Role of Streptococcus gordonii Amylase-Binding Protein A in Adhesion to Hydroxyapatite, Starch Metabolism, and Biofilm Formation (original) (raw)
Abstract
Interactions between bacteria and salivary components are thought to be important in the establishment and ecology of the oral microflora. α-Amylase, the predominant salivary enzyme in humans, binds to Streptococcus gordonii, a primary colonizer of the tooth. Previous studies have implicated this interaction in adhesion of the bacteria to salivary pellicles, catabolism of dietary starches, and biofilm formation. Amylase binding is mediated at least in part by the amylase-binding protein A (AbpA). To study the function of this protein, an erythromycin resistance determinant [erm(AM)] was inserted within the abpA gene of S. gordonii strains Challis and FAS4 by allelic exchange, resulting in abpA mutant strains Challis-E1 and FAS4-E1. Comparison of the wild-type and mutant strains did not reveal any significant differences in colony morphology, biochemical metabolic profiles, growth in complex or defined media, surface hydrophobicity, or coaggregation properties. Scatchard analysis of adhesion isotherms demonstrated that the wild-type strains adhered better to human parotid-saliva- and amylase-coated hydroxyapatite than did the AbpA mutants. In contrast, the mutant strains bound to whole-saliva-coated hydroxyapatite to a greater extent than did the wild-type strains. While the wild-type strains preincubated with purified salivary amylase grew well in defined medium with potato starch as the sole carbohydrate source, the AbpA mutants did not grow under the same conditions even after preincubation with amylase. In addition, the wild-type strain produced large microcolonies in a flow cell biofilm model, while the abpA mutant strains grew much more poorly and produced relatively small microcolonies. Taken together, these results suggest that AbpA of S. gordonii functions as an adhesin to amylase-coated hydroxyapatite, in salivary-amylase-mediated catabolism of dietary starches and in human saliva-supported biofilm formation by S. gordonii.
Saliva-bacterium interactions are thought to be of key importance in the establishment and maintenance of the oral microflora to form dental plaques, which are responsible for dental caries and periodontal diseases (37). Salivary components may influence the formation of dental plaques through a number of mechanisms. The binding of salivary constituents to microorganisms may diminish plaque formation by facilitating their physical clearance from the oral cavity or through bacteriostatic or bactericidal mechanisms. Conversely, salivary pellicles may foster microbial colonization of host surfaces by serving as adhesin receptors or by providing nutrients through the enzymatic breakdown of salivary or dietary constituents.
Amylase is the most abundant salivary enzyme in humans (1) and catalyzes the hydrolysis of α-1,4-glucosidic linkages in dietary starch. The primary products of this enzymatic activity are glucose, maltose, and maltodextrins, which are all fermentable substrates for many oral bacterial species. Amylase has also been identified as an abundant constituent of the acquired enamel pellicle (32, 33), the film composed primarily of salivary components that selectively adhere to clean teeth. In this case, amylase may act as a receptor for bacterial adhesion to the tooth surface. Amylase has also been detected in dental plaque (42) and binds with high avidity to a number of the oral streptococci (11, 38). Bacterium-bound amylase retains approximately half of its enzymatic activity (12, 39), suggesting a potential role in starch catabolism.
Amylase binds to several species of dental plaque streptococci, including Streptococcus gordonii (5, 10, 42). Amylase-binding bacteria constitute a substantial proportion of the total cultivable flora from human teeth (41, 46) and appear only to colonize the mouths of animals having salivary amylase activity (41). These findings suggest that amylase binding is advantageous and aids in bacterial colonization of the oral cavity.
Previous studies have shown greater adhesion of S. gordonii to human parotid saliva (HPS)- and amylase-coated hydroxyapatite (HAP) compared to non-amylase-binding oral streptococci (43). It has also been shown that incubation of S. gordonii in the presence of maltotriose increases adhesion to experimental pellicles formed on HAP (43). A role for amylase binding in streptococcal carbohydrate catabolism has also been suggested by the observation that bound amylase continues to catalyze the hydrolysis of dietary starches, resulting in the liberation of fermentable saccharides. Support for this hypothesis is provided by studies showing that strains of oral streptococci able to bind amylase exhibited functional enzyme on their surface and produced acid from the products of amylolytic degradation (13, 39).
Amylase binds to S. gordonii through high-affinity receptors that appear to cluster around cell division sites on the surfaces of actively dividing cells (40). Previous studies suggested that proteins of 20 and 82 kDa mediated amylase binding to S. gordonii (12, 16, 40). The gene encoding the 20-kDa amylase-binding protein (AbpA) of S. gordonii Challis has been cloned and sequenced (34). Mutants lacking AbpA were found to bind amylase at levels similar to those seen for non-amylase-binding organisms, suggesting that AbpA is involved in amylase binding to S. gordonii. In addition, the expression of abpA is at least partially controlled by a carbon catabolite repression regulatory mechanism (34a). Finally, a recent study has suggested that disruption of abpA yields mutants defective in biofilm formation (26).
The available data suggest that amylase binding by the oral streptococci is involved in adhesion and biofilm formation by the bacteria to oral surfaces and aids in growth of the organisms in the presence of dietary starch, the enzymatic substrate of amylase. The following study was performed to evaluate the potential role of amylase binding by S. gordonii in the adhesion to HAP, in biofilm formation using a flow cell model, and in bacterial growth in the presence of starch.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The streptococcal and Escherichia coli strains and plasmids used in this study are listed in Table 1. Streptococci were maintained on blood agar or tryptic soy broth supplemented with 0.5% yeast extract (TSBY) (Difco, Detroit, Mich.) agar. The streptococci were routinely cultured in a defined medium (FMC [45]) with either 2% glucose or 2% potato starch as the carbon source, or in brain heart infusion (Difco) medium, and were incubated for 12 to 16 h at 37°C without shaking in a candle jar. Where indicated, erythromycin (5 μg/ml) was used in the selection of recombinant strains of S. gordonii. E. coli strains were grown under aerobic conditions with shaking for 12 to 16 h at 37°C in Luria-Bertani (LB) broth and maintained on LB agar. E. coli strains containing recombinant clones were plated on LB agar supplemented as required with ampicillin (100 μg/ml) or erythromycin (300 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5 | F− φ_80dlacZΔM15 Δ_(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rkv,/_m_k+) phoA supE44+ thi-1 gyrA96 relA1 | Gibco-BRL |
| S. gordonii Challis | Wild-type reference strain | University at Buffalo |
| S. gordonii G9B | Wild-type reference strain | University at Buffalo |
| S. gordonii Blackburn | Wild-type reference strain | University at Buffalo |
| S. gordonii NCTC 7865 | Wild-type reference strain | University at Buffalo |
| S. gordonii FAS4 | Wild-type reference strain | University at Buffalo |
| S. gordonii Challis-E1 | Challis Δ_abpA_::erm(AM); Emr | This study |
| S. gordonii Challis-E2 | Challis Δ_abpA_::erm(AM); Emr (single crossover) | This study |
| S. gordonii FAS4-E1 | FAS4 Δ_abpA_::erm(AM); Emr | This study |
| S. oralis C104 | Reference strain | 23 |
| S. oralis 34 | Reference strain | 23 |
| Streptococcus sp. strain SM PK509 | Reference strain | 7 |
| S. sanguis ATCC 10556 | Reference strain | University at Buffalo |
| A. naeslundii PK947 | Reference strain | 7 |
| A. naeslundii T14V | Reference strain | 23 |
| Plasmids | ||
| pTS19E | erm(AM); Emr Amr | 50 |
| pCR2KB-7 | abpA; Amr Kmr | 21 |
| pCR2KB-E1 | abpA::erm(AM); Amr Kmr Emr | This study |
| pCR2KB-E2 | abpA::erm(AM); Kmr Emr | This study |
DNA manipulations.
Standard methodologies were used for the isolation and manipulation of S. gordonii genomic DNA (34, 36). Southern blot analysis was carried out using biotinylated probes labeled using the Photogene Nucleic Acid Detection System (Gibco-BRL) according to the manufacturer's protocol. Plasmid DNA was isolated as previously described (3).
Insertional inactivation of the S. gordonii Challis and FAS4 abpA genes.
Mutation of the abpA genes of S. gordonii strains Challis and FAS4 was obtained by allelic exchange using the following strategy. The erm(AM) gene (GenBank no. K00551) from pTS19E (50) was restricted with _Pvu_II, the 2-kb erm(AM) fragment was gel purified on 1% agarose, and the blunt-ended fragment was cloned into the _Sna_BI site of the abpA gene in pCR2KB-7 (34). The resulting plasmid was designated pCR2KB-E1. To avoid introduction of ampicillin resistance into S. gordonii, a Bsa_I/Sca_I double digest of pCR2KB-E1 was used to cleave an internal 415-bp portion of the ampicillin gene from the plasmid. The remaining 7.5-kb fragment containing the abpA::erm(AM) construct was purified on 0.8% agarose gels, excised, religated, and designated pCR2KB-E2. pCR2KB-E2 was transformed into E. coli DH5α, and erythromycin-resistant (300 μg/ml) transformants were selected. Colonies were then replica plated to LB agar supplemented with ampicillin (100 μg/ml) to ensure that the ampicillin determinant had been inactivated. pCR2KB-E2 was linearized by restriction with _Asp_I (located at position 3525 and within the kanamycin resistance open reading frame) to increase the probability of integration into the chromosome by a double-crossover event. Genetic competence of S. gordonii Challis was induced as previously described (2). Transformation of S. gordonii FAS4 was accomplished using electrocompetent cells prepared as previously described (25). Transformation reaction mixtures were incubated for 2 h with shaking (120 rpm) at 37°C before aliquots were plated on TSBY agar supplemented with erythromycin (5 μg/ml). The plates were incubated at 37°C overnight in a candle jar. The mutant strains were compared to their wild-type parental strains by Gram staining, evaluation of colony morphology on blood agar and mitis salivarius agar, and biochemical metabolic profiles obtained using the API Rapid Strep kit (bioMerieux Vitek) according to the manufacturer's instructions.
PCR screening of transformants.
PCR was used to distinguish between mutants arising from single- or double-crossover events. Primers specific for the 5′ (5′-TGATGAAGCTACTGATGC-3′) and 3′ (5′-ATCACTGAACCAATGGCC-3′) ends of abpA were used to amplify products from genomic DNA isolated from the wild-type and mutant strains of S. gordonii. Following an initial denaturation period of 4 min at 94°C, 30 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C were executed, followed by an extension time of 10 min at 72°C.
Amylase-binding assays.
A solid-phase amylase ligand-binding assay was used to determine the presence of AbpA in culture supernatants as previously described (12). Briefly, streptococcal extracts or supernatants in phosphate-buffered saline (PBS) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred to Immobilon-P membranes or nitrocellulose. Blots were incubated with lyophilized HPS adjusted to 1 mg ml−1 in 3% (wt/vol) nonfat dry milk in 10 mM Tris-buffered normal saline containing 0.05% Tween 20 (TBST) for 1 h, washed, and incubated with antibody to purified human salivary amylase (40) in 1% (wt/vol) nonfat dry milk in TBST for 30 min. The blots were then washed, incubated with goat anti-rabbit immunoglobulin G conjugated to alkaline phosphatase (Promega) or to horseradish peroxidase-conjugated swine anti-rabbit immunoglobulin G and developed with appropriate reagents. The binding of soluble 125I-labeled amylase to whole cells was evaluated as described previously (38). In all cases, the wild-type parental strains of S. gordonii served as positive amylase-binding controls, and Streptococcus sanguis 10556 served as the nonbinding negative control. Whole saliva, HPS, and purified salivary amylase were prepared as previously described (38).
Coaggregation and cell surface hydrophobicity assays.
S. gordonii strains Challis, Challis-E1, FAS4, and FAS4-E1 were evaluated for the ability to coaggregate with each of the following partners: Streptococcus oralis strains C104 and 34, Streptococcus sp. strain SM PK509, and Actinomyces naeslundii strains PK947 and T14V. The visual coaggregation assay, the coaggregation scoring system used, and the coaggregating characteristics of the partner strains with S. gordonii have been previously described (47). Cell surface hydrophobicity was measured by hexadecane partitioning (18).
HAP binding studies.
Adhesion of bacteria to experimental pellicles was performed as previously described (43), except that buffered KCl (15) served as the adhesion buffer. Experimental pellicles were prepared by incubating 30 mg of HAP (Bio-Rad) with fresh whole saliva (1 ml/aliquot of HAP), fresh HPS (1 ml/aliquot of HAP), pool 3 of Bio-Gel P-60 chromatographed HPS containing primarily the glycosylated isoenzyme of amylase (500 μl of a 1-mg/ml solution), or pool 4 from a Bio-Gel P-60 fractionation of HPS containing predominantly nonglycosylated amylase (500 μl of a 1-mg/ml solution) (38).
Amylase-facilitated growth studies.
Approximately 106 cells of wild-type or mutant strains of S. gordonii were incubated with a sterile solution of purified, nonglycosylated amylase (1 mg/ml) for 30 min on a rotary mixer, washed three times with PBS, and then resuspended in 10 ml of FMC with 2% potato starch or 2% glucose as the primary carbon source. Cells were incubated for 24 h at 37°C in a candle jar. Samples were monitored spectrophotometrically at a wavelength of 600 nm. In addition, growth of all S. gordonii strains was also done without prior incubation with amylase. S. sanguis 10556 served as the non-amylase-binding negative control.
Biofilm growth of wild-type S. gordonii and the abpA mutant.
Biofilm culture methods were performed as described previously (33a). Briefly, two-track flow cells (parallel-plate flow chambers with a working volume of 250 μl/track) were constructed using a microscope slide as the bottom and a no. 1.5 coverglass as the top (22). The flow cells were acid washed overnight, rinsed with several changes of distilled water over a period of 3 h, and then autoclaved. Sterile 25% stimulated human whole saliva was prepared according to a modification (33a) of the method of de Jong and van der Hoeven (9).
Overnight, anaerobically (N2-CO2-H2, 95:5:5) grown, static liquid (BHI; Difco) cultures of S. gordonii wild-type or abpA mutant bacteria were transferred to fresh medium and regrown statically in an aerobic incubator for 2 h at 37°C to reestablish exponential growth. The cells were pelleted, washed three times with 25% saliva, and resuspended in 25% saliva to an _A_600 of 0.04.
All the following manipulations were performed in a 34°C aerobic incubator. Flow cells were conditioned by exposure to 25% saliva for 20 min prior to the introduction of bacteria. Bacterial suspensions (0.5 ml) were injected into two flow cells: in each flow cell, one track received the wild-type cells and other track received the abpA mutant cells. The flow cells were inverted and the bacteria were permitted to attach for 20 min, after which the flow cells were returned to the original orientation and flow of 25% saliva through the system at 0.0125 mm s−1 (200 μl min−1) was begun. After 20 min of salivary flow, one flow cell was removed from the incubator, stained with BacLight Live/Dead (Molecular Probes, Eugene, Oreg.) (0.5 ml of a 1:1 mixture of the two dyes diluted 1,000-fold in 25% saliva), and observed with a Leica TCS 4D confocal microscope (Leica LaserTechnik, Heidelberg, Germany) to document initial attachment levels. The settings on the microscope were interactively adjusted so that the specimen appearance when viewed through the oculars using a 515 LP filter was reproduced in the true-color red-green-blue overlay display. Three randomly selected fields of view were imaged in each track, and optical sections were collected at 0.5-μm intervals through the biofilms using a 40× (1.0 numerical aperture) oil immersion lens at an Airy disk value of 1. After an overnight (approximately 18-h) period of salivary flow, the remaining flow cell was removed from the incubator, stained, and imaged. This experiment was repeated twice.
Statistical analysis.
Analysis of the adhesion data and determination of significance were performed using the nonparametric Mann-Whitney test (data are presented as medians with the range in parentheses). Differences were considered significant when a P value of <0.05 was obtained.
RESULTS
Construction of abpA::erm(AM) mutants in S. gordonii
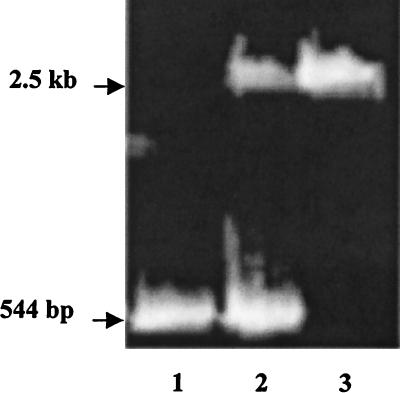
The genetic competence of five strains of S. gordonii previously shown to have abpA (5) was evaluated. With the exception of Challis, natural competence was not observed for any of the strains of S. gordonii using the conditions previously developed for the Challis strain (2). Electroporation of S. gordonii FAS4, a more recent clinical amylase-binding isolate (38), resulted in transformation and insertion of Tn_916_. The Challis and FAS4 strains were thus selected for inactivation of the abpA gene. Linearized pCR2KB-E2 was transformed into the naturally competent Challis and the electrocompetent FAS4 strains of S. gordonii, resulting in the mutant strains Challis-E1 and FAS4-E1. Confirmation of insertion of the erm(AM) gene into abpA was determined using PCR, with the results shown in Fig. 1. The abpA gene alone would be expected to yield a product of 544 bp (positive control), the presence of abpA and abpA::erm(AM) would result in amplicons of 544 bp and 2.5 kb (potential outcome of a single-crossover event), while a double-crossover event would yield a single 2.5-kb PCR product [abpA::erm(AM)]. Only the larger product was observed in transformants arising from a double-crossover event as seen in recombinant strains Challis-E1 (Fig. 1, lane 3) and FAS4-E1, indicating that no intact copies of abpA were present. Results obtained from a single-crossover mutant (Challis-E2) that produced amplicons of 544 bp (abpA) and 2,500 bp [abpA::erm(AM)] are shown for comparison (Fig. 1, lane 2). Southern blot analysis of the mutant S. gordonii Challis-E1 and FAS4-E1 _Hin_dIII-digested chromosomal DNA showed identical hybridization patterns when probed with either the erm(AM) or the abpA gene (results not shown). The erm(AM) probe failed to hybridize with genomic DNA from the wild-type strains. These data confirmed that erm(AM) had integrated into abpA in the transformants Challis-E1 and FAS4-E1 yielding a double-crossover mutation.
FIG. 1.
PCR screening of abpA::erm(AM) recombinants of S. gordonii. Lane 1, abpA (544-bp) positive control amplified from Challis; lane 2, recombinant in S. gordonii Challis-E2 resulting from the duplication (single crossover) of abpA (544 bp) and insertion of abpA::erm(AM) (2.5 kb); lane 3, recombinant in S. gordonii Challis-E1 resulting from allelic exchange (double crossover) of abpA::erm(AM) (2.5-kb band only).
Characterization of S. gordonii Challis-E1 and FAS4-E1 strains.
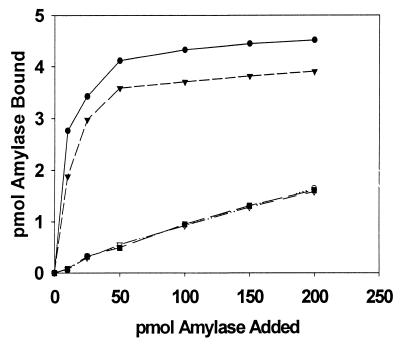
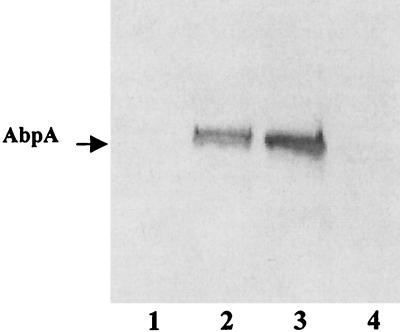
The amylase-binding capacity of the mutant strains of S. gordonii was compared to that of their wild-type progenitors (Fig. 2). In each case, the mutant strain bound salivary amylase only at low levels similar to that seen for S. sanguis 10556, the non-amylase-binding negative control. A solid-phase amylase ligand-binding assay (16) demonstrated no detectable levels of AbpA in the culture supernatants from either S. gordonii Challis-E1 or S. gordonii FAS4-E1 (Fig. 3, lanes 1 and 4). These results demonstrated that disruption of the abpA gene with erm(AM) had resulted in the loss of expression of AbpA. Comparisons of the wild-type and mutant strains of S. gordonii failed to reveal any differences regarding doubling time in nutrient-rich or chemically defined culture media. Additionally, the wild-type and mutant strains were indistinguishable with respect to colony morphology on blood agar or mitis salivarius agar or biochemical profiles as determined by the API Rapid Strep assay.
FIG. 2.
Binding of 125I-labeled amylase to S. gordonii strains Challis (●), Challis-E1 (○), FAS4 (▾), FAS4-E1 (▿), and S. sanguis 10556 (▪). Data points represent the mean values of duplicate samples from two experiments.
FIG. 3.
Comparison of culture supernatant levels of AbpA from wild-type and AbpA mutants of S. gordonii. An amylase ligand-binding assay was used. Lanes are loaded with equal amounts of protein (25 μg) from culture supernatants of S. gordonii Challis-E1 (lane 1), Challis (lane 2), FAS4 (lane 3), and FAS4-E1 (lane 4).
Coaggregation and hydrophobic properties of S. gordonii Challis-E1 and FAS4-E1 strains.
In order to determine if disruption of abpA affected interbacterial interactions, coaggregation of the wild-type and mutant strains of S. gordonii with a number of well-characterized coaggregation partners of S. gordonii (47) were determined. No differences were noted between the amylase-binding and non-amylase-binding pairs (data not shown). These results suggest that AbpA is not an important contributor to the coaggregation of S. gordonii with the organisms tested. Cell surface hydrophobicity also was not affected by the insertional inactivation of abpA as determined by hexadecane partitioning (data not shown). The results suggest that any difference in adhesion to experimental pellicles by the mutant strains is not due to alterations in surface hydrophobicity, a property previously shown to affect adhesion (18).
HAP-binding studies.
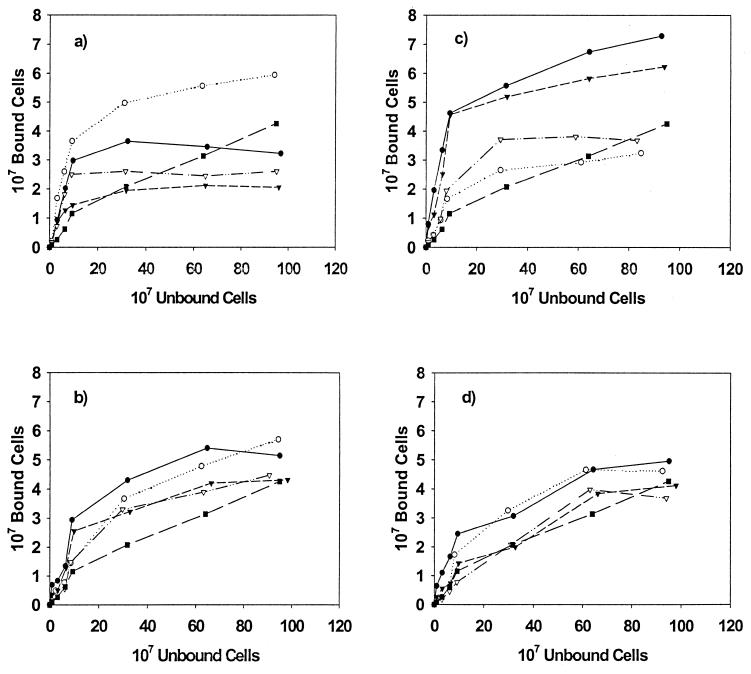
The role of the amylase-binding phenotype in the adhesion of S. gordonii strains to experimental pellicles was determined using coated HAP, and the resulting binding isotherms are summarized in Fig. 4. All of the strains tested demonstrated saturation of binding to whole-saliva-coated HAP (Fig. 4a). No significant differences between strains were noted for binding to uncoated HAP (data not shown). In the case of binding to adhesion buffer-coated HAP, saturation was not achieved suggesting nonspecific binding in that system. Comparison of the binding isotherms for S. gordonii strains Challis and Challis-E1 (Fig. 4a) show that the mutant strain, Challis-E1, bound better to whole-saliva-coated HAP than did the wild-type strain. A similar, though less pronounced, difference was noted for the mutant and wild-type strains of S. gordonii FAS4. In both cases, AbpA mutants bound better to whole-saliva-coated HAP than did the wild-type organisms. Interestingly, although there were pronounced differences in the total number of cells bound, saturation was achieved for strains Challis, FAS4, and FAS4-E1 when approximately 108 cells were added to the system. Approximately threefold more cells were required to reach saturation of 30 mg of whole-saliva-coated HAP with the non-amylase-binding mutant strain Challis-E1. While binding of all strains to whole-saliva-coated HAP exceeded that seen for binding to adhesion buffer-coated HAP at lower concentrations of cells, the nonspecific binding seen in the adhesion buffer-coated HAP system did exceed that seen for S. gordonii Challis, FAS4, and FAS4-E1 at higher cell input levels.
FIG. 4.
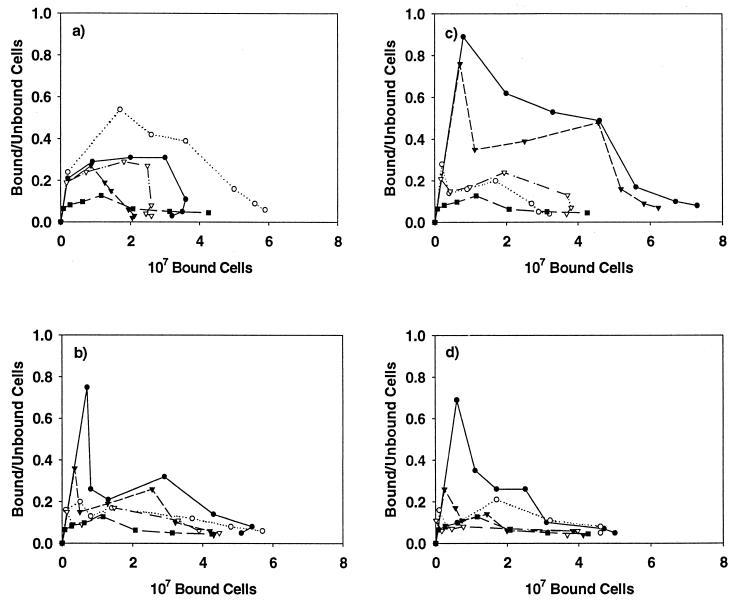
Binding isotherms for the adherence of S. gordonii strains to coated HAP. (a) Whole-saliva-coated HAP; (b) HPS-coated HAP; (c) glycosylated amylase-coated HAP; (d) nonglycosylated amylase-coated HAP. Data for the following S. gordonii strains are shown: Challis (●), Challis-E1 (○), FAS4 (▾), FAS4-E1 (▿), and S. sanguis 10556 (▪).
These findings were reversed when the binding of S. gordonii strains to HPS-coated HAP was assessed (Fig. 4b). Similar binding patterns were seen for each of the mutant and wild-type pairs. Small differences were noted in the binding isotherms at lower concentrations of cells, with greater numbers of the wild-type organisms adhering to the parotid-saliva-coated HAP than were found for the mutant strains. The greatest differences in binding were noted when wild-type and mutant strains were compared for adhesion to HAP coated with the glycosylated isoenzyme of human salivary amylase (Fig. 4c). The binding of S. gordonii Challis and FAS4 to glycosylated amylase-coated HAP was approximately twofold better than that of their mutant counterparts. In contrast to what was seen for adhesion to whole-saliva- and HPS-coated HAP, S. gordonii Challis and FAS4 strains demonstrated very similar adhesion patterns. The binding isotherms for HAP coated with the nonglycosylated isoenzyme of amylase were nearly identical for both the wild-type and mutant strains, with the Challis–Challis-E1 pair binding at higher levels than the FAS4–FAS4-E1 counterpart (Fig. 4d).
Results for Scatchard analysis of the adhesion data for whole-saliva-coated HAP (Fig. 5a) are characterized by initial positive slopes for both the mutant and wild-type strains suggestive of positive cooperativity (29). This is especially evident for the Challis-E1 strain and may explain the markedly greater adhesion of this organism to whole-saliva-coated HAP. The Scatchard plot describing binding to adhesion buffer-coated HAP is consistent with nonspecific binding interactions. No statistically significant differences were noted for adhesion to whole-saliva-coated HAP when comparing the ratios of bound versus unbound cells (Table 2).
FIG. 5.
Scatchard plots for adherence of S. gordonii strains to coated HAP. (a) Whole-saliva-coated HAP; (b) HPS-coated HAP; (c) glycosylated amylase-coated HAP; (d) nonglycosylated amylase-coated HAP. Data for the following S. gordonii strains are shown: Challis (●), Challis-E1 (○), FAS4 (▾), FAS4-E1 (▿), and S. sanguis 10556 (▪).
TABLE 2.
S. gordonii cells bound to coated HAP
| Cells bound/cells unbound to HAP coated withg: | |||||
|---|---|---|---|---|---|
| Strain | Adhesion buffer | Whole salivaa | HPSb | Amylased (glycosylated) | Amylasee (nonglycosylated) |
| Challis | 0.09 (0.07–0.12) | 0.21 (0.19–0.24) | 0.75 (0.71–0.80)c | 0.89 (0.85–0.91) | 0.69 (0.66–0.70) |
| Challis-E1 | 0.11 (0.10–0.13) | 0.24 (0.23–0.27) | 0.16 (0.13–0.18) | 0.28 (0.24–0.31) | 0.16 (0.14–0.17) |
| FAS4 | 0.08 (0.06–0.10) | 0.19 (0.18–0.22) | 0.36 (0.33–0.37) | 0.76 (0.72–0.79) | 0.26 (0.23–0.27)f |
| FAS4-E1 | 0.10 (0.09–0.10) | 0.19 (0.16–0.22) | 0.15 (0.13–0.16) | 0.21 (0.18–0.23) | 0.11 (0.08–0.12) |
The Scatchard plot shown in Fig. 5b illustrates the magnitude of the differences in adhesion to HPS at low cell concentrations when comparing the wild-type and mutant strains. The wild-type strains (Challis and FAS4) demonstrated a marked initial positive slope that was absent from the mutant strains when lower numbers of cells were present, again suggestive of positive cooperativity. These differences were statistically significant for the Challis (P < 0.001) and FAS4 (P < 0.05) strains compared to Challis-E1 and FAS4-E1, respectively (Table 2). In addition, a second smaller positive slope was associated with the wild-type organisms. Overall binding to HPS-coated HAP was greater than that observed for whole-saliva-coated HAP except for Challis-E1, which showed a decrease in overall adhesion. The trend for greater adhesion of the Challis and Challis-E1 strains over the FAS4 and FAS4-E1 strains remained.
Scatchard analysis of the binding to glycosylated amylase demonstrated a very strong positive slope associated with adhesion at lower concentrations of cells for the wild-type strains (Fig. 5c), but differences in the amplitude of the slope were not statistically significant (Table 2). The initial positive slope of the FAS4 strain was followed by a steep negative slope and then a gradual, second positive slope suggestive of alternating positive and negative cooperativity. The positive slope S. gordonii Challis, on the other hand, was followed by a relatively broad, gradual negative slope. When compared at saturating amounts of cells, the Challis (P < 0.005) and FAS4 (P < 0.005) strains bound significantly better than did the mutant strains (Table 2). Scatchard analysis of binding to nonglycosylated amylase again revealed that relatively large differences in binding did occur at lower cell concentrations (Fig. 5d). These differences were lost at concentrations greater than 107 cells. A slope consistent with positive cooperativity remained evident for the Challis strain and was present, although less so, for S. gordonii FAS4. The ratio of bound versus unbound cells was significantly greater for S. gordonii Challis than for the Challis-E1 (P < 0.005), FAS4 (P < 0.05), and FAS4-E1 (P < 0.005) strains (Table 2). In addition, a significant difference (P < 0.05) was noted between the FAS4 and FAS4-E1 strains.
Taken together, these findings suggest that AbpA most efficiently binds to HAP coated with the glycosylated isoenzyme of amylase, followed by the nonglycosylated isoenzyme. This adhesion is obscured when the amount of amylase in the pellicle is decreased (as would occur in pellicles prepared from whole saliva).
Role of AbpA in growth.
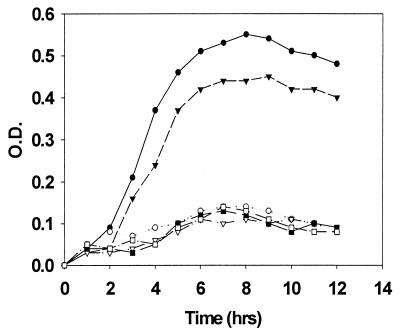
Recent experiments have suggested that the regulation of abpA gene expression is under the control of a catabolite repression mechanism (34a). This implies that AbpA may play a role in the catabolic activities of S. gordonii. We investigated this potential role by culturing the wild-type and mutant strains of S. gordonii in a chemically defined medium (FMC) with either 2% glucose or 2% potato starch as the primary carbon source. S. sanguis 10556 was used as a non-amylase-binding control. The strains were first incubated in a sterile solution of human salivary nonglycosylated amylase in PBS. After washing to remove unbound amylase, approximately 106 cells were inoculated into 10 ml of defined medium, and growth was monitored spectrophotometrically, with the results shown in Fig. 6. In all cases, only organisms with the amylase-binding phenotype were able to grow appreciably in the starch-containing FMC medium and only after preincubation of the cells with salivary amylase. Strains expressing the amylase-binding phenotype failed to grow in the starch-containing medium without prior incubation with amylase, while all strains grew equally well in the glucose-supplemented medium. This suggests that AbpA facilitates the growth of these organisms under conditions under which starch is the predominant carbon source.
FIG. 6.
Amylase-facilitated growth of amylase-binding and nonbinding streptococcal strains. Bacteria were grown in a defined medium with 2% potato starch as the primary carbon source, and growth was monitored spectrophotometrically at 600 nm. All cells were incubated with amylase prior to inoculation into the culture medium. Data for the following S. gordonii strains are shown: Challis (●), Challis-E1 (○), FAS4 (▾), FAS4-E1 (▿), and S. sanguis 10556 (▪). Data represent mean values of three experiments, each done in duplicate.
Role of AbpA in biofilm formation.
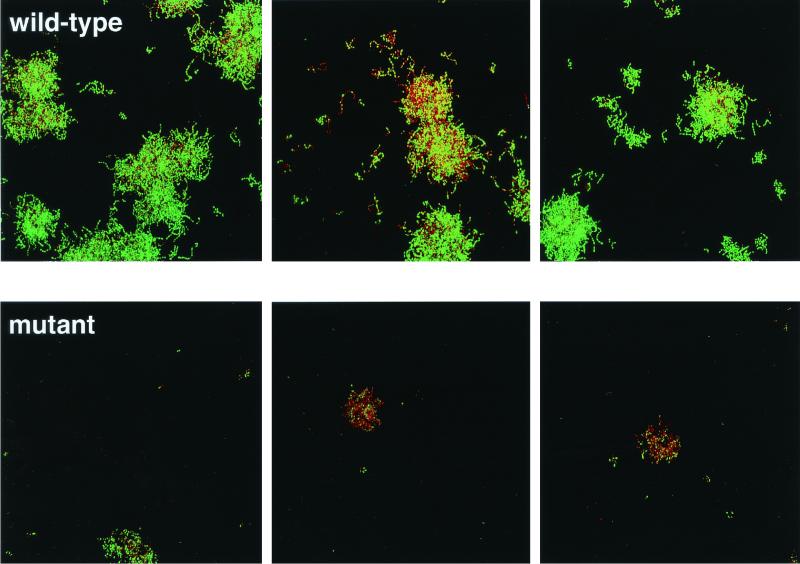
Images of biofilms of the wild-type (Challis) and abpA mutant (Challis-E1) strains of S. gordonii acquired immediately after the initial 20 min of salivary flow demonstrated that colonization was initially sparse. The cells were present primarily as short chains composed of two or three cells, and the vast majority of cells stained green with Live/Dead, thus showing that the cells were metabolically active. After overnight growth on 25% saliva, major differences between the two strains became obvious (Fig. 7). The wild-type strain produced large microcolonies that were approximately 10 μm high and in which the majority of cells were stained green to yellow-green; only one field of view showed many red- or orange-stained cells. In sharp contrast, the abpA mutant strain grew much more poorly than did the wild-type. The microcolonies were relatively small and were about 5 μm high. Red- and orange-stained cells were predominant in two of the three fields. A repeat of this experiment yielded essentially identical results, which indicated that the absence of the abpA gene product reduces the ability of S. gordonii to grow as a biofilm on human saliva and that this reduction in growth is linked to a reduction in overall vitality as measured by the membrane integrity assay (Live/Dead stain).
FIG. 7.
Live/Dead-stained biofilms of three random fields of view from the flow cell containing S. gordonii Challis (top row) and three random fields of view of the abpA mutant Challis-E1 (bottom row) after overnight growth on 25% saliva. Image dimensions are 250 by 250 μm. Maximum microcolony height is about 10 μm (wild type) and about 5 μm (mutant). The vast majority of cells stained green in the wild-type biofilm, but the majority of the cells were red to orange in the mutant biofilm.
DISCUSSION
The observed twofold increase in the adhesion of amylase-binding strains to amylase-coated HAP compared to the AbpA mutants suggests a role for AbpA as an adhesin. This result supports previous data showing that amylase promoted the adhesion of S. gordonii to experimental pellicles formed on HAP (43). S. gordonii produces over 30 polypeptides on its cell surface (19, 48). A number of these proteins (adhesins) promote adhesion of this bacterium to oral surfaces, including antigen I/II, LraI, or the PAAP antigen (14, 17). The adhesion to amylase appears to involve AbpA, which is distinct from previously described adhesins since it does not share extensive nucleotide or amino acid sequence homology with any known proteins (34). Unlike members of the antigen I/II, LraI, or PAAP adhesins, AbpA possesses a truncated cell wall-anchoring consensus sequence, suggesting transient association between AbpA and the cell wall. Previous studies have shown the localization of the protein to cell wall division sites (40). The rapid appearance of AbpA in culture supernatants support the hypothesis that AbpA is a secreted protein.
There have been conflicting reports in the literature regarding the contribution of amylase binding to the process of adhesion. Douglas found no correlation between the amylase-binding phenotype of a number of streptococcal strains and the ability to adhere to saliva-coated HAP (11). In addition, a more recent study found no association between salivary amylase concentration and adhesion of S. gordonii to microtiter plates coated with human saliva (35). Note that in both of these studies, adhesion to whole-saliva-coated HAP rather than HPS- or amylase-coated HAP was evaluated. This distinction is important since amylase accounts for approximately half of the protein content of HPS while representing only <5% of whole-saliva total protein. Other studies have shown that the composition of saliva, and therefore the composition of the oral pellicles, differs throughout the oral cavity (6). Caution must also be used when comparing results from studies that assess adhesion to HAP versus microtiter plates, as these two model systems may not be comparable (35, 43). The comparison of the wild type with its isogenic amylase-deficient mutants clearly supports a role for AbpA as an adhesin to amylase-coated HAP.
Adhesion isotherms of S. gordonii to whole-saliva-coated HAP demonstrated that the mutant strains were bound in markedly greater numbers than their wild-type progenitors. The increase in adhesion of the amylase-deficient mutant strains to whole-saliva-coated HAP suggests that other adhesins may become redistributed on the surface of the amylase-deficient mutants to increase the likelihood of their binding to receptors in the pellicle. This trend was reversed when adhesion to HPS- or amylase-coated HAP was measured, indicating that AbpA is important in adhesion when amylase is in higher salivary concentrations within the pellicle. It is also possible that the nonrandom spatial location of AbpA at cell division sites may also reduce the activity of AbpA as an adhesin (19). This would be especially apparent in the presence of multiple ligands found in more complex fluids such as whole saliva. There may also be substantial redundancy among the various salivary receptors for S. gordonii. Redundancy refers to the observation that many salivary proteins exhibit similar functions (24). Thus, although AbpA may serve as an amylase adhesin, the display of multiple adhesins on the surface of S. gordonii may serve to diminish the relative contribution of AbpA in adhesion to complex pellicles.
In Scatchard analysis, data are weighted for greatest sensitivity at low concentrations of ligand or high-affinity regions (44). This was clearly seen in the coated HAP studies, in which differences were obvious between the mutant and wild-type organisms at lower cell concentrations that were not as evident from the binding isotherms. The Challis and FAS4 strains showed pronounced positive slopes under those conditions that were absent in Scatchard plots of their mutant counterparts. Scatchard plots that show nonlinearity can be diagnostic of mechanistic features of the attachment reaction (44). Indeed, the Scatchard plots of S. gordonii adhesion to amylase-coated pellicles suggested the presence of positive cooperativity in ligand binding (29). This implies that the first few ligands bind with a lower affinity than the subsequent ligands to the macromolecule (8). The maximum in a Scatchard plot relates cooperativity and the degree of saturation. The early maximums seen in these studies correlate well with the relatively rapid saturation depicted on the binding isotherms.
While inactivation of abpA does not appear to alter the expression of other cell surface proteins (34), surface hydrophobicity, or coaggregation with the organisms tested, adhesion to saliva-coated HAP is affected by the loss of AbpA. Reduced adhesion to parotid-saliva-coated HAP was also observed in Streptococcus mutans mutants that lacked the major cell surface-associated protein P1 (4). Unlike AbpA, P1 also appears to affect the hydrophobicity of the cell (21). In contrast, mutant strains of S. gordonii that did not express surface SspA or SspB demonstrated diminished coaggregation with A. naeslundii and adhesion to parotid salivary agglutinin glycoprotein without concomitant alterations in cell surface hydrophobicity. These mutations also did not affect adhesion to whole-saliva- or HPS-coated HAP. Isogenic mutants of S. gordonii which did not express cell surface CshA or CshB were also deficient in adhesion to A. naeslundii, S. oralis, and immobilized human fibronectin (28). Recent studies have shown that the cell wall-anchored CshA polypeptide forms surface fibrils that confer hydrophobic and adhesive properties (27). Fap1, an adhesin involved in the assembly of S. parasanguis FW213 fimbriae, has also been shown to play an important role in adhesion to saliva-coated HAP (49). It is of particular interest that each of these proteins appears to affect multiple cell surface properties and may in fact be multifunctional (20).
Based on the diversity of binding specificities of the adhesins described for the oral streptococci, consideration must be given to the possibility that these adhesins may help determine the initial microbial composition of dental plaque within different parts of the mouth. Saliva from the major salivary glands is not distributed evenly throughout the oral cavity, and these compositional variations in the resulting pellicles may be important in the establishment of microflora and tooth-related disease patterns in various parts of the dentition (6). For instance, the ability to bind amylase-coated HAP predicts that S. gordonii and other amylase-binding bacteria would preferentially colonize the buccal surfaces of maxillary posterior teeth due to their proximity to the parotid ducts, since HPS is rich in amylase. The relatively greater amounts of amylase bound to the tooth surface adjacent to the parotid ducts may aid in the adhesion of the organism during the early phases of plaque formation.
In addition to its role as an adhesin, our results validate a second function for AbpA in dietary carbohydrate catabolism. Thus, AbpA mediates the binding of amylase to the cell surface of S. gordonii, and the bound amylase hydrolyzes starch to fermentable substrates. This hypothesis is supported by the observation that both AbpA and amylase are required for maximal growth in defined media with potato starch as the predominant carbon source. Poor growth was observed when either one of these components was absent from the cultures. This is in agreement with earlier reports that provided indirect evidence for this phenomenon (12, 39). The lack of growth by the wild-type strains on starch in the absence of amylase also suggests that S. gordonii Challis does not produce significant quantities of an endogenous amylase.
The study of the wild-type and abpA mutant strains in a biofilm model also yielded interesting insights into the potential role of this protein in S. gordonii colonization of the oral cavity. Growth of single oral bacterial strains using saliva as the sole carbon and nitrogen source has been relatively ignored because it has been thought not to be relevant to the microbial ecology of the oral cavity. It is known that multigenus oral bacterial consortia can grow directly on unamended saliva (9), presumably through their ability to metabolize high-molecular-weight glycoproteins. However, in the initial colonization of the tooth surface, we propose that nascent bacterial communities form and acquire the metabolic cooperation necessary to break down salivary glycoproteins. Isolated clusters of cells must first adhere and metabolize the suite of nutrients present in saliva. Therefore, the ability to grow and form biofilms on saliva alone is a selective factor that has been shown to influence biofilm composition in vitro and may play a role in vivo (33a). The present results suggest that AbpA is important in this regard. The mutant strain lacking functional AbpA grew much more poorly in the flow cell biofilm model than did the wild-type strain. Also, when forced to utilize saliva as the sole carbon and nitrogen source, the physiological state of the mutant cells, but not of the wild-type cells, was impaired as assessed by the Live/Dead stain. Thus, it appears that the ability to bind salivary amylase may play an important role in S. gordonii colonization. Perhaps amylase-binding species are able to first adhere to amylase receptors and then form a biofilm utilizing saliva as the sole carbon and nitrogen source. Thus, these species appear well adapted as initial colonizers in vivo. Those species that lack abpA may be predicted to be less likely to produce significant biomass during early stages of tooth colonization. S. oralis strain 34 lacks abpA (5), and biofilm formation of strain 34 was poor in the same saliva-supported in vitro biofilm model used here (33a). Streptococci constitute 60 to 90% of initial colonizers (30, 31), and a large proportion of these isolates bind amylase (41). The interaction between early colonizing amylase-binding streptococcal species and those species lacking the ability to bind amylase could be critical to retention of the latter and their subsequent growth in early dental plaque. It is possible that the amylase-binding species contribute to the plaque community by efficiently metabolizing dietary starch and providing nearby nonbinding species with metabolizable by-products from starch.
In summary, the results from this study suggest that the AbpA protein of S. gordonii appears to play a role in the adhesion of this bacterium to amylase receptors in the acquired enamel pellicle. Amylase bound to the bacterial cell surface may also serve to liberate fermentable carbohydrates through the hydrolysis of dietary starches. Finally, AbpA may further function in initial biofilm formation and maturation. Together, these properties may facilitate colonization of this species to oral surfaces. The relative contribution of AbpA to adhesion, carbon catabolism, and biofilm formation awaits further in vivo studies comparing the wild-type and abpA strains of S. gordonii.
ACKNOWLEDGMENTS
This work was supported by grant DE 09838 (F.A.S.) and a Dentist Scientist Award DE 00158 (J.D.R.), both from the National Institute of Dental and Craniofacial Research.
We are grateful to Howard K. Kuramitsu and Elaine M. Haase for helpful discussions throughout the course of this work.
REFERENCES
- 1.Aguirre A, Levine M J, Cohen R E, Tabak L A. Immunochemical quantitation of α-amylase and secretory IgA in parotid saliva from people of various ages. Arch Oral Biol. 1987;32:297–301. doi: 10.1016/0003-9969(87)90024-0. [DOI] [PubMed] [Google Scholar]
- 2.Behnke D. Plasmid transformation of Streptococcus sanguis (Challis) occurs by circular and linear molecules. Mol Gen Genet. 1981;182:490–497. doi: 10.1007/BF00293940. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowen W H, Schilling K, Giertsen E, Pearson S, Lee S F, Bleiweis A, Beeman D. Role of a cell surface-associated protein in adherence and dental caries. Infect Immun. 1991;59:4606–4609. doi: 10.1128/iai.59.12.4606-4609.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown A E, Rogers J D, Haase E M, Zelasko P M, Scannapieco F A. Prevalence of the amylase binding protein A gene (abpA) in oral streptococci. J Clin Microbiol. 1999;37:4081–4085. doi: 10.1128/jcm.37.12.4081-4085.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlen A, Borjesson A C, Nikdel K, Olsson J. Composition of pellicles formed in vivo on tooth surfaces in different parts of the dentition, and in vitro on hydroxyapatite. Caries Res. 1998;32:447–455. doi: 10.1159/000016486. [DOI] [PubMed] [Google Scholar]
- 7.Clemans D L, Kolenbrander P E. Isolation and characterization of coaggregation-defective (Cog-) mutants of Streptococcus gordonii DL1 (Challis) J Ind Microbiol. 1995;15:193–197. doi: 10.1007/BF01569825. [DOI] [PubMed] [Google Scholar]
- 8.Dahlquist F W. The meaning of Scatchard and Hill plots. Methods Enzymol. 1978;48:270–299. doi: 10.1016/s0076-6879(78)48015-2. [DOI] [PubMed] [Google Scholar]
- 9.de Jong M H, van der Hoeven J S. The growth of bacteria on saliva. J Dent Res. 1987;66:498–505. doi: 10.1177/00220345870660021901. [DOI] [PubMed] [Google Scholar]
- 10.Douglas C W I. Bacterial-protein interactions in the oral cavity. Adv Dent Res. 1994;8:254–262. doi: 10.1177/08959374940080021901. [DOI] [PubMed] [Google Scholar]
- 11.Douglas C W I. The binding of human salivary α-amylase by oral strains of streptococcal bacteria. Arch Oral Biol. 1983;28:567–573. doi: 10.1016/0003-9969(83)90003-1. [DOI] [PubMed] [Google Scholar]
- 12.Douglas C W I. Characterization of the α-amylase receptor of Streptococcus gordonii NCTC 7868. J Dent Res. 1990;69:1746–1752. doi: 10.1177/00220345900690110701. [DOI] [PubMed] [Google Scholar]
- 13.Douglas C W I, Heath J, Gwynn J P. Enzymic activity of salivary amylase when bound to the surface of oral streptococci. FEMS Microbiol Let. 1992;92:193–198. doi: 10.1016/0378-1097(92)90511-l. [DOI] [PubMed] [Google Scholar]
- 14.Erickson P R, Herzberg M C. The Streptococcus sanguis platelet-associated protein. Identification and characterization of the minimal platelet-interactive domain. J Biol Chem. 1993;268:1646–1649. [PubMed] [Google Scholar]
- 15.Gibbons R J, Hay D I. Human salivary acidic proline-rich proteins and statherin promote the attachment of Actinomyces viscosus LY7 to apatitic surfaces. Infect Immun. 1988;56:439–445. doi: 10.1128/iai.56.2.439-445.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gwynn J P, Douglas C W I. Comparison of amylase-binding proteins in oral streptococci. FEMS Microbiol Lett. 1994;124:373–380. doi: 10.1111/j.1574-6968.1994.tb07311.x. [DOI] [PubMed] [Google Scholar]
- 17.Jenkinson H F. Cell surface protein receptors in oral streptococci. FEMS Microbiol Lett. 1994;121:133–140. doi: 10.1111/j.1574-6968.1994.tb07089.x. [DOI] [PubMed] [Google Scholar]
- 18.Jenkinson H F. Novobiocin-resistant mutants of Streptococcus sanguis with reduced cell hydrophobicity and defective in coaggregation. J Gen Microbiol. 1987;133:1909–1918. doi: 10.1099/00221287-133-7-1909. [DOI] [PubMed] [Google Scholar]
- 19.Jenkinson H F, Lamont R J. Streptococcal adhesion and colonization. Crit Rev Oral Biol Med. 1997;8:175–200. doi: 10.1177/10454411970080020601. [DOI] [PubMed] [Google Scholar]
- 20.Jenkinson H F, Terry S D, McNab R, Tannock G W. Inactivation of the gene encoding surface protein SspA in Streptococcus gordonii DL1 affects cell interactions with human salivary agglutinin and oral actinomyces. Infect Immun. 1993;61:3199–3208. doi: 10.1128/iai.61.8.3199-3208.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koga T, Okahashi N, Takahashi I, Kanamoto T, Asakawa H, Iwaki M. Surface hydrophobicity, adherence and aggregation of cell surface protein antigen mutants of Streptococcus mutans serotype c. Infect Immun. 1990;58:289–296. doi: 10.1128/iai.58.2.289-296.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolenbrander P E, Andersen R N, Kazmerzak K, Wu R, Palmer R J., Jr Spatial organization of oral bacteria in biofilms. Methods Enzymol. 1999;310:322–332. doi: 10.1016/s0076-6879(99)10026-0. [DOI] [PubMed] [Google Scholar]
- 23.Kolenbrander P E, Andersen R N, Moore L V H. Intrageneric coaggregation among strains of human oral bacteria: potential role in primary colonization of the tooth surface. Appl Environ Microbiol. 1990;56:3890–3894. doi: 10.1128/aem.56.12.3890-3894.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine M J. Development of artificial salivas. Crit Rev Oral Biol Med. 1993;4:279–286. doi: 10.1177/10454411930040030401. [DOI] [PubMed] [Google Scholar]
- 25.Loimaranta V, Tenovuo J, Koivisto L, Karp M. Generation of bioluminescent Streptococcus mutans and its usage in rapid analysis of the efficacy of antimicrobial compounds. Antimicrob Agents Chemother. 1998;42:1906–1910. doi: 10.1128/aac.42.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loo C Y, Corliss D A, Ganeshkumar N. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J Bacteriol. 2000;182:1374–1382. doi: 10.1128/jb.182.5.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNab R, Forbes H, Handley P S, Loach D M, Tannock G W, Jenkinson H F. Cell wall-anchored CshA polypeptide (259 kilodaltons) in Streptococcus gordonii forms surface fibrils that confer hydrophobic and adhesive properties. J Bacteriol. 1999;181:3087–3095. doi: 10.1128/jb.181.10.3087-3095.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McNab R, Holmes A R, Clarke J M, Tannock G W, Jenkinson H F. Cell surface polypeptide CshA mediates binding of Streptococcus gordonii to other oral bacteria and to immobilized fibronectin. Infect Immun. 1996;64:4204–4210. doi: 10.1128/iai.64.10.4204-4210.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nesbitt W E, Doyle R J, Taylor K G, Staat R H, Arnold R R. Positive cooperativity in the binding of Streptococcus sanguis to hydroxylapatite. Infect Immun. 1982;35:157–165. doi: 10.1128/iai.35.1.157-165.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nyvad B, Kilian M. Comparison of the initial streptococcal microflora on dental enamel in caries active and caries inactive individuals. Caries Res. 1990;24:267–272. doi: 10.1159/000261281. [DOI] [PubMed] [Google Scholar]
- 31.Nyvad B, Kilian M. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand J Dent Res. 1987;95:369–380. doi: 10.1111/j.1600-0722.1987.tb01627.x. [DOI] [PubMed] [Google Scholar]
- 32.Orstavik D, Kraus F W. The acquired pellicle: enzyme and antibody activities. Scand J Dent Res. 1974;82:202–205. doi: 10.1111/j.1600-0722.1974.tb00381.x. [DOI] [PubMed] [Google Scholar]
- 33.Orstavik D, Kraus F W. The acquired pellicle: immunofluorescent demonstration of specific proteins. J Oral Pathol. 1973;2:68–76. doi: 10.1111/j.1600-0714.1973.tb01675.x. [DOI] [PubMed] [Google Scholar]
- 33a.Palmer R J, Jr, Kazmerzak K, Hansen M C, Kolenbrander P E. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect Immun. 2001;69:5794–5804. doi: 10.1128/IAI.69.9.5794-5804.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogers J D, Haase E M, Brown A B, Douglas C W I, Gwynn J P, Scannapieco F A. Identification and analysis of a gene (abpA) essential for amylase-binding to Streptococcus gordonii. Microbiology. 1998;144:1223–1233. doi: 10.1099/00221287-144-5-1223. [DOI] [PubMed] [Google Scholar]
- 34a.Rogers J D, Scannapieco F A. RegG, a CcpA homolog, participates in regulation of amylase-binding protein A gene (abpA) expression in Streptococcus gordonii. J Bacteriol. 2001;183:3521–3525. doi: 10.1128/JB.183.11.3521-3525.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudney J D, Hickey K L, Ji Z. Cumulative correlations of lysozyme, lactoferrin, peroxidase, S-IgA, amylase, and total protein concentrations with adherence of oral viridans streptococci to microplates coated with human saliva. J Dent Res. 1999;78:759–768. doi: 10.1177/00220345990780030801. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Scannapieco F A. Saliva-bacterium interactions in oral microbial ecology. Crit Rev Oral Biol Med. 1994;5:203–248. doi: 10.1177/10454411940050030201. [DOI] [PubMed] [Google Scholar]
- 38.Scannapieco F A, Bergey E J, Reddy M S, Levine M J. Characterization of salivary α-amylase binding to Streptococcus sanguis. Infect Immun. 1989;57:2853–2863. doi: 10.1128/iai.57.9.2853-2863.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scannapieco F A, Bhandary K, Ramasubbu N, Levine M J. Structural relationship between the enzymatic and streptococcal binding sites of human salivary α-amylase. Biochem Biophys Res Commun. 1990;173:1109–1115. doi: 10.1016/s0006-291x(05)80900-3. [DOI] [PubMed] [Google Scholar]
- 40.Scannapieco F A, Haraszthy G G, Cho M I, Levine M J. Characterization of an amylase-binding component from Streptococcus gordonii G9B. Infect Immun. 1992;60:4726–4733. doi: 10.1128/iai.60.11.4726-4733.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scannapieco F A, Solomon L, Wadenya R O. Emergence in human dental plaque and host distribution of amylase-binding streptococci. J Dent Res. 1994;73:1627–1635. doi: 10.1177/00220345940730100701. [DOI] [PubMed] [Google Scholar]
- 42.Scannapieco F A, Torres G, Levine M J. Salivary α-amylase: role in dental plaque and caries formation. Crit Rev Oral Biol Med. 1993;4:301–307. doi: 10.1177/10454411930040030701. [DOI] [PubMed] [Google Scholar]
- 43.Scannapieco F A, Torres G I, Levine M J. Salivary amylase promotes adhesion of oral streptococci to hydroxyapatite. J Dent Res. 1995;74:1360–1366. doi: 10.1177/00220345950740070701. [DOI] [PubMed] [Google Scholar]
- 44.Staat R H, Peyton J C. Adherence of oral streptococci: evidence for nonspecific adsorption to saliva-coated hydroxylapatite surfaces. Infect Immun. 1984;144:653–659. doi: 10.1128/iai.44.3.653-659.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terleckyj B, Willet N P, Schockman G D. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect Immun. 1975;11:649–655. doi: 10.1128/iai.11.4.649-655.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tseng C C, Scannapieco F A, Levine M J. Use of a replica plate assay for the rapid assessment of salivary protein-bacteria interactions. Oral Microbiol Immunol. 1992;7:53–56. doi: 10.1111/j.1399-302x.1992.tb00021.x. [DOI] [PubMed] [Google Scholar]
- 47.Whittaker C J, Clemans D L, Kolenbrander P E. Insertional inactivation of an intrageneric coaggregation-relevant adhesin locus from Streptococcus gordonii DL1 (Challis) Infect Immun. 1996;64:4137–4142. doi: 10.1128/iai.64.10.4137-4142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whittaker C J, Klier C M, Kolenbrander P E. Mechanisms of adhesion by oral bacteria. Annu Rev Microbiol. 1996;50:513–552. doi: 10.1146/annurev.micro.50.1.513. [DOI] [PubMed] [Google Scholar]
- 49.Wu H, Fives-Taylor P M. Identification of dipeptide repeats and a cell wall sorting signal in the fimbriae-associated adhesin, Fap1, of Streptococcus parasanguis. Mol Microbiol. 1999;34:1070–1081. doi: 10.1046/j.1365-2958.1999.01670.x. [DOI] [PubMed] [Google Scholar]
- 50.Yamashita Y, Takehara T, Kuramitsu H K. Molecular characterization of a Streptococcus mutans mutant altered in environmental stress responses. J Bacteriol. 1993;175:6220–6228. doi: 10.1128/jb.175.19.6220-6228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]