Spontaneous tumorigenesis in mice defective in the MTH1 gene encoding 8-oxo-dGTPase (original) (raw)
Abstract
Oxygen radicals, which can be produced through normal cellular metabolism, are thought to play an important role in mutagenesis and tumorigenesis. Among various classes of oxidative DNA damage, 8-oxo-7,8-dihydroguanine (8-oxoG) is most important because of its abundance and mutagenicity. The MTH1 gene encodes an enzyme that hydrolyzes 8-oxo-dGTP to monophosphate in the nucleotide pool, thereby preventing occurrence of transversion mutations. By means of gene targeting, we have established MTH1 gene-knockout cell lines and mice. When examined 18 months after birth, a greater number of tumors were formed in the lungs, livers, and stomachs of _MTH1_-deficient mice, as compared with wild-type mice. The _MTH1_-deficient mouse will provide a useful model for investigating the role of the MTH1 protein in normal conditions and under oxidative stress.
Oxygen radicals are produced through normal cellular metabolism, and formation of such radicals is enhanced further by ionizing radiation and by various chemicals (1). The oxygen radicals attack nucleic acids and generate various modified bases in DNA (2, 3). Among them, 8-oxo-7,8-dihydroguanine (8-oxoG) is the most abundant, and seems to play a critical role in carcinogenesis and in aging (4). 8-OxoG can pair with both cytosine and adenine during DNA synthesis, and as a result, G:C to T:A transversions are induced (5). Oxidation of guanine also occurs in the cellular nucleotide pool. 8-Oxo-dGTP, when formed, is a potent mutagenic substrate for DNA synthesis: it is equally incorporated opposite adenine and cytosine in DNA (6), resulting in both A:T to C:G and G:C to T:A transversions (7).
Studies with Escherichia coli mutator mutants revealed that cells possess elaborate mechanisms which prevent mutations caused by oxidation of the guanine base, in both DNA and free-nucleotide forms. In DNA, 8-oxoG residues are removed by the enzyme encoded by the_mutM_ gene, whereas the mutY gene product removes adenine from an adenine:8-oxoG mismatch (8, 9). Thus, two proteins, MutM and MutY, act consecutively at the site of the oxidized guanine residue in DNA to prevent the occurrence of mutations in E. coli (9, 10). On the other hand, mutations owing to misincorporation of 8-oxo-dGTP can be prevented by the mutT gene product, which hydrolyzes 8oxo-dGTP to 8-oxo-dGMP (6). Mutation in mutT specifically induces A:T to C:G transversions (11). This mutational specificity occurs through the concerted actions of the MutM and MutY proteins (12).
8-OxoG-related mutagenesis may account for a considerable number of spontaneous mutations in mammalian cells. Enzyme activities similar to those of the E. coli proteins were identified in mammalian cells (13–15). Among them, the MutT-related protein has been studied most extensively (16). Based on the partial amino acid sequence determined with the purified 18-kDa protein bearing 8-oxo-dGTPase activity, cDNA and the gene for the human enzyme were isolated and named as MTH1 (mutT homolog-1; refs. 17 and 18). Expression of the human MTH1 cDNA in _E. coli mutT_− cells significantly suppressed the frequency of spontaneous mutation in _mutT_− cells (17, 18). Additionally, more striking suppressive effects were observed when mouse or rat cDNA was expressed in the_mutT_− cells (19, 20). These observations imply that the mammalian proteins may have the same antimutagenic capacity as E. coli MutT protein and may act to sanitize the nucleotide pools in these organisms.
It is interesting to note that the E. coli and the mammalian enzymes have some different spectra in their substrate specificity. Human MTH1 protein hydrolyzes 2-hydroxydeoxyadenosine triphosphate but acts only slightly on 8-oxoguanosine triphosphate, a ribonucleotide counterpart of 8-oxo-dGTP. In contrast, E. coli MutT protein exhibits opposite actions on these nucleotides (21, 22). Based on the substrate specificity of human MTH1 protein, the name “oxidized purine nucleoside triphosphatase” has been proposed recently (23).
To ascertain the exact role of MTH1 protein, particularly in the case of spontaneous carcinogenesis, we carried out targeted disruption of the MTH1 gene in mice. Experiments with the MTH1-null mice generated here showed their disposition to develop tumors during a normal lifespan, and provided an important insight into the role of this nucleotide sanitization enzyme in terms of spontaneous tumorigenesis as well as mutagenesis caused by the oxygen-induced DNA damage.
Materials and Methods
Construction of the Targeting Vector and Isolation of Targeted Embryonic Stem (ES) Cell Clones.
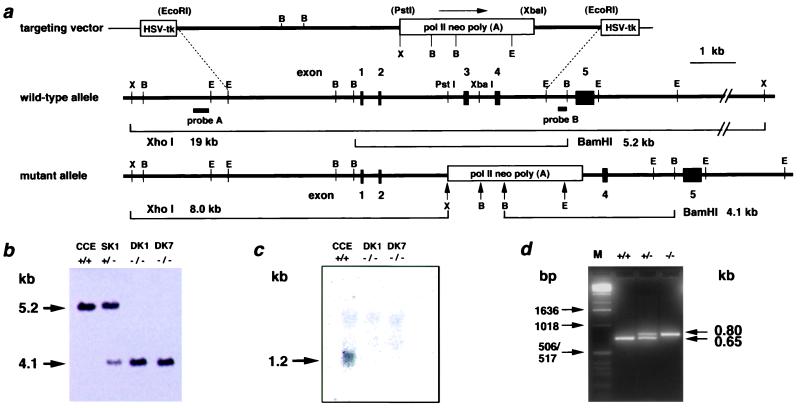
The procedures for gene targeting in ES cells were essentially as described (24). Briefly, the targeting vector contained a 7.0-kb genomic sequence interrupted by a_polII_-_neo_-poly(A) cassette and flanked by a pair of herpes simplex virus thymidine kinase cassettes for negative selection. Insertion of the neo cassette was accompanied by deletion of a 680-bp _Pst_I-Xba_I fragment of_MTH1 gene, containing 165 nucleotides of exon 3, 370 nucleotides of intron 2, and 145 nucleotides of intron 3 (Fig.1a). Colonies doubly resistant to G418 (250 μg/ml) and ganciclovir (5 μM) were selected, and homologous recombinants were identified by Southern blot analysis. The DNA (8 μg) was cleaved with _Xho_I or Bam_HI, subjected to agarose gel electrophoresis, blotted onto Hybond N+ membrane (Amersham Pharmacia), and hybridized to the 3′-flanking probe (Fig. 1a). Expected sizes of hybridized bands for wild-type and mutant MTH1 alleles were 5.2 kb and 4.1 kb, respectively. To ensure targeted disruption of the_MTH1 gene, the DNA was digested with _Xho_I, followed by hybridization with 5′-flanking probe (Fig. 1a). To isolate the homozygously targeted ES cells, cells that were identified as those heterozygous for the MTH1 sequence were cultured in the presence of a higher dose of G418 (1.5 mg/ml), as described (25).
Figure 1.
Targeted disruption of the MTH1 gene by homologous recombination. (a) Targeting of the MTH1 gene. The upper lines represent targeting vector and wild-type_MTH1_ allele, whereas the lower line shows mutated_MTH1_ allele. The thick lines show genomic sequences with exons (filled boxes), whereas a thin line shows the bacterial plasmid portion. Open boxes, a positive [pol II neo poly (A)] or a negative (HSV-tk) selection cassette; lettered bars, 5′-flanking probe A (0.1-kb _Apa_I-_Eco_RI fragment) and 3′-flanking probe B (0.1-kb _Pst_I-_Bam_HI fragment). The observed sizes of the diagnostic restriction fragments, used to distinguish the wild-type and mutant alleles, correspond to their expected sizes. The restriction enzyme sites are abbreviated as B for _Bam_HI, E for Eco_RI, and X for_Xho_I. The restriction enzyme sites in parentheses are lost in the process of targeting vector construction. (b) Southern blot analysis of_Bam_HI-digested genomic DNA from_MTH1+/+(CCE),MTH1+/−(SK1), and two_MTH1_−/−(DK1, DK7) ES cell clones, using the external 3′ probe. (c) Northern blot analysis of poly(A)+ RNA from MTH1+/+ and two _MTH1_−/− ES cell clones, using the 503-bp _Nco_I/_Bam_HI fragment of mouse cDNA as a probe that detects an ≈1.2-kb band corresponding to the size of MTH1 transcript. Each lane contained 3 μg of poly(A)+ RNA. (d) Genotype analysis of DNAs from tails from MTH1+/− intercrosses by PCR. PCR amplification of the wild-type MTH1 allele produces a 0.65-kb DNA fragment (bottom band), whereas amplification of the mutated MTH1 allele produces a 0.80-kb DNA fragment (upper band). Lane 1, marker DNA fragments (M); lane 2,MTH1+/+; lane 3,MTH1+/−; and lane 4,_MTH1_−/−. Sizes of marker fragments are indicated Left.
Generation of Chimeric, Heterozygous, and Homozygous Mice.
Mutant ES cells were injected into C57BL/6J blastocysts, and resulting male chimeras were mated with female BDF1 mice. Germ-line transmission of mutant MTH1 allele to F1 mice was confirmed by Southern blot analysis. Genotyping of F2 offspring was performed by a PCR-based method (26). A combination of a pair of primers was used to detect wild-type and mutant alleles: a pair of 5′-CTCTCCAGCCCTTGTTCAAGTTC-3′ as forward MTH1 primer (MT5) and 5′-CCTACTCTCTTGGGCTTCATCC-3′ as reverse MTH1 primer (MT3) for the wild-type allele, and a pair of MT5 and 5′-GAACCTGCGTGCAATCCATCTTGT-3′ as reverse neo primer (MTN) for the mutated allele. All experiments were conducted in accordance with institutional guidelines for Kyushu University.
Detection of the MTH1 Protein.
Precleared extracts (100 μg of protein) from livers and 1 ng of purified mouse MTH1 protein were reacted with anti-MTH1 (5 μg) overnight, and the immunocomplexes were subjected to Western blotting as described (19). 8-Oxo-dGTPase activity was assayed by measuring the hydrolysis of 8-oxo-dGTP to 8-oxo-dGMP, as described (17).
Mutation Analysis.
Of each independent colony, 15 were isolated from two_MTH1_−/− cell lines, DK1 and DK7, and 24 independent colonies were isolated from_MTH1_+/+ cells. Cells of each colony were expanded and plated onto two 100-mm dishes (1.0 × 106 cells per dish). After 2 days of incubation, ES medium (27) containing 6-thioguanine (6-TG; 1 μg/ml) was supplied to one of the two dishes, and the number of colonies was counted after 10 days of incubation with 6-TG (25). By using the rest of the dishes, the number of viable cells at the time of 6-TG treatment was determined.
Results
Generation of _MTH1_-Deficient Cells.
The mouse MTH1 gene is composed of five exons and spans about 10 kb (28). By using the isogenic genomic DNA fragment, the whole part of the third exon containing the initiation codon and the adjacent intron regions were replaced by a neo cassette (Fig.1a). The resulting construct was electroporated into ES cells, and cells showing resistance to both G418 and ganciclovir were selected. Clones were screened by Southern blot hybridization, using 3′- and 5′-flanking probes with a combination of restriction enzymes_Bam_HI and _Xho_I. Approximately 14% of the resistant cells (11 of 78) carried the expected structure of the mutated allele (Fig. 1a).
To isolate the double knock-out cells, targeted clones (MTH1+/−) were grown in the presence of 1.5 mg/ml of G418, a concentration which is 6 times higher than that used for isolation of MTH1+/− cells. All of the resistant colonies, examined after 8 days of incubation, were shown to be mutated in both of the MTH1 alleles. DK1 and DK7 are double knock-out cells, derived from the different singly targeted clones (Fig. 1b). Northern blot analysis was done to determine the level of expression of the MTH1 gene in ES cells. A band corresponding to 1.2-kb MTH1 mRNA was detected only in the wild-type ES cell (Fig. 1c).MTH1_−/− cells thus established grew normally in ES medium, and the average doubling times of_MTH1+/+ and_MTH1_−/− cells at 37°C were essentially the same (9.2 h).
Mutation Frequency of _MTH1_−/− Cells.
Mutations in the Hprt gene, located on the X chromosome in the mouse genome, render cells resistant to 6-TG, and this forward mutation assay was adopted for examining the effect of MTH1 deficiency on spontaneous mutagenesis. Mutation rates toward 6-TG resistance were determined by a fluctuation test using the two cell lines with_MTH1_+/+ and_MTH1_−/− background. With these data, a mutation rate can be estimated either from the fraction of replicate cultures in which no resistant cells occurred [P(0)] (29) or from the median frequency of resistant cells per replicate cultures, using the method of Lea and Coulson (30). An approximately 2-fold higher mutation rate was observed in two independently isolated_MTH1_−/− cells (DK1 and DK7), compared with the value of MTH1+/+ cells (Table1). MTH1 may have a potential to prevent the occurrence of mutations under the normal growth conditions.
Table 1.
Mutation rates determined by fluctuation tests
| Name of cell line (genotype) | No. of replicate cultures | Final no. of cells per replicate | No. of replicates with 6-TGR | No. of 6-TGR colonies per replicate | Mutation rate, × 10−8 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N colonies, N = | Range | Mean | Variance | P(0) | P(0) calculation* | Median calculation* | |||||||
| 0 | 1 | 2 | 3–5 | 6–10 | |||||||||
| CCE (+/+) | 24 | 3.0 × 107 ± 0.4 | 12 | 5 | 4 | 3 | 0 | 0–4 | 1.00 | 1.58 | 0.50 | 1.65 (1.00) | 1.43 (1.00) |
| DK1 (−/−) | 15 | 3.0 × 107 ± 0.2 | 4 | 6 | 3 | 0 | 2 | 0–10 | 1.93 | 7.39 | 0.27 | 3.06 (1.85) | 2.55 (1.78) |
| DK7 (−/−) | 15 | 3.0 × 107 ± 0.3 | 3 | 2 | 0 | 9 | 1 | 0–7 | 3.06 | 4.73 | 0.20 | 3.72 (2.25) | 3.65 (2.55) |
Generation of Mice with Mutated MTH1 Alleles.
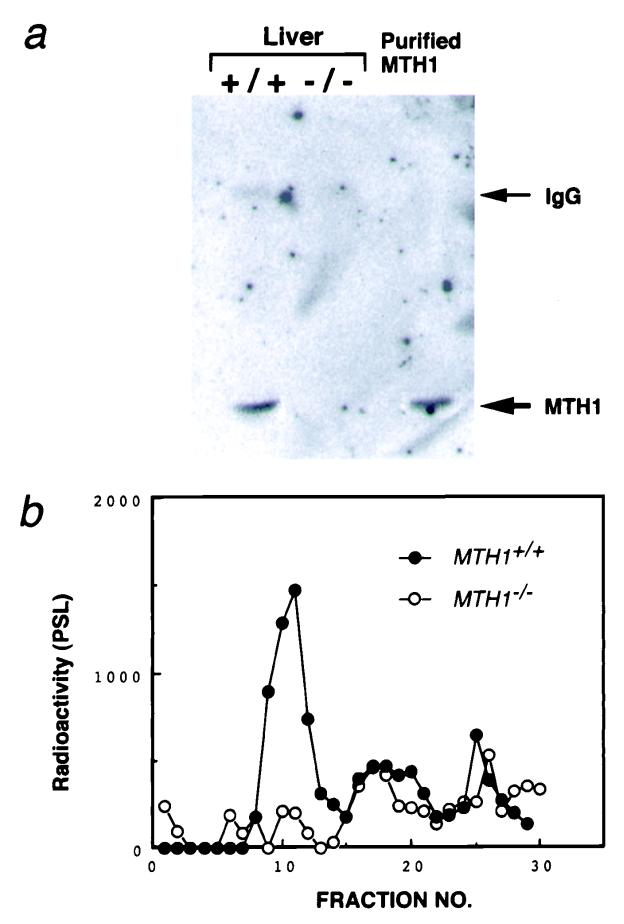
MTH1 +/− ES cell clones were microinjected into C57BL/6J blastocysts and 8 chimeras (6 males and 2 females) were obtained. Two independent germ-line chimeras transmitted mutant MTH1 alleles to their agouti offspring. By crossing the MTH1+/− mice, homozygous mutant mice (_MTH1_−/−) were obtained, according to Mendel's law (25 of 98). The _MTH1_−/− mice appeared normal. Both copies of the exon 3 segment were disrupted in the _MTH1_−/− mice (Fig. 1d). To ensure that modification of the MTH1 gene resulted in a null mutation, we examined the liver of wild-type and_MTH1_−/− mice for MTH1 protein. Western blot analysis detected normal MTH1 protein levels in wild-type mice, whereas no MTH1 protein was detected in_MTH1_−/− mice (Fig.2a). Further, assay of 8-oxo-dGTPase activity in the liver sample confirmed this finding (Fig.2b).
Figure 2.
Absence of MTH1 protein in MTH1_-deficient mouse. (a) Western blot analysis of extracts of liver from_MTH1+/+ and_MTH1_−/− mice, with Abs against purified MTH1 protein. Lane 1, MTH+/+; lane 2,_MTH1_−/−; and lane 3, purified MTH1 protein (1 ng). Arrows (Right) indicate the positions of normal rabbit IgG heavy-chain and purified MTH1 protein, respectively. (b) Assay of 8-oxo-dGTPase activity in the liver. Crude extracts of liver (1 g) prepared from MTH+/+ and _MTH1_−/− mice loaded on a HiTrap Q column and eluted by a linear gradient of 0–0.5 M NaCl. Radioactivities of 8-oxo-dGMP produced were measured by PSL (photo-stimulated luminescence).
Tumorigenesis of MTH1-Defective Mice.
Offspring of F2 heterozygotes were used for analyzing the susceptibility for spontaneous tumorigenesis. Briefly, the F2 heterozygotes were intercrossed and resulting F3 heterozygote offspring were then intercrossed to other heterozygote F3 offspring to generate F4 animals with 0, 1, or 2 germ-line null MTH1 alleles. These animals were ≈50% C57BL/6J, 25% 129/Sv, and 25% DBA/2, respectively, and monitored for tumor development as described below. Groups of wild-type and null mice (≈50 males and females), each containing almost the same number of F3 and F4 generations, were kept under specific pathogen-free (SPF) conditions for an extended period. No distinct difference in survival rates of these_MTH1_+/+ and_MTH1_−/− males and females was observed. After 1.5 years of observation, all of the animals were necropsied. Systematic pathological examination revealed a significant difference in the incidence of tumors in the two types of mice. Tumors were found in lung, liver, and stomach. Here, the term “tumor” is used without distinction between benign and malignant neoplasms.
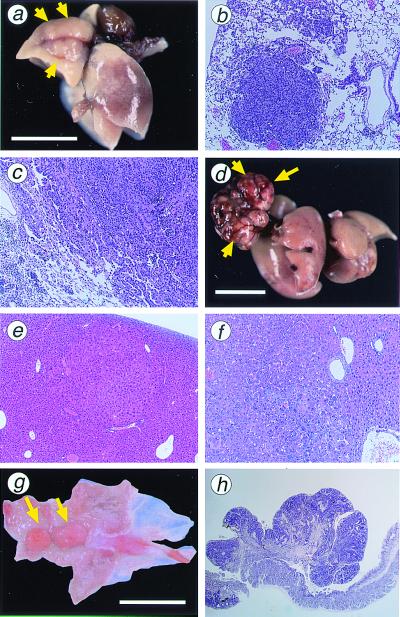
Tumors of the lungs were grayish-white nodules with a maximum diameter of 1–7 mm (Fig. 3a). They are mostly solitary. Histologically, lung tumors were diagnosed as adenoma or adenocarcinoma (Table 2). Adenomas were relatively small spherical lesions composed of basophilic tumor cells simulating alveolar pattern with little cellular atypism (Fig.3b). Adenocarcinomas were composed of irregular-sized tumor cells, densely arranged in a glandular pattern and showing peripheral infiltration in some parts (Fig. 3c). Both tumor types developed in MTH1_−/− and_MTH1+/+ mice with no apparent sex predisposition. However, when tumor yield was viewed with respect to the total number of tumors, more lung tumors were formed in_MTH1_−/− mice (15 of 93 mice) than in_MTH1_+/+ mice (4 of 90 mice,P = 0.014; Table 2). Both adenomas and adenocarcinomas developed in the liver of MTH1_−/− and_MTH1+/+ mice (Fig. 3 d_–_f). More liver tumors were likely to be formed in _MTH1_−/− mice (18 of 93) than in MTH1+/+ mice (6 of 90), and these values are statistically significant (P = 0.015). High susceptibility of male mice to liver tumorigenic events was evident, compared with the female counterparts. As for the tumor formation in liver, 38% of MTH1_−/− male and 13% of_MTH1+/+ male mice developed tumors, whereas only 3.9% of MTH1_−/− female and 0% of_MTH1+/+ female mice yielded tumors.
Figure 3.
Tumors in lung, liver, and glandular stomach developed in_MTH1_−/− mice. (a) Adenocarcinoma of the lung developed in_MTH1_−/− mouse; 7 mm in maximum diameter protruding on the surface of a lung lobe (arrows). (b) Histologic section of a lung adenoma developed in_MTH1_−/− mice. (c) Histologic section of a lung adenocarcinoma. Papillary/alveolar proliferation of basophilic cells is the common feature of both tumor types. (d) Large carcinoma nodule of the liver developed in _MTH1_−/− mouse; 16 mm in maximum diameter (arrows). (e) Histologic section of an adenoma developed in liver of _MTH1_−/− mice. (f) Histologic section of a hepatocellular carcinoma developed in _MTH1_−/− mouse showing typical trabecular pattern of malignant hepatocytes. (g) Elevated lesion of the pyloric mucosa of_MTH1_−/− mouse (arrows) diagnosed as well differentiated adenocarcinoma. (h) Histologic section of adenocarcinoma of the stomach in MTH1_−/− mouse, cut perpendicular to the mucosal surface, consisted of irregular proliferation of dysplastic cells arranged in tubular structure. [Bar = 10 mm (a, d, and_g).] Paraffin-embedded sections (4 mm) were stained with hematoxylin and eosin. Magnification: b, ×100;c, ×100; e, ×40; f, ×100; h, ×20.
Table 2.
Spontaneous tumorigenesis in wild-type and MTH1-deficient mice
| Organ | Type of tumor | Male | Female | ||
|---|---|---|---|---|---|
| −/− | +/+ | −/− | +/+ | ||
| Lung | Carcinoma | 2 | 1 | 3 | 1 |
| Adenoma | 5 | 2 | 5 | 0 | |
| No. of mice with tumors (%) | 7 (17) | 3 (7) | 8 (16) | 1 (2) | |
| Liver | Hepatocellular carcinoma | 10 | 2 | 0 | 0 |
| Adenoma | 6 | 4 | 2 | 0 | |
| No. of mice with tumors (%) | 16 (38) | 6 (13) | 2 (4) | 0 (0) | |
| Stomach | Carcinoma | 2 | 1 | 0 | 0 |
| Adenoma | 4 | 1 | 2 | 0 | |
| No. of mice with tumors (%) | 6 (14) | 2 (4) | 2 (4) | 0 (0) | |
| Total no. of mice examined | 42 | 46 | 51 | 44 |
It is of interest to note that 13 of 93_MTH1_−/− mice had elevated lesions in glandular stomach, whereas only 4 of 90_MTH1_+/+ mice had similar lesions during the same observation period. Most lesions were grayish-white in color, and showed polypoid growths in the pyloric mucosa. Histologically, they were diagnosed as adenomatous hyperplasia, adenoma, or adenocarcinoma. The incidence of adenomatous hyperplasia (not shown) was found in 2_MTH1_+/+ male mice and 5 of_MTH1_−/− mice (2 male and 3 female mice). Examples of stomach tumors developed in_MTH1_−/− mice are shown (Fig.3g). Polypoid carcinomas consisted of irregular proliferation of dysplastic cells arranged in tubular structure (Fig.3h). Of 13 gastric polyps, 2 that developed in_MTH1_−/− mice were classified into well differentiated adenocarcinomas, whereas 1 adenocarcinoma was found of 4 gastric polyps developed in MTH1+/+ mice. It should be noted that the 3 of these 4 gastric polyps in wild-type mice were siblings.
In summary, more tumors were formed in the three internal organs (lungs, livers, and stomachs) of _MTH1_−/− mice than in those of the wild-type mice. When statistical analysis was carried out comparing the total number of mice bearing tumors between the MTH1_−/− and_MTH1+/+ groups, there was significant difference between these two groups: 34 of 93 (36%)MTH1_−/− mice compared with 10 of 90 (11%) MTH1+/+ mice (P < 0.001). Other tumors observed in the_MTH1_−/− mice included skin sarcomas (two male cases) and two cervical lymphomas, one intestinal lymphoma and one thymoma in females. However, only one cervical lymphoma was found in an_MTH1+/+ female mouse in this experiment.
Discussion
It has been proposed that one early step in the progression of human tumors is an elevation of the rate of spontaneous mutation (i.e., to induce a mutator phenotype). This argument is based on the finding that the progression of many human tumors is accompanied by accumulation of a large number of mutations (31–34). Thus, if changes in spontaneous mutation rates are indeed involved in carcinogenesis, it is important to define pathways that influence spontaneous mutation rates in mammalian cells. Experiments with the _MTH1_-null cell lines generated here will provide some clues as to the role of elimination of oxidized forms of DNA precursors.
Degrees of increase in spontaneous mutation frequency, because of the loss of MutT-related functions, differ considerably in E. coli and mouse cells. As shown in the present study, the increases in Hprt mutations detected in mouse_MTH1_−/− cells is 2-fold compared with_MTH1_+/+ cells, whereas the value of mutation frequency for _E. coli mutT_− cells is 100 times more than that of wild-type cells (12). Several hypotheses to explain this difference may be considered, among which the most plausible is a possibility that mammalian cells may possess an enzyme capable of degrading 8-oxo-dGTP in addition to MTH1. This notion was strengthened by the recent finding that E. coli, despite its predominant MutT activity, possesses an additional enzyme activity that degrades 8-oxo-dGTP. GTP cyclohydrolase II, encoded by the ribA gene of E. coli, can hydrolyze 8-oxo-dGTP to the corresponding nucleoside monophosphate. In the _mutT_− background,ribA_− cells showed 2 times higher spontaneous mutation frequencies as compared with_ribA+ cells (35). It is unlikely that a GTP cyclohydrolase II-type enzyme is present in mammalian cells because the biosynthesis of riboflavin, in which this type of enzyme functions, does not take place in animals. Mammalian cells may possess another type of enzyme(s) that is capable of degrading 8-oxo-dGTP. In support of this idea, we have identified recently a mouse cDNA clone that considerably suppresses the high mutability of _E. coli mutT_− cells (J.-P. Cai, Y. Takagi, and M.S., unpublished result). The protein encoded by this cDNA carries the MutT box, an amino acid sequence highly conserved through MutT and MTH1 family proteins (36), and could act as an MTH1 redundancy factor.
MTH1 homozygous mutant mice were found to have a normal physical appearance. Pathological examination revealed a statistically significant difference in the incidence of tumors in_MTH1_+/+ and_MTH1_−/− mice. Around 18 months after birth, many tumors were found in lungs and livers of the_MTH1_-deficient mice, but there were a few in_MTH1_+/+ mice. The elevated incidence of tumor formation in the liver of _MTH1_−/− mice was well correlated with the highest content of MTH1 protein in this organ of the wild-type mouse (19). As generally observed in spontaneous and carcinogen-induced hepatocarcinogenesis in rodents (37), there is a high susceptibility of male mice to liver tumorigenic events, compared with their female counterparts. It has been suggested that the hormonal environment of the host affects the development of many types of tumors, especially those in liver. More tumors tend to form in the stomach of _MTH1_−/− mice, as compared with MTH1+/+ mice, although the statistical significance is rather weak. Indeed, as there are differing profiles of antioxidant enzymes, such as superoxide dismutases and catalases among organs, different susceptibility to tumorigenesis among organs may reflect the metabolic balance of oxidative stress, inducing lesions in DNA as well as in the nucleotide pool. Altogether, we concluded that the intracellular level of MTH1 protein is an important factor in determining susceptibility of mice to tumor induction by endogenous oxidative DNA damage. These studies with MTH1-null mutant mice will provide an important insight into the role of this nucleotide sanitization enzyme in terms of spontaneous tumorigenesis.
To prevent occurrence of cancers, organisms are equipped with multiple defense mechanisms, including DNA repair systems, apoptotic cell death, and immune systems. Indeed, in_MSH2-_deficient mice, where one of the mismatch repair enzymes is absent (38, 39), and p53_-deficient mice, where appropriate apoptotic cell death is disturbed (40), tumors frequently occur during early life. Although it is generally conceived that blockage of DNA replication by DNA damage somehow acts as a signal for inducing apoptosis, some type of DNA lesions that do not prevent progression of the replication fork also cause apoptosis. Indeed, O6-methylguanine, produced by alkylating agents, does not prevent DNA replication but induces efficient apoptosis (25). Evidence has been presented that mismatch recognition proteins, such as MLH1, are involved in recognition of O6-methylguanine in the DNA as a signal for induction of apoptosis (41). Accumulation of 8-oxoG paired with adenine may be recognized by the mismatch-protein complex (42). If cells carrying 8-oxoG were eliminated through apoptosis, the incidence of mutation and tumors in_MTH1_−/− mice might be reduced. Moreover, another pathway, such as nucleotide excision repair pathway, may also participate in repairing the mutagenic lesion, 8-oxoG (43). It is interesting to note that recent works with Ogg1 (Mmh)-deficient mice exhibited significantly elevated spontaneous mutation rates but did not show an increased incidence of tumor formation (44, 45). No such marked pathological changes observed with the 8-oxoG DNA glycosylase-deficient mice might be attributed to their observation period (not beyond a 1-year term), because histopathological examination of both_MTH1+/+ and_MTH1_−/− mice killed at around 12 months of age did not reveal clear abnormalities.
More definitive studies designed to evaluate the role of this oxidation-induced DNA damage-defense system in the intact animal will benefit from double- or triple-mutant mice, which are defective in other pathways of DNA repair.
Acknowledgments
We thank Drs. T. Kakuma, K. Sakumi, M. Furuichi, Y. Fujii, and Mr. N. Kinoshita for their assistance and helpful discussion, and Drs. D. E. Rancourt and M. A. Wride for comments on the manuscript. This work was supported, in part, by Kyushu University Interdisciplinary Programs in Education and Projects in Research Development, a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and by a research grant from Fukuoka Cancer Association.
Abbreviations
8-oxoG
8-oxo-7,8-dihydroguanine
ES cells
embryonic stem cells
6-TG
6-thioguanine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ames B N, Gold L S. Mutat Res. 1991;250:3–16. doi: 10.1016/0027-5107(91)90157-j. [DOI] [PubMed] [Google Scholar]
- 2.Boiteux S, Gajewski E, Laval J, Dizdaroglu M. Biochemistry. 1992;31:106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- 3.Gajewski E, Rao G, Nackerdien Z, Dizdaroglu M. Biochemistry. 1990;29:7876–7882. doi: 10.1021/bi00486a014. [DOI] [PubMed] [Google Scholar]
- 4.Kasai H, Crain P F, Kuchino Y, Nishimura S, Ootsuyama A, Tanooka H. Carcinogenesis. 1986;7:1849–1851. doi: 10.1093/carcin/7.11.1849. [DOI] [PubMed] [Google Scholar]
- 5.Shibutani S, Takeshita M, Grollman A P. Nature (London) 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 6.Maki H, Sekiguchi M. Nature (London) 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 7.Cheng K C, Cahill D S, Kasai H, Nishimura S, Loeb L A. J Biol Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- 8.Au K G, Cabrera M, Miller J H, Modrich P. Proc Natl Acad Sci USA. 1988;85:9163–9166. doi: 10.1073/pnas.85.23.9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michaels M L, Cruz C, Grollman A P, Miller J H. Proc Natl Acad Sci USA. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tchou J, Grollman A P. Mutat Res. 1993;299:277–287. doi: 10.1016/0165-1218(93)90104-l. [DOI] [PubMed] [Google Scholar]
- 11.Yanofsky C E, Cox E C, Horn V. Proc Natl Acad Sci USA. 1966;55:274–281. doi: 10.1073/pnas.55.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tajiri T, Maki H, Sekiguchi M. Mutat Res. 1995;336:257–267. doi: 10.1016/0921-8777(94)00062-b. [DOI] [PubMed] [Google Scholar]
- 13.Mo J-Y, Maki H, Sekiguchi M. Proc Natl Acad Sci USA. 1992;89:11021–11025. doi: 10.1073/pnas.89.22.11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bessho T, Tano K, Kasai H, Ohtsuka E, Nishimura S. J Biol Chem. 1993;268:19416–19421. [PubMed] [Google Scholar]
- 15.Yeh Y-C, Chang D-Y, Masin J, Lu A-L. J Biol Chem. 1991;266:6480–6484. [PubMed] [Google Scholar]
- 16.Sekiguchi M. Genes to Cells. 1996;1:139–145. doi: 10.1046/j.1365-2443.1996.d01-232.x. [DOI] [PubMed] [Google Scholar]
- 17.Sakumi K, Furuichi M, Tsuzuki T, Kakuma T, Kawabata S, Maki H, Sekiguchi M. J Biol Chem. 1993;268:23524–23530. [PubMed] [Google Scholar]
- 18.Furuichi M, Yoshida M C, Oda H, Tajiri T, Nakabeppu Y, Tsuzuki T, Sekiguchi M. Genomics. 1994;24:485–490. doi: 10.1006/geno.1994.1657. [DOI] [PubMed] [Google Scholar]
- 19.Kakuma K, Nishida J, Tsuzuki T, Sekiguchi M. J Biol Chem. 1995;270:25942–25948. doi: 10.1074/jbc.270.43.25942. [DOI] [PubMed] [Google Scholar]
- 20.Cai J-P, Kakuma T, Tsuzuki T, Sekiguchi M. Carcinogenesis. 1995;16:2343–2350. doi: 10.1093/carcin/16.10.2343. [DOI] [PubMed] [Google Scholar]
- 21.Fujikawa K, Kamiya H, Yakushiji H, Fujii Y, Nakabeppu Y, Kasai H. J Biol Chem. 1999;274:18201–18205. doi: 10.1074/jbc.274.26.18201. [DOI] [PubMed] [Google Scholar]
- 22.Hayakawa H, Hofer A, Thelander L, Kitajima S, Cai Y, Oshiro S, Yakushiji H, Nakabeppu Y, Kuwano M, Sekiguchi M. Biochemistry. 1999;38:3610–3614. doi: 10.1021/bi982361l. [DOI] [PubMed] [Google Scholar]
- 23.Nakabeppu Y. Mutat Res. 2001;477:59–70. doi: 10.1016/s0027-5107(01)00096-3. [DOI] [PubMed] [Google Scholar]
- 24.Tsuzuki T, Sakumi K, Shiraishi A, Kawate H, Igarashi H, Iwakuma T, Tominaga Y, Zhang S, Shimizu S, Ishikawa T, et al. Carcinogenesis. 1996;17:1215–1220. doi: 10.1093/carcin/17.6.1215. [DOI] [PubMed] [Google Scholar]
- 25.Tominaga Y, Tsuzuki T, Shiraishi A, Kawate H, Sekiguchi M. Carcinogenesis. 1997;18:889–896. doi: 10.1093/carcin/18.5.889. [DOI] [PubMed] [Google Scholar]
- 26.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robertson E J. Teratocarcinomas and Embryonic Stem Cells: A Practical Approach. Oxford: IRL; 1987. [Google Scholar]
- 28.Igarashi H, Tsuzuki T, Kakuma T, Tominaga Y, Sekiguchi M. J Biol Chem. 1997;272:3766–3772. doi: 10.1074/jbc.272.6.3766. [DOI] [PubMed] [Google Scholar]
- 29.Luria S E, Delbrück M. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lea D E, Coulson C A. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 31.Ionov Y, Peinado M A, Malkhosyan S, Shibata D, Perucho M. Nature (London) 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 32.Loeb L A. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 33.Vogelstein B, Fearon E R, Hamilton S R, Kern S E, Preisinger A C, Leppert M, Nakamura Y, White R, Smits A M, Bos J L. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 34.Stoler D L, Chen N, Basik M, Kahlenberg M S, Rodriguez-Bigas M A, Petrelli N J, Anderson G R. Proc Natl Acad Sci USA. 1999;96:15121–15126. doi: 10.1073/pnas.96.26.15121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi M, Ohara-Nemoto Y, Kaneko M, Hayakawa H, Sekiguchi M, Yamamoto K. J Biol Chem. 1998;273:26394–26399. doi: 10.1074/jbc.273.41.26394. [DOI] [PubMed] [Google Scholar]
- 36.Fujii Y, Shimokawa H, Sekiguchi M, Nakabeppu Y. J Biol Chem. 1999;274:38251–38259. doi: 10.1074/jbc.274.53.38251. [DOI] [PubMed] [Google Scholar]
- 37.Poole T M, Drinkwater N R. Carcinogenesis. 1996;17:191–196. doi: 10.1093/carcin/17.2.191. [DOI] [PubMed] [Google Scholar]
- 38.Reitmair A H, Schmits R, Ewel A, Bapat B, Redson M, Mitri A, Waterhouse P, Mittrucher H W, Wakeham A, Liu B, et al. Nat Genet. 1995;11:64–70. doi: 10.1038/ng0995-64. [DOI] [PubMed] [Google Scholar]
- 39.de Wind N, Dekker M, Berna A, Radman M, te Riele H. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 40.Harvey M, McArthur M J, Montgomery C A, Jr, Bradley A, Donehower L A. FASEB J. 1993;7:938–943. doi: 10.1096/fasebj.7.10.8344491. [DOI] [PubMed] [Google Scholar]
- 41.Kawate H, Sakumi K, Tsuzuki T, Nakatsuru Y, Ishikawa T, Takahashi S, Takano H, Noda T, Sekiguchi M. Proc Natl Acad Sci USA. 1998;95:5116–5120. doi: 10.1073/pnas.95.9.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeWeese T L, Shipman J M, Larrier N A, Buckley N M, Kidd L R, Groopman J D, Cutler R G, te Riele H, Nelson W G. Proc Natl Acad Sci USA. 1998;95:11915–11920. doi: 10.1073/pnas.95.20.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reardon J T, Bessho T, Kung H C, Bolton P H, Sancar A. Proc Natl Acad Sci USA. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes D E. Proc Natl Acad Sci USA. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minowa O, Arai T, Hirano M, Monden Y, Nakai S, Fukuda M, Itoh M, Takano H, Hippou Y, Aburatani H, et al. Proc Natl Acad Sci USA. 2000;97:4156–4161. doi: 10.1073/pnas.050404497. . (First Published March 21, 2000; 10.1073/pnas.050404497) [DOI] [PMC free article] [PubMed] [Google Scholar]