Viral Late Domains (original) (raw)
Domains have been identified in the Gag proteins of a number of retroviruses, and in the matrix proteins of the rhabdoviruses and filoviruses, that play a critical role in the pinching off of virus particles from the plasma membrane. These sequences are collectively termed late or L domains to reflect their function late in the virus budding process. One of the intriguing features of these L domains is that they all contain highly conserved motifs known to mediate protein-protein interactions between cellular proteins. Three classes of motifs have been defined in viral L domains: PTAP, PPXY, and YXXL. In each case, the integrity of these motifs appears to be essential for L domain activity, suggesting that L domains function by interacting with a host factor(s). This minireview summarizes the present state of our knowledge concerning viral L domain function and describes recent tantalizing clues regarding the identity of the cellular partners with which L domains interact.
Retroviral Gag proteins are synthesized as polyprotein precursors that are cleaved by the viral protease (PR) during or shortly after release from the cell. Although sequence conservation between the Gag proteins of different retroviral genera is very limited, all retroviruses, with the exception of the spumaretroviruses, encode three major Gag domains: matrix (MA), capsid (CA), and nucleocapsid (NC) (for reviews, see references 15, 16, and 74). In addition to these well-characterized domains, which are invariably ordered MA-CA-NC in the Gag precursor, a variety of additional domains and spacer peptides with diverse functions are encoded by the gag genes of individual retroviruses. Expression of retroviral Gag precursor proteins is both necessary and sufficient for the assembly and release of noninfectious, virus-like particles; Gag processing by PR is not required for particle production.
Retroviruses have evolved several mechanisms for particle assembly and release (for reviews, see references 21 and 74). For those viruses that use the so-called C-type pathway (e.g., alpharetroviruses such as Rous sarcoma virus [RSV], gammaretroviruses such as murine leukemia virus [MLV], and lentiviruses such as human immunodeficiency virus type 1 [HIV-1]), particle assembly occurs at the plasma membrane. In contrast, betaretroviruses (e.g., mouse mammary tumor virus and Mason-Pfizer monkey virus [M-PMV]) use the B/D-type pathway, in which particles assemble in the cytosol and are then transported to the plasma membrane. For both C- and B/D-type retroviruses, budding occurs from the plasma membrane and L domains have been defined for both classes of retroviruses. Intracytoplasmic A-type particles and spumaretroviruses bud primarily at intracellular membranes and will not be discussed further, since L domains for these viruses have not been characterized. Regardless of the assembly pathway, virus budding from the plasma membrane requires a membrane fusion or pinching-off event. As is the case for the membrane fusion step in virus entry, this fusion reaction likely involves a complex interplay between viral and host factors.
IDENTIFICATION OF L DOMAINS BY MUTAGENESIS
HIV-1: the PTAP prototype.
In 1991, Gottlinger and colleagues reported that deletion of the 6-kDa C-terminal domain of HIV-1 Gag caused a marked defect in virus particle production in transfected COS cells (22). Intriguingly, electron microscopy (EM) indicated that the mutant particles failed to detach from virus-expressing cells but rather remained attached to the plasma membrane by a thin tether. These observations suggested that p6 plays a critical role in virus release. Subsequent studies, using a variety of cell lines and Gag expression systems, failed to observe a virus release defect upon p6 truncation (33, 35, 39, 46, 58, 63, 68) (see below). However, by using a full-length molecular clone expressed in human (HeLa) cells, the requirement for p6 in virus release was eventually confirmed (36). A highly conserved Pro-Thr-Ala-Pro (PTAP) motif located near the N terminus of p6 was identified as playing a crucial role in the L domain activity of p6 (36). Even subtle mutations in this motif cause a severe defect in virus particle budding in HeLa cells, whereas mutations outside this small sequence are well tolerated (13, 36). The L domain function of p6 does not depend on residues toward the C terminus of p6, as truncation four residues downstream of the PTAP motif has no effect on virus release in HeLa cells (13).
RSV: the PPXY prototype.
A detailed mutational analysis of the RSV Gag protein revealed that the small peptide p2b, located between MA and p10 (Fig. 1), plays an important role in virus particle production (80, 82). This activity maps to the Pro-rich motif Pro-Pro-Pro-Tyr (PPPY). Subsequently, PPPY motifs in other retroviruses, namely in pp16 of M-PMV (84) and in p12 of MLV (86, 87), were demonstrated to be required for efficient budding. In the M-PMV and MLV studies (84, 86), EM analyses revealed a tethering defect similar to that previously observed for HIV-1 p6 mutants (22, 36).
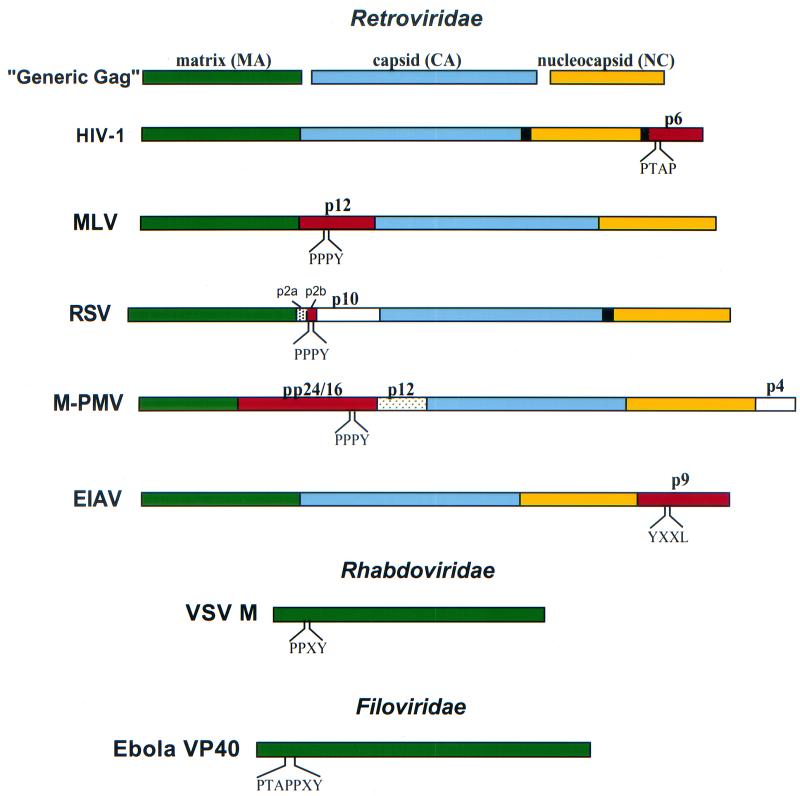
FIG. 1.
Location of L domains in retroviral Gag proteins and in the matrix proteins of the rhabdoviruses and filoviruses. The retroviral Gag proteins that harbor L domains are shown in red; matrix, capsid, and nucleocapsid proteins are depicted in green, blue, and yellow, respectively. L domain motifs (PTAP, PPXY, and YXXL) are indicated.
The matrix proteins of members of both the Rhabdoviridae (e.g., vesicular stomatitis virus [VSV]) and Filoviridae (e.g., Ebola virus), which are known as M and VP40, respectively, play central roles in the assembly and release of these viruses (for reviews, see references 62 and 64). Interestingly, a PPXY motif in the rhabdovirus M protein was demonstrated to possess L domain activity (10, 26, 37). The Ebola virus VP40 protein has overlapping PTAP and PPXY motifs (sequence PTAPPXY; Fig. 1); both motifs appear to contribute to particle release (25, 47). These findings suggest that highly divergent enveloped viruses use a similar strategy to bud off from their host cells.
EIAV: requirement for a YXXL motif.
To address the question of whether the C-terminal domain of equine infectious anemia virus (EIAV) Gag, like that of HIV-1, encodes an L domain, Parent et al. (55) introduced the entire EIAV p9 coding region (Fig. 1) into an RSV Gag deletion mutant lacking p2b. Remarkably, p9 was able to provide L domain activity to the L domain-deficient RSV clone. Subsequent mutational analysis (59) mapped the p9 L domain not to a Pro-rich motif but rather to a Tyr-X-X-Leu (YXXL) sequence.
A diversity of phenotypes.
As mentioned above, early attempts to define the role of the HIV-1 L domain in virus release were complicated by the fact that a number of studies failed to observe a requirement for p6 in efficient virus production (33, 35, 39, 46, 58, 63, 68). Although the reasons for these disparate findings are not entirely clear, a variety of factors appear to influence HIV-1 L domain function. (i) Several of the studies that did not find a role for p6 in virus release used high-level vaccinia virus and baculovirus expression systems in which Gag is synthesized at superphysiological concentrations. It has been observed in 293T cells that by using a high-efficiency transfection procedure an effect of p6 deletion on virus release is not observed, whereas with a low-efficiency transfection method p6 mutations markedly reduce virus production (H. Göttlinger, personal communication). Thus, very high levels of Gag expression per cell can “swamp out” the requirement for p6 in virus budding, perhaps by driving the release of tethered chains of virus particles. Consistent with this hypothesis, RSV PPPY mutants do not display a virus release defect in the baculovirus overexpression system (V. Vogt and M. Johnson, personal communication). (ii) Huang et al. (36) observed that the effect of p6 mutation on virus release was less pronounced in the context of a molecular clone lacking a functional PR. These results suggested a relationship between PR activity and p6 L domain function. This observation is relevant to the differences in p6 phenotypes mentioned above, since in many of the studies that failed to observe a role for p6 in virus budding, only Gag was expressed. It should be noted, however, that the presence of an active PR is not an absolute requirement for p6-induced stimulation of virus release (1). (iii) Demirov et al. performed a detailed characterization of the phenotype of p6 mutations in diverse cell types (13). These authors observed that while p6 mutations significantly impaired virus release in adherent cell lines (e.g., HeLa, 293T, COS-7, and CV-1) and in primary human macrophages, the mutations had little or no effect on the efficiency of virus production in T-cell lines and in primary T cells. However, EM analysis revealed that p6-mutant virus produced from T cells exhibited a defect in virion-virion detachment such that long chains of tethered particles were produced (13) (Fig. 2). Thus, although L domain mutations induce a defect in membrane detachment in a wide diversity of cell types, including those that are relevant for HIV-1 infection in vivo (i.e., primary lymphocytes and macrophages), the manner in which this defect is manifested differs between cell types. Interestingly, heterokaryons formed between HeLa cells and a T-cell line displayed the T-cell phenotype with respect to the release of p6 mutants (13).
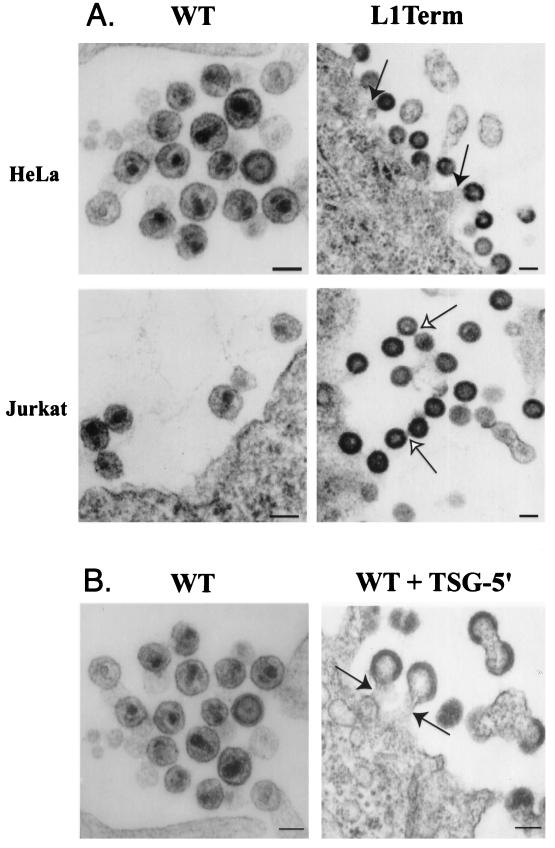
FIG. 2.
EM analysis of HIV-1 L domain defects. (A) Effect of HIV-1 L domain mutation on virus budding. Virus expression was directed by WT pNL4-3 and a p6 mutant derivative truncated at p6 amino acid 1 (L1Term [36]). Budding from HeLa cells and the Jurkat T-cell line is shown. Adapted from reference 13. (B) Effect of TSG-5′ overexpression on HIV-1 budding. HeLa cells were transfected with pNL4-3 alone (WT) or cotransfected with pNL4-3 plus TSG-5′ (WT + TSG-5′). Adapted with permission from reference 12. In panels A and B, closed arrows indicate virus particles tethered to the plasma membrane; open arrows highlight virion-virion tethers. Bar, 100 nm.
In addition to promoting virus release, several additional activities have been attributed to L domain-containing retroviral Gag proteins. The effects of HIV-1 p6 mutations seem to be particularly pleiotropic. Several reports demonstrated that p6 L domain mutations caused defects in Gag processing (13, 22, 36) and reduced levels of the _pol_-encoded enzymes (i.e., reverse transcriptase and integrase) in virions (13, 14, 85). The increased levels of Gag processing intermediates and the reduced virion-associated reverse transcriptase and integrase are particularly striking when p6-mutant virions are produced at high levels (i.e., from T-cell lines) (13). As a result of the Gag processing defect, p6-mutant virions released by T-cell lines exhibit a predominantly immature morphology (Fig. 2) (13). Truncations in p6 that have no effect on virus release in HeLa cells can block virus replication in T-cell lines (13). It has also been reported that p6 L domain mutations increase virion size, as determined by velocity gradient centrifugation (17, 18); normal size is restored by introducing the L domains of RSV and EIAV (17). This apparent increase in particle size appears to result from the virion-virion tethering phenotype mentioned above (R. Craven, personal communication). Finally, p6 possesses a function not associated with its role in virus release: a Leu repeat motif near the C terminus of p6 is essential for the incorporation of the accessory protein Vpr into virus particles (38, 42, 58). p6 is also required for the packaging of the HIV-2/simian immunodeficiency virus SIVsm/SIVmac accessory protein Vpx into virions (81).
The L domain-containing proteins of other retroviruses also appear to serve multiple functions in the virus replication cycle. Mutations in the p12 protein of MLV not only reduce virus budding but also impair Gag processing and virus infectivity (87). In addition, PR-mediated cleavage of the cytoplasmic tail of the MLV envelope glycoprotein is inhibited by p12 mutations, again consistent with a defect in PR activity (87). Remarkably, insertion of peptides containing the RSV and HIV-1 L domains restored not only virus release but also infectivity to p12 mutants lacking the PPPY motif (86). As observed with HIV-1 (13), truncations have been reported in EIAV p9 that block virus replication without significantly affecting virus release (7).
Intriguingly, a number of L domain-containing proteins, including those of M-PMV (6), MLV (67), HIV-1 (49), and VSV (9) are phosphorylated. At this time, a role for phosphorylation in L domain activity remains to be defined.
HOST FACTORS AND L DOMAIN FUNCTION
Evidence of a role for host factors.
As L domains were identified and characterized, several observations suggested that these domains might function by interacting with host factors. For example, L domains contain motifs that are crucial for protein-protein interactions between cellular proteins. Also, L domains are quite small and can in some cases still function when moved from one virus to another and when placed at nonnative positions within the Gag precursor. Their small size, interchangeability, and positional independence are consistent with L domains functioning as modules that bind and recruit host factors to the site of budding.
(i) Dependence on protein-protein interaction signals.
The three types of motifs that play central roles in L domain activity (PXXP, PPXY, and YXXL) are well-recognized protein-protein interaction modules. PXXP motifs in a diversity of proteins interact with Src homology 3 (SH3) domains (for reviews, see references 11 and 48). SH3 domains, which are in the range of 50 to 70 amino acids long, are found in dozens of proteins that function as cytoskeletal elements, GTPases, lipases, phosphatases, and adapter proteins. Two classes of SH3-binding motifs have been identified; these are referred to as class I (consensus Arg-X-X-Pro-X-X-Pro) and class II (consensus Pro-X-X-Pro-X-Arg). The L domain of HIV-1 p6 fits the class I consensus; however, the Arg at the N terminus of this motif (amino acid 4 of HIV-1 p6) is dispensable for virus budding (13, 36). Thus, either the p6 virus budding function does not involve SH3 domain binding or the p6 L domain constitutes a novel class of SH3-binding element.
PPXY sequences comprise a class of Pro-rich ligands distinct from PXXP motifs. These motifs interact with proteins containing WW domains, so named because they contain two Trp residues separated by approximately 40 amino acids (reviewed in reference 72). Frequently found in multiple copies per protein, WW domains were initially identified in the Yes-associated protein Yap, which interacts with the product of the yes oncogene during signaling (8). WW domains have since been identified in a number of proteins that perform a wide variety of functions.
The YXXL motif, which is found in the EIAV L domain, has been shown to interact with the medium-chain subunits of the heterotetrameric, clathrin-associated adapter protein complexes AP-1 and AP-2 (50). AP-1 and AP-2 are associated with clathrin-coated vesicles in the trans-Golgi network and at the plasma membrane, respectively. YXXL motifs are frequently located in the cytoplasmic tails of proteins and function in sorting and endocytosis (50).
(ii) Functional interchangeability and positional independence.
The ability of one type of L domain to functionally replace another in a chimeric Gag protein was first demonstrated by Parent et al. (55). The L domain defect displayed by RSV Gag mutants containing deletions that spanned p2b could be restored by placing HIV-1 p6 or EIAV p9 at the C terminus of Gag. A 12-amino-acid peptide that included the p6 L domain could also restore L domain function when exchanged for p2b. Moreover, the p2b L domain activity could function when moved from its native location (Fig. 1) to other positions within RSV Gag. Similar experiments were also performed with MLV (86); the p12 L domain can be moved to several places within Gag, and HIV-1 and RSV L domains can provide budding activity when substituted for the native PPPY p12 L domain. Accola et al. (1) also showed, using so-called minimal Gag constructs that contain large deletions but are nevertheless assembly competent, that RSV p2b can replace HIV-1 p6 L domain activity. This L domain exchangeability is not limited to swaps between retroviruses; the L domain peptide from Ebola virus VP40 can enhance budding of HIV-1 minimal Gag constructs lacking p6 (1), and the rhabdovirus M protein functionally replaces the L domain activity of RSV p2b (10).
(iii) Binding proteins.
Several early reports directly supported the hypothesis that retroviral L domains interact with cellular proteins. Glutathione-S-transferase (GST) fusion proteins containing HIV-1, RSV, and EIAV L domains were tested for their ability to bind Yap-derived WW domains in a filter binding assay (19). The PPPY-containing RSV L domain bound, whereas the HIV-1 and EIAV L domains scored negative in the binding assay. Similar methods were later used to demonstrate that peptides spanning the rhabdovirus PPXY L domain could interact with specific WW domains (26). To investigate whether the YXXL-containing p9 L domain from EIAV could associate with its predicted partner, Puffer et al. (60) tested the ability of a GST-p9 fusion protein to bind the medium chain (AP-50) subunit of the AP-2 complex. Binding was observed, whereas similar fusions derived from RSV or HIV-1 L domains failed to interact. In addition, colocalization between EIAV p9 and AP-2 complexes could be visualized by immunofluorescence (60). Although the functional relevance of the interactions observed in these studies was not determined, these reports clearly indicated that L domains are capable of interacting with the predicted cellular proteins.
The ubiquitin (Ub) connection.
A number of independent lines of evidence suggest that viral L domains interact with the host ubiquitination machinery (78). While it is beyond the scope of this minireview to describe in detail the cellular ubiquitination pathway, a brief mention of the major players is warranted. Excellent reviews of ubiquitination are available elsewhere (28, 76). The ubiquitination process begins when Ub, a 76-amino-acid protein, is covalently linked via a high-energy thioester bond to a Cys residue on a Ub-activating (also known as E1) enzyme. Ub is then transferred to a Ub-conjugating (E2 or UBC) enzyme. Finally, with the assistance of a Ub ligase (E3), Ub is transferred to a Lys residue on a target protein. The mammalian cell expresses a large family of E2s and E3s having different specificities, allowing the ubiquitination of a great diversity of cellular proteins. What determines the temporal and spatial control of the ubiquitination process is presently under active investigation. Interestingly, a family of proteins exists in the cell whose members bear a high degree of sequence and predicted structural similarity with bona fide E2s. However, these proteins, known variously as E2-like, UBC-like, or UEV (for ubiquitin E2 variant), lack the Cys at the catalytic site and are therefore presumably incapable of directly conjugating Ub. Although the function of these enzymatically inactive proteins is not fully understood, they appear to play a regulatory role in modulating or inhibiting the ubiquitination reaction (34, 44).
Ubiquitination can influence the fate of Ub-modified proteins in a variety of ways. The best-characterized outcome of ubiquitination is degradation in the 26S proteasome; this generally involves attachment of a chain of Ubs to the target protein and recognition of the multi-Ub chain by the proteasome. Perhaps more a propos of the topic of L domain function, it is now well appreciated that mono-Ub can serve as a signal for protein internalization from the plasma membrane (30) or sorting from either the endocytic or biosynthetic pathway into the multivesicular body (MVB)/late endosome (29). As has been pointed out previously (20, 57), the topological similarity between budding of vesicles into the MVB/late endosome and budding of virus particles from the plasma membrane (away from the cytoplasm) makes the sorting function of Ub seem particularly relevant for L domain activity.
A connection between retrovirus assembly and Ub was first suggested by the finding that RSV particles contain approximately 100 molecules of free Ub (61). Since that initial report, a number of additional lines of evidence have argued for a link between L domain function and cellular ubiquitination machinery. (i) The L domain-containing proteins of HIV and SIV (p6), MLV (p12), and EIAV (p9) are ubiquitinated at a low level (2 to 5%) (53, 54). It should be noted that these studies probably underestimate the degree to which Gag is ubiquitinated, since ubiquitin hydrolases readily catalyze a deubiquitination reaction (79). (ii) Attachment of L domains to minimal Gag constructs not only stimulates their ability to bud but also induces their ubiquitination (70). (iii) Treating virus-producing cells with proteasome inhibitors, which depletes intracellular levels of free Ub, inhibits retrovirus and rhabdovirus budding (24, 57, 66, 70). (iv) The L domains of RSV p2b (41), Ebola virus VP40 (25), and rhabdovirus M (24) interact with members of the Nedd4 family of Ub ligases (27). (v) Finally, the TSG101 protein, which contains at its N terminus an E2-like domain, interacts with HIV-1 p6 (12, 20, 77).
Although the connection between L domain function and ubiquitination is compelling, exactly what role Ub plays in the budding process is not yet clear. Some evidence suggests that Gag itself is the relevant target for ubiquitination. As mentioned above, the L domains of MLV, HIV, and EIAV are ubiquitinated. Furthermore, fusing Ub to the C terminus of RSV Gag causes the release of the modified Gag to be partially resistant to proteasome inhibitors (57). However, other data argue against Gag, or at least the L domain itself, being the primary target. For example, amino acid substitutions (52) or truncations (13) in HIV-1 Gag that prevent p6 from being ubiquitinated do not disrupt virus budding. This latter observation does not rule out the possibility that Lys residues outside p6 could be ubiquitinated. Thus, it is not yet firmly established that ubiquitination of Gag itself is critical for virus release; ubiquitination of Gag could simply be a by-product of the L domain-mediated recruitment of ubiquitination machinery to the site of budding. Interestingly, although EIAV p9 is reportedly ubiquitinated (54), EIAV budding is relatively insensitive to proteasome inhibitors (54, 56). This insensitivity can be transferred to RSV by constructing RSV-p9 chimeric Gags (56). It has been suggested that p9 contains a motif that resembles a small portion of Ub, perhaps explaining the insensitivity of EIAV release to Ub depletion (56). Although this potential Ub-like motif is not required for EIAV budding or infectivity in culture (7), it is conceivable that EIAV has evolved a redundant mechanism in which either p9 ubiquitination or the p9 Ub-like motif can assist in promoting virus release.
The results in the studies described above are consistent with proteasome inhibitors disrupting virus release by depleting intracellular pools of free Ub, thereby preventing the ubiquitination of Gag and/or a host factor(s) involved in the budding process. However, the observation that proteasome inhibitors disrupt virus release before intracellular pools of free Ub are depleted (U. Schubert and D. Ott, personal communication) suggests that proteasome inhibitors may interfere with virus release by a mechanism other than Ub depletion. One such mechanism is based on the defective ribosomal products (DRIP) hypothesis (65), which postulates that a significant percentage of proteins synthesized in the cell are misfolded and are rapidly degraded in the proteasome. According to this model, proteasome inhibitors would cause the accumulation of misfolded Gag molecules which would in some way impair virus budding (66). Why the accumulation of misfolded Gags would specifically inhibit virus budding rather than assembly remains to be determined.
It is clear that further studies will be needed to elucidate fully the relationship between Ub and virus budding. To this end, it is informative to consider the role ubiquitination plays in cellular processes, particularly endocytosis and endosomal sorting. The internalization of a number of cell-surface receptor proteins requires the recruitment of host ubiquitination machinery. Although in some cases ubiquitination of the receptor itself is required for internalization, in other cases it is not. Following growth hormone (GH) binding, the GH receptor undergoes ubiquitination on multiple Lys residues in its cytoplasmic tail. The GH receptor is subsequently internalized and degraded in the lysosome. While an intact ubiquitin-conjugating system is required for GH receptor internalization (71), ubiquitination of the receptor itself is reportedly not essential for this process (23). In contrast, internalization of the Saccharomyces cerevisiae G protein-coupled receptor Ste2p requires both the presence of ubiquitin-conjugating enzymes and the ubiquitination of Ste2p (31). It is noteworthy that not only are some membrane receptors ubiquitinated following ligand binding but also that components of the endocytic machinery may undergo this modification. This has been demonstrated for the AP-2-binding protein EPS15, which becomes ubiquitinated following cell stimulation with epidermal growth factor (75). One could imagine that the recruitment of ubiquitination enzymes by L domains might induce the ubiquitination of components of the cellular endocytic or endosomal sorting machinery.
Endosomal sorting, endocytosis, and L domains.
Increasing evidence suggests that pathways involved in endosomal sorting and endocytosis play a central role in the function of all three classes of L domains (i.e., those containing PTAP, PPXY, or YXXL motifs). As discussed above, a connection between endocytosis and L domain function was suggested initially by the observation of binding and colocalization between the EIAV p9 and AP-2 complexes (60). More recently, functional associations of RSV and Ebola virus L domains with Nedd4, and HIV-1 p6 with TSG101, have been reported.
(i) Nedd4.
Nedd4 and Nedd4-like proteins form a large family of Ub ligases that regulate a variety of cellular functions (for a review, see reference 27). These proteins have three domains in common (Fig. 3): an N-terminal lipid-binding (C2) domain; WW modules, present in multiple copies; and a C-terminal Hect (for homologous to E6-associated protein C terminus) domain that contains the Ub ligase activity. One of the principal functions of human Nedd4 appears to be the Ub-dependent regulation of cell-surface expression of the epithelial sodium channel (EnaC) (69). The yeast Nedd4 homolog, Rsp5, controls the Ub-mediated internalization of at least three membrane proteins. A role for the Nedd4 family of Ub ligases in RSV L domain function was suggested by the finding that a p2b-derived peptide hybridized to the WW domain region of a chicken Nedd4 homolog (41). p2b peptides containing substitutions known to inactivate L domain activity failed to bind. Furthermore, RSV Gag and a Nedd4-like protein could be coimmunoprecipitated from COS cell extracts. The functional relevance of the interaction between p2b and the Nedd4-like protein was supported by the finding that overexpression of the WW domain region from the Nedd4-like protein inhibited RSV particle production. Although it was not demonstrated that a budding block was responsible for the impaired virus production, the specific interaction between the L domain and the Nedd4-like protein suggests that L domain activity (i.e., budding) was likely the step affected.
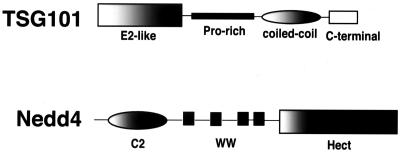
FIG. 3.
Schematic organization of TSG101 and Nedd4. Domains (described in the text) are indicated. Diagram is not intended to be to scale.
A link between Ebola virus and rhabdovirus L domains and Nedd4 has also been reported. Far-Western blotting analysis demonstrated binding between Ebola virus VP40 and WW domain-containing proteins, including Nedd4 (25). VP40 bound the yeast Nedd4 homolog Rsp5 in vitro; an L domain mutation disrupted this interaction. The PPXY-containing region of rhabdovirus M bound Nedd4 in a cDNA library screen (24). Finally, Rsp5 was able to ubiquitinate Ebola virus VP40 and rhabdovirus M proteins in an in vitro ubiquitination assay (24, 25).
(ii) TSG101.
As discussed above, TSG101 is a member of the family of proteins whose members bear an enzymatically inactive E2 domain. Originally identified in a random RNA knockout screen for tumor suppressor genes, a number of cellular functions have been proposed for TSG101. It has been implicated in genome stability, mitotic spindle formation (83), gene transcription (32, 73), and regulation of the ubiquitination and stability of the Ub ligase MDM2 and the tumor suppressor p53 (44). Perhaps more relevant from the perspective of virus budding, TSG101 and the yeast homolog Vps23 function in the sorting of proteins into the endosomal pathway (2, 5, 40, 45). Deficient TSG101/Vps23 expression leads to defects in the sorting and maturation of vacuolar hydrolases, the downregulation of activated G protein-coupled receptors in yeast cells and of epidermal growth factor receptors in mammalian cells, and abnormal redistribution of the mannose-6-phosphate receptor to the cell surface in mammalian cells (2, 40, 45). TSG101/Vps23 forms part of a high-molecular-weight complex, termed ESCRT-1, containing at least two other proteins involved in endosomal sorting (2, 5, 40). TSG101 is a multidomain protein (Fig. 3); its coiled-coiled and C-terminal domains are required for interaction with other subunits of the ESCRT-1 complex (5, 40). Interestingly, although the E2-like domain of TSG101 cannot directly conjugate Ub (since it lacks the active-site Cys), this domain nevertheless interacts with ubiquitinated cargo proteins during the sorting process. Substitution of the ubiquitinated Lys residues in the cytoplasmic tail of carboxypeptidase S prevents its recognition by Vps23 (40), and the E2-like domain of TSG101 binds Ub weakly in vitro (20). Thus, ubiquitination apparently plays a role in the recognition of proteins whose sorting into the endosomal/MVB pathway is regulated by TSG101/Vps23.
What is the evidence that TSG101 is involved in virus budding? Two groups (20, 77) identified TSG101 as a p6-interacting protein by yeast two-hybrid analysis. Mutations in the PTAP motif disrupt this interaction. Garrus et al. (20) used small interfering RNAs to show that inhibition of TSG101 expression markedly impairs HIV-1 virus production. Using a complementary approach, Demirov et al. (12) demonstrated that overexpression of the N-terminal domain of TSG101 (termed TSG-5′) dramatically reduces virus release. In both cases, EM analysis indicated that the block is at the level of virus budding (Fig. 2B) (12, 20). TSG101-underexpressing and TSG-5′-overexpressing cells exhibit a striking increase in the number of tethered particles at the plasma membrane (Fig. 2B), a phenotype very reminiscent of the pinching-off defect observed for p6 mutants (Fig. 2A) (13, 22, 36). The block is specific for HIV-1, as MLV release is unaffected by either TSG101 underexpression or TSG-5′ overexpression. TSG-5′ is incorporated into wild-type (WT) but not PTAP-mutant virus particles, confirming the p6-specific Gag/TSG101 interaction in virus-expressing cells (12). TSG101 was shown to colocalize with Ebola virus VP40 at the plasma membrane, and recruitment of TSG101 to the site of HIV-1 budding replaced the need for an intact L domain in particle release (47).
The role of p6 ubiquitination in TSG101's virus release function remains to be defined. Garrus et al. (20) observed in experiments using a surface plasmon resonance biosensor that binding between the TSG101 E2-like domain and p6 was 10-fold higher if p6 contained a Ub fused to its C terminus. In contrast, VerPlank et al. (77) reported that mutating the Ub-acceptor sites in p6 had no effect on Gag/TSG101 binding in yeast two-hybrid assays. Demirov et al. found that removal of the sites of p6 ubiquitination by truncating p6 several residues downstream of the PTAP motif only modestly reduced the efficiency of TSG-5′ incorporation into virions (12). Thus, p6 ubiquitination may promote, but is not required for, its interaction with TSG101. As noted above, these data do not address a possible contribution of ubiquitinated Lys residues outside p6.
What role Nedd4 and TSG101 play in retrovirus budding remains to be elucidated. Both are involved in regulating the sorting of proteins from the cell surface or the trans-Golgi network into the endosomal pathway. Recruitment of these proteins, and the partners with which they associate, to the plasma membrane could alter the protein composition at the site of budding in such a way that pinching off is stimulated. The recruitment of Nedd4 or TSG101 to the site of budding could modulate the ubiquitination of Gag itself or of an associated host factor(s). The recent observation of a potential link between HIV-1 L domain function and plasma membrane rafts (51), together with the finding that Nedd4 can associate with rafts (43), suggests that interaction between L domains and host factors may occur in these microdomains. While the studies described above argue that Nedd4 and TSG101 play key roles in the budding of viruses bearing PPXY and PTAP L domains, respectively, it seems likely that these Gag-binding factors function by recruiting additional players involved in protein sorting and internalization. In support of this hypothesis, when TSG101 is recruited to the site of budding via a direct HIV-1 Gag-TSG101 fusion, the C-terminal half of TSG101, which contains a binding site(s) for other components of the ESCRT-1 complex (5, 40), is sufficient to provide L domain activity (47).
While PTAP, PPXY, and YXXL L domains appear to interact with different host factors during the budding process, there is some evidence suggesting that retroviruses containing PTAP and PPXY L domains may interface with a common pathway. Several groups have demonstrated that ATPase-defective mutants of the vacuolar sorting protein Vps4 disrupt the protein complex required for endosomal sorting and induce the formation of large, aberrant endosomes (3, 4). Garrus et al. (20) and Demirov and Freed (D. Demirov and E. Freed, unpublished) have observed that Vps4 ATPase-defective mutants potently inhibit both HIV-1 and MLV budding. Similar observations have also been made for RSV (J. Wills and R. Craven, personal communication). These results suggest that these divergent retroviruses all use components of the endosomal sorting pathway to promote their release. It should be noted, however, that Vps4 ATPase mutants induce a diversity of defects in protein sorting and also disrupt cholesterol trafficking (3, 4), suggesting that caution should be used in interpreting the virus release data.
A number of established or putative L domain-containing proteins (e.g., those of M-PMV, human T-cell leukemia virus type 1, Ebola virus, and VSV) contain both PTAP and PPXY motifs, and in some cases these sequences overlap (Fig. 1) (70). The relative importance to virus budding of PTAP versus PPXY sequences in these viruses and their role in host factor binding remain to be determined. Dual motifs could provide a built-in redundancy and could perhaps enable the L domain to interact with multiple host factors in a sequential fashion.
CONCLUDING REMARKS
Over the course of the past decade, our understanding of virus budding has progressed from the initial identification of a domain in HIV-1 Gag that promotes particle release to an appreciation of the fact that analogous motifs are present in a wide range of retroviruses and in the filoviruses and rhabdoviruses as well. In recent years, it has become widely accepted that L domains function by interacting with host factors and several candidates have been identified. Additional work will clearly be required to elucidate the fascinating interplay between ubiquitination, internalization, endosomal sorting, and virus budding. Future L domain research will be exciting not only from the point of view of virology but because it is also likely to contribute important information to several key areas of cell biology.
Acknowledgments
I thank John Wills, Rebecca Craven, Heinrich Göttlinger, Volker Vogt, Marc Johnson, Ulrich Schubert, and David Ott for generously sharing unpublished data and Dimiter Demirov and Jan Orenstein for assistance with the EM data presented in Fig. 2. I am grateful to Dimiter Demirov, Akira Ono, and Ritu Goila for critical reviews of the manuscript. I particularly thank John Wills for insightful discussions and careful review of the manuscript.
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Göttlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74**:**5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babst, M., G. Odorizzi, E. J. Estepa, and S. D. Emr. 2000. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 1**:**248-258. [DOI] [PubMed] [Google Scholar]
- 3.Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17**:**2982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop, N., and P. Woodman. 2000. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol. Biol Cell. 11**:**227-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop, N., and P. Woodman. 2001. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J. Biol. Chem. 276**:**11735-11742. [DOI] [PubMed] [Google Scholar]
- 6.Bradac, J., and E. Hunter. 1984. Polypeptides of Mason-Pfizer monkey virus. I. Synthesis and processing of the gag-gene products. Virology 138**:**260-275. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C., F. Li, and R. C. Montelaro. 2001. Functional roles of equine infectious anemia virus Gag p9 in viral budding and infection. J. Virol. 75**:**9762-9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, H. I., and M. Sudol. 1995. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc. Natl. Acad. Sci. USA 92**:**7819-7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinton, G. M., B. W. Burge, and A. S. Huang. 1979. Phosphoproteins of vesicular stomatitis virus: identity and interconversion of phosphorylated forms. Virology 99**:**84-94. [DOI] [PubMed] [Google Scholar]
- 10.Craven, R. C., R. N. Harty, J. Paragas, P. Palese, and J. W. Wills. 1999. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 73**:**3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalgarno, D. C., M. C. Botfield, and R. J. Rickles. 1997. SH3 domains and drug design: ligands, structure, and biological function. Biopolymers 43**:**383-400. [DOI] [PubMed] [Google Scholar]
- 12.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99**:**955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76**:**105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dettenhofer, M., and X. F. Yu. 1999. Proline residues in human immunodeficiency virus type 1 p6Gag exert a cell type-dependent effect on viral replication and virion incorporation of Pol proteins. J. Virol. 73**:**4696-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freed, E. O. 1998. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology 251**:**1-15. [DOI] [PubMed] [Google Scholar]
- 16.Freed, E. O., and M. A. Martin. 2001. HIVs and their replication, p. 1971-2041. In D. M. Knipe and P. M. Howley (ed.). Fields virology, 4th ed. Lippincott, Philadelphia, Pa.
- 17.Garnier, L., L. J. Parent, B. Rovinski, S. X. Cao, and J. W. Wills. 1999. Identification of retroviral late domains as determinants of particle size. J. Virol. 73**:**2309-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garnier, L., L. Ratner, B. Rovinski, S. X. Cao, and J. W. Wills. 1998. Particle size determinants in the human immunodeficiency virus type 1 Gag protein. J. Virol. 72**:**4667-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garnier, L., J. W. Wills, M. F. Verderame, and M. Sudol. 1996. WW domains and retrovirus budding. Nature 381**:**744-745. [DOI] [PubMed] [Google Scholar]
- 20.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107**:**55-65. [DOI] [PubMed] [Google Scholar]
- 21.Goff, S. 2001. Retroviridae: the retroviruses and their replication, p. 1871-1940. In D. M. Knipe and P. M. Howley (ed.). Fields virology, 4th ed. Lippincott, Philadelphia, Pa.
- 22.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88**:**3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Govers, R., T. ten Broeke, P. van Kerkhof, A. L. Schwartz, and G. J. Strous. 1999. Identification of a novel ubiquitin conjugation motif, required for ligand-induced internalization of the growth hormone receptor. EMBO J. 18**:**28-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harty, R. N., M. E. Brown, J. P. McGettigan, G. Wang, H. R. Jayakar, J. M. Huibregtse, M. A. Whitt, and M. J. Schnell. 2001. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 75**:**10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97**:**13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73**:**2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harvey, K. F., and S. Kumar. 1999. Nedd4-like proteins: an emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol. 9**:**166-169. [DOI] [PubMed] [Google Scholar]
- 28.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67**:**425-479. [DOI] [PubMed] [Google Scholar]
- 29.Hicke, L. 2001. A new ticket for entry into budding vesicles-ubiquitin. Cell 106**:**527-530. [DOI] [PubMed] [Google Scholar]
- 30.Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2**:**195-201. [DOI] [PubMed] [Google Scholar]
- 31.Hicke, L., and H. Riezman. 1996. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84**:**277-287. [DOI] [PubMed] [Google Scholar]
- 32.Hittelman, A. B., D. Burakov, J. A. Iniguez-Lluhi, L. P. Freedman, and M. J. Garabedian. 1999. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 18**:**5380-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hockley, D. J., M. V. Nermut, C. Grief, J. B. Jowett, and I. M. Jones. 1994. Comparative morphology of Gag protein structures produced by mutants of the gag gene of human immunodeficiency virus type 1. J. Gen. Virol. 75**:**2985-2997. [DOI] [PubMed] [Google Scholar]
- 34.Hofmann, R. M., and C. M. Pickart. 1999. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96**:**645-653. [DOI] [PubMed] [Google Scholar]
- 35.Hoshikawa, N., A. Kojima, A. Yasuda, E. Takayashiki, S. Masuko, J. Chiba, T. Sata, and T. Kurata. 1991. Role of the gag and pol genes of human immunodeficiency virus in the morphogenesis and maturation of retrovirus-like particles expressed by recombinant vaccinia virus: an ultrastructural study. J. Gen. Virol. 72**:**2509-2517. [DOI] [PubMed] [Google Scholar]
- 36.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69**:**6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jayakar, H. R., K. G. Murti, and M. A. Whitt. 2000. Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J. Virol. 74**:**9818-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jenkins, Y., O. Pornillos, R. L. Rich, D. G. Myszka, W. I. Sundquist, and M. H. Malim. 2001. Biochemical analyses of the interactions between human immunodeficiency virus type 1 Vpr and p6Gag. J Virol. 75**:**10537-10542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jowett, J. B., D. J. Hockley, M. V. Nermut, and I. M. Jones. 1992. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J. Gen. Virol. 73**:**3079-3086. [DOI] [PubMed] [Google Scholar]
- 40.Katzmann, D. J., M. Babst, and S. D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106**:**145-155. [DOI] [PubMed] [Google Scholar]
- 41.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98**:**11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kondo, E., F. Mammano, E. A. Cohen, and H. G. Göttlinger. 1995. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J. Virol. 69**:**2759-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lafont, F., and K. Simons. 2001. Raft-partitioning of the ubiquitin ligases Cbl and Nedd4 upon IgE- triggered cell signaling. Proc. Natl. Acad. Sci. USA 98**:**3180-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li, L., J. Liao, J. Ruland, T. W. Mak, and S. N. Cohen. 2001. A TSG101/MDM2 regulatory loop modulates MDM2 degradation and MDM2/p53 feedback control. Proc. Natl. Acad. Sci. USA 98**:**1619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li, Y., T. Kane, C. Tipper, P. Spatrick, and D. D. Jenness. 1999. Yeast mutants affecting possible quality control of plasma membrane proteins. Mol. Cell. Biol. 19**:**3588-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu, Y. L., P. Spearman, and L. Ratner. 1993. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J. Virol. 67**:**6542-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7**:**1313-1319. [DOI] [PubMed] [Google Scholar]
- 48.Mayer, B. J., and M. J. Eck. 1995. SH3 domains. Minding your p's and q's. Curr. Biol. 5**:**364-367. [DOI] [PubMed] [Google Scholar]
- 49.Muller, B., T. Patschinsky, and H. G. Krausslich. 2002. The late-domain-containing protein p6 is the predominant phosphoprotein of human immunodeficiency virus type 1 particles. J. Virol. 76**:**1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohno, H., J. Stewart, M. C. Fournier, H. Bosshart, I. Rhee, S. Miyatake, T. Saito, A. Gallusser, T. Kirchhausen, and J. S. Bonifacino. 1995. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science 269**:**1872-1875. [DOI] [PubMed] [Google Scholar]
- 51.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 98**:**13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278**:**111-121. [DOI] [PubMed] [Google Scholar]
- 53.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder II, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72**:**2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ott, D. E., L. V. Coren, R. C. Sowder, I. I., J. Adams, K. Nagashima, and U. Schubert. 2002. Equine infectious anemia virus and the ubiquitin-proteasome system. J. Virol. 76**:**3038-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69**:**5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patnaik, A., V. Chau, F. Li, R. C. Montelaro, and J. W. Wills. 2002. Budding of equine infectious anemia virus is insensitive to proteasome inhibitors. J. Virol. 76**:**2641-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97**:**13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paxton, W., R. I. Connor, and N. R. Landau. 1993. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of Gag and mutational analysis. J. Virol. 67**:**7229-7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71**:**6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Puffer, B. A., S. C. Watkins, and R. C. Montelaro. 1998. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J. Virol. 72**:**10218-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Putterman, D., R. B. Pepinsky, and V. M. Vogt. 1990. Ubiquitin in avian leukosis virus particles. Virology 176**:**633-637. [DOI] [PubMed] [Google Scholar]
- 62.Rose, J. K., and M. A. Whitt. 2001. Rhabdoviridae: the viruses and their replication, 1221-1244. In D. M. Knipe and P. M. Howley (ed.). Fields virology, 4th ed. Lippincott, Philadelphia, Pa.
- 63.Royer, M., M. Cerutti, B. Gay, S. S. Hong, G. Devauchelle, and P. Boulanger. 1991. Functional domains of HIV-1 gag-polyprotein expressed in baculovirus-infected cells. Virology 184**:**417-422. [DOI] [PubMed] [Google Scholar]
- 64.Sanchez, A., A. S. Khan, S. R. Zaki, G. J. Nabel, T. G. Ksiazek, and C. J. Peters. 2001. Filoviridae: Marburg and Ebola viruses, 1279-1304. In D. M. Knipe and P. M. Howley (ed.). Fields virology, 4th ed. Lippincott, Philadelphia, Pa.
- 65.Schubert, U., L. C. Anton, J. Gibbs, C. C. Norbury, J. W. Yewdell, and J. R. Bennink. 2000. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404**:**770-774. [DOI] [PubMed] [Google Scholar]
- 66.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97**:**13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sen, A., C. J. Sherr, and G. J. Todaro. 1977. Phosphorylation of murine type C viral p12 proteins regulates their extent of binding to the homologous viral RNA. Cell 10**:**489-496. [DOI] [PubMed] [Google Scholar]
- 68.Spearman, P., J. J. Wang, N. Vander Heyden, and L. Ratner. 1994. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J. Virol. 68**:**3232-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Staub, O., S. Dho, P. Henry, J. Correa, T. Ishikawa, J. McGlade, and D. Rotin. 1996. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 15**:**2371-2380. [PMC free article] [PubMed] [Google Scholar]
- 70.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97**:**13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Strous, G. J., P. van Kerkhof, R. Govers, A. Ciechanover, and A. L. Schwartz. 1996. The ubiquitin conjugation system is required for ligand-induced endocytosis and degradation of the growth hormone receptor. EMBO J. 15**:**3806-3812. [PMC free article] [PubMed] [Google Scholar]
- 72.Sudol, M. 1996. Structure and function of the WW domain. Prog. Biophys. Mol. Biol. 65**:**113-132. [DOI] [PubMed] [Google Scholar]
- 73.Sun, Z., J. Pan, W. X. Hope, S. N. Cohen, and S. P. Balk. 1999. Tumor susceptibility gene 101 protein represses androgen receptor transactivation and interacts with p300. Cancer 86**:**689-696. [DOI] [PubMed] [Google Scholar]
- 74.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.). Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 75.van Delft, S., R. Govers, G. J. Strous, A. J. Verkleij, and P. M. van Bergen en Henegouwen. 1997. Epidermal growth factor induces ubiquitination of Eps15. J. Biol. Chem. 272**:**14013-14016. [DOI] [PubMed] [Google Scholar]
- 76.Varshavsky, A. 1997. The ubiquitin system. Trends Biochem. Sci. 22**:**383-387. [DOI] [PubMed] [Google Scholar]
- 77.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 98**:**7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vogt, V. M. 2000. Ubiquitin in retrovirus assembly: actor or bystander? Proc. Natl. Acad. Sci. USA 97**:**12945-12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wilkinson, K. D. 2000. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin. Cell. Dev. Biol. 11**:**141-148. [DOI] [PubMed] [Google Scholar]
- 80.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68**:**6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu, X., J. A. Conway, J. Kim, and J. C. Kappes. 1994. Localization of the Vpx packaging signal within the C terminus of the human immunodeficiency virus type 2 Gag precursor protein. J. Virol. 68**:**6161-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70**:**5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xie, W., L. Li, and S. N. Cohen. 1998. Cell cycle-dependent subcellular localization of the TSG101 protein and mitotic and nuclear abnormalities associated with TSG101 deficiency. Proc. Natl. Acad. Sci. USA 95**:**1595-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72**:**4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu, X. F., L. Dawson, C. J. Tian, C. Flexner, and M. Dettenhofer. 1998. Mutations of the human immunodeficiency virus type 1 p6Gag domain result in reduced retention of Pol proteins during virus assembly. J. Virol. 72**:**3412-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74**:**7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18**:**4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]