Role for radA/sms in Recombination Intermediate Processing in Escherichia coli (original) (raw)
Abstract
RadA/Sms is a highly conserved eubacterial protein that shares sequence similarity with both RecA strand transferase and Lon protease. We examined mutations in the radA/sms gene of Escherichia coli for effects on conjugational recombination and sensitivity to DNA-damaging agents, including UV irradiation, methyl methanesulfonate (MMS), mitomycin C, phleomycin, hydrogen peroxide, and hydroxyurea (HU). Null mutants of radA were modestly sensitive to the DNA-methylating agent MMS and to the DNA strand breakage agent phleomycin, with conjugational recombination decreased two- to threefold. We combined a radA mutation with other mutations in recombination genes, including recA, recB, recG, recJ, recQ, ruvA, and ruvC. A radA mutation was strongly synergistic with the recG Holliday junction helicase mutation, producing profound sensitivity to all DNA-damaging agents tested. Lesser synergy was noted between a mutation in radA and recJ, recQ, ruvA, ruvC, and recA for sensitivity to various genotoxins. For survival after peroxide and HU exposure, a radA mutation surprisingly suppressed the sensitivity of recA and recB mutants, suggesting that RadA may convert some forms of damage into lethal intermediates in the absence of these functions. Loss of radA enhanced the conjugational recombination deficiency conferred by mutations in Holliday junction-processing function genes, recG, ruvA, and ruvC. A radA recG ruv triple mutant had severe recombinational defects, to the low level exhibited by recA mutants. These results establish a role for RadA/Sms in recombination and recombinational repair, most likely involving the stabilization or processing of branched DNA molecules or blocked replication forks because of its genetic redundancy with RecG and RuvABC.
The radA/sms gene was initially identified in a screen for radiation-sensitive mutants of Escherichia coli (13). radA mutants showed a modest decrease in survival after UV or X-irradiation exposure and in repair of DNA breaks (40). This phenotype is growth medium dependent: in minimal medium, strains are less resistant and have no _radA_-dependent component of survival. This may be related to the fact that in rich growth medium, E. coli cells have multiple sister chromosomes that can interact by recombination to effect repair. The sms gene was defined as an open reading frame downstream of, and coregulated with, serB, whose inactivation caused slight sensitivity to methyl methanesulfonate (MMS) (39). Later, sms and radA were identified as the same gene (46).
The RadA/Sms predicted protein sequence is a composite of three characteristic regions. It contains a putative zinc finger at its N terminus with a CXXC-Xn-CXXC motif. Its middle region is related to the RecA strand exchange protein and the DnaB replicative DNA helicase (24), containing both Walker A and Walker B boxes characteristic of ATPases. The highly conserved motif among prokaryotic RadA orthologs, KNRFG, is found at the C-terminal edge of this RecA-related region. The C-terminal 150 amino acids of RadA/Sms is related to Lon protease, an ATP-dependent serine protease that binds to DNA (51) and that regulates capsular polysaccharide synthesis and the SOS response (16). The active-site serine is present in E. coli RadA/Sms, but this residue is replaced by alanine in many of the eubacterial radA/sms orthologs. Orthologs of radA/sms carrying all three sequence motifs are ubiquitous among the eubacteria and can be seen in the genomes of at least 40 eubacterial genera to date. The radA gene of archaea, despite its name, is not related to the radA/sms gene of eubacteria, except that both have similarity to recA of prokaryotes and RAD51 of eukaryotes. There is one bona fide eukaryotic radA/sms form in Arabidopsis thaliana, which may have entered the plant genome via the chloroplast of eubacterial origin.
A role for radA/sms in recombination is suggested by the facts that the repair pathways affected by radA are recA dependent (13) and that recombination mutants have concomitant defects in repair of radiation-induced DNA lesions. However, radA/sms does not affect survival as severely as do other mutations that result in defects in double-strand-break repair, such as recA, recB, recC, or recN (40). As determined by conjugational recombination, radA mutants are not appreciably affected in the ability to recombine (13). However, conjugational recombination differs significantly from repair recombination, and several mutations resulting in strong defects in recombinational repair (such as recJ, recN, recFOR, and ruvABC) have only minor effects on conjugational recombination frequencies (reviewed in reference 27). A radA/sms ortholog mutant in Bacillus subtilis has been reported to be defective in transformational inheritance (23).
We examined here the role of radA in DNA damage survival and conjugational recombination. In addition, we constructed double mutants with mutations in both radA and other known recombination function genes to reveal properties of radA that may be redundant to those of other genes. Sensitivity to various types of DNA-damaging agents, such as UV irradiation, MMS, hydrogen peroxide, hydroxyurea (HU), and phleomycin, were assayed. The radA/sms gene does indeed play a role in recombination and DNA repair. Its effects on repair were revealed most strongly in mutants deficient in postsynaptic DNA processing, especially those lacking the RecG branch migration helicase, suggesting that RadA may also play a role in this type of branched-structure processing.
MATERIALS AND METHODS
Strains and plasmids.
Strains were grown at 37°C as previously described on Luria-Bertani (LB) medium, consisting of 1% Bacto Tryptone, 0.5% yeast extract, 0.5% sodium chloride, and, for plates, 1.5% agar (50). For transductions and preparation of P1 phage lysates, cultures were grown in LCG medium, consisting of LB medium supplemented with 1% glucose, 2 mM calcium chloride, and, for plates, 1% agar. LCG top agar contained 0.7% agar. The following antibiotics at the indicated concentrations were used: ampicillin and streptomycin, 100 μg/ml each; kanamycin, 30 to 60 μg/ml; tetracycline and chloramphenicol, 15 μg/ml each. Isogenic strains in an AB1157 background were constructed by P1 transduction (37) and are listed in Table 1.
TABLE 1.
Escherichia coli K-12 strains
| Strain | Relevant genotype | Origin or reference |
|---|---|---|
| AB1157 and derivatives used in survival and recombination assaysa | ||
| AB1157 | Wild typea | (1) |
| CS140 | ruvC53 | R. G. Lloyd |
| JC10287 | (srlR-recA)Δ_304_ | A. J. Clark (11) |
| JC12123 | recJ284::Tn_10_ | (28) |
| N2096 | ruvA_Δ_63 | R. G. Lloyd |
| N2101 | recB268::Tn_10_ | B. Michel (26) |
| N4452 | recG_Δ_265::cat | R. G. Lloyd |
| SR776 | radA100 | N. Sargentini (13) |
| STL1548 | recQ1802::Tn_3_ | Apr transductant P1 RDK16980 (32) × AB1157 |
| STL4799 | radA1::_kan (srlR-recA)Δ_304 | Cys+ transductant P1 JC10298 × STL4759 |
| STL5037 | ruvA_Δ_63 radA1::kan | Kmr transductant P1 STL1815 × N2096 |
| STL5042 | recJ284::Tn_10 radA1_::kan | Kmr transductant P1 STL1815 × JC12123 |
| STL5046 | ruvC53 radA1::kan | Kmr transductant P1 STL1815 × CS140 |
| STL5048 | recQ1802::Tn_3 radA1_::kan | Kmr transductant P1 STL1815 × STL1548 |
| STL5280 | radA1::kan | Kmr transductant P1 STL1815 × AB1157 |
| STL5480 | recB268::Tn_10 radA1_::kan | Tcr transductant P1N2101 × STL5280 |
| STL6430 | radA_Δ_3::FRT | Plasmid-cured Kms Aps pCP20 transformant of STL6036 |
| STL6571 | recG_Δ_265::cat ruvA_Δ_63 | Cmr transductant P1 STL5130 × N2096 |
| STL6586 | recG_Δ_265::cat ruvC53 | Cmr transductant P1 STL5130 × CS140 |
| STL6588 | recG_Δ_265::cat radA1::kan | Kmr transductant P1 STL1815 × STL5130 |
| STL6592 | recG_Δ_265::cat ruvA_Δ_63 radA1::kan | Kmr Cmr transductant P1 STL1815 × STL6571 |
| STL6640 | recG_Δ_265::cat ruvC53 radA1::kan | Kmr Cmr transductant P1 STL1815 × STL6586 |
| Other strains | ||
| STL1542a | radA1::kan helD104 uvrD517am srjD7 recB21 recC22 sbcA23 | Kmr disruptant of STL941 (32) |
| STL1815a | radA1::kan (Cys−)b | Kmr transductant P1 STL1542 × AB1157 |
| STL4759 | radA1::kan cysC95::Tn_10_ | Kmr transductant P1 STL1815 × STL700 (29) |
| STL5821c | radA_Δ_3::FRT kan | Kmr gene disruptant of BW26308 (12) |
| STL6036a | radA_Δ_3::FRT kan | Kmr transductant P1 STL5821 × AB1157 |
| STL6804 | Hfr PO1 radA1::kan serA6 thi-1 relA1 lac122 | Kmr transductant P1 1815 × JC158 (6) |
| STL7130 | Hfr PO1 radA1::kan serA6 thi-1 relA1 lac122 λ+ | Kmr transductant P1 1815 × RDK1911 (30) |
Construction of radA mutant alleles.
The radA gene was amplified by PCR using Taq polymerase (Promega, Inc.) with the primers 5′ CTGAATTCAG AAGTAATTGCTCGCCCG and 5′ ATCCGGCACG GTCGGCTGCTGCGACAT from chromosomal DNA derived from wild-type E. coli K-12 strain MG1655. The PCR product was digested with _Eco_RI and _Bst_BI and ligated into vector pBS SK− (Stratagene, Inc.) that had been cut with _Eco_RI and _Cla_I, producing plasmid pSTL307. The radA1::kan allele was constructed by insertion of a Bam_HI fragment of mini-Tn_10-kan (48) into the unique Bgl_II site of radA, producing plasmid pSTL310. This plasmid was digested with Hin_cII and Bgl_I restriction endonucleases and transformed by electroporation (14) into strain STL941 (31), with selection for kanamycin resistance (Kmr) and screening for ampicillin sensitivity (Aps). A P1 transducing lysate from the resulting disruptant, STL1542, was used to convert AB1157 to Kmr, yielding STL1815. The location of the kan insertion within radA was confirmed by its linkage in transductional crosses with CAG18429 (zjh-606::Tn_10), CAG18430 (zji-202::Tn_10), and CAG18442 (thr-34::Tn_10) (43). Because STL1815 developed an auxotrophy for cysteine in an uncharacterized gene, all experiments employed a Kmr transductant of STL1815 into AB1157, strain STL5280, which does not carry this additional auxotrophy. The _radA3_Δ::FRT allele with a precise deletion of the entire radA coding region was constructed by the method of Datsenko and Wanner (12) with PCR primers 5′ CCGCCATCCTGCGGGCGGCACAGCATTAACGAGGTACACCTGTAGGCTGGAGCTGCTTCG and 5′ TCAGGTAATCAAATGACGACATATCTCCCTCCGTATATCTCATATGAATATCCTCCTTAG to amplify the kan gene of plasmid pKD4 flanked by FRT site-specific recombination sites and _radA_-specific sequences. The resultant PCR fragment carried a 40-bp homologous region where the allele was substituted by recombination into the chromosome of strain BW26308, producing STL5821. From this strain, the allele was transduced into AB1157 by using P1, with selection for Kmr (producing strain STL6036); subsequent deletion of the kan gene at the flanking FRT sites was accomplished by transformation with FLP recombinase-encoding plasmid pCP20, selecting for Apr at 30°C. After streaking STL6036 onto LB medium at 42°C to cure the plasmid, deletion strain STL6430 was produced. Genetic crosses with CAG18442 (42) confirmed the appropriate genetic location, and Southern blotting and PCRs confirmed deletion of the radA coding sequence.
DNA damage survival assays.
For UV survival determinations, cells were grown in LB liquid medium to exponential stage (optical density at 600 nm [OD600], 0.4 to 0.6), serially diluted in 56/2 buffer (50), and plated on LB agar plates. The plates were immediately irradiated with various doses of UV (254 nm) and incubated at 37°C in the dark overnight. The total viable cells in serially diluted unirradiated cells were determined. Resistance to MMS was determined for at least eight independent cultures in 2 ml of LB medium after overnight growth, and resistance to mitomycin C (MMC), phleomycin, and HU was determined for at least eight independent cultures in 2 ml of LB medium after growth to exponential phase (OD600, 0.4 to 0.6). To score for survival of cells to these DNA-damaging agents, the cultures were serially diluted in 56/2 buffer and plated directly onto LB plates containing 0.1% MMS, 1 μg of MMC/ml, 0.5 μg of phleomycin/ml, or 10 mM HU. Chemicals were purchased from Sigma, Inc. Plates containing MMS were used within 48 h. Hydrogen peroxide sensitivity was measured with exponentially growing liquid cultures, with hydrogen peroxide added to cultures to a final concentration of 5 mM. After incubation of shaking cultures at 37°C for 20 min, 50 μg of catalase per ml was added directly to the cultures to inactivate the peroxide. The cultures were serially diluted in 56/2 buffer and plated directly onto LB plates. Total viable cells were determined by serial dilution with 56/2 buffer followed by plating on LB medium. Cell counts were determined on all platesafter overnight growth at 37°C.
Assay for conjugational recombination.
All strains were assayed for conjugational inheritance in parallel with the radA+ control strain. Matings were performed using a 10:1 recipient-to-donor ratio, with recipient cells grown to an OD600 of 0.4 and donor cells grown to an OD600 of 0.3. The matings were allowed to proceed for 30 min at 37°C, and then the cultures were shaken vigorously and serially diluted in 56/2 buffer. STL6804 was used as the Hfr donor in recombination proficiency tests. (The radA gene was mutated in the Hfr donor to prevent complementation of radA in the zygote due to early transfer of radA+ in the cross.) The Leu+ Ser+ Smr recombinants were selected on streptomycin-minimal medium plates lacking leucine after serial dilution and plating. As a control for the efficiency of conjugal transfer, the strains were also crossed with STL7130, a similar Hfr donor lysogenic for lambda at attB. Production of zygotically induced infective centers was assayed as previously described (30). Although the data are not reported, none of the strains listed below (see Table 3) had any defects in conjugational transfer of lambda. Viable counts were determined by plating serially diluted unmated cultures on LB plates. All plates were counted after 1 or 2 days of growth at 37°C.
TABLE 3.
Conjugational inheritance in radA mutantsa
| Strain | Relevant genotype | Relative Leu+ Ser+ Smr recombination frequency |
|---|---|---|
| AB1157 | radA+ | 1 |
| SR776 | radA100 | 7.4 × 10−1 |
| STL5280 | radA1::kan | 3.8 × 10−1 |
| STL6430 | radA_Δ_3 | 4.8 × 10−1 |
| JC10287 | recA_Δ_304 | 2.7 × 10−4 |
| STL4799 | recA_Δ_304 radA1::kan | 6.9 × 10−4 |
| N4452 | recG_Δ_265::cat | 8.3 × 10−2 |
| STL6588 | recG_Δ_265::cat radA1::kan | 2.6 × 10−2 |
| N2096 | ruvA_Δ_63 | 1.2 × 10−1 |
| STL5037 | ruvA_Δ_63 radA1::kan | 5.1 × 10−2 |
| CS140 | ruvC53 | 3.8 × 10−2 |
| STL5046 | ruvC53 radA1::kan | 1.6 × 10−2 |
| STL6571 | recG_Δ_265::cat ruvA_Δ_63 | 3.2 × 10−2 |
| STL6592 | recG_Δ_265::cat ruvA_Δ_63 radA1::kan | 3.8 × 10−4 |
| STL6586 | recG_Δ_265::cat ruvC53 | 2.2 × 10−3 |
| STL6640 | recG_Δ_265::cat ruvC53 radA1::kan | <3.0 × 10−4 |
RESULTS
A modest effect of single radA mutations on DNA survival and recombination.
The original allele of radA is radA100, which encodes a mutation in one of the conserved cysteine residues in the putative zinc finger of the RadA protein (46). We generated two additional mutant alleles: radA1::kan, a simple insertion, and _rad3_Δ::FRT, a complete and precise deletion of the entire open reading frame. We tested the survival of these three single mutants to a group of DNA-damaging agents to which recA and other recombination mutants of E. coli K-12 are sensitive.
Unlike the results of a previous report (13), we were unable to demonstrate any consistent defect in survival after UV light exposure for the radA1 single mutant (Fig. 1 and 2), although slight sensitivity was evident in one of the six experiments (Fig. 1A). This is not an allele effect, since we were also unable to demonstrate UV sensitivity for the other radA alleles, radA100 and _radA3_Δ (B. Levinson, C. E. Beam, and S. T. Lovett, unpublished results). As reported earlier (39), these mutants did show sensitivity to the methylating agent MMS (Table 2), although their decrease in survival was much less than the more than four orders of magnitude of killing seen for the recA mutant of E. coli at the same MMS concentration. None of the mutants was sensitive to oxidative damage in the presence of hydrogen peroxide, and all showed a weak sensitivity to the cross-linking agent MMC. The presumed null alleles radA1 and radA3, but not the radA100 point mutation, conferred modest sensitivity to phleomycin, a compound related to the antitumor agent bleomycin, that induces strand breaks in DNA (4, 44). Conversely, HU, an inhibitor of deoxynucleotide synthesis, was an effective killer of the radA100 point mutant but had much less effect on the presumed null mutants carrying radA1 or radA3. The last results suggest that the radA100 putative zinc finger mutation has specific and limited effects on RadA function in vivo. The radA single mutants had modest, if any, defects in conjugational inheritance (Table 3), results in agreement with a previously published report (13).
FIG. 1.
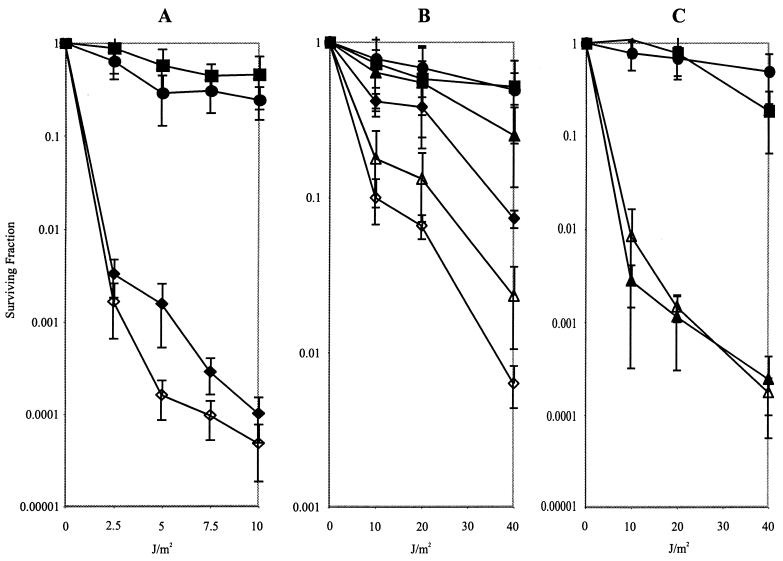
Synergy of radA with mutations affecting recombinational UV repair. Shown are UV survival curves for AB1157-derived strains both singly and doubly deficient for radA, recA, recJ, recQ, and recB. (A) AB1157, rec+ (▪); STL5280, radA1::kan (•); JC10287, recA_Δ (⧫); STL4799, recA_Δ radA1::kan (◊); (B) AB1157, rec+ (▪); STL5280, radA1::kan (•); STL1548, recQ1802::Tn_3 (▴); JC12123, recJ284::Tn_10 (⧫); STL5048, recQ1802::Tn_3 radA1_::kan (Δ); STL5042, recJ284::Tn_10 radA1_::kan (◊); (C) AB1157, rec+ (▪); STL5280, radA1::kan (•); N2101, recB268::Tn_10_ (▴); STL5480, recB268::Tn_10 radA1_::kan (Δ). Error bars indicate standard errors of the determinations.
FIG. 2.
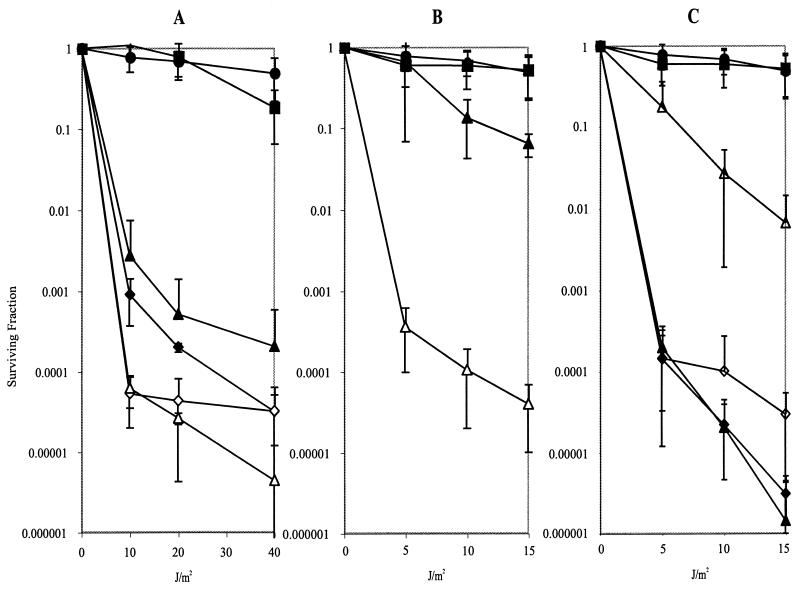
RadA is synergistic with Holliday junction-processing genes in UV repair. UV survival curves are shown for E. coli strains both singly and multiply deficient for radA, recG, ruvA, and ruvC. All strains assayed for UV survival were derived from the AB1157 background. (A) AB1157, rec+ (▪); STL5280, radA1::kan (•); N2096, ruvAΔ63 (▴); CS140, ruvC53 (⧫); STL5046, ruvC53 radA1::kan (◊); STL5037, ruvAΔ63 radA1::kan (Δ); (B) AB1157, rec+ (▪); STL5280, radA1::kan (•); N4452, recGΔ265::cat (▴); STL6588, recGΔ265::cat radA1::kan (Δ); (C) AB1157, rec+ (▪); STL5280, radA1::kan (•); STL6586, recGΔ265::cat ruvC53 (◊); STL6640, recGΔ265::cat radA1::kan ruvC53 (⧫); STL6571, recGΔ265::cat ruvAΔ63(Δ); STL6592, recGΔ265::cat radA1::kan ruvAΔ63 (▴). Error bars indicate standard errors of the determinations.
TABLE 2.
Relative survival of radA and rec mutant strains to DNA-damaging agentsa
| Strain | Genotype | Relative survivalb to: | ||||
|---|---|---|---|---|---|---|
| MMS | MMC | Phleomycin | H2O2 | HU | ||
| radA single mutants | ||||||
| AB1157 | Wild type | 1 | 1 | 1 | 1 | 1 |
| SR776 | radA100 | 2.1 × 10−2 | 3.5 × 10−1 | 4.6 | 1.1 | 3.8 × 10−4 |
| STL5280 | radA1::kan | 6.6 × 10−2 | 7.5 × 10−1 | 2.2 × 10−2 | 1.1 | 1.4 |
| STL6430 | radA_Δ_3 | 1.3 × 10−1 | 4.0 × 10−1 | 9.5 × 10−2 | 9.6 × 10−1 | 2.9 × 10−1 |
| Holliday junction-processing mutants | ||||||
| N4452 | recG | 1.9 × 10−4 | 1.1 × 10−1 | 2.0 × 10−1 | 4.1 × 10−1 | 2.1 × 10−4 |
| STL6588 | recG radA1 | 4.5 × 10−7 | 1.8 × 10−4 | 4.4 × 10−4 | 5.9 × 10−3 | 3.9 × 10−5 |
| N2096 | ruvA | 6.4 × 10−5 | 8.6 × 10−6 | 7.1 × 10−4 | 6.7 × 10−2 | 5.1 × 10−5 |
| STL5037 | ruvA radA1 | 7.6 × 10−6 | 3.5 × 10−7 | 1.9 × 10−4 | 5.3 × 10−3 | 1.9 × 10−5 |
| CS140 | ruvC | 2.8 × 10−6 | 2.6 × 10−6 | 6.9 × 10−4 | 7.3 × 10−2 | 1.2 × 10−4 |
| STL5046 | ruvC radA1 | 1.1 × 10−5 | 7.4 × 10−7 | 1.8 × 10−4 | 1.1 × 10−2 | 1.4 × 10−4 |
| Mutants in other recombination functions | ||||||
| JC12123 | recJ | 3.2 × 10−2 | 1.8 × 10−1 | 1.8 | 4.2 × 10−1 | 6.6 × 10−1 |
| STL5042 | recJ radA1 | 6.8 × 10−3 | 2.2 × 10−1 | 2.7 × 10−1 | 7.3 × 10−1 | 4.1 × 10−1 |
| STL1528 | recQ | 4.4 × 10−1 | 5.3 × 10−1 | 3.2 × 10−3 | 4.8 × 10−1 | 1.4 |
| STL5048 | recQ radA1 | 2.8 × 10−2 | 4.4 × 10−1 | 2.8 × 10−4 | 9.8 × 10−1 | 1.9 |
| N2101 | recB | 2.7 × 10−2 | 4.9 × 10−3 | 5.3 × 10−3 | 3.1 × 10−2 | 9.9 × 10−5 |
| STL5480 | recB radA1 | 2.5 × 10−2 | 2.7 × 10−3 | 2.9 × 10−4 | 1.0 × 10−1 | 7.3 × 10−2 |
| JC10287 | recA | 1.9 × 10−5 | 1.2 × 10−5 | 1.6 × 10−3 | 1.7 × 10−3 | 1.1 × 10−3 |
| STL4799 | recA radA1 | 1.7 × 10−6 | 3.0 × 10−6 | 2.9 × 10−4 | 1.4 × 10−2 | 6.0 × 10−1 |
Effect of radA when combined with other recombination mutations.
Recombination and recombinational repair are mediated in E. coli by several genetically distinct pathways (7, 21, 27). In addition, some processing events in recombination are redundant; for this reason, mutational effects are sometimes not manifest unless other functions are also mutated. We therefore placed the radA1 insertion allele in combination with mutations in genes encoding several known recombination functions (reviewed in reference 22), including recA (DNA strand exchange protein), recB (component of DNA nuclease/helicase complex), recJ (single-strand DNA exonuclease), recQ (DNA helicase), recG (branch migration helicase), ruvA (component of branch migration helicase), and ruvC (Holliday junction endonuclease).
With respect to UV survival, the radA mutation exacerbated the UV sensitivity of several mutants, including the recA, recJ, recQ (Fig. 1A and B), recG, ruvA, and possibly ruvC mutants (Fig. 2A and B). The effect was especially great with recG: although neither single mutant showed pronounced survival defects, the double mutant was severely affected. The recG radA ruv triple mutants were as sensitive (Fig. 2C), if not more so, than the recA mutants (Fig. 1A) that are defective in all pathways of homologous recombination and in induction of the SOS response. No effect on the recB mutants was afforded by addition of the radA mutation (Fig. 1C). The RecG, RuvA, and RuvC proteins play a role in the processing of branched recombinational intermediates, such as Holliday junctions, and therefore act at a late step in recombination pathways (42, 49). The RecQ and RecJ proteins in concert may reveal single-strand DNA to initiate recombination (reviewed in reference 21). There is some evidence that these proteins also play a role in processing of replication forks stalled by UV lesions (9). The RecJ exonuclease also has a postsynaptic role in stabilization of joint molecules, both in vitro and in vivo (8, 15)
In other DNA damage survival assays (Table 2), genetic synthetic effects were again strongest between radA and recG, where radA was found to sensitize recG mutants to all agents tested 5- to 500-fold. The sensitivity of the double mutants approached or exceeded the sensitivity of recA mutants that are deficient in homologous recombination and induction of the SOS response. For MMC, phleomycin, and hydrogen peroxide, although both recG and radA single mutants were only modestly sensitive, the recG radA double mutant showed profound defects in survival. This genetic effect is consistent with the idea that recG and radA/sms share some redundant role in DNA damage tolerance or repair.
Mutants in the Holliday junction helicase and resolvase, RuvA and RuvC, were very sensitive to all agents tested. The addition of a mutation in radA exacerbated DNA damage sensitivity of ruvA or ruvC mutants to all agents except the replication inhibitor HU. Strains with mutations in recJ (encoding exonuclease) and recQ (encoding helicase), like those with mutations in radA, were only modestly sensitive to the array of DNA-damaging agents, with the strongest killing effects demonstrated by either MMS or phleomycin. A mutation in radA was found to sensitize recJ and recQ mutants to the killing effect of MMS and recQ and recB mutants to that of phleomycin.
An unexpected but consistent result was the suppression of sensitivity of recA and recB mutants to hydrogen peroxide and to HU by mutations in radA. This contrasts to the slightly enhancing effect of a radA mutation on the extreme sensitivity to UV light (Fig. 1), MMS, MMC, and phleomycin (Table 2) conferred by recA. The suppressive result seems to suggest that functional RadA may convert some kinds of DNA damage, such as that after oxidation or replication inhibition, into a form that is lethal in the absence of RecABCD-dependent repair.
Several different double and triple mutant combinations were tested for effects on conjugational recombination (Table 3). Although conjugational recombination was reduced by approximately 4 orders of magnitude by a mutation in recA, mutations in radA, recG, ruvA, or ruvC reduced recombination no more than about 10-fold. A mutation in radA substantially reduced residual recombination of recG or ruv mutants, and the radA recG ruv triple mutants achieved recombination deficiency comparable to that of recA mutants. Again, this result is consistent with the idea that RadA plays a role in recombination that is redundant to that of the Holliday junction-processing proteins RecG and RuvABC.
DISCUSSION
The ubiquity of the radA/sms gene in bacterial genomes suggests that it has played an important role in the promotion of cell growth or survival. We show here that the radA/sms gene is required for efficient repair of certain forms of DNA damage and is required for genetic recombination in a step that is apparently redundant to that provided by the Holliday junction-processing proteins RecG and RuvABC. For promotion of survival after DNA damage, in particular, RadA/Sms has genetic effects that are often highly redundant to those of the RecG helicase, such that only the loss of both functions results in severe sensitivity to genotoxins and radiation.
Recombinational repair processes can, in various ways, alleviate blocks to DNA replication imposed by genotoxic assaults and restore chromosome integrity (for reviews, see references 10, 20, 21, 36). Lesions in DNA, such as those produced by UV light or by the cross-linking agent MMC, can block DNA polymerases during replication, leading to the accumulation of single-strand DNA gaps or double-strand breaks. Other agents can directly or indirectly via processing enzymes cause the accumulation of single-strand nicks or double-strand breaks. Recombination can restore broken chromosomes or broken replication forks in a manner dependent on the double-strand-break-processing helicase/nuclease RecBCD and the strand invasion protein RecA. In contrast, single-strand DNA gaps are filled by a recombinational mechanism involving RecA and the RecFOR proteins (45).
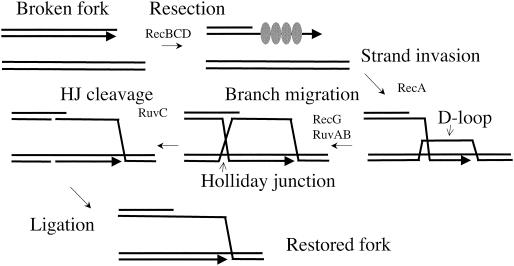
Recombinational reactions involve the formation of branched intermediate structures that are processed by the RecG helicase and by the RuvAB helicase in concert with the Holliday junction endonuclease RuvC. RecG and the Ruv proteins have redundant effects on genetic recombination and DNA repair (25), such that loss of either function causes a modest reduction whereas the loss of both has a severe effect. We show here that RadA/Sms joins this redundant team of processing enzymes; loss of all three functions produces a strain with severe conjugational recombination defects that is comparable to recA strand transferase mutants of E. coli. Since conjugational recombination occurs primarily via the RecBCD-dependent pathway (7, 27), RadA/Sms is implicated in recombination initiated from double-strand ends (Fig. 3).
FIG. 3.
Double-strand-break (DSB)-mediated recombination. A broken fork can be repaired by recombinational reactions. (Likewise, ends of conjugative DNA or transducing fragments can be integrated via this mechanism.) Double-strand ends are resected by RecBCD nuclease; RecBCD also assists in loading of RecA onto single-strand DNA. The RecA-single-strand DNA filament promotes strand invasion into a homologous duplex molecule (the sister chromosome), forming a D-loop intermediate. Branch migration helicases can extend the region of pairing to form a Holliday junction (HJ), which can be resolved by cleavage mediated by Holliday junction endonucleases such as RuvC. Ligation of strand scissions restores an intact recombinant chromosome. RadA may participate in recombination by stabilizing any of these joint intermediates or by mediating branch migration or cleavage.
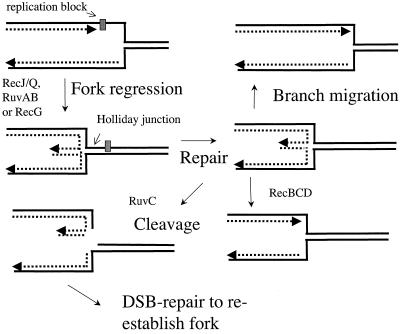
In addition to a role in genetic recombination, the Holliday junction-processing proteins of E. coli may contribute to the process of replication fork regression (Fig. 4). When a fork is stalled, perhaps by lesions or tightly bound proteins, the nascent strands can anneal to one another, producing a four-way branched or “chicken foot” structure. This junction can be cleaved by Holliday junction endonuclease RuvC in vivo (40). Fork regression serves two important purposes in DNA repair (18): (i) it reconverts parental template DNA in the fork region into a double strand so that excision repair can remove replication-blocking lesions, and (ii) it allows a template switch reaction so that one nascent strand can provide a template for the other, overcoming blocks to replication on the leading strand parental template. As both RuvAB and RecG have been implicated in the regression reaction (34, 35, 41) along with RecJ/RecQ (9), it is possible that RadA plays a similar and partially redundant role in the processing of regressed forks that accounts for its effects on DNA damage survival. One explanation for the suppressive effect that radA mutations have on the sensitivity of recA and recB mutants to peroxide and HU is that RadA converts damaged or stalled forks into double-strand breaks, which recA and recB mutants are unable to process. In the absence of RadA, these lesions can be channeled into other repair pathways. Such an effect may also explain similar suppressive effects of various Bacillus subtilis recombination mutants with mutation in radA/sms (5).
FIG. 4.
Fork regression and repair. Lesions can stall DNA polymerase. Fork regression catalyzed by RecG or RuvAB helicase activity can move the lesion into double-strand DNA, where it can be repaired. 5′ to 3′ degradation of the lagging strand by RecJ/RecQ may facilitate repair when the lagging strand has moved ahead of the leading strand. The regressed fork forms a Holliday junction, which can be cleaved by RuvC. Double-strand break repair can then restore integrity of the fork. Alternatively, RecBCD degradation of the double-strand arm or reversed branch migration can restore a fork structure.
RadA has an unusual motif structure, with a putative Zn finger N-terminal region, a region similar to RecA, including Walker A and Walker B boxes conserved in ATPases, and a C-terminal domain similar to that of Lon protease. Despite its similarity to RecA, we have no evidence that RadA can replace the RecA protein; on the contrary, most RadA-dependent genetic functions require RecA as well. In this way, RadA resembles the family of Rad51 (and RecA-related) paralogs in eukaryotes that function in concert with the Rad51 strand exchange protein (which plays a role comparable to RecA in prokaryotes) (33). In Saccharomyces cerevisiae, two of the paralogs, Rad55 and Rad57, act as “mediator” proteins to load Rad51 onto presynaptic filaments in vitro (47). The Dmc1 protein, also homologous to Rad51, is a meiosis-specific factor that works in concert with Rad51 to promote meiotic recombination (3) and has weak strand exchange properties itself (17, 19, 38). In sequence comparisons, RadA and the eukaryotic Rad51-related proteins show the most extensive similarity to Dmc1. Sequence alignment of the vertebrate Dmc1 protein extends through the putative Zn finger region of the RadA protein, with two of the four cysteines conserved and the other two replaced by lysine in the Dmc1 sequence (Fig. 5); two flanking invariant glycines are also shared between RadA and Dmc1.
FIG. 5.
Alignment of E. coli RadA/Sms with human Dmc1 showing similarity through the putative Zn finger region. Arrows indicate the conserved cysteines of the Zn finger.
Zn fingers can act as motifs promoting DNA binding or protein interactions (2). The function of RadA's putative Zn finger is important in repair, since the original radA100 mutation results in a cysteine-to-tyrosine change at one of the invariant residues (46). In our DNA damage survival assays, the radA100 mutant was as sensitive as two null mutants in radA to the alkylating agent MMS. However, in assays of survival after exposure to other agents, the radA100 mutant behaved differently than the null mutants. Unlike the null mutants, the radA100 mutant was not sensitive to the DNA breakage antibiotic phleomycin. Moreover, the radA100 mutant showed extreme sensitivity to the replication inhibitor HU, whereas neither null mutant was appreciably sensitive, suggesting that RadA processing in the absence of the Zn finger is worse for the HU-exposed cell than the absence of RadA altogether. These results must mean that Zn finger is essential for only one of multiple functions of the RadA protein, which is not unexpected given its predicted multiple-domain structure.
The role of the Lon protease domain of RadA is unknown. Although many radA/sms open reading frames are annotated in some genome databases as encoding “probable” or “predicted” ATP-dependent proteases, no protease activity has ever been experimentally demonstrated for RadA/Sms. The active-site serine of the comparable Lon domain is converted to alanine in many of the RadA bacterial forms (although E. coli RadA/Sms does retain the active-site serine), which in our opinion makes its improbable that a Lon-like protease activity contributes to its biological function. Assays detecting Lon protease have failed to demonstrate protease activity of purified RadA protein (A. Long, S. T. Lovett, and L. Hedstom, unpublished results). However, this domain is conserved among the RadA/Sms family, and truncations of this domain fail to complement radA (D. Resnicow and S. T. Lovett, unpublished results). This suggests that the Lon protease domain of RadA/Sms plays an important function, perhaps by protein or nucleic acid interactions similar to those of the Lon protease.
Acknowledgments
This work was supported by U.S. Public Health Service grants T32 GM07122 (to C.E.B. and C.J.S.) and RO1 GM51753 from the National Institute of General Medical Sciences.
We thank M. Berlyn of the E. coli genetic stock center and A. J. Clark, R. Kolodner, R. G. Lloyd, B. Michel, N. Sargentini, and B. Wanner for providing strains used in this study. We acknowledge D. Resnicow and B. Levinson for their work on the complementation analysis and cloning of radA mutants.
REFERENCES
- 1.Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd., vol. 2. ASM Press, Washington, D.C.
- 2.Berg, J. M. 1986. Potential metal-binding domains in nucleic acid binding proteins. Science 232**:**485-487. [DOI] [PubMed] [Google Scholar]
- 3.Bishop, D. K., D. Park, L. Xu, and N. Kleckner. 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69**:**439-456. [DOI] [PubMed] [Google Scholar]
- 4.Byfield, J. E., Y. C. Lee, L. Tu, and F. Kulhanian. 1976. Molecular interactions of the combined effects of bleomycin and x-rays on mammalian cell survival. Cancer Res. 36**:**1138-1143. [PubMed] [Google Scholar]
- 5.Carrasco, B., S. Fernandez, K. Asai, N. Ogasawara, and C. Alonso. 2002. Effect of the recU suppressors sms and subA on DNA repair and homologous recombination in Bacillus subtilis. Mol. Genet. Genomics 266**:**899-906. [DOI] [PubMed] [Google Scholar]
- 6.Clark, A. J. 1963. Genetic analysis of a “double male” strain of Escherichia coli K-12. Genetics 48**:**105-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark, A. J., and S. J. Sandler. 1994. Homologous genetic recombination: the pieces begin to fall into place. Crit. Rev. Microbiol. 20**:**125-142. [DOI] [PubMed] [Google Scholar]
- 8.Corrette-Bennett, S. E., and S. T. Lovett. 1995. Enhancement of RecA strand-transfer activity by the RecJ exonuclease of Escherichia coli. J. Biol. Chem. 270**:**6881-6885. [DOI] [PubMed] [Google Scholar]
- 9.Courcelle, J., and P. C. Hanawalt. 1999. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol. Gen. Genet. 262**:**543-551. [DOI] [PubMed] [Google Scholar]
- 10.Cox, M. M. 2001. Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu. Rev. Genet. 35**:**53-82. [DOI] [PubMed] [Google Scholar]
- 11.Csonka, L. N., and A. J. Clark. 1979. Deletions generated by the transposon Tn_10_ in the srl recA region of the Escherichia coli K-12 chromosome. Genetics 93**:**321-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97**:**6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diver, W. P., N. J. Sargentini, and K. C. Smith. 1982. A mutation (radA100) in Escherichia coli that selectively sensitizes cells grown in rich medium to X- or u.v.-radiation, or methyl methanesulphonate. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 42**:**339-346. [DOI] [PubMed] [Google Scholar]
- 14.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16**:**6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman-Ohana, R., and A. Cohen. 1998. Heteroduplex joint formation in Escherichia coli recombination is initiated by pairing of a 3′-ending strand. Proc. Natl. Acad. Sci. USA 95**:**6909-6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottesman, S. 1989. Genetics of proteolysis in Escherichia coli. Annu. Rev. Genet. 23**:**163-198. [DOI] [PubMed] [Google Scholar]
- 17.Gupta, R. C., E. Golub, B. Bi, and C. M. Radding. 2001. The synaptic activity of HsDmc1, a human recombination protein specific to meiosis. Proc. Natl. Acad. Sci. USA 98**:**8433-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins, N. P., K. Kato, and B. Strauss. 1976. A model for replication repair in mammalian cells. J. Mol. Biol. 101**:**417-425. [DOI] [PubMed] [Google Scholar]
- 19.Hong, E. L., A. Shinohara, and D. K. Bishop. 1912. 2001. Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J. Biol. Chem. 276**:**41906-41912. [DOI] [PubMed] [Google Scholar]
- 20.Kogoma, T. 1996. Recombination by replication. Cell 85**:**625-627. [DOI] [PubMed] [Google Scholar]
- 21.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25**:**156-165. [DOI] [PubMed] [Google Scholar]
- 22.Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Lauder, and W. M. Rehrauer. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58**:**401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kruger, E., T. Msadek, S. Ohlmeier, and M. Hecker. 1997. The Bacillus subtilis clpC operon encodes DNA repair and competence proteins. Microbiology 143**:**1309-1316. [DOI] [PubMed] [Google Scholar]
- 24.Leipe, D. D., L. Aravind, N. V. Grishin, and E. V. Koonin. 2000. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res. 10**:**5-16. [PubMed] [Google Scholar]
- 25.Lloyd, R. G. 1991. Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J. Bacteriol. 173**:**5414-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lloyd, R. G., C. Buckman, and F. E. Benson. 1987. Genetic analysis of conjugational recombination in Escherichia coli K12 strains deficient in RecBCD enzyme. J. Gen. Microbiol. 133**:**2531-2538. [DOI] [PubMed] [Google Scholar]
- 27.Lloyd, R. G., and G. J. Sharples. 1992. Genetic analysis of recombination in prokaryotes. Curr. Opin. Genet. Dev. 2**:**683-690. [DOI] [PubMed] [Google Scholar]
- 28.Lovett, S. T., and A. J. Clark. 1984. Genetic analysis of the recJ gene of Escherichia coli K-12. J. Bacteriol. 157**:**190-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lovett, S. T., P. T. Drapkin, V. A. Sutera, Jr., and T. J. Gluckman-Peskind. 1993. A sister-strand exchange mechanism for _recA_-independent deletion of repeated DNA sequences in Escherichia coli. Genetics 135**:**631-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovett, S. T., C. Luisi-DeLuca, and R. D. Kolodner. 1988. The genetic dependence of recombination in recD mutants of Escherichia coli. Genetics 120**:**37-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lovett, S. T., and V. A. Sutera, Jr. 1995. Suppression of RecJ exonuclease mutants of Escherichia coli by alterations in DNA helicases II (uvrD) and IV (helD). Genetics 140**:**27-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luisi-DeLuca, C., S. T. Lovett, and R. D. Kolodner. 1989. Genetic and physical analysis of plasmid recombination in recB recC sbcB and recB recC sbcA Escherichia coli K-12 mutants. Genetics 122**:**269-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masson, J. Y., and S. C. West. 2001. The Rad51 and Dmc1 recombinases: a non-identical twin relationship. Trends Biochem. Sci. 26**:**131-136. [DOI] [PubMed] [Google Scholar]
- 34.McGlynn, P., and R. G. Lloyd. 2001. Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc. Natl. Acad. Sci. USA 98**:**8227-8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGlynn, P., R. G. Lloyd, and K. J. Marians. 2001. Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc. Natl. Acad. Sci. USA 98**:**8235-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michel, B. 2000. Replication fork arrest and DNA recombination. Trends Biochem. Sci. 25**:**173-178. [DOI] [PubMed] [Google Scholar]
- 37.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 38.Nara, T., F. Hamada, S. Namekawa, and K. Sakaguchi. 2001. Strand exchange reaction in vitro and DNA-dependent ATPase activity of recombinant LIM15/DMC1 and RAD51 proteins from Coprinus cinereus. Biochem. Biophys. Res. Commun. 285**:**92-97. [DOI] [PubMed] [Google Scholar]
- 39.Neuwald, A. F., D. E. Berg, and G. V. Stauffer. 1992. Mutational analysis of the Escherichia coli serB promoter region reveals transcriptional linkage to a downstream gene. Gene 120**:**1-9. [DOI] [PubMed] [Google Scholar]
- 40.Sargentini, N. J., and K. C. Smith. 1986. Quantitation of the involvement of the recA, recB, recC, recF, recJ, recN, lexA, radA, radB, uvrD, and umuC genes in the repair of X-ray-induced DNA double-strand breaks in Escherichia coli. Radiat. Res. 107**:**58-72. [PubMed] [Google Scholar]
- 41.Seigneur, M., V. Bidnenko, S. D. Ehrlich, and B. Michel. 1998. RuvAB acts at arrested replication forks. Cell. 95**:**419-430. [DOI] [PubMed] [Google Scholar]
- 42.Shinagawa, H., and H. Iwasaki. 1996. Processing the Holliday junction in homologous recombination. Trends Biochem. Sci. 21**:**107-111. [PubMed] [Google Scholar]
- 43.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichai coli. Microbiol. Rev. 53**:**1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sleigh, M. J., and G. W. Grigg. 1976. The mechanism of sensitivity to phleomycin in growing Escherichia coli cells. Biochem. J. 155**:**87-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, K. C., T. V. Wang, and R. C. Sharma. 1987. _recA_-dependent DNA repair in UV-irradiated Escherichia coli. J. Photochem. Photobiol. B 1**:**1-11. [DOI] [PubMed] [Google Scholar]
- 46.Song, Y., and N. J. Sargentini. 1996. Escherichia coli DNA repair genes radA and sms are the same gene. J. Bacteriol. 178**:**5045-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sung, P. 1997. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 11**:**1111-1121. [DOI] [PubMed] [Google Scholar]
- 48.Way, J. C., M. A. Davis, D. Morisato, D. E. Roberts, and N. Kleckner. 1988. New Tn_10_ derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32**:**369-379. [DOI] [PubMed] [Google Scholar]
- 49.West, S. C. 1997. Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 31**:**213-244. [DOI] [PubMed] [Google Scholar]
- 50.Willetts, N. S., A. J. Clark, and B. Low. 1969. Genetic location of certain mutations conferring recombination deficiency in Escherichia coli. J. Bacteriol. 97**:**244-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zehnbauer, B. A., E. C. Foley, G. W. Henderson, and A. Markowitz. 1981. Identification and purification of the Lon+ (capR+) gene product, a DNA-binding protein. Proc. Natl. Acad. Sci. USA 78**:**2043-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]