Physical and Functional Interactions of Human DNA Polymerase η with PCNA (original) (raw)
Abstract
Human DNA polymerase η (hPolη) functions in the error-free replication of UV-damaged DNA, and mutations in hPolη cause cancer-prone syndrome, the variant form of xeroderma pigmentosum. However, in spite of its key role in promoting replication through a variety of distorting DNA lesions, the manner by which hPolη is targeted to the replication machinery stalled at a lesion site remains unknown. Here, we provide evidence for the physical interaction of hPolη with proliferating cell nuclear antigen (PCNA) and show that mutations in the PCNA binding motif of hPolη inactivate this interaction. PCNA, together with replication factor C and replication protein A, stimulates the DNA synthetic activity of hPolη, and steady-state kinetic studies indicate that this stimulation accrues from an increase in the efficiency of nucleotide insertion resulting from a reduction in the apparent K m for the incoming nucleotide.
DNA polymerase η (Polη) is unique among eukaryotic DNA polymerases in its proficient ability to replicate through distorting DNA lesions. Both in yeast and in humans, Polη functions in the error-free replication of UV-damaged DNA (19, 26, 34, 39), and mutations in human Polη (hPolη) result in cancer-prone syndrome, the variant form of xeroderma pigmentosum (XP-V) (17, 25). Interestingly, both yeast Polη and hPolη replicate through a _cis_-syn thymine-thymine (TT) dimer with the same efficiency and accuracy as they replicate through undamaged T's (18, 21, 37). Also, genetic studies with yeast have indicated a role for Polη in the error-free bypass of cyclobutane pyrimidine dimers that are formed at 5′-TC-3′ and 5′-CC-3′ sites (40). Polη also promotes replication through a (6-4) TT photoproduct, a highly distorting DNA lesion, by preferentially inserting a G residue opposite the 3′ T of the photoproduct. Subsequently, Polζ efficiently promotes extension from the G residue by inserting the correct nucleotide, A, opposite the 5′ T of the lesion (16). Although the insertion of a G opposite the 3′ T of the (6-4) TT photoproduct would cause 3′ T→C substitutions, it was previously suggested that a similar insertion of G by Polη opposite the 3′ C of the 5′-TC-3′ and 5′-CC-3′ (6-4) photoproducts, followed by extension by Polζ by the insertion of the correct nucleotide opposite the 5′ residue of this lesion, would lead to error-free bypass of the DNA lesion (16). Since (6-4) photoproducts are formed much more frequently at TC and CC sites than at TT sites (4, 6), Polη would largely contribute to the error-free bypass of (6-4) lesions as well. Yeast Polη and hPolη also efficiently replicate through other DNA lesions, such as 8-oxoguanine (15) and _O_6-methylguanine (13).
The ability of Polη to replicate through distorting DNA lesions has suggested that the active site of Polη is tolerant of geometric distortions introduced into DNA by these lesions. As a consequence, Polη is a low-fidelity enzyme, and on undamaged DNA, the yeast and human enzymes misincorporate nucleotides with a frequency of 10−2 to 10−3 (21, 38). In sharp contrast, replicative DNA polymerases exhibit a much higher fidelity, misinserting nucleotides with a frequency of 10−4 to 10−7 (3, 8, 33). However, because of their enhanced sensitivity to geometric distortions in DNA (9), these polymerases are unable to replicate through DNA lesions.
Although hPolη plays a critical role in the error-free replication of UV-damaged DNA and thus prevents the formation of sunlight-induced skin cancers, the manner by which this polymerase gains access to the replication machinery stalled at a lesion site is not known. Here, we examine the role of proliferating cell nuclear antigen (PCNA) in promoting the access of hPolη to the replication machinery. PCNA, a ring-shaped homotrimeric protein, forms a sliding clamp at the primer-template junction. PCNA is loaded onto DNA by the multiprotein clamp loader, replication factor C (RFC), which couples ATP hydrolysis to open and close the PCNA ring around the DNA. Replication protein A (RPA) binds single-stranded DNA, and after the loading of PCNA, RFC stays on DNA via its interaction with RPA (2, 22, 41). The replicative DNA polymerase, Polδ, then assembles with the PCNA ring, and this association endows the polymerase with a high processivity (30, 32). However, because of its inability to replicate through DNA lesions such as cyclobutane pyrimidine dimers, Polδ stalls at such lesion sites (18), necessitating the action of a translesion synthesis polymerase, such as Polη. Here, we provide evidence for the physical interaction of hPolη with PCNA and show that PCNA, together with RFC and RPA, stimulates the DNA synthetic activity of hPolη. These studies identify PCNA as a crucial element for the assembly of hPolη into the replication machinery.
MATERIALS AND METHODS
Proteins.
Human PCNA (hPCNA), RFC, and RPA were purified as described previously (5, 10, 24). Six-His-tagged hPCNA, used for interaction studies, was overexpressed in Escherichia coli and purified as described previously (23). Wild-type and mutant hPolη proteins fused with glutathione S-transferase (GST) were purified as described previously (16, 21); for the DNA synthesis studies (see Fig. 3 and 4), the GST portion was removed by treatment with PreScission protease (Amersham Pharmacia Biotech).
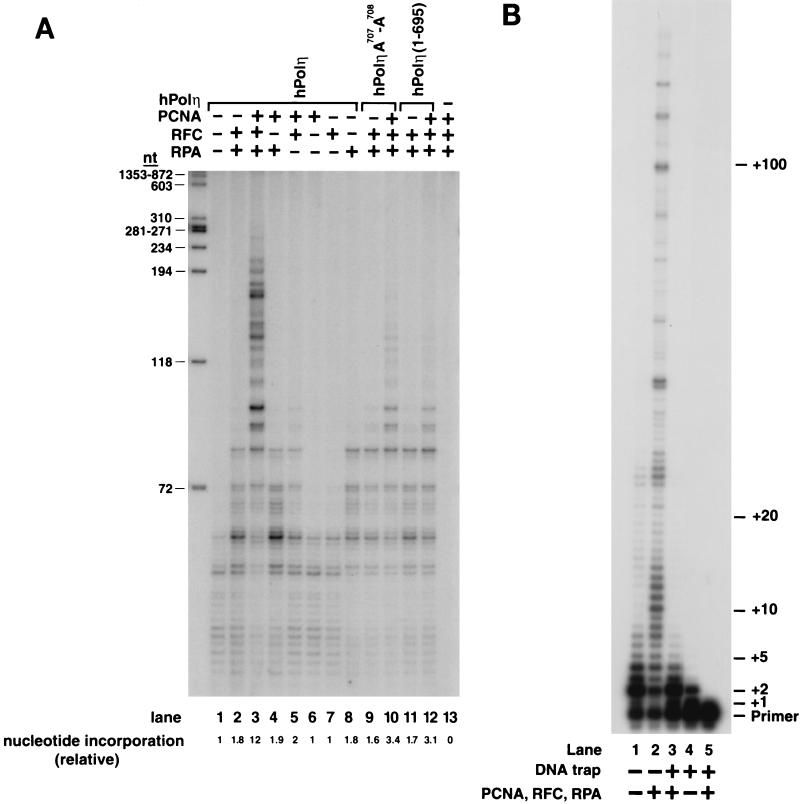
FIG. 3.
Stimulation of DNA synthetic activity of hPolη by PCNA. (A) DNA synthesis by the wild-type or PCNA binding site mutant hPolη proteins in the presence or absence of PCNA, RFC, and RPA. The reaction mixtures contained either the wild type hPolη protein (lanes 1 to 8) or the mutant hPolη A707-A708 (lanes 9 and 10) or hPolη (1-695) (lanes 11 and 12) proteins (10 ng each) along with singly primed M13 single-stranded DNA (25 ng), all four dNTPs (100 μM each), [α-32P]dATP, PCNA (100 ng), RFC (50 ng), or RPA (250 ng) or combinations of these proteins. No hPolη was added in lane 13. The amount of DNA synthesis is indicated at the bottom as the relative nucleotide (nt) incorporation. _Hae_III-digested ϕX174 DNA labeled with polynucleotide kinase is shown on the left as a molecular size marker. (B) Processivity of hPolη in the presence of PCNA, RFC, and RPA. hPolη (10 ng) alone (lanes 1 and 4) or in the presence of PCNA (100 ng), RFC (50 ng), and RPA (250 ng) (lanes 2 and 3) was preincubated with a circular single-stranded M13 template DNA (50 ng) singly primed with a 5′ 32P-labeled oligonucleotide for 5 min at 37°C. Primer extension reactions were initiated by adding all four dNTPs (500 μM each) (lanes 1 and 2) or all four dNTPs and excess sonicated herring sperm DNA (0.5 mg/ml) as a trap (lanes 3 and 4). After incubation for10 min at 37°C , samples were quenched and run on a 10% polyacrylamide gel. To demonstrate the effectiveness of the trap, hPolη, along with PCNA, RFC, and RPA, was preincubated with the trap DNA and the primer-template substrate before the addition of dNTPs (lane 5).
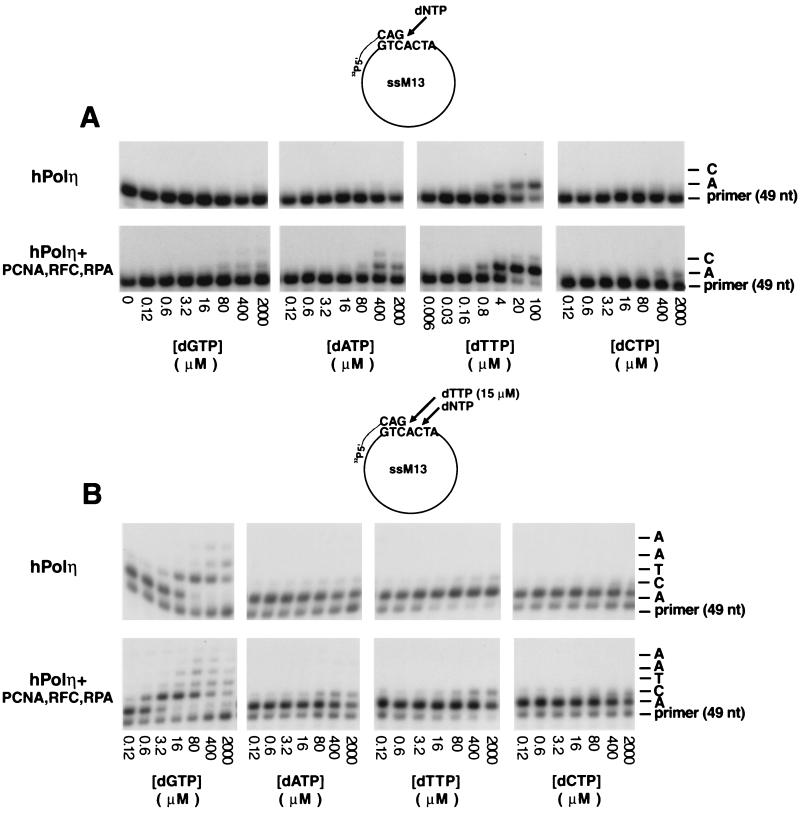
FIG. 4.
PCNA stimulates deoxynucleotide incorporation by hPolη. (A) Steady-state kinetics of deoxynucleotide incorporation opposite an A residue by hPolη in the presence or absence of PCNA, RFC, and RPA in a standing-start reaction. A portion of the DNA substrate is shown at the top. Polη (10 ng) was incubated with a singly primed circular single-stranded M13 (ssM13) DNA substrate (25 ng) and increasing concentrations of a single deoxynucleotide in the absence or presence of PCNA (100 ng), RFC (50 ng), and RPA (250 ng). The nucleotide (nt) incorporation rate was plotted against the dNTP concentration, and the data were fit to the Michaelis-Menten equation describing a hyperbola. The apparent K m and _V_max values were obtained from the fit and used to calculate the efficiency of deoxynucleotide incorporation (_V_max/K m). (B) Steady-state kinetics of deoxynucleotide incorporation opposite a template C residue by hPolη in the presence or absence of PCNA, RFC, and RPA in a running-start reaction. Reactions were carried out as described for panel A, except that each reaction also included the addition of dTTP (15 μM).
Generation of hrad30A mutations.
To generate the _hrad30A F707_-F708 → _A707_-A _70_8 and the hrad30A (_1_-695) mutations, a portion of the 3′ end of the hRAD30A gene was amplified by PCR using oligonucleotide N4919 (5′-GGGGTGTCGA AGCTAGAAG AATCCTCTA AAGCAACTCC-3′ (17) and mutagenic oligonucleotides N7819 (5′-CCTGGGATCC TAATGTGTTA ATGGCTTAG CAGCTGATTC CAATGTTTG CATGCCC-3′) and N7820 (5′-CTTGGGATCC TAGCGTTTAT TAGTGCAGGC CAAAGGGCTC-3′), respectively. The hrad30A (1-695) mutant gene encodes only amino acid residues 1 to 695. A 223-bp _Asp_718/_Bam_HI PCR fragment containing the _hRAD30A F707_-F708 → _A707_-A _70_8 mutation and a 169-bp _Asp_718/_Bam_HI PCR fragment containing the hrad30A (_1_-695) mutation were cloned into plasmid YIplac211, generating plasmids pBJ835 and pBJ840, respectively. The cloned PCR fragments in pBJ835 and pBJ840 were sequenced to confirm the presence of the mutations. Subsequently, an _Asp_718 DNA fragment containing the rest of the hRAD30A gene was cloned into plasmids pBJ835 and pBJ840; the hRAD30A open reading frame was restored, but either the _hrad30A A707_-A708 or the hrad30A (_1_-695) mutation was retained. Each hrad30A mutant gene was cloned in frame with the GST gene under the control of the galactose-inducible phosphoglycerate kinase promoter in pBJ842, generating plasmids pBJ867 and pBJ868, respectively. For the yeast two-hybrid analysis, the hrad30A (_1_-695) and _hrad30A A707_-A708 mutant genes and the wild-type hRAD30A gene were cloned in frame with the GAL4 DNA binding domain (BD) (amino acid residues 1 to 147) in plasmid pAS1, and the resulting plasmids were designated pR30.219, pR30.220, and pR30.218, respectively. Also, for these studies, hPCNA was cloned in frame with the GAL4 activation domain (AD) in plasmid pPCNA1.32.
DNA polymerase assays.
The circular DNA substrate used for some of the DNA synthesis studies (see Fig. 3A) was a 7.2-kb M13mp18 single-stranded DNA primed with a nonlabeled 36-nucleotide oligomer spanning nucleotides 6330 to 6294. For the processivity assays shown in Fig. 3B and the kinetic studies shown in Fig. 4, we used a single-stranded M13-derived (M13mp7L2) DNA primed with a 5′ 32P-labeled oligomer primer, LP-097, 5′-GGGTAACGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAG-3′. The standard DNA polymerase reaction mixture (10 μl) contained 40 mM Tris-HCl (pH 7.5); 8 mM MgCl2; 150 mM NaCl; 1 mM dithiothreitol; 100 μg of bovine serum albumin/ml; 500 μM ATP; and 100 μM each dGTP, dATP, dTTP, and dCTP. For reactions with a circular DNA substrate primed with a nonlabeled oligonucleotide (see Fig. 3A), α-32P-labeled dATP (final dATP concentration, ∼200 to 500 cpm/pmol) was added. When needed, wild-type or mutant hPolη (10 ng), PCNA (100 ng), RFC (50 ng), and/or RPA (250 ng) was incubated with 25 ng of M13 DNA substrate. Assays were assembled on ice and incubated at 37°C for 10 min, and the reaction was stopped by the addition of loading buffer (40 μl) containing EDTA (20 mM), 95% formamide, 0.3% bromphenol blue, and 0.3% cyanol blue. The reaction products were resolved on 10% polyacrylamide gels containing 8 M urea. Quantitation of the results was done using a Molecular Dynamics STORM PhosphorImager and ImageQuant software.
Processivity assays.
hPolη (10 ng) was preincubated with a circular M13 primer-template DNA substrate (50 ng) in standard reaction buffer, which contained no deoxynucleotides, for 5 min at 37°C. Reactions were initiated by adding all four deoxynucleoside triphosphates (dNTPs) (500 μM each) or all four dNTPs plus excess sonicated herring sperm DNA (0.5 mg/ml) as a trap. To demonstrate the effectiveness of the trap, hPolη was preincubated with the DNA trap and the primer-template substrate before the addition of dNTPs.
Steady-state kinetic analyses.
Steady-state kinetic analyses for deoxynucleotide incorporation opposite an A or a C site were performed as described previously (7, 11). Briefly, hPolη alone or in the presence of PCNA, RFC, and RPA was incubated with increasing concentrations of a single dNTP for 10 min under standard reaction conditions. In the running-start assay, each reaction included dTTP (15 μM). Gel band intensities of the substrates and products were quantitated with a PhosphorImager, and the percentage of primer extension was plotted as a function of dNTP concentration. The data were fit by nonlinear regression, using SigmaPlot 5.0, to the Michaelis-Menten equation describing a hyperbola, v = (_V_max × [dNTP])/(_Km_+ [dNTP]). Apparent Km and Vmax steady-state parameters were obtained from the fit and used to calculate the efficiency of deoxynucleotide incorporation (_V_max/Km).
Physical interaction of hPolη with PCNA.
To make complexes, wild-type or mutant GST-hPolη proteins (4 μg) were mixed with six-His–hPCNA (4 μg) in 75 μl of buffer I [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Tris(2 carboxyethyl)-phosphine–HCl, 0.01% Nonidet P-40, 10% glycerol] and incubated for 30 min at 4°C followed by 10 min at 25°C. Subsequently, to 25 μl of these samples, either 10 μl of glutathione-Sepharose (Pharmacia) beads or 10 μl of Ni-nitrilotriacetic acid (NTA) (Qiagen) beads was added to bind GST-Polη or six-His–PCNA and their complexes, respectively. The samples were further incubated with rocking for 30 min at 4°C. The glutathione-Sepharose and Ni-NTA beads were washed five times each with buffer I, followed by elution of the bound proteins with buffer I containing 40 mM glutathione and 500 mM imidazole, respectively. All protein samples, including the protein mixture before the addition of affinity beads, the flowthrough plus wash fractions, and the eluted proteins, were precipitated with 5% trichloroacetic acetic acid (TCA) and separated on a sodium dodecyl sulfate–12% polyacrylamide gel followed by Coomassie blue R-250 staining.
Two-hybrid analyses.
The HF7c yeast cell line was transformed with the GAL4 BD-hPolη and theGAL4 AD-hPCNA fusion constructs. Transformants harboring both the GAL4 BD-hPolη and the GAL4 AD-hPCNA fusion constructs were grown on synthetic complete media lacking leucine and tryptophan. β-Galactosidase activity was examined to determine the interaction between hPolη and PCNA as described in the Clontech Yeast Protocols Handbook (PT3024-1; chapter VI). Experiments were performed at least three times with triplicate samples.
RESULTS
Generating mutations in the PCNA binding motif of hPolη.
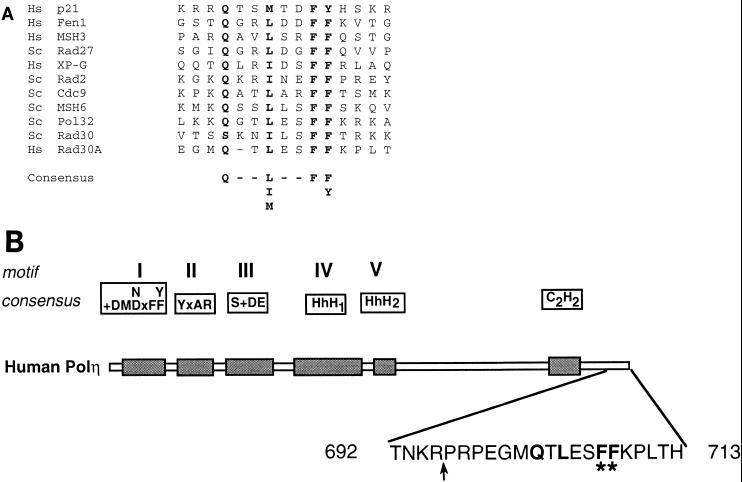
Many proteins involved in DNA replication and repair contain a consensus PCNA binding motif, QXX(I, L, or M)XXF(F or Y) (22, 35), and structural and mutational studies have indicated the involvement of the conserved hydrophobic residues within this motif in the interaction with PCNA (27, 31, 36). In the protein sequence of hPolη, the product of the human RAD30A gene, a putative PCNA binding sequence, Q-TLESFF, is located at the extreme C terminus and encompasses residues 702 to 708 of the 713-amino-acid protein (Fig. 1). To test if this motif was involved in the interaction of hPolη with PCNA, two mutations in the hRAD30A gene were generated, rad30A (_1_-695) and _rad30A A707_-A708. In the hPolη (1-695) protein, the C-terminal 18 amino acids, including the conserved F707 and F708 residues of the putative PCNA binding motif (Fig. 1B), have been removed, whereas in the hPolη A707-A708 protein, both of these phenylalanine residues have been changed to alanines (Fig. 1B). The wild-type and mutant hPolη proteins were expressed in yeast as GST fusion proteins. During purification, the mutant hPolη proteins displayed the same chromatographic properties as the wild-type protein, and the proteins were at least 95% pure, as judged from Coomassie blue staining (data not shown). To rule out the possibility that the mutations caused improper folding, the DNA polymerase activities of the wild-type and mutant hPolη proteins were compared. Running start-DNA synthesis reactions were carried out using a linear DNA substrate either containing or not containing a _cis_-syn TT dimer. The wild-type and mutant hPolη (1-695) and hPolη A707-A708 proteins displayed identical DNA polymerase and TT dimer bypass activities (data not shown).
FIG. 1.
PCNA binding motif of hPolη. (A) C-terminal amino acids 699 to 712 of hPolη are aligned with the PCNA binding motifs identified in various PCNA binding proteins. The highly conserved residues are shown in bold. Hs, human; Sc, S. cerevisiae. (B) Mutations made in the PCNA binding motif of hPolη. In the schematic representation of hPolη, the five highly conserved motifs (I to V) shared among different members of the Polη/UmuC/DinB protein family are indicated, and the C2H2 motif conserved in the Polη family is shown (20). The amino acid residues present in the extreme C-terminal region are shown; the amino acids highly conserved in the consensus PCNA binding motif are shown in bold. In the hPolη (1-695) mutant protein, the last 18 amino acids from the C terminus were deleted (indicated by an arrow). In the hPolη A707-A708 mutant protein, the F residues at positions 707 and 708 (indicated by asterisks) were changed to A residues.
Interaction of hPolη with PCNA—two-hybrid analysis.
We used the yeast two-hybrid system to examine the interaction of hPolη and hPCNA proteins in vivo. In one of the plasmids, the GAL4 BD was fused with either wild-type RAD30A or mutant rad30 (_1_-695) or _rad30 A707_-A708 open reading frames, and in the other plasmid, the GAL4 AD was fused with hPCNA. The HF7c yeast reporter strain harboring the GAL4 AD-hPCNA plasmid was transformed with one of the GAL4 BD-hPolη plasmids. The expression of GAL4 BD-hPolη fusion proteins was confirmed by immunoblotting using anti-hPolη antibodies (data not shown). The interaction of the wild-type and mutant hPolη proteins with PCNA in these transformants was analyzed by a β-galactosidase liquid assay, and the results are summarized in Table 1. Compared to the low level of β-galactosidase activity detected with the wild-type GAL4 BD-hPolη protein and the GAL4 AD protein, the wild-type GAL4 BD-hPolη protein showed a strong interaction with PCNA bound to GAL4 AD, resulting in 21-fold higher level of β-galactosidase activity. Deletion or point mutations in the conserved PCNA binding site in the hPolη (1-695) and hPolη A707-A708 proteins strongly reduced the interaction between hPolη and PCNA, yielding only a small increase in β-galactosidase activity. These results establish an interaction of hPolη with PCNA in vivo and show that the PCNA binding motif at the C terminus of hPolη plays an important role in mediating this interaction.
TABLE 1.
Interaction of hPolη with hPCNA in the yeast two-hybrid system
| BD fusiona | AD fusion | Mean ± SD β-galactosidase activity | Fold activation |
|---|---|---|---|
| GAL4 BD-hPolη (WT) | GAL4 AD | 0.82 ± 0.05 | 1 |
| GAL4 BD-hPolη (WT) | GAL4 AD-hPCNA | 17.6 ± 0.6 | 21 |
| GAL4 BD-hPolη A707-A708 | GAL4 AD-hPCNA | 1.75 ± 0.03 | 2 |
| GAL4 BD-hPolη (1-695) | GAL4 AD-hPCNA | 2.71 ± 0.05 | 3 |
Physical interaction of hPolη with PCNA.
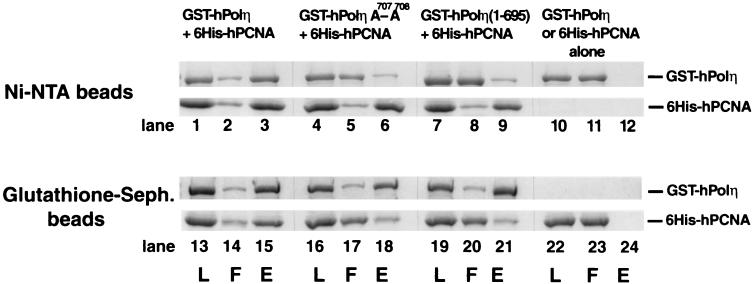
Next, we examined if purified hPolη physically interacts with purified hPCNA in vitro. Wild-type GST-hPolη or mutant GST-hPolη (1-695) or GST-hPolη A707-A708 proteins were incubated with six-His–PCNA, and a pull down assay was carried out using Ni-NTA or glutathione-Sepharose affinity beads (Fig. 2). As expected, the Ni-NTA beads bound only six-His–PCNA and not GST hPolη (Fig. 2, lanes 10 to 12) and the gluthathione-Sepharose beads bound GST-hPolη but not six-His–PCNA (Fig. 2, lanes 22 to 24). Hence, the two proteins could be pulled down together only if they interacted with one another.
FIG. 2.
Polη forms a complex with PCNA. Six-His–hPCNA (4 μg) was mixed either with wild-type GST-hPolη protein (lanes 1 to 3 and 13 to 15) or with mutant GST-hPolη A707-A708 (lanes 4 to 6 and 16 to 18) or GST-hPolη (1-695) (lanes 7 to 9 and 19 to 21) protein (4 μg each). As controls, the same amounts of GST-hPolη (lanes 10 to 12) or six-His–hPCNA (lanes 22 to 24) protein were used alone. After incubation, samples were bound to Ni-NTA (lanes 1 to 12) or glutathione-Sepharose (Seph.) (lanes 13 to 24) beads, followed by washing and elution of the bound proteins by imidazole- or glutathione-containing buffer, respectively. Aliquots of each sample before loading on the beads (L), the flowthrough and wash (F), and the eluted proteins (E) were precipitated by TCA and analyzed on a sodium dodecyl sulfate–12% polyacrylamide gel stained with Coomassie blue. The positions of GST-hPolη and six-His–hPCNA are indicated on the right.
When six-His–PCNA was bound to the Ni-NTA beads, a large proportion of wild-type hPolη remained bound to PCNA (Fig. 2, lanes 1 to 3); similarly, when GST-hPolη was bound to the glutathione-Sepharose beads, PCNA was retained on the beads via an interaction with hPolη (Fig. 2, lanes 13 to 15). However, the interaction of hPolη with PCNA was greatly reduced for both the hPolη A707-A708 and the hPolη (1-695) mutant proteins. For example, when six-His–PCNA was bound to the Ni-NTA beads, almost all of each mutant hPolη protein was recovered in the flowthrough (Fig. 2, lanes 5 and 8); conversely, when the mutant hPolη proteins were bound to the glutathione-Sepharose beads, the majority of PCNA was recovered in the flowthrough (Fig. 2, lanes 17 and 20). Thus, hPolη interacts with PCNA in vitro, and mutations in the conserved PCNA binding motif of hPolη reduce this interaction very substantially.
PCNA cooperates with RFC and RPA to enhance the DNA synthetic activity of hPolη.
Next, we examined if PCNA stimulates the DNA synthetic activity of hPolη. Stimulation of the synthetic activity of the replicative DNA polymerase, Polδ, by PCNA requires the action of RFC and RPA as well. Therefore, we examined the effect of PCNA on the DNA synthetic activity of hPolη in the presence of RFC and RPA by using a single-stranded M13 template DNA primed at a unique site (Fig. 3A). The DNA synthetic activity of hPolη is enhanced ∼12-fold upon the addition of PCNA, RFC, and RPA (Fig. 3A, compare lanes 1 and 3). This stimulation requires PCNA, since in the absence of PCNA, RFC and RPA increased the DNA synthetic activity of hPolη only ∼2-fold (Fig. 3A, compare lanes 1 and 2). This weak enhancement could be attributed to RPA, since the addition of RPA alone also resulted in ∼2-fold stimulation (Fig. 3, lane 8). This effect probably stems from a reduction in the nonspecific binding of hPolη to single-stranded DNA by the presence of RPA. No stimulation of DNA synthesis occurred with PCNA or RFC alone (Fig. 3, lanes 6 and 7). As no significant stimulation of DNA synthesis occurs unless all three proteins are present (Fig. 3A, lanes 1 to 8), PCNA, RFC, and RPA cooperate to stimulate the activity of hPolη. In contrast, the hPolη A707-A708 and hPolη (1-695) mutant proteins were greatly impaired in their ability to be stimulated by PCNA in the presence of RFC and RPA (Fig. 3A, compare lanes 9 and 10 or lanes 11 and 12).
Effect of PCNA on the processivity of hPolη.
Both yeast Polη and hPolη are low-processivity enzymes, incorporating only a few nucleotides per DNA binding event (21, 38). A low processivity is desirable for this enzyme, in order to limit its activity to synthesizing only short stretches of DNA, thereby preventing the high mutation rates that would otherwise occur if this low-fidelity polymerase were to synthesize long tracts of DNA. However, the possibility existed that an increase in processivity was in fact responsible for stimulation of the activity of hPolη by PCNA. To test if PCNA, together with RFC and RPA, increases the processivity of hPolη, we used a circular single-stranded M13 template DNA primed singly with a 5′ 32P-labeled oligonucleotide primer. To ensure that we were observing deoxynucleotide incorporation resulting from a single DNA binding event, we monitored DNA synthesis in the presence of an excess of nonradiolabeled, sonicated herring sperm DNA as a trap (Fig. 3B). The reactions were performed by first preincubating hPolη in the absence (Fig. 3B, lanes 1 and 4) or in the presence (Fig. 3B, lanes 2 and 3) of PCNA, RFC, and RPA with the DNA substrate. All four dNTPs (Fig. 3B, lanes 1 and 2) or a mixture of excess herring sperm DNA and all four dNTPs (Fig. 3B, lanes 3 and 4) was then added to initiate the reaction. In the presence of the DNA trap, all hPolη molecules that dissociate from the labeled DNA substrate will be bound by the excess of nonradiolabeled herring sperm DNA. The effectiveness of the trap was verified by first preincubating hPolη with the DNA substrate together with the excess herring sperm DNA before the addition of nucleotides (Fig. 3B, lane 5). The lack of any DNA synthesis in this sample shows that the excess herring sperm DNA (about 100-fold) was sufficient to trap all hPolη molecules. Despite the strong stimulation of hPolη activity by PCNA, RFC, and RPA in the reactions containing no DNA trap (Fig. 3B, lane 2), in samples in which single-hit conditions were provided by excess herring sperm DNA, PCNA together with RFC and RPA stimulated the processivity of hPolη only weakly; processivity remained low, at ∼4 nucleotides per DNA binding event (Fig. 3B, compare lanes 3 and 4).
Kinetic analysis of the DNA synthetic activity of hPolη in the presence of PCNA.
To identify the mechanism by which PCNA stimulates the activity of hPolη, we examined the steady-state kinetic parameters Km and _V_max for nucleotide insertion by hPolη in the presence of PCNA, RFC, and RPA. Using a circular single-stranded M13 substrate DNA primed singly with a 5′ 32P-labeled oligonucleotide primer, we examined the kinetics of insertion of a single deoxynucleotide opposite an A residue in a standing-start reaction (Fig. 4A) or opposite a C residue in a running-start reaction (Fig. 4B) under steady-state conditions. From the kinetics of deoxynucleotide incorporation, the steady-state apparent Km and _V_max values for each deoxynucleotide were obtained from the curve fitted to the Michaelis-Menten equation. These Km and _V_max values and the efficiencies of nucleotide incorporation (_V_max/Km) for hPolη in the presence or absence of PCNA, RFC, and RPA are summarized in Table 2.
TABLE 2.
Kinetic parameters for nucleotide insertion reactions catalyzed by hPolη
| Reaction | Insertion | Sitea | dNTP added | Km (μM)b | _V_max (%/min)b | _V_max/Km | _f_incc |
|---|---|---|---|---|---|---|---|
| Standing start | Opposite an A by hPolη | 5′----CAG | dTTP | 23.6 ± 1.3 | 5.6 ± 0.15 | 0.24 | ND |
| ----GTCACT | |||||||
| Opposite an A by hPolη + PCNA, RFC, and RPA | 5′----CAG | dGTP | 70 ± 12 | 1.1 ± 0.05 | 0.016 | 5 × 10−3 | |
| ----GTCACT | dATP | 83 ± 28 | 2.3 ± 0.2 | 0.027 | 8.5 × 10−3 | ||
| dTTP | 2.7 ± 0.3 | 8.5 ± 0.3 | 3.15 | 1 | |||
| dCTP | 102 ± 19 | 0.98 ± 0.08 | 0.0096 | 3 × 10−3 | |||
| Running start | Opposite a C by hPolη | 5′----CAG | dGTP | 14.3 ± 1.5 | 6.6 ± 0.45 | 0.46 | ND |
| ----GTCACT | |||||||
| Opposite a C by hPolη + PCNA, RFC, and RPA | 5′----CAG | dGTP | 1 ± 0.09 | 7.6 ± 0.43 | 7.6 | 1 | |
| ----GTCACT | dATP | 82 ± 16 | 2.5 ± 0.41 | 0.03 | 3.9 × 10−3 | ||
| dTTP | 238 ± 51 | 2.8 ± 0.3 | 0.012 | 1.5 × 10−3 | |||
| dCTP | 380 ± 95 | 0.78 ± 0.45 | 0.002 | 2.7 × 10−4 |
As indicated by the _V_max/Km values, hPolη incorporates the correct nucleotide about 15-fold more efficiently in the presence of PCNA, RFC, and RPA than in the absence of these proteins (Table 2). The incorporation of incorrect nucleotides is also stimulated by PCNA, RFC, and RPA (Fig. 4); however, because of the low _V_max values for the insertion of incorrect nucleotides in the absence of these proteins, we did not quantitate this enhancement. Importantly, PCNA, together with RFC and RPA, promotes nucleotide insertion by hPolη primarily via an ∼10- to 14-fold reduction in the apparent Km for the nucleotide. In contrast, there was only a slight increase in the _V_max when PCNA, RFC, and RPA were present. As judged from the comparison of the _V_max/Km values for the incorporation of correct and incorrect nucleotides, the fidelity of nucleotide insertion of hPolη in the presence of PCNA, RFC, and RPA remains low, ranging from 8.5 × 10−3 to 2.7 × 10−4.
DISCUSSION
Polη plays a key role in the error-free replication of UV-damaged DNA, and inactivation of Polη in humans results in the cancer-prone syndrome XP-V. However, the mechanism by which this important translesion synthesis DNA polymerase is recruited to the stalled replication machinery in humans has remained unclear so far. Here, we identify a PCNA binding motif in the C terminus of Polη and provide both in vivo and in vitro evidence for the physical interaction of hPolη with PCNA. Mutations in the PCNA binding motif greatly reduce the affinity of hPolη for PCNA, indicating a role for this motif in PCNA binding.
PCNA, together with RFC and RPA, stimulates the DNA synthetic activity of hPolη ∼12-fold. However, this increase does not result from an increase in the processivity of the enzyme. We find that even in the presence of PCNA, RFC, and RPA, hPolη processivity remains low, at three or four nucleotides per DNA binding event. The effect of PCNA on hPolη processivity stands in sharp contrast to the large increase in processivity that occurs for Polδ in the presence of PCNA, RFC, and RPA (30, 32). However, since Polη misincorporates nucleotides at a higher rate than Polδ, any increase in Polη processivity would have conferred high mutagenicity.
Steady-state kinetic analyses showed that PCNA, RFC, and RPA increase the efficiency of hPolη for inserting the correct nucleotide by ∼15-fold, and this increase is achieved primarily by a reduction in the apparent Km for the nucleotide. Even though the processivity of hPolη is not significantly increased, the stimulation of its synthetic activity in the presence of PCNA, RFC, and RPA may be due to an increased affinity of the enzyme for the primer 3′ end, as has been suggested for the stimulation in synthesis by E. coli PolV that occurs with RecA, single-stranded DNA binding protein, and the β,γ-complex (28). However, the possibility that PCNA, RFC, and RPA stimulate the activity of hPolη by increasing the affinity of the enzyme for the incoming nucleotide cannot be ruled out.
Saccharomyces cerevisiae Polη and hPolη resemble one another in their damage bypass ability, and they promote the error-free replication of UV-damaged DNA. Similar to the results reported here for hPolη, evidence was recently provided for physical and functional interactions of yeast Polη (Polη) with PCNA (12). Like that of hPolη, the DNA synthetic activity of yeast Polη is stimulated ∼15-fold in the presence of PCNA, RFC, and RPA; processivity, however, is not affected. Both yeast Polη and hPolη are highly inefficient at inserting a nucleotide opposite an abasic site (14). However, yeast PCNA, together with yeast RFC and yeast RPA, greatly stimulates the ability of yeast Polη to insert a nucleotide opposite an AP site; by comparison to the ∼14-fold increase in the efficiency of G insertion opposite the template C, the ability of yeast Polη to insert a G opposite an AP site is stimulated over 350-fold in the presence of these protein factors (12). Additionally, genetic studies with the yeast Polη mutant proteins unable to bind PCNA have shown that an interaction with PCNA is indispensable for the in vivo function of Polη. From these studies, we infer a crucial role for PCNA in the targeting of Polη to the replication machinery stalled at a lesion site and in promoting the efficient bypass of DNA lesions.
Polδ, required for the replication of both the leading and the lagging strands, stalls at DNA lesion sites, such as cyclobutane pyrimidine dimers. While an interaction with PCNA would promote the targeting of Polη to the replication machinery stalled at a lesion site, the question remains as to how Polη displaces stalled Polδ and gains access to the template-primer junction. Studies of human DNA replication have revealed that the Polα-to-Polδ switch is coordinated via competition for RPA (41). First, Polα binds RPA for firm attachment to the primed site. After Polα has synthesized the primer, RFC is able to bind RPA at the primed template junction and competes with Polα for RPA, resulting in the release of Polα from DNA. RFC then loads PCNA onto DNA. Next, Polδ binds both PCNA and RPA and competes with RFC for these two proteins. This process results in the displacement of RFC from the 3′ terminus and in the binding of Polδ to the 3′ terminus. RFC, however, remains bound to DNA via its interaction with RPA. The Polδ-to-Polη switch differs from the Polα-to-Polδ switch in that whereas both Polδ and Polη bind PCNA, Polα does not. The Polδ-to-Polη switch could occur in any of the following ways. First, Polη may compete with Polδ for the 3′ terminus opposite a lesion site, perhaps because Polη binds such a terminus more tightly than does Polδ, resulting in the release of Polδ from the replication complex. Alternatively, the displacement of Polδ may be a more active process, requiring the action of the Rad6-Rad18 complex, which is essential for the replication of damaged DNA (29) and which comprises ubiquitin-conjugating and DNA binding activities (1). Conjugation of ubiquitin to a stalled Polδ subunit may destabilize the Polδ interaction with PCNA as well as its binding to the 3′ terminus. Finally, it is possible that Polη displaces Polδ from the 3′ terminus but that both polymerases remain in the replication ensemble via their binding to different monomers of the homotrimeric PCNA ring. This scenario raises the possibility of physical and functional interactions of Polη with Polδ that may further affect the fidelity, processivity, or damage bypass ability of Polη.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants GM19261 and GM38559.
REFERENCES
- 1.Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J Biol Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 2.Bambara R A, Murante R S, Henricksen L A. Enzymes and reactions at the eukaryotic DNA replication fork. J Biol Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 3.Bloom L B, Chen X, Fygenson D K, Turner J, O'Donnell M, Goodman M F. Fidelity of Escherichia coli DNA polymerase III holoenzyme The effects of β, γ complex processivity proteins and ɛ proofreading exonuclease on nucleotide misincorporation efficiencies. J Biol Chem. 1997;272:27919–27930. doi: 10.1074/jbc.272.44.27919. [DOI] [PubMed] [Google Scholar]
- 4.Brash D E. Sunlight and the onset of skin cancer. Trends Genet. 1997;13:410–414. doi: 10.1016/s0168-9525(97)01246-8. [DOI] [PubMed] [Google Scholar]
- 5.Cai J, Gibbs E, Uhlmann F, Phillips B, Yano N, O'Donnell M, Hurwitz J. A complex consisting of human replication factor C, p40, p37, and p36 subunits is a DNA-dependent ATPase and an intermediate in the assembly of the holoenzyme. J Biol Chem. 1997;272:18974–18981. doi: 10.1074/jbc.272.30.18974. [DOI] [PubMed] [Google Scholar]
- 6.Canella K A, Seidman M M. Mutation spectra in supF: approaches to elucidating sequence context effects. Mutat Res. 2000;450:61–73. doi: 10.1016/s0027-5107(00)00016-6. [DOI] [PubMed] [Google Scholar]
- 7.Creighton S, Bloom L B, Goodman M F. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- 8.Creighton S, Goodman M F. Gel kinetic analysis of DNA polymerase fidelity in the presence of proofreading using bacteriophage T4 DNA polymerase. J Biol Chem. 1995;270:4759–4774. doi: 10.1074/jbc.270.9.4759. [DOI] [PubMed] [Google Scholar]
- 9.Echols H, Goodman M F. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 10.Gibbs E, Kelman Z, Gulbis J M, O'Donnell M, Kuriyan J, Burgers P M, Hurwitz J. The influence of the proliferating cell nuclear antigen-interacting domain of p21 (CIP1) on DNA synthesis catalyzed by the human and Saccharomyces cerevisiae polymerase delta holoenzymes. J Biol Chem. 1997;272:2373–2381. doi: 10.1074/jbc.272.4.2373. [DOI] [PubMed] [Google Scholar]
- 11.Goodman M F, Creighton S, Bloom L B, Petruska J. Biochemical basis of DNA replication fidelity. Crit Rev Biochem Mol Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 12.Haracska L, Kondratick C M, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase η function. Mol Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 13.Haracska L, Prakash S, Prakash L. Replication past O6-methylguanine by yeast and human DNA polymerase η. Mol Cell Biol. 2000;20:8001–8007. doi: 10.1128/mcb.20.21.8001-8007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haracska L, Washington M T, Prakash S, Prakash L. Inefficient bypass of an abasic site by DNA polymerase η. J Biol Chem. 2001;276:6861–6866. doi: 10.1074/jbc.M008021200. [DOI] [PubMed] [Google Scholar]
- 15.Haracska L, Yu S-L, Johnson R E, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 16.Johnson R E, Haracska L, Prakash S, Prakash L. Role of DNA polymerase η in the bypass of a (6-4) TT photoproduct. Mol Cell Biol. 2001;21:3558–3563. doi: 10.1128/MCB.21.10.3558-3563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson R E, Kondratick C M, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 18.Johnson R E, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 19.Johnson R E, Prakash S, Prakash L. Requirement of DNA polymerase activity of yeast Rad30 protein for its biological function. J Biol Chem. 1999;274:15975–15977. doi: 10.1074/jbc.274.23.15975. [DOI] [PubMed] [Google Scholar]
- 20.Johnson R E, Washington M T, Prakash S, Prakash L. Bridging the gap: a family of novel DNA polymerases that replicate faulty DNA. Proc Natl Acad Sci USA. 1999;96:12224–12226. doi: 10.1073/pnas.96.22.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson R E, Washington M T, Prakash S, Prakash L. Fidelity of human DNA polymerase η. J Biol Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 22.Kelman Z, Hurwitz J. Protein-PCNA interactions: a DNA-scanning mechanism? Trends Biol Sci. 1998;23:236–238. doi: 10.1016/s0968-0004(98)01223-7. [DOI] [PubMed] [Google Scholar]
- 23.Kelman Z, Yao N, O'Donnell M. Escherichia coli expression vectors containing a protein kinase recognition motif, His6-tag and hemagglutinin epitope. Gene. 1995;166:177–178. doi: 10.1016/0378-1119(95)00556-7. [DOI] [PubMed] [Google Scholar]
- 24.Lee S H, Eki T, Hurwitz J. Synthesis of DNA containing the simian virus 40 origin of replication by the combined action of DNA polymerases alpha and delta. Proc Natl Acad Sci USA. 1989;86:7361–7365. doi: 10.1073/pnas.86.19.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 26.McDonald J P, Levine A S, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakanishi M, Robetorye R S, Pereira-Smith O M, Smith J R. The C-terminal region of p21SD11/WAF1/CIP1 is involved in proliferating cell nuclear antigen binding but does not appear to be required for growth inhibition. J Biol Chem. 1995;270:17060–17063. doi: 10.1074/jbc.270.29.17060. [DOI] [PubMed] [Google Scholar]
- 28.Pham P, Bertram J G, O'Donnell M, Woodgate R, Goodman M F. A model for SOS-lesion-targeted mutations in Escherichia coli. Nature. 2001;409:366–370. doi: 10.1038/35053116. [DOI] [PubMed] [Google Scholar]
- 29.Prakash L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol Gen Genet. 1981;184:471–478. doi: 10.1007/BF00352525. [DOI] [PubMed] [Google Scholar]
- 30.Prelich G, Kostura M, Marshak D R, Matthews M B, Stillman B. The cell-cycle regulated proliferating cell nuclear antigen is required for SV40 DNA replication in vitro. Nature. 1987;326:471–475. doi: 10.1038/326471a0. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds N, Warbrick E, Fantes P A, MacNeill S A. Essential interaction between the fission yeast DNA polymerase δ subunit Cdc27 and Pcn1 (PCNA) mediated through a C-terminal p21Cip1-like PCNA binding motif. EMBO J. 2000;19:1108–1118. doi: 10.1093/emboj/19.5.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan C K, Castillo C, So A G, Downey K M. An auxiliary protein for DNA polymerase δ from fetal calf thymus. J Biol Chem. 1986;261:12310–12316. [PubMed] [Google Scholar]
- 33.Thomas D C, Roberts J D, Sabatino R D, Myers T W, Tan C-K, Downey K M, So A G, Bambara R A, Kunkel T A. Fidelity of mammalian DNA replication and replicative DNA polymerases. Biochemistry. 1991;30:11751–11759. doi: 10.1021/bi00115a003. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y-C, Maher V M, Mitchell D L, McCormick J J. Evidence from mutation spectra that the UV hypermutability of xeroderma pigmentosum variant cells reflects abnormal, error-prone replication on a template containing photoproducts. Mol Cell Biol. 1993;13:4276–4283. doi: 10.1128/mcb.13.7.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warbrick E. PCNA binding through a conserved motif. Bioessays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 36.Warbrick E, Lane D P, Glover D M, Cox L S. A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21WAF1 and proliferating cell nuclear antigen. Curr Biol. 1995;5:275–282. doi: 10.1016/s0960-9822(95)00058-3. [DOI] [PubMed] [Google Scholar]
- 37.Washington M T, Johnson R E, Prakash S, Prakash L. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc Natl Acad Sci USA. 2000;97:3094–3099. doi: 10.1073/pnas.050491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Washington M T, Johnson R E, Prakash S, Prakash L. Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase η. J Biol Chem. 1999;274:36835–36838. doi: 10.1074/jbc.274.52.36835. [DOI] [PubMed] [Google Scholar]
- 39.Waters H L, Seetharam S, Seidman M M, Kraemer K H. Ultraviolet hypermutability of a shuttle vector propagated in xeroderma pigmentosum variant cells. J Investig Dermatol. 1993;101:744–748. doi: 10.1111/1523-1747.ep12371686. [DOI] [PubMed] [Google Scholar]
- 40.Yu S-L, Johnson R E, Prakash S, Prakash L. Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol Cell Biol. 2001;21:185–188. doi: 10.1128/MCB.21.1.185-188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuzhakov A, Kelman Z, Hurwitz J, O'Donnell M. Multiple competition reactions for RPA order the assembly of the DNA polymerase δ holoenzyme. EMBO J. 1999;18:6189–6199. doi: 10.1093/emboj/18.21.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]