A PP2C-type phosphatase dephosphorylates the PII signaling protein in the cyanobacterium Synechocystis PCC 6803 (original) (raw)
Abstract
The family of the PII signal transduction proteins contains the most highly conserved signaling proteins in nature. The cyanobacterial PII-homologue transmits signals of the cellular nitrogen status and carbon status through phosphorylation of a seryl-residue. To identify the enzyme responsible for dephosphorylation of the phosphorylated PII protein in_Synechocystis_ PCC 6803, prospective phosphatase encoding genes were inactivated by targeted insertion of kanamycin resistance cassettes. Disruption of ORF sll1771 generates a mutant unable to dephosphorylate PII under various experimental conditions. On the basis of conserved signature motifs, the sll1771 product (termed PphA) is a member of the protein phosphatase 2C (PP2C) superfamily, which is characterized by Mg2+/Mn2+-dependent catalytic activity. Biochemical analysis of overexpressed and purified PphA confirms its PP2C-type enzymatic properties and demonstrated its reactivity toward the phosphorylated PII protein. Thus, PphA is the first protein phosphatase in Synechocystis PCC 6803 for which the physiological substrate and function is known.
Growth of cyanobacteria is based on oxygenic photosynthesis and the assimilation of simple inorganic nutrients, a lifestyle that allows these organisms to inhabit nearly all illuminated environments. They employ highly efficient sensing and control mechanisms to acclimate to the various challenges posed by physical and chemical parameters of their environment. An important part of this homeostatic control system are the bacterial two-component regulatory systems, and the genome of the cyanobacterium_Synechocystis_ PCC 6803 has an abundance of genes encoding such systems (1). During the past years, it has become increasingly evident that in addition to this signaling mode, protein O-phosphorylation, previously thought to be only important in eukaryotic signal transduction, occurs in various groups of Gram-negative and Gram-positive bacteria (2, 3) and is particularly widespread in cyanobacteria (4). Surveys of the 3.6-Mbp genome of_Synechocystis_ PCC 6803 revealed more than 20 putative serine/threonine and tyrosine-specific protein kinases (PK) and protein phosphatases (PP) (4, 5).
The mechanism of signal transduction through histidine (sensor) kinases and response regulators is highly conserved and the function and physiological role of various two-component systems is well established (6). By contrast, our knowledge of the molecular networks of bacterial signal transduction systems involving protein O-phosphorylation and their function in cell regulation is still preliminary (7). For example, stress response and sporulation in Bacillus subtilis involves a sophisticated network of sigma-factor/anti-sigma-factor regulation, which is controlled by protein O-phosphorylation (8). The icfG gene cluster in_Synechocystis_ PCC 6803 also has been shown to encode protein kinases and phosphatases homologous to the _Bacillus_sigma-factor regulators (9). Furthermore, a putative protein kinase is involved in motility of Synechocystis cells (10), and in the filamentous cyanobacterium Anabaena PCC7120, PK, and PP activities play a role in the differentiation of nitrogen-fixation-competent heterocysts (11).
Through the analysis of in vivo 32P-labeled proteins, a highly phosphorylated 13-kDa protein in the unicellular cyanobacterium_Synechococcus_ PCC 7942 could be identified as a homologue of the PII signal protein (12), which is a key regulator in nitrogen control in Escherichia coli (13). PII homologues are now known to be one of the most conserved signaling protein families in nature, and they have been subject to detailed studies in recent years (reviewed in refs. 14 and 15). In contrast to most bacterial PII proteins, which are reversibly uridylylated at a conserved tyrosine residue (Tyr-51) by a bifunctional uridylyltransferase/uridylyl-removing enzyme, the cyanobacterial PII protein is phosphorylated at seryl-residue 49 (16). Apart from this difference, biochemical studies have shown a remarkable degree of structural and functional conservation between the_Synechococcus_ PII protein and its proteobacterial counterpart (14, 17, 18). Both proteins are trimers, composed of 12.3-kDa polypeptides, and when coexpressed in E. coli the_Synechococcus_ and E. coli PII polypeptides form heterotrimers. Moreover, both proteins bind ATP and 2-oxoglutarate in a synergistic manner. In vivo, the phosphorylation state of the Synechococcus PII protein responds to the nitrogen and carbon supply to the cells (19). In the presence of excess ammonium, the trimeric PII is unphosphorylated (PII0), whereas in nitrogen-starved cells it is highly phosphorylated (two or three phosphorylated subunits, PII2 or PII3, respectively). In nitrate-grown cells, the phosphorylation state of PII is intermediate and increases with increasing CO2 supply, whereas inhibition of CO2 fixation leads to dephosphorylation of phospho-PII (PII-P). Through in vitro analysis of PII-kinase and phosphatase activities, we suggested that these activities are localized on different polypeptides, and that both PII phosphorylation and dephosphorylation are regulated by the metabolite-binding status of PII with its ligands 2-oxoglutarate and ATP (20). Moreover, we were able to characterize a Mg2+-dependent PII-P phosphatase activity in partially purified preparations of_Synechococcus_ PCC 7942 extracts (20).
According to catalytic and effector properties, the serine/threonine-specific protein phosphatases were originally grouped into four distinct families. On a structural basis, they comprise two superfamilies, the large PPP superfamily with phosphatases PP1, PP2A, and PP2B and the smaller PPM superfamily with the Mg2+/Mn2+-dependent protein phosphatase 2C (PP2C) family, characterized by 11 conserved signature motifs in the catalytic domain (21, 22). Previous inspections of the Synechocystis PCC 6803 genome revealed eight genes for prospective members of the PP2C family, but only one for a putative PPP-type phosphatase (4, 5). Because Synechocystis PCC 6803 possesses a highly homologous PII system to _Synechococcus_PCC 7942 (23, 24), we intended to create mutants in prospective PP encoding genes in order to identify PII-specific phosphatase(s). Here we describe the identification of a gene whose product is the cellular PII-P phosphatase of Synechocystis PCC 6803.
Materials and Methods
Culture Conditions.
Cells of Synechocystis sp. strain PCC 6803 T (25) with enhanced transformation efficiency were grown photoautotrophically at a constant illumination of 50 μmol photons s−1⋅m−2 and at 25°C in liquid BG11-medium (26) with nitrate as the nitrogen source. Aeration of the cultures was obtained by chicane flasks with foamed plastic caps, shaken at 100 rpm. For low-carbon conditions, cells were grown in Allan and Arnon's medium (26) at an irradiance of 30 μmol photons s−1⋅m−2 and the cultures were not stirred.
DNA and RNA Manipulations.
Manipulations and analyses of DNA were performed according to standard protocols (27). Restriction enzymes were purchased from New England Biolabs. Taq polymerase from Roche Diagnostics was used for PCR amplification of DNA. Total RNA extraction and Northern blot analysis were performed as described (28). A 762-bp fragment, containing the whole ORF of pphA, was used as a probe to identify pphA transcripts in 30 μg of total RNA loaded on each lane. The 5′ end of the pphA transcript was determined by 5′ rapid amplification of cDNA ends (RACE) assay and sequence analysis of the resulting PCR product by using the 5′/3′ RACE kit (Roche Diagnostics) under the conditions recommended by the supplier.
The primers used were as follows. Primer 1: 5′-GTACCCATATCCCGCCGTTCCA; primer 2: 5′-CATTGGCATCCATTAGTGCATCCC; primer 3: 5′-CCGAGGTAATCTCCGATTGCCAG.
Generation of Insertion Mutants.
The coding regions and neighboring sequences of selected genes were amplified by PCR and the ≈2-kbp fragments were cloned into suitable cloning vectors (Table 1). The sequences for designing the PCR primers were taken from the complete genomic sequence of Synechocystis PCC 6803 (1). The _aph_II gene (aminoglycoside 3′ phosphotransferase II) conferring kanamycin resistance (KmR) was inserted into unique restriction sites within the coding regions by using the kanamycin-resistance cartridges from pUC4K-derived vectors (Amersham Pharmacia). The constructs were checked by sequencing and were then used to transform Synechocystis cells (29). Transformants were first streaked on 30 μg/ml kanamycin-containing BG11 plates, which were solidified with Gelrite (Roth, Karlsruhe, Germany). Segregation of the mutation was achieved by restreaking two to three times on plates supplemented with 50 μg/ml of kanamycin. Liquid cultures of the mutants were supplemented with 50 μg/ml of kanamycin. Complete segregation of all mutants was checked by PCR and by Southern blot analysis.
Table 1.
Primers, vectors, and kanamycin resistance cartridges used to inactivate genes in _Synechocystis_PCC 6803
| ORF | Primer | Enzyme | Vector | KmRcassette | Insertion site |
|---|---|---|---|---|---|
| slr0114 | 5′; 5′-CTAAGCGGTACCGAATGTCC-3′ | KpnI | pBlueIIKS | KIND | NheI |
| 3′; 5′-GCTAACATGAGCTCCACGTC-3′ | SacI | ||||
| slr1983 | 5′; 5′-CATCTAGCCCGGGCATATCA-3′ | XmaI | pBlueIIKS | KAPA | EcoRI |
| 3′; 5′-CCAAGAAAATGGTACCAC-3′ | KpnI | ||||
| sll0602 | 5′; 5′-GGCCATGGTACCGCTTTTATCG-3′ | KpnI | pBlueIIKS | KAPA | ApaI |
| 3′; 3′-GCTAGCGGAGCTCTCATCTAAAC-3′ | SacI | ||||
| sll1033 | 5′; 5′-GGGGATGGTGTTCTAGAATTAGCG-3′ | XbaI | pUC19 | KISS | KpnI |
| 3′; 5′-CCTACCGAGCTCGTGGAAACAG-3′ | SacI | ||||
| sll1365 | 5′; 5′-GCTGGTACCATTGATTTTGCCC-3′ | KpnI | pBlueIIKS | KIXX | SmaI |
| 3′; 5′-CCTAGAGCTCTACGGCAGTG-3′ | SacI | ||||
| sll1771 | 5′; 5′-CGATCAAGCTTTTCCGTTTCC-3′ | HindIII | pUC19 | KISS | KpnI |
| 3′; 5′-TTACCACTGGCGGAGCTCTCAT-3′ | SacI | ||||
| sll1387 | 5′; 5′-GCCTACTGGCACCACCAAAA-3′ | HincII | pBlueIIKS | KIXX | BamHI |
| 3′; 5′-GGTCGAAACTTCCGCAAGCT-3′ | HindIII | ||||
| sll1329 | 5′; 5′-GCCATTCCCAAGGGAGAAAAAG-3′ | XmaI | pUC19 | KIXX | (SfiI) blunt/SmaI |
| 3′; 5′-GCACGTAGAGCAAACTGAATTC-3′ | SalI | ||||
| sll1770 | 5′; 5′-CCGGTACCGTGTTCACAGCGGGCATTATG-3′ | KpnI | pBlueIIKS | KIXX | SmaI |
| 3′; 5′-GCTCTAGACGTCCCTCTGGATCAAGGTA-3′ | XbaI |
Determination of the Phosphorylation State of PII.
The phosphorylation state of PII in vivo and in vitro was determined by nondenaturing gel electrophoresis followed by immunoblot analysis of PII as described (30). PII0, PII1, PII2, and PII3 correspond to the trimeric PII protein containing none, one, two, or three phosphorylated subunits, respectively.
Cloning and Purification of Overexpressed PphA Protein (sll1771 Gene Product).
The ORF sll 1771 of Synechocystis PCC 6803 was PCR amplified by using the following primer combination: 5′-CCA ATT TTC TAA GGC ATA TGA CA-3′ with an _Nde_I site and 5′-CTT CCC GGG TAC TAA ATC TAT-3′ with an _Sma_I site. After restriction with these enzymes, the fragment was cloned into _Nde_I- and _Sma_I-restricted vector pT7–7 vector (31) and the construct was transformed into E. coli_BL21 (DE3) cells. Overexpression of sll 1771 was induced by addition of 1 mM IPTG. After 4 h induction, cells from 1-l cultures were harvested by centrifugation. All subsequent steps were carried out at 0–4°C. The pellet was resuspended in 6 ml of lysis buffer (20 mM Tris⋅Cl, pH 7.4/50 mM KCl/5 mM MgCl2/0.5 mM EDTA/3 mM DTT/1 mM benzamidine/0.2 mM phenylmethylsulfonyl-fluoride) and the cells were broken by sonification. Cell debris and insoluble material was removed by three consecutive centrifugations (15 min at 10,000 ×_g = S10; 20 min at 30,000 × g = S30; 1 h at 100,000 × g = S100). Ammonium sulfate was added to the final supernatant to a saturation of 50%. The precipitate was collected by centrifugation, resuspended in 2 ml of buffer II (20 mM Tris⋅Cl, pH 7.4/50 mM KCl/5 mM MgCl2/0.5 mM EDTA/3 mM DTT/1 mM benzamidine), dialyzed against the same buffer, and applied to a 23-ml DEAE Sepharose Fast Flow column (Amersham Pharmacia) equilibrated in buffer II. Elution was performed with a 50-ml linear gradient from 50–550 mM KCl. PphA-containing fractions were pooled and were concentrated by ammonium sulfate precipitation (50% saturation). After resuspending the precipitate in 1.5 ml of buffer II, the proteins were dialyzed against the same buffer and applied to a 1.3-ml UnoQ anion exchange column (Bio-Rad). Proteins were eluted with a 35-ml linear gradient from 200–550 mM KCl. PphA-containing fractions were precipitated with ammonium sulfate and the pellet was resuspended in 2 ml of buffer III (20 mM Tris⋅Cl, pH 7.4/50 mM NaCl/5 mM MgCl2/0.5 mM EDTA/3 mM DTT/1 mM benzamidine/30% saturated with ammonium sulfate) and dialyzed against the same buffer. The solution was applied to a 5-ml Methyl HIC Econo-Pac cartridge (Bio-Rad). Elution was performed with a 50-ml linear gradient in buffer III from 30% to 0% ammonium sulfate saturation. PphA-containing fractions were pooled and precipitated with ammonium sulfate. The sediment was resuspended in 800 μl of buffer IV (20 mM Tris⋅Cl, pH 7.4/150 mM NaCl/5 mM MgCl2/0.5 mM EDTA/3 mM DTT/1 mM benzamidine) and applied to a Superdex 200 gelfiltration column (HiLoad 16/60, Amersham Pharmacia) equilibrated in buffer IV. Pure PphA, which eluted as a single peak, was dialyzed against buffer V (20 mM Tris⋅Cl, pH 7.4/50 mM NaCl/5 mM MgCl2/0.5 mM EDTA/50% glycerol) and stored at −20°C. To determine the Mg2+/Mn2+-dependence of PphA activity, purified PphA was dialyzed against buffer V without MgCl2 and stored as above.
In Vitro Assays of PphA Reactivity Toward PII-P.
Phosphorylated PII was purified as described (20) with the following modifications: (i) The cells were grown in modified BG11 medium (containing only 1 mM sodium nitrate) to an optical density (OD750) of 0.9, which caused initial nitrogen chlorosis, and thus, a maximal degree of PII phosphorylation (28); (ii) the heparin Econo-Pac cartridge was developed with a 200-ml gradient. The PII protein obtained was highly phosphorylated with almost exclusively the form PII3 and a small amount of PII2. To assay the reactivity of PphA toward PII-P, 14 ng of phosphorylated PII was incubated with the indicated amount of PphA in 10 μl of 10 mM Tris⋅Cl, pH 7.4, 50 mM NaCl, 1 mM DTT, 10 μg BSA, 0.05% Nonidet P-40, and MgCl2 or MnCl2, as indicated. The reaction mixtures were incubated at 37°C; after a certain time, the reactions were stopped by chilling on ice followed by the addition of ice-cold native gel-loading buffer (30). Subsequently, the status of PII phosphorylation was determined as described above.
Results
Inactivation of Protein Phosphatase-Encoding Genes in_Synechocystis_ PCC 6803.
Of the eight ORFs encoding putative PP2C-type phosphatases, six (slr0114, slr1983, sll0602, sll1033, sll1365, and sll1771) were inactivated by gene-disruption through the integration of a kanamycin-resistance cartridge (apaII) in the ORF as described in Materials and Methods. A slr2031 mutant (slr2031 encodes the PP2C-type phosphatase RsbU-homologue) was provided by M. Hagemann (32) and a slr1860 (icfG) mutant was investigated in the lab of C. C. Zhang (Universite Aix-Marseille II, Marseille, France). In addition to the PP2C homologues, ORF sll1387, encoding a putative PPP-type phosphatase, and ORF sll1329, encoding a monophosphatase homologue, were inactivated by using the same procedure. Complete segregation of the mutant alleles could be achieved in all but the slr1983 construct; this strain was therefore omitted from the following investigation. The other mutants (slr0114, sll0602, sll1033, sll1365, sll1771, slr2031, sll1387, and sll1329) were analyzed for their ability to dephosphorylate phospho-PII. For this purpose, cells were grown in nitrate-supplemented medium under photoautotrophic conditions, which results in an intermediate phosphorylation state of PII (19). To exponentially growing cells, 5 mM ammonium chloride (final concentration) was added and after different times, the phosphorylation state of PII was analyzed by immunoblot analysis of nondenaturing gels. In all but the mutant in sll1771, dephosphorylation of PII-P was unaffected (data not shown).
PII-P Dephosphorylation in sll1771-Mutant Cells.
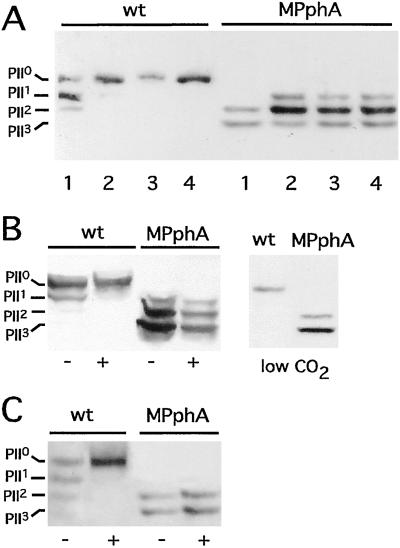
Disruption of ORF sll1771 resulted in a mutant that was unable to dephosphorylate PII. Fig. 1 shows the results obtained with the mutant, in which aphII was inserted in the opposite orientation to sll1771 (the mutant with_aphII_ in the same direction yielded identical results). Wild-type cells completely dephosphorylate PII-P within 5 min after ammonium shock, whereas the phosphorylation state of PII in the sll1771 mutant remained high, and even after 30 min no unphosphorylated PII appeared. Note that the mutant already displays a significantly higher degree of PII phosphorylation in untreated nitrate-grown cells. Other conditions known to cause PII-P dephosphorylation in nitrate-grown cells are low CO2 supply, inhibition of CO2 fixation (19), or oxidation of the photosystem I acceptor site by methylviologen treatment, which was suggested to generate a redox signal that acts on PII (24). In contrast to the wild type, the sll1771 mutant retains a high degree of PII phosphorylation under all conditions (Fig. 1). These results implied that sll1771 encodes a phosphatase that is essential for dephosphorylation of PII-P not only in response to ammonium treatment but also to carbon or redox signals. Because this is the first protein phosphatase in Synechocystis PCC 6803 for which a precise_in vivo_ function can be assigned, we termed the gene_pphA_ (_p_rotein ph_osphatase_A) and the corresponding mutant strain MPphA.
Figure 1.
Phosphorylation state of PII in wild-type (wt) and sll1771 inactivated (MPphA) cells of Synechocystis PCC6803. (A) Response to ammonium treatment; cells were first grown in BG11-medium (lane 1), then NH4Cl was added (5 mM final concentration) and after 5 min (2), 15 min (3), and 30 min (4) aliquots were removed and the phosphorylation state of PII was analyzed. (B) Response to different carbon conditions: (Left) Cells grown in BG11-medium were treated with an inhibitor of CO2-fixation,d,l-glyceraldehyde (d,l-GA). Negative control (−) and after 5 min in the presence of 30 mM d,l-GA (+); (Right) cells grown under CO2-poor conditions in Allan and Arnon's medium, which is devoid of carbonate and bicarbonate. (C) Response toward methylviologen (MV): to BG11-grown cells 0.1 mM MV was added and, after 5 min, the phosphorylation state of PII was analyzed.
Chromosomal Localization of sll1771 (pphA).
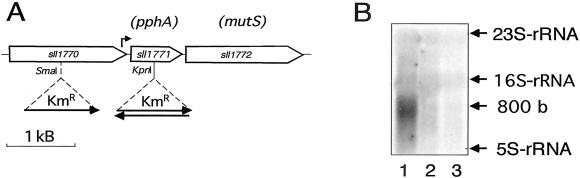
The pphA gene is flanked by two ORFs that are transcribed in the same orientation (Fig.2A). ORF sll1770 is localized upstream of pphA, spaced by a 54-bp intergenic region, and ORF sll1772 (encoding an homologue of the DNA-repair enzyme MutS) starts 29 bp downstream of the pphA stop codon. To elucidate whether pphA is transcribed in an operon with one of its flanking genes, Northern blot analysis was carried out with a_pphA_-specific probe. As shown in Fig. 2B,Synechocystis wild-type cells display a signal at about 800 nucleotides, which is absent in the pphA_-inactivated mutants. This size is in good agreement with that expected for a monocistronic pphA transcript. The 5′ end of the_pphA transcript was analyzed by a 5′ rapid amplification of cDNA ends (RACE) assay, which yielded “A −121” relative to the GTG start codon as a possible transcriptional start site. A putative −10 promoter box (TATTAT) is located at the canonical 7-bp distance from the probable 5′ end. Furthermore, a hexamer (TTGTAA) resembling the −35 promoter box can be found 27 bp upstream of the −10 element. A 29-bp spacing between the −10 and −35 boxes has been described in_Synechocystis_ promoters recently (33). From this it appears that pphA is transcribed monocistronically from a promoter, which is located within the 3′ end of the preceding ORF sll1770. Interestingly, the predicted sll1770 product has been described as a putative protein kinase in a search for “eukaryote-like” protein serine/threonine kinases in prokaryotes (4). To test whether the sll1770 product may be a PII kinase, a mutant was created in which this gene was disrupted (Fig. 2A). After complete segregation of the mutant allele, phosphorylation of PII was analyzed in nitrogen-starved cells of the mutant. However, the mutant could still phosphorylate PII (data not shown).
Figure 2.
(A) Organization of the DNA region containing sll1770, sll1771, and sll1772 according to the gene map from the Cyanobase database (1) and position of the inserted kanamycin resistance cassette (KmR) in mutants of sll1770 and sll1771. Mutants in sll1771 were generated with the KmR cassette in both orientations, indicated by the orientation of the arrows. The bent arrow indicates the putative transcriptional start site, as deduced from 5′ RACE assay. (B) Northern blot analysis of_pphA_-transcripts from Synechocystis PCC 6803 (lane 1) and the sll1771 mutants with the KmR cassette in the same orientation (lane 2) or opposite (lane 3) to_pphA_. RNA was prepared from nitrate-grown cells and was probed with the pphA gene.
pphA Encodes a PP2C Homologue.
The deduced gene product of pphA contains 254 aa, comprising a polypeptide with a calculated molecular mass of 28.3 kDa. This size corresponds to that of the catalytic domain of PP2C phosphatases, with its 11 conserved motifs (22) scattered over the PphA sequence (4). A BLASTP homology search revealed the highest degree of amino acid identity with hypothetical proteins from other cyanobacterial genomes (Anabaena sp. PCC 7120 ORF 2136 and_Nostic punctiforme_ ORF 3387) and with a hypothetical protein from Deinococcus radiodurans (55%, 54%, or 42% amino acid identity, respectively). Furthermore, 30 other putative PP2C homologues, which are found in various bacterial genomes, show clear similarities throughout their sequence (summarized in ref. 34). By contrast, the similarity to the eukaryotic PP2C homologues and to the previously characterized B. subtilis PP2C homologues SpoIIE (8) and relatives is restricted to highly conserved signature sequences (4). A phylogenetic tree of the _Synechocystis_PP2C-homologues was constructed, which shows that most of these (the products of slr0114, slr1860, slr1983, slr2031, and sll1365) belong to the SpoIIE-class of PP2C-homologues, whereas pphA is a member of the bacterial PP2C-phosphatases, designated “group I” in ref. 34 (Fig. 3).
Figure 3.
Phylogenetic analysis of PphA and the prospective serine/threonine protein phosphatases of Synechocystis PCC 6803 together with selected PP2C members from bacterial and eukaryotic origin. The predicted size (amino acids) of the proteins is given in brackets. Sequence homology searches were carried out by using the BLAST analysis (37). The dendrogram was constructed with the programs PHYLIP Version 3.572 and CLUSTALX (38). Accession nos. of the sequences (from top to bottom): BAB08417;P35813; BAA10651; BAA18661; L35574; BAA17283; P37979; BAA18228; P37475;BAA10367; CAB65437; BAA16771; AAF73659; E71538; AF223364; BAB06224;G69878; CAB60748; CAA10712; RAN02136; RNPU03387; BAA17671; H75265;A83438.
In Vitro Reactivity of PphA Toward Phospho-PII.
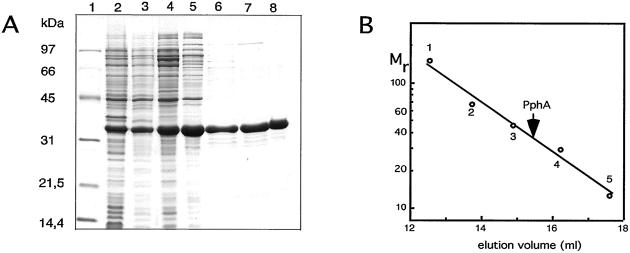
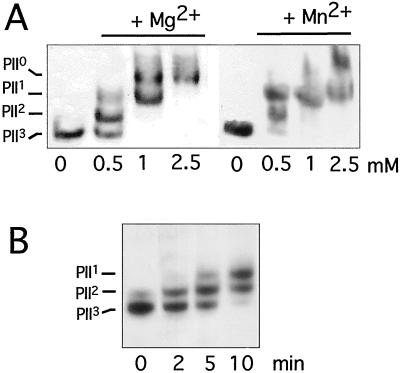
To test the reactivity of PphA toward PII-P in vitro, the_pphA_ gene was cloned into the E. coli expression plasmid pT7–7, which overexpresses the cloned gene devoid of a tagging sequence and avoids a possible interference of foreign sequences in PphA with PII-P recognition. After overproduction of native PphA in_E. coli_, the protein was purified to near homogeneity by conventional chromatographic procedures (Fig.4A). The purified protein migrates with a size of 33 kDa in gel-filtration columns, suggesting that it is monomeric (Fig. 4B). Purified PphA was incubated under various conditions with highly phosphorylated PII protein. The phosphorylation state of PII in the reaction mixtures was determined by immunoblot analysis of nondenaturing PAGE. PphA displays a Mg2+-dependent reactivity toward PII3 and is able to dephosphorylate it completely. Reactivity toward phospho-PII increases with increasing Mg2+ concentration and saturates at about 2.5 mM. Mn2+ has a similar effect than Mg2+ on PII dephosphorylation (Fig.5A). No reactivity of PphA toward _p_-nitrophenylphosphate, a frequently used standard substrate for a wide range of phosphatases, could be detected by using the PII-Pase assay conditions (data not shown). From kinetic experiments, as shown in Fig. 5B, it can be estimated that at a PII concentration of 38 nM (trimers), the turn-over rate of PphA is ≈1.3 PII-P monomers per minute. The reaction can be inhibited by the broad-range phosphatase inhibitor NaF and the specific inhibitor of protein phosphatase 2C, EDTA, but is insensitive toward okadaic acid, which is a potent inhibitor of the PPP family members PP1 and PP2A (not shown).
Figure 4.
(A) Purification of overproduced PphA shown by Coomassie-blue-stained 12.5% SDS/PAGE. The lanes correspond to the different purification steps as described in Materials and Methods. 1, molecular weight marker; 2, S10; 3, S100; 4, first (NH4)2SO4 precipitate; 5, DEAE Sepharose Fast Flow-pool; 6, UnoQ-pool; 7, Methyl HIC-pool; 8, Superdex 200-pool. (B) Determination of the native molecular size of PphA by gel-permeation chromatography. PphA and proteins used as size standards (1, alcohol dehydrogenase, 150 kDa; 2, BSA, 67 kDa; 3, ovalbumin, 43 kDa; 4, carbonic anhydrase, 29 kDa; 5, cytochrome C, 12.4 kDa) were passed separately through a Superdex S200 h 10/30 column (Amersham Pharmacia) equilibrated in 20 mM Tris⋅Cl (pH 7.4), 150 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, 2 mM DTT, 1 mM benzamidine, and 1 mM ɛ-aminocaproic acid.
Figure 5.
In vitro reactivity of PphA toward purified phosphorylated PII. (A) Mg2+/Mn2+ dependence of PII-P dephosphorylation. Mg2+/Mn2+-free PphA (10 ng) was incubated with PII-P in the presence of 0, 0.5, 1, or 2.5 mM MgCl2 or MnCl2 as indicated. After 30 min the phosphorylation state of PII was determined. (B) Time course of in vitro PII-P dephosphorylation. Standard reactions (with 10 mM MgCl2) were started by the addition of 3 ng PphA. After 2, 5, and 10 min the reactions were stopped and analyzed.
Discussion
Here we report the identification and initial characterization of the phosphatase responsible for dephosphorylation of the PII signaling protein in the cyanobacterium Synechocystis PCC 6803. The phosphatase, which we designate PphA, is the product of ORF sll1771 in the genome of Synechocystis PCC 6803. It contains the 11 conserved motifs of the catalytic domain of the PP2C family (4, 22) and displays a Mg2+- or Mn2+-dependent enzymatic activity typical for PP2C phosphatases. Because complete segregation of the mutant allele could be achieved for all ORFs encoding PP homologues except for slr1983, it appears that the corresponding phosphatases are not essential to the Synechocystis cells under laboratory conditions. None of the phosphatase mutants displayed a significant growth deficiency in terms of growth rate and cell yields in BG11 medium. These gene products may be required for specific stress conditions or for fine-tuning and optimization of cellular processes, which provide no apparent growth advantage under noncompetitive laboratory conditions. Several phosphatases may be functionally redundant, so that single knockout mutations would not reveal a phenotype. PphA appears to be the only phosphatase in the_Synechocystis_ cell able to dephosphorylate PII-P; a mutation in sll1860 (icfG) does not impair dephosphorylation of PII either (C. C. Zhang and S. Bedut, personal communication). Future studies will address the basis of the specificity of PII-P dephosphorylation by PphA.
The nature of the PII kinase remains elusive: the putative kinase gene sll1770, which is located upstream of, but not cotranscribed with_pphA_, is not essential for PII phosphorylation. Either there are multiple PII kinases that are functionally redundant, or the gene for a specific PII kinase is located distantly on the chromosome.
In addition to nitrogen and carbon signals, redox-signals also were shown to elicit PII dephosphorylation in Synechocystis PCC 6803 (24). So far, it has not been clear whether the different stimuli are transmitted through separate signaling pathways to PII. The fact that PphA is required for PII dephosphorylation under all conditions suggests that the different signals have to converge at the level of the PphA–PII-P interaction. With a size of 28.5 kDa, PphA is just as large as the catalytic domain of a PP2C phosphatase and lacks sequences for a putative regulatory domain. PphA shares the small size with most of the thirty closely related bacterial PP2C homologues. Lacking a regulatory domain, it is unclear how their activity is regulated. Recently, three of these related gene products, Pseudomonas aeruginosa Stp1 (35), Myxococcus xanthus Pph1 (34), and_Bacillus subtilis_ PrpC (36) were shown _in vitro_to display Mg2+/Mn2+-dependent activities toward standard phosphorylated assay substrates. A recent investigation of the related phosphatase Pph1 from Myxococcus xanthus suggested that these newly discovered bacterial PP2C homologues, which by phylogenetic analysis group close to the root, should be classified as a distinct phylogenetic group (compare Fig. 4; ref. 8). PphA would be the first member of this group of PP2C phosphatases for which the physiological substrate is known.
It has been widely assumed that the occurrence of eukaryote-like protein phosphatases and kinases in bacteria is related to complex cellular processes in these organisms, like sporulation or formation of fruiting bodies. However, as shown by PII phosphorylation, these enzymes may also be involved in the regulation of central metabolic processes. Signaling by the chemically stable protein-O-phosphorylation mode, as opposed to the chemically labile protein phosphorylation mode in bacterial two-component systems, might be favorable when signal integration over prolonged time periods is required.
Acknowledgments
We thank Dr. A. Treuner-Lange for expert help with the computational analysis and for helpful discussions, Dr. G. Sawers for critically reading the manuscript, and Dr. C. C. Zhang for communicating his unpublished results. Excellent technical assistance by Ulrike Ruppert and Noelia Pohl is gratefully acknowledged. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (Fo 195/2 and Fo 195/4).
Abbreviation
PP
protein phosphatases
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 2.Kenelly P, Potts M. J Bacteriol. 1996;178:4759–4764. doi: 10.1128/jb.178.16.4759-4764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang C C. Mol Microbiol. 1996;20:9–15. doi: 10.1111/j.1365-2958.1996.tb02483.x. [DOI] [PubMed] [Google Scholar]
- 4.Shi L, Potts M, Kenelly M. FEMS Microbiol Rev. 1998;22:229–253. doi: 10.1111/j.1574-6976.1998.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 5.Zhang C C, Gonzalez L, Phalip V. Nucleic Acids Res. 1998;26:3619–3625. doi: 10.1093/nar/26.16.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stock A M, Robinson V L, Goudreau P N. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 7.Bakal C J, Davies J E. Trends Cell Biol. 2000;10:32–38. doi: 10.1016/s0962-8924(99)01681-5. [DOI] [PubMed] [Google Scholar]
- 8.Adler E, Donella-Deana A, Arigoni F, Pina L A, Stragier P. Mol Microbiol. 1997;23:57–62. doi: 10.1046/j.1365-2958.1997.1801552.x. [DOI] [PubMed] [Google Scholar]
- 9.Shi L, Bischoff K M, Kennelly P J. J Bacteriol. 1999;181:4761–4767. doi: 10.1128/jb.181.16.4761-4767.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamei A, Yuasa T, Orikawa K, Geng X X, Ikeuchi M. J Bacteriol. 2001;183:1505–1510. doi: 10.1128/JB.183.5.1505-1510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang C C, Friry A, Peng L. J Bacteriol. 1998;180:2616–2622. doi: 10.1128/jb.180.10.2616-2622.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsinoremas N F, Castets A M, Harrison M A, Allen J F, Tandeau de Marsac N. Proc Natl Acad Sci USA. 1991;88:4565–4569. doi: 10.1073/pnas.88.11.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stadtman E R. Methods Enzymol. 1990;182:793–809. doi: 10.1016/0076-6879(90)82062-7. [DOI] [PubMed] [Google Scholar]
- 14.Ninfa A J, Atkinson M R. Trends Microbiol. 2000;8:172–179. doi: 10.1016/s0966-842x(00)01709-1. [DOI] [PubMed] [Google Scholar]
- 15.Arcondeguy T, Jack R, Merrick M. Microbiol Mol Biol Rev. 2001;65:80–105. doi: 10.1128/MMBR.65.1.80-105.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forchhammer K, Tandeau de Marsac N. J Bacteriol. 1995;177:5812–5817. doi: 10.1128/jb.177.20.5812-5817.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forchhammer K, Hedler A. Eur J Biochem. 1997;244:869–875. doi: 10.1111/j.1432-1033.1997.00869.x. [DOI] [PubMed] [Google Scholar]
- 18.Forchhammer K, Hedler A, Strobel H, Weiss V. Mol Microbiol. 1999;33:338–349. doi: 10.1046/j.1365-2958.1999.01477.x. [DOI] [PubMed] [Google Scholar]
- 19.Forchhammer K, Tandeau de Marsac N. J Bacteriol. 1995;177:2033–2040. doi: 10.1128/jb.177.8.2033-2040.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irmler A, Sanner S, Dierks H, Forchhammer K. Mol Microbiol. 1997;26:81–90. doi: 10.1046/j.1365-2958.1997.5521918.x. [DOI] [PubMed] [Google Scholar]
- 21.Cohen P T W. Adv Protein Phosphatases. 1994;8:371–376. [Google Scholar]
- 22.Bork P, Brown N P, Hegyi H, Schulz J. Protein Sci. 1996;5:1421–1425. doi: 10.1002/pro.5560050720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Dominguez M, Florencio J F. Plant Mol Biol. 1997;35:723–734. doi: 10.1023/a:1005846626187. [DOI] [PubMed] [Google Scholar]
- 24.Hisbergues M, Jeanjean R, Joset F, Tandeau de Marsac N, Bedut S. FEBS Lett. 1999;463:216–220. doi: 10.1016/s0014-5793(99)01624-5. [DOI] [PubMed] [Google Scholar]
- 25.Griegorieva G, Shestakov S V. FEMS Microbiol Lett. 1982;13:367–370. [Google Scholar]
- 26.Rippka R. Methods Enzymol. 1988;167:3–27. doi: 10.1016/0076-6879(88)67004-2. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 28.Sauer J, Görl M, Forchhammer K. Arch Microbiol. 1999;172:247–255. doi: 10.1007/s002030050767. [DOI] [PubMed] [Google Scholar]
- 29.Golden S S, Brusslaan J, Haselkorn R. Methods Enzymol. 1987;153:215–231. doi: 10.1016/0076-6879(87)53055-5. [DOI] [PubMed] [Google Scholar]
- 30.Forchhammer K, Tandeau de Marsac N. J Bacteriol. 1994;176:85–91. doi: 10.1128/jb.176.1.84-91.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tabor S, Richardson C C. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huckauf J, Nomura C, Forchhammer K, Hagemann M. Microbiology. 2000;146:2877–2889. doi: 10.1099/00221287-146-11-2877. [DOI] [PubMed] [Google Scholar]
- 33.Figge R M, Cassier-Chauvat C, Chauvat F, Cerff R. Mol Microbiol. 2000;36:44–54. doi: 10.1046/j.1365-2958.2000.01806.x. [DOI] [PubMed] [Google Scholar]
- 34.Treuner-Lange A, Ward M J, Zusman D R. Mol Microbiol. 2001;39:1–16. doi: 10.1046/j.1365-2958.2001.02362.x. [DOI] [PubMed] [Google Scholar]
- 35.Mukhopadhyay S, Kapatral V, Xu W, Chakrabarty A M. J Bacteriol. 1999;181:6615–6622. doi: 10.1128/jb.181.21.6615-6622.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Obuchowski M, Madec E, Delattre D, Boel G, Iwanicki A, Foulger D, Seror S J. J Bacteriol. 2000;182:5634–5638. doi: 10.1128/jb.182.19.5634-5638.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul S F, Madden T L, Schaeffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]