 L-domains, respectively, were inserted into the original location of the PPPY motif, could both synthesize linear and circular viral DNAs, although the total amount of viral DNA was somewhat less than that of the wild-type control (Fig. 5). Most of the other mutants could direct the synthesis of linear DNA to various extents, but none of them could make detectable levels of circular viral DNA (Fig. 5).
FIG. 5.
Southern blot detection of linear and circular viral DNAs synthesized in infected NIH 3T3 cells. Supernatants from transfected 293T cells were used to acutely infect NIH 3T3 cells. The virus titers in the supernatants were normalized for RT activity. Low-molecular-weight DNAs were extracted by the Hirt method, and the viral DNAs were detected by hybridization with a radiolabeled virus-specific DNA probe. The same membrane was stripped and rehybridized with a mitochondrial DNA probe to monitor the recovery of DNA loaded in each lane. The positions and sizes of linear and two circular viral DNAs are marked. WT, wild type.
Mutants PY-NC and P2-NC, containing insertions of the wild-type PPPY motif or p2b into NC, resulted in even less viral DNA production than the DPY mutant (Fig. 5). The NC protein plays an important role in the early events of the life cycle, particularly reverse transcription (5, 12, 18), and these insertions into NC may have affected its function during viral DNA synthesis.
Viability of insertion mutants.
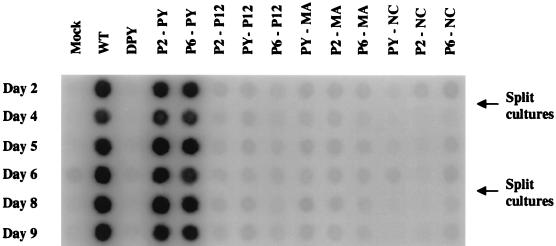
Since some of the insertion mutants showed substantially normal levels of virion production and viral DNA synthesis, it was possible that a subset of these viruses could be fully replication competent. To test for viral replication, culture supernatants of transfected 293T cells were used to infect fresh NIH 3T3 cells and virion yields in the cultures were monitored by RT assays daily. As expected, the wild-type virus could efficiently infect and spread in the culture, and strong RT activity could be detected within 2 days after infection. The DPY mutant remained replication defective (Fig. 6). Mutants P2-PY and P6-PY produced progeny virus as quickly as the wild-type virus did (Fig. 6). These data suggest that the heterologous retroviral L-domains can fully replace the M-MuLV L-domain when placed in the original location of the PPPY motif, even though the HIV-1 p6 L-domain does not bear primary sequence similarity to the L-domain of M-MuLV. As described above, Gag and TM protein processing for mutant P2-PY was similar to that for the wild type. However, mutant P6-PY exhibited partial defects in virion production and in Gag and TM protein processing (Fig. 2 to 4). Despite these defects, this mutant virus was still infectious. Therefore, the modest defects in assembly and protein processing exhibited by this mutant were not limiting in the replication of the mutant virus.
FIG. 6.
Virus infectivity. Equal volumes of culture medium from transfected 293T cells were collected 48 h after transfection and used to infect naive NIH 3T3 cells. Culture supernatants were harvested every day up to 17 days, and the virus yield was monitored by the RT assay. The RT assay results from day 2 to day 9 are shown. The days when cultures were split are marked. WT, wild type.
None of the remaining mutants were infectious, even though the particle release and protein processing of mutants PY-P12, PY-MA, P2-P12, P2-MA, and P6-NC were comparable to those of the wild-type virus (Fig. 3 to 6). For mutants PY-P12 and PY-MA, low levels of RT activity were detected after 13 days. Whether these signals were due to extremely limited replication of the mutant or to the appearance of revertants is not known. Similar results were obtained when Rat2-2 cells were used instead of NIH 3T3 cells (data not shown).
Analysis of virion proteins and viral DNA sequences of the viable mutants.
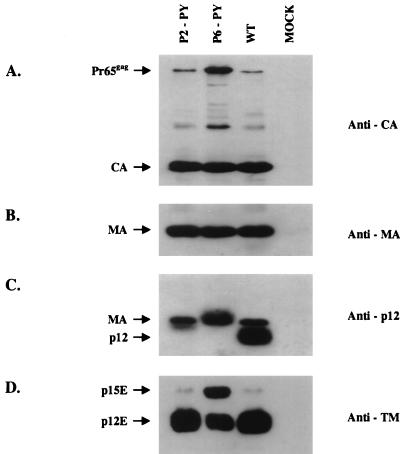
To ensure that the apparent viability of mutants P2-PY and P6-PY could not be attributed to contamination from wild-type virus, recombination with endogenous DNAs, or reversion, the culture supernatants from infected NIH 3T3 cells were collected on day 6 and the virions were pelleted by ultracentrifugation. The viral particles were lysed and subjected to Western blot analysis. Protein processing of mutant P2-PY was similar to that of wild-type virus, but mutant P6-PY remained partially defective for both Gag and TM protein cleavage (Fig. 7). These phenotypes were identical to those of mutant viruses obtained from transient transfection (Fig. 3, 4 and 7). As expected, the p12 proteins of both mutants migrated slower than that of wild-type virus on an SDS-PAGE gel because of the insertions (Fig. 7C).
FIG. 7.
Western blot analysis of virion proteins in the replication-competent chimeric mutants. The culture supernatants of infected NIH 3T3 cells in Fig. 6 were collected on day 6. The viral particles were pelleted by ultracentrifugation, lysed, and analyzed by Western blotting. The membranes were probed with anti-CA (A), anti-MA (B), anti-p12 (C) polyclonal antiserum. (D) In addition, a specific monoclonal antibody was used to detect TM envelope protein. The anti-p12 polyclonal antiserum also showed contaminating reactivity with MA. The positions of Pr65_gag_, CA, MA, and p12 are indicated. WT, wild type.
After the infected NIH 3T3 cells were cultured for 17 days, the supernatants of these two mutant viruses were harvested and used to infect fresh NIH 3T3 cells. The viral DNAs were isolated 24 h after infection by using the Hirt method. The viral DNA fragments between the _Bsr_GI and _Xho_I sites were amplified by PCR and cloned into a plasmid vector. DNA sequencing demonstrated that the original insertions of the p2b and p6 L-domains into the PPPY location remained intact in the P2-PY and P6-PY mutants. No other mutations were found in the insertion region between _Bsr_GI and _Xho_I. These data strongly suggest that the infectious chimeric viruses replicating in the NIH 3T3 cells were the original P2-PY and P6-PY mutants, although we cannot completely rule out the possibility that suppressor mutations could have occurred outside the insertion region.
Quantification of the infectivity of DPY, P2-PY, and P6-PY in a single round of infection.
Although the above experiments suggest that two chimeric viruses can replicate, they do not provide an estimate of the efficiency of infection. To quantitatively measure the efficiency of a single round of replication, the ability of the DPY, P2-PY, and P6-PY viruses to provide helper function in trans for the transduction of a retroviral vector was tested. 293T cells were transfected with the helper DNAs plus a vector carrying the luciferase reporter gene, virus was collected, and the efficiency of transfer was assessed by measuring the luciferase levels present in extracts of Rat2-2 cells after infection. The DPY virus failed to mediate transduction of cells by the retroviral vector (the luciferase specific activity was 0.5% of that present after infection mediated by wild-type helper). In contrast, the P2-PY and P6-PY viruses transduced cells with approximately 10 and 20% of the efficiency of the wild-type virus, respectively. These results demonstrate that the RSV p2 and the HIV-1 p6 sequences partially rescued the infectivity of the DPY virus.
EM analysis of particles released by mutants.
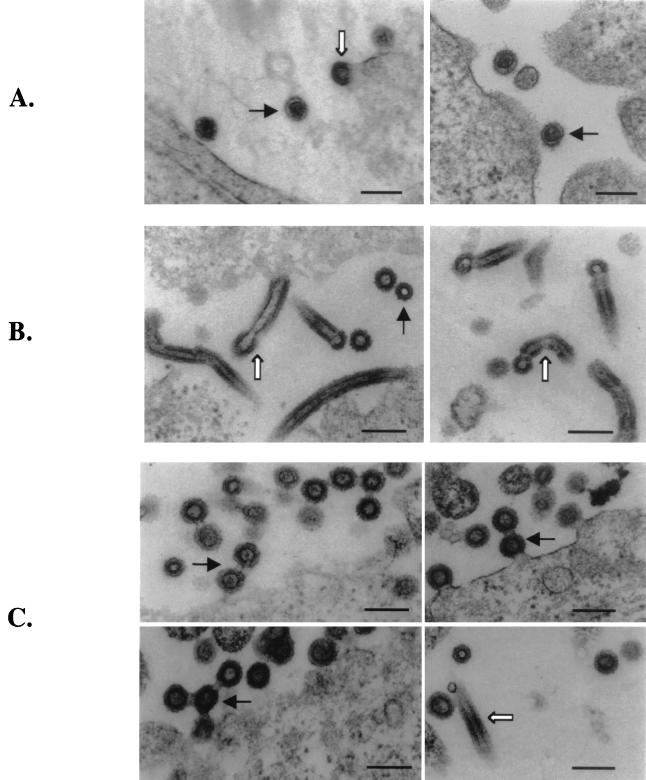
Deletion of the entire p12 protein or the PPPY motif of M-MuLV leads to significant reductions in virion production and viral protein processing (38). However, the exact stage in the process of assembly or budding that is affected by the mutation is not clear. To determine the effects of the mutations on virion morphology, producer cells were examined by EM. 293T cells were transfected with the wild-type or mutant (Del-p12 and DPY) proviruses and fixed 48 h after transfection. The samples were embedded, sectioned, and stained, and sections were examined under the electron microscope. The wild-type mature viruses appeared as round particles with a condensed, concentric core. In the immature budding viruses, the particles were characterized by a spherical structure with an electron-lucent center, giving a doughnut-shaped appearance (Fig. 8A). Surprisingly, cells expressing the Del-p12 mutant produced abnormal particles with elongated tube-like structures (Fig. 8B). The tubes were generally 300 to 500 nm in length and two-thirds to three-quarters of the diameter of normal immature virus. Most of the rods were still attached to the cell membrane and contained a head or cap at one end. The heads of the tubes showed a structure similar to that of the immature budding viruses. There were also low levels of spherical immature virions but no mature viral particles (Fig. 8B). It is not clear whether the few “immature particles” are simply sections through the “heads” at the ends of the tubes.
FIG. 8.
EM images of 293T cells expressing wild-type and mutant viral genomes. (A) Wild-type virus particle release. The mature viruses are marked by black arrows, and the immature viruses are marked by white arrows. (B) Particle release from the Del-p12 mutant. The tube-like structures are indicated by white arrows. A black arrow points to a cross-section of a tube. (C) Particle release from the DPY mutant. The multiple virus chain structures are marked by black arrows, and tubes are indicated by white arrows. Bars, 200 nm.
The DPY mutant assembled chain-like structures containing several spherical immature viruses connected by bridges (Fig. 8C). In the multiple-chain structures, the immature virions appeared to emanate from the cell membrane one after another to form a linear chain or branches. The virions were connected with each other by a narrow neck; typically two to five virions were chained together (Fig. 8C). There were also occasional tube-like structures and no mature virions. These data were consistent with observations that the Del-p12 and DPY mutants were defective in viral particle release and protein maturation and that the defects in the Del-p12 mutant were more severe than those in the DPY mutant. The fact that multiple virions in the DPY mutants could bud out from the same location on the cell surface and remain connected to form a “daisy chain” structure suggests that there may be some preferred sites at the plasma membrane for virion budding.
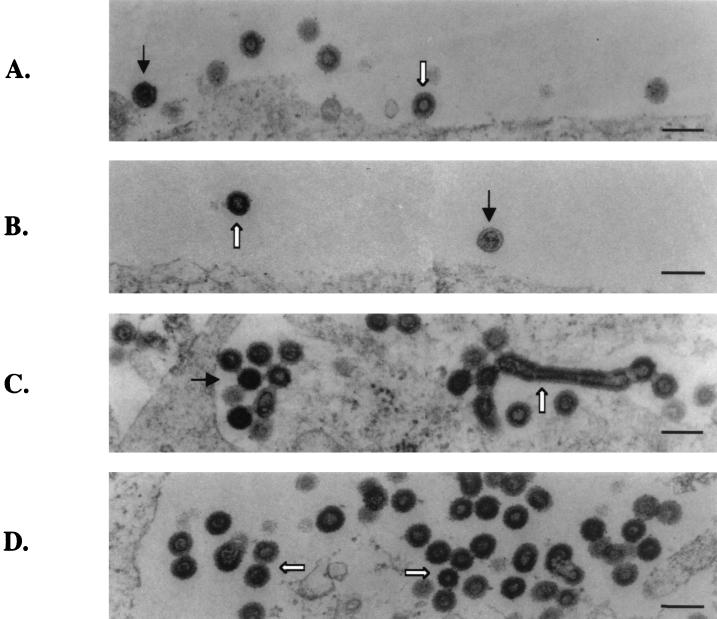
Several of the insertion mutants were also analyzed by EM. The two viable mutants (P2-PY and P6-PY) released virion particles indistinguishable from those of the wild type (Fig. 9A and B). Mutants P6-P12 and P6-MA contained all three virus types seen with the DPY mutants, including the immature viruses, chain structures, and very occasionally tube-like structures (Fig. 9C and D). Therefore, these two mutants failed to correct the virion release defects of the PPPY deletion mutant. All the other mutants examined (PY-P12, PY-MA, P2-P12, and P2-MA) released wild-type-like virions (data not shown). However, these mutants were noninfectious, and no circular viral DNA could be detected in infected NIH 3T3 cells (Fig. 5 and 6). Therefore, although the particles showed normal morphology, these mutant viruses must be blocked during the early events of the life cycle, suggesting that other Gag functions were disrupted by the insertions (Fig. 2 to 5).
FIG. 9.
EM images of virions released from 293T cells transfected by the insertion mutants. (A) Mutant P2-PY. (B) Mutant P6-PY. In these panels, black arrows point to mature virions and white arrows point to immature virions (C) Mutant P6-P12. (D) Mutant P6-MA. In these two panels, the chain structures are indicated by white arrows and the tube structures are indicated by black arrows. Scale bars, 200 nm.
DISCUSSION
The results presented here show that the PPPY motif contained in the M-MuLV p12 protein serves as an L-domain that is functionally interchangeable with the RSV and HIV-1 L-domains. The most compelling observation is that the insertion of the RSV p2b or HIV-1 p6 L-domain into the original location of the PPPY motif fully restored viral replication. Insertions of L-domains at several different locations of Gag protein were able to fully or partially restore normal particle release.
L-domains were first identified during mutational analyses of the RSV p2b protein (34). The crucial motif of the domain was subsequently identified as a polyproline motif, PPPY, conserved in the Gag precursors of many retroviruses (34). Mutation or deletion of this motif resulted in defects late in the course of virus assembly and virion budding (34, 36–38). The L-domains of the lentiviruses were distinctive: they are located in a small protein (p6 for HIV-1 and p9 for EIAV) at the C terminus of Gag proteins and do not contain a PPPY motif. Instead, the core sequence of these L-domains consists of a PTAPP motif for HIV-1 and a YXXL motif for EIAV (19, 22). In spite of these different sequences, the L-domains of RSV, HIV-1, and EIAV are functionally interchangeable for virus budding (21, 22, 36). Data presented here demonstrate that the RSV and HIV-1 L-domains can also restore viral assembly to an M-MuLV PPPY mutant. Therefore, all these different retroviral L-domains may function through a similar mechanism.
The position of insertion of the various L-domains had some effect on the ability of the domain to rescue virion assembly. The RSV and M-MuLV L-domains restored the assembly function more efficiently when inserted in MA or p12 than when inserted in NC. In contrast, the HIV-1 p6 functioned better in NC than in MA or p12 (Fig. 2). These data suggest that although the function of the L-domain is largely position independent, the L-domain may function best when located near its original, native location (relative to the N and C termini of the Gag protein).
Although previous reports have shown that swapping L-domains among different retroviruses results in variant Gag proteins that are competent for assembly, the infectivity of these chimeric viruses was not determined (21, 22, 36). This report demonstrates that the insertion of the RSV or HIV-1 L-domain at the location of the original PPPY motif not only restored normal assembly to the M-MuLV L-domain mutant but also generated a fully replication-competent virus. The recovery of infectivity did not always require complete restoration of efficient assembly; for example, the P6-PY mutant was infectious in spite of modest defects in virion release and protein processing. Other mutants, such as PY-P12, PY-MA, P2-P12, and P2-MA, showed efficient budding and protein cleavage but were still noninfectious. One explanation here could be that deletion of the PPPY motif in these mutants disrupted the functions of p12 needed in an early phase of the viral life cycle. Alternatively, insertions in MA or another position of p12 might interfere with their functions during early events.
The mature wild-type M-MuLV virion assembles into a spherical particle with an electron-dense core. Deletion of the L-domain in HIV-1 leads to retention of immature virions on the cell surface, but no dramatic changes in the overall morphology of the virion were observed (13). In contrast, an M-MuLV mutant lacking the entire p12 directed the formation of elongated tubes or cylinders, which projected perpendicularly outward from the plasma membrane, as the predominant particle form. These cylindrical particles were reproducibly but less frequently observed when only the PPPY motif was deleted. Presumably, the proteins of the Gag precursor are somehow affected so that tubes rather than spherical structures form. The virions are apparently surrounded by membrane, since their buoyant density is at least grossly similar to that of the wild type. Similar cylindrical particles were previously observed when HIV-1 Gag deletion mutants (missing p6 and the C-terminal half of NC) were overexpressed in insect cells (17) and when certain M-PMV mutants with alterations in the CA domain were expressed in mammalian cells (28). Other viruses can also form rods under certain circumstances; influenza viruses, for example, form rods in vivo, and the elongated forms are thought to be harder for the host to remove from pulmonary airways than are the spherical variants (9, 26, 27). It is unclear whether the retroviral structures seen here reflect a similar capacity for formation of filamentous virions under particular conditions.
Rod-like cylindrical structures have also been observed in assembly studies in vitro using only the CA domain of HIV-1 (10, 15) or the CA-NC fragment of RSV and HIV-1 Gag (6). More relevant to the present study is the observation that deletion of the p10 domain in RSV, which is positionally analogous to p12 in M-MuLV, resulted in the formation of cylindrical particles in vitro. From this in vitro work with RSV, it was concluded that in addition to the M-, I-, and L-domains, the p10 domain of RSV Gag acts as an S-domain (shape-determining domain) (7). The results presented here suggested that the p12 domain of M-MuLV is also a shape-determining domain. It is not clear whether the shape-determining properties of p12 are inherent within Gag or whether an interaction with a cellular factor is required. In vitro assembly studies using M-MuLV Gag proteins are currently in progress to address this question.
The chain-structure particles assembled by the DPY mutant are even more striking. These structures provide direct evidence that the PPPY motif may function during the final stages of virion release. The presence of the chains also suggests that the virions may bud out one after another at the same site on the plasma membrane. It is possible that virions may not assemble at random positions on the plasma membrane but that some preferred exit points exist. Alternatively, budding of one virion at a certain site of the plasma membrane could promote or “seed” the formation of other virions at that same site. This process may be coupled with recruitment of host cytoskeletal proteins to the site of assembly. It would be interesting to determine whether reagents affecting the cytoskeletal protein can affect these budding phenotypes.
Mutations in the PPPY motif partially block Gag protein cleavage and completely block TM protein processing (38). These defects could be corrected by insertions of the M-MuLV L-domain or by insertions of other retroviral L-domains in different locations of the Gag protein. As was true for viral assembly, inserting the L-domains near their original locations resulted in better recovery of the protein-processing function (Fig. 3 and 4). It is possible that the viral protease activity or the accessibility of its substrate is influenced by the L-domain. How the L-domain and the viral protease work together is still unclear. Studies of protease-inactivated viruses have revealed different results with different viruses. In HIV-1, inactivation of the viral protease alleviated the requirement of p6 for budding (19). In contrast, in both RSV and M-PMV, protease inactivation did not alter the requirement of the late domains for budding (34, 37). The L-domain may activate the protease or may somehow allow the viral protease to recognize its substrates, including the TM protein. It is also possible that host proteins may interact with the L-domain and be involved in the activation of the protease function.
Host proteins have long been suggested to participate in the L-domain function, particularly since the PPPY motif can act as the binding site for the WW motif, an important protein-protein interaction motif found in many signal transduction and cytoskeletal proteins (29, 30; L. Garnier, J. W. Wills, M. F. Verderame, and M. Sudol, Letter, Nature **381:**744–745, 1996). One or more host proteins containing the WW motif may interact with the PPPY motif of the retroviral L-domain and facilitate virion release. Further studies are needed to identify which of the host proteins that bind to the L-domain are important for its function. One puzzle is that there is no primary sequence similarity among the HIV-1 p6, EIAV p9, and M-MuLV L-domains (23). It is possible that three different host proteins are used by these different viruses; alternatively, one host protein may somehow bind all three motifs by using different contacts.
ACKNOWLEDGMENTS
We are grateful to Jason Gonsky for advice and for providing the pNCSB plasmid, to Matthew Evans for preparing anti-p15E antiserum, and to Andrew Yueh, Baojie Li, and Ari Fassati for helpful discussions. We thank Theodora Hatziioannou, Nicole Bouvier, and Sharon Boast for reading the early drafts. We also thank Kunio Nagashima for excellent EM technical support.
This work was partially supported by Public Health Service grant CA 30488 from the National Cancer Institute. It was also partially sponsored by the National Cancer Institute, Department of Health and Human Services, under contract with ABL. S.P.G. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.An D S, Koyanagi Y, Zhao J Q, Akkina R, Bristol G, Yamamoto N, Zack J A, Chen I S. High-efficiency transduction of human lymphoid progenitor cells and expression in differentiated T cells. J Virol. 1997;71:1397–1404. doi: 10.1128/jvi.71.2.1397-1404.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arcement L, Karshin W, Naso R, Jamjoom G, Arlinghaus R. Biosynthesis of Rauscher leukemia viral proteins: presence of p30 and envelope p15 sequences in precursor polypeptides. Virology. 1976;69:763–774. doi: 10.1016/0042-6822(76)90504-3. [DOI] [PubMed] [Google Scholar]
- 3.Arcement L J, Karshin W I, Naso R B, Arlinghaus R B. “Gag” polyprotein precursors of Rauscher murine leukemia virus. Virology. 1977;81:284–297. doi: 10.1016/0042-6822(77)90145-3. [DOI] [PubMed] [Google Scholar]
- 4.Barnes W M. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthoux L, Pechoux C, Ottmann M, Morel G, Darlix J L. Mutations in the N-terminal domain of human immunodeficiency virus type 1 nucleocapsid protein affect virion core structure and proviral DNA synthesis. J Virol. 1997;71:6973–6981. doi: 10.1128/jvi.71.9.6973-6981.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell S, Vogt V M. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colicelli J, Goff S P. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988;199:47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- 9.Cox J C, Hampson A W, Hamilton R C. An immunofluorescence study of influenza virus filament formation. Arch Virol. 1980;63:275–284. doi: 10.1007/BF01315033. [DOI] [PubMed] [Google Scholar]
- 10.Ehrlich L S, Agresta B E, Carter C A. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J Virol. 1992;66:4874–4883. doi: 10.1128/jvi.66.8.4874-4883.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao G, Goff S P. Somatic cell mutants resistant to retrovirus replication: intracellular blocks during the early stages of infection. Mol Biol Cell. 1999;10:1705–1717. doi: 10.1091/mbc.10.6.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorelick R J, Chabot D J, Ott D E, Gagliardi T D, Rein A, Henderson L E, Arthur L O. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc-coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J Virol. 1996;70:2593–2597. doi: 10.1128/jvi.70.4.2593-2597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Granowitz C, Berkowitz R D, Goff S P. Mutations affecting the cytoplasmic domain of the Moloney murine leukemia virus envelope protein: rapid reversion during replication. Virus Res. 1996;41:25–42. doi: 10.1016/0168-1702(95)01278-8. [DOI] [PubMed] [Google Scholar]
- 15.Gross I, Hohenberg H, Krausslich H G. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 16.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 17.Hockley D J, Nermut M V, Grief C, Jowett J B, Jones I M. Comparative morphology of Gag protein structures produced by mutants of the gag gene of human immunodeficiency virus type 1. J Gen Virol. 1994;75:2985–2997. doi: 10.1099/0022-1317-75-11-2985. [DOI] [PubMed] [Google Scholar]
- 18.Housset V, De Rocquigny H, Roques B P, Darlix J L. Basic amino acids flanking the zinc finger of Moloney murine leukemia virus nucleocapsid protein NCp10 are critical for virus infectivity. J Virol. 1993;67:2537–2545. doi: 10.1128/jvi.67.5.2537-2545.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang M, Orenstein J M, Martin M A, Freed E O. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leis J, Baltimore D, Bishop J M, Coffin J, Fleissner E, Goff S P, Oroszlan S, Robinson H, Skalka A M, Temin H M, Vogt V. Standardized and simplified nomenclature for proteins common to all retroviruses. J Virol. 1988;62:1808–1809. doi: 10.1128/jvi.62.5.1808-1809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puffer B A, Parent L J, Wills J W, Montelaro R C. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol. 1997;71:6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puffer B A, Watkins S C, Montelaro R C. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J Virol. 1998;72:10218–10221. doi: 10.1128/jvi.72.12.10218-10221.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ragheb J A, Anderson W F. Uncoupled expression of Moloney murine leukemia virus envelope polypeptides SU and TM: a functional analysis of the role of TM domains in viral entry. J Virol. 1994;68:3207–3219. doi: 10.1128/jvi.68.5.3207-3219.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rein A, Mirro J, Haynes J G, Ernst S M, Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol. 1994;68:1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts P C, Compans R W. Host cell dependence of viral morphology. Proc Natl Acad Sci USA. 1998;95:5746–5751. doi: 10.1073/pnas.95.10.5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts P C, Lamb R A, Compans R W. The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology. 1998;240:127–137. doi: 10.1006/viro.1997.8916. [DOI] [PubMed] [Google Scholar]
- 28.Strambio-de-Castillia C, Hunter E. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J Virol. 1992;66:7021–7032. doi: 10.1128/jvi.66.12.7021-7032.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sudol M. The WW module competes with the SH3 domain? Trends Biochem Sci. 1996;21:161–163. [PubMed] [Google Scholar]
- 30.Sudol M. From Src Homology domains to other signaling modules: proposal of the ‘protein recognition code’. Oncogene. 1998;17:1469–1474. doi: 10.1038/sj.onc.1202182. [DOI] [PubMed] [Google Scholar]
- 31.Swanstrom R, Wills J W. Synthesis, assembly, and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 32.Telesnitsky A, Blain S, Goff S P. Assays for retroviral reverse transcriptase. Methods Enzymol. 1995;262:347–362. doi: 10.1016/0076-6879(95)62029-x. [DOI] [PubMed] [Google Scholar]
- 33.Vogt V M, Eisenman R, Diggleman H. Generation of avian myeloblastosis virus structural proteins by proteolytic cleavage of a precursor polypeptide. J Mol Biol. 1975;96:471–493. doi: 10.1016/0022-2836(75)90174-6. [DOI] [PubMed] [Google Scholar]
- 34.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wills J W, Craven R C. Form, function, and use of retroviral gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 36.Xiang Y, Cameron C E, Wills J W, Leis J. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yasuda J, Hunter E. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J Virol. 1998;72:4095–4103. doi: 10.1128/jvi.72.5.4095-4103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan B, Li X, Goff S P. Mutations altering the moloney murine leukemia virus p12 gag protein affect virion production and early events of the virus life cycle. EMBO J. 1999;18:4700–4710. doi: 10.1093/emboj/18.17.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]