Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics (original) (raw)
Abstract
Antibiotics such as erythromycin and rifampicin, at low concentrations, alter global bacterial transcription patterns as measured by the stimulation or inhibition of a variety of promoter–lux reporter constructs in a Salmonella typhimurium library. Analysis of a 6,500-clone library indicated that as many as 5% of the promoters may be affected, comprising genes for a variety of functions, as well as a significant fraction of genes with no known function. Studies of a selection of the reporter clones showed that stimulation varied depending on the nature of the antibiotic, the promoter, and what culture medium was used; the response differed on solid as compared with liquid media. Transcription was markedly reduced in antibiotic-resistant hosts, but the presence of mutations deficient in stress responses such as SOS or universal stress did not prevent antibiotic-induced modulation. The results show that small molecules may have contrasting effects on bacteria depending on their concentration: either the modulation of bacterial metabolism by altering transcription patterns or the inhibition of growth by the inhibition of specific target functions. Both activities could play important roles in the regulation of microbial communities. These studies indicate that the detection of pharmaceutically useful natural product inhibitors could be effectively achieved by measuring activation of transcription at low concentrations in high-throughput assays using appropriate bacterial promoter–reporter constructs.
Microbes such as bacteria and fungi produce a bewildering array of low-molecular-weight organic molecules that have many biological activities; their roles in nature are largely unknown, although many have been suggested (1–4). In addition, it is well established that bacteria are exposed to and respond to many different extracellular signals in the environment (5). Antibiotics are the most extensively studied of these molecules, and their use in the therapy of infectious disease since the 1940s has revolutionized medicine, leading to many life-saving treatments. Numerous small molecules (SMs) with other biological activities (e.g., antiviral, antifungal, antitumor, and immunosuppressive) have also been isolated, creating an enormous market for natural products as therapeutics (6). However, there have been comparatively few studies of the potential roles of SMs in nature, apart from the recent identification of a diversity of molecules as autoinducers of quorum sensing, a process in which a specific chemical signal (autoinducer) triggers a variety of biological functions when microbial populations attain certain cell densities (7). It is popularly assumed that the majority of SMs with inhibitory (antibiotic) activity are important as weapons in intermicrobial competition. Nonetheless, many antibiotics have been shown to possess biological activities other than inhibition (3, 8), and this prompted us to examine the possibility that they might act as chemical signals to modulate metabolic processes in bacteria at low concentrations (9, 10). We demonstrate that SMs, at concentrations below the minimal inhibitory concentrations (MICs), stimulate or depress bacterial gene expression at the transcription level, as detected by their effects on bacterial promoter–reporter constructs.
Materials and Methods
Bacterial Strains and Growth Conditions.
Strains used in the study are listed in Table 1. Cultures were grown aerobically in Luria–Bertani (LB) broth at 30 or 37°C. When appropriate, kanamycin (50 μg/ml), tetracycline (20 μg/ml), erythromycin (50 and 500 μg/ml), and rifampicin (50 and 200 μg/ml) were added. All antibiotics were obtained from Sigma or from the laboratory collection.
Table 1.
Bacterial strains and plasmids employed in this study
| Strain or plasmid | Characteristics | Source and/or ref. |
|---|---|---|
| Escherichia coli | ||
| N281 | Erythromycin-resistant mutant rplV of AB301 | A. E. Dahlberg (11) |
| K802NR | Nalidixic acid (gyrA), rifampicin-resistant (rpoB) mutant of K802 | J.D. |
| 7120 | lexA3, tifsf1A1, ind− derivative of W3110 | C. A. Gross |
| CA8306 | Δcya derivative of CA8000 | R. J. Redfield (12) |
| 13703 | ΔdnaKJ derivative of MG1655 | C. A. Gross (13) |
| 13751 | dnaJ313 derivative of MG1655 | C. A. Gross (13) |
| CF1652 | ΔrelA261:kan derivative of MG1655 | M. Cashel (14) |
| CF1693 | ΔspoT207:cml derivative of CF1652 | M. Cashel (14) |
| RH90 | rpoS359:Tn_10_ derivative of MC4100 | R. Hengge-Aronis (15) |
| RH100 | Δ(nlpD-rpoS)360 zfi3251:Tn_10_ derivative of MC4100 | R. Hengge-Aronis (15) |
| S. typhimurium | ||
| WG | Rifampicin-resistant (rpoB) derivative of 14028 | J.D. |
| SA2386 | pyrE123, recAI derivative of 14028 | Salmonella Stock Center |
| SA3670 | recA, srl202:Tn_10_ derivative of 14028 | Salmonella Stock Center |
| Plasmids | ||
| pSB401 | luxI′:luxCDABE, luxR, _tc_R | B. Ahmer (16) |
| pSB536 | ahyI′:luxCDABE, ahyR, _ap_R | B. Ahmer (17) |
| pSB1075 | lasI′:luxCDABE, lasR, _ap_R | B. Ahmer (16) |
| pBA428 | rck:luxCDABE | B. Ahmer (18) |
| pCS26 | Low-copy cloning vector | M.G.S. (19) |
Solid Media Assay.
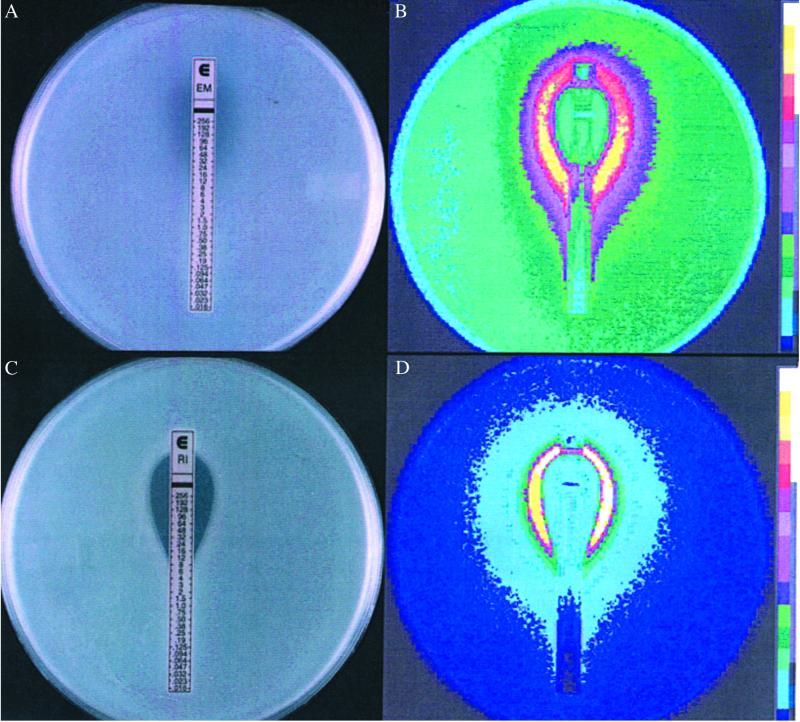
Overnight LB (BD Biosciences, Sparks, MD) cultures from single colonies of reporter strains were diluted 100-fold, inoculated into 0.7% agar, and overlaid on LB plates. Etest strips (AB Biodisk, Solna, Sweden), Sensi-discs (BD Biosciences), or antibiotic-sensitivity discs made in our laboratory were placed on the overlay. Etest strips contain precisely graduated concentrations of antibiotics that permit the accurate determination of MICs where the lower end of the inhibition zone intersects the strip (see Fig. 1 A and C). Plates were incubated at 30°C or 37°C overnight and luminescence (relative light units) was detected with a Berthold USA (Oakridge, TN) LB980 photon camera.
Fig 1.
Comparison between growth inhibition and promoter activation (light production) by erythromycin (A and B) and rifampicin (C and D) over a range of different concentrations as measured with Etest strips placed on bacterial cell overlays. (B and D) Luminescence. The _lux_–reporter constructions used are listed in Table 1. The bottom of the inhibition zone (where it intersects with the Etest strip) gives a measure of the MICs, which are 32 μg/ml (erythromycin) and 12 μg/ml (rifampicin).
Liquid Media Assay.
Two-fold serial dilutions of antibiotics were made in the wells of black clear-bottom or white 96-well plates (Thermo Labsystems, Helsinki). Overnight liquid cultures of reporter strains were diluted from 1:100 to 1:300 in LB and added to the wells containing antibiotics. OD620 and luminescence from each well were recorded at 37°C in a Wallac 1420 Victor multilabel counter (Perkin–Elmer) or a Tecan SpectraFluor Plus (Tecan, Durham, NC).
Screening for Promoters Activated by Subinhibitory Concentrations of Antibiotics.
Salmonella enterica serovar Typhimurium (Salmonella typhimurium) strain ATCC 14028 was used in this study. A random promoter library was constructed by cloning genomic restriction endonuclease fragments into the expression vector pCS26 upstream of a promoterless luxCDABE operon (J. Bjarnason, C. M. Southward, and M.G.S., unpublished data). The library consisted of 6,528 clones (17 × 384 microtiter plates) exhibiting promoter activity under different growth conditions. Salmonella clones were cultured aerobically at 37°C in LB containing kanamycin (25 μg/ml). Erythromycin was added to selected cultures at a concentration of 1–30 μg/ml, and rifampicin at a concentration of 0.2–5 μg/ml. Screening was conducted by using black 384-well solid-bottom plates. A 384-pin replicator (V&P Scientific, San Diego) was used to inoculate 384-well plates from overnight cultures. The plates were incubated at 37°C and light production was measured in a multilabel counter at 6 and 24 h. Clones showing differential expression of 3× or greater were chosen and rearrayed into 384-well plates. A second round of screening was done in a similar manner, except that light readings were taken at 2, 4, 6, 8, and 24 h; additional readings of OD620 were taken to account for possible growth effects. Clones showing differential expression of 3× or more were rearrayed into 96-well plates. These clones give a consistent positive antibiotic response when reassayed. As with any high-throughput method, some false positives were selected initially; however, these were identified and eliminated by screening potential positives by using a more rigorous second screen. In the screens reported here the initial number of false positives was between 5% and 10%. Consistent, reproducible responses were obtained for true positives in the final rescreening. (Details of library construction and screening methodology will be published elsewhere.) Plasmid DNA was isolated from positive clones by using the Concert Miniprep system (Life Technologies, Rockville, MD) and sequenced by using a vector primer pZE06 5′-AATCATCACTTTCGGGAA-3′ (Qiagen Operon, Alameda, CA). Sequencing was carried out by the Marine Biotechnology Lab, National Research Council of Canada (Halifax, NS). The promoters were identified by comparison to the GenBank database by using the National Center for Biotechnology Information (NCBI) standard nucleotide-nucleotide blastn program (www.ncbi.nlm.nih.gov/BLAST/) and analyzed by using vector nti software (Informax, Bethesda).
Results
Activation of lux Genes Using Quorum-Sensing Promoters.
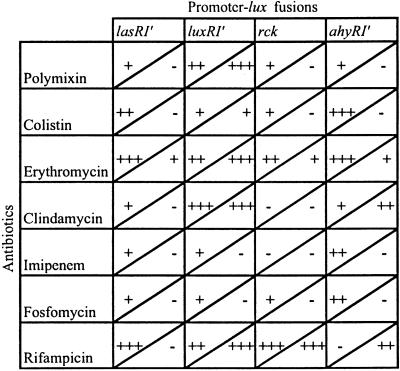
Initially, the effects of antibiotics on transcription from different plasmid-borne quorum-sensing promoters of the luxI type (Table 1) were examined by using lux reporter constructs with and without the luxR element. This testing was done by placing Etest strips on bacterial lawns and examining light production in a luminometer; the use of Etest strips provided a simultaneous indication of the concentration dependence of promoter activation and MIC (20). As shown in Fig. 1 A and B, erythromycin, an antibiotic inhibitor of bacterial protein synthesis, activated transcription (as measured by light production) at concentrations significantly lower than the MIC. A similar result was obtained with the RNA polymerase-inhibitor rifampicin (Fig. 1 C and D). A variety of other antibiotics were found to be active in stimulating different promoter–lux constructs in similar Etest studies. Patterns of activation differed in liquid as compared with solid media, depending on the strain and the antibiotic being used (Table 2). Antibiotics with distinct modes of action (inhibition of transcription, translation, cell-wall synthesis, or metabolic reactions) were active, suggesting that transcriptional modulation is a common bacterial response to SMs. However, not all antibiotics were active in these tests; for example, a number of _β_-lactams (including penicillin), certain protein synthesis inhibitors (some aminoglycosides), and the gyrase B inhibitors, coumermycin and novobiocin, showed no response. However, the “inactive” compounds may well be active against other promoters or hosts that have not been tested. It should be noted that inhibition of the growth of the tester strain was not a requirement for transcriptional modulation by SMs, as indicated by both Etest and growth curves in liquid media (see Fig. 6); thus strong responses were detected with SMs that had little or no inhibitory activity against the bacterial strains tested.
Table 2.
Activation of promoter–lux fusions by different antibiotics on solid and in liquid media
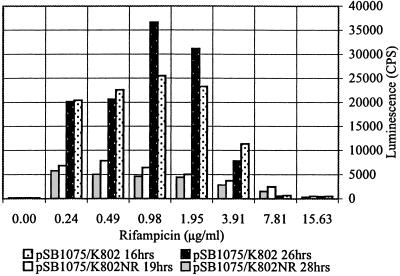
Fig 6.
Concentration dependence of promoter activation (_plasI_′:luxCDABE) in a rifampicin-sensitive (K802; MIC 12 μg/ml) and a rifampicin-resistant (K802NR; MIC >256 μg/ml) E. coli host.
Antibiotic Activation on a Global Scale.
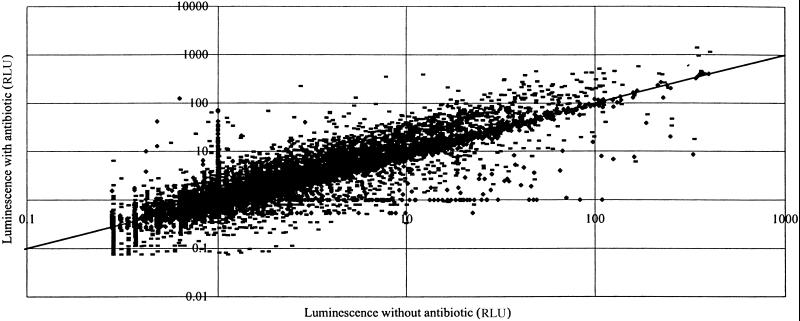
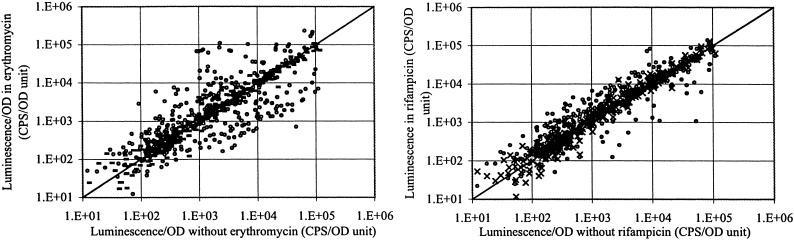
The global effects of antibiotics on transcription in bacteria were examined against 6,500 isolates of a “promoter-clone” library of S. typhimurium, which was constructed by cloning _Sau_3A fragments of S. typhimurium DNA into a plasmid vector upstream of a promoterless luxCDABE cluster and screened automatically by using different antibiotic concentrations in liquid medium (see Materials and Methods). The results of two such surveys using erythromycin and rifampicin are shown as scatter plots in Figs. 2 and 3. The patterns show clearly that these two antibiotics, at subinhibitory concentrations, activate (points above the diagonal) or inhibit (below the diagonal) many different promoters in S. typhimurium. Nucleotide sequence analyses showed that many different genes are activated by low concentrations of antibiotics, including those involved in transport, virulence, DNA repair, and numerous unidentified functions. In addition, different classes of antibiotics modulate the function of different promoters, presumably by a variety of mechanisms, as discussed below.
Fig 2.
Combined scatter plot of the actions of rifampicin at 1 μg/ml and erythromycin at 5 μg/ml determined by using a 6,500-clone S. typhimurium random promoter–lux library. RLU, relative light units. Incubation in microtiter plate liquid cultures was for 24 h at 37°C. Points above the diagonal indicate promoter-activated strains and points below the diagonal indicate clones in which promoter activity was repressed. (-, erythromycin; ♦, rifampicin.)
Fig 3.
(Left) Scatter plots of the reassay of selected clones activated by erythromycin at 1 μg/ml (-) and 30 μg/ml (•). (Right) Reassay of selected clones activated by rifampicin at 0.2 μg/ml (×) and 2.5 μg/ml (•). CPS, counts per second. OD is at 620 nm; 1.E+01 = 101, etc.
Response of Antibiotic-Resistant Hosts.
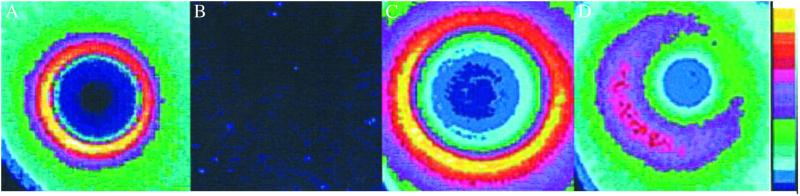
To obtain information on the mechanism of induction, a number of bacterial strains resistant to the active antibiotics were examined. In Fig. 4 we show the responses of erythromycin-resistant and rifampicin-resistant mutants, in which the inhibitory targets of the drugs are altered. For erythromycin, mutants in ribosomal protein L22 with attenuated binding of the antibiotic to the 50S ribosome were used (11). For rifampicin, mutants in the rpoB gene for the RNA polymerase _β_-subunit (the binding site for the antibiotic), were used (13). Significantly reduced activation of transcription by erythromycin was seen in eryR mutants. The rifampicin-resistant strain showed no response to rifampicin. As expected, a rifR mutation had no effect on activation by erythromycin and the eryR mutants responded normally to rifampicin.
Fig 4.
The effect of mutations to resistance to erythromycin (rplV) and to rifampicin (rpoB) in S. typhimurium on antibiotic inhibition and promoter activation on solid media. (A) Rifampicin-sensitive. (B) Rifampicin-resistant. (C) Erythromycin-sensitive. (D) Erythromycin-resistant.
Mutants Defective in Stress Responses Show Antibiotic-Induced Modulation.
Numerous cell stress responses are mediated through the association of alternative σ factors with the transcription complex (21). We introduced several “responsive” promoter–lux constructs (including luxR promoters) into bacterial hosts defective in a variety of stress responses and examined the modulation effects of antibiotics. Mutations in rec and lex (SOS), dnaJ and Δ_dnaKJ_ (heat shock response), and Δ_relA_ Δ_spoT_ (universal stress response) did not significantly reduce the antibiotic stimulatory effects. We also examined the effect of mutations in rpoS, which is considered to be the general regulator of a variety of stress responses (22). No influence on antibiotic activation was observed (results not shown). In addition, SM activation was normal in a strain lacking cAMP (cya), a global regulatory molecule in bacteria.
Characterization of Promoter Responses.
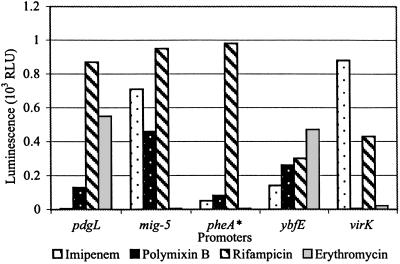
The above studies show that promoters may differ in their response to antibiotics. This finding was confirmed by testing a selection of the active S. typhimurium promoter–lux reporters for their activity in the presence of different structural classes of antibiotics, with different modes of action. The results in Fig. 5 indicate that any given promoter may be activated to a different extent, depending on the antibiotic being used (we assume that this is true for those promoters that are repressed). These variations appear to be a function of the antibiotic mode of action. In some cases the discrimination is subtle, because 14- and 16-membered macrolide inhibitors of translation, which are known to block the peptide exit tunnel of the ribosome, activate different promoters (results not shown).
Fig 5.
The antibiotic specificity of the transcription activation of different promoters. Overlays of S. typhimurium promoter–lux fusions on LB agar were exposed to discs containing antibiotics (10 μg of imipenem, 10 μg of polymixin B, 10 μg of rifampicin, or 15 μg of erythromycin). Light production was measured as described in Materials and Methods. RLU, relative light units. pheA* is a presumptive identification.
The kinetics and concentration dependence of promoter activation were examined in liquid cultures, and Fig. 6 shows that positive responses can be detected in most cases at antibiotic concentrations lower than the MIC for the E. coli strain being used. The stimulatory activity reaches a maximum level near the MIC, where transcription levels increased some 2- to 10-fold. At concentrations greater than the MIC, promoter activation was considerably reduced. The activity of different antibiotics was also dependent on the phase of bacterial growth.
Discussion
We show that antibiotics with different chemical structures and modes of inhibitory action activate or repress a wide variety of promoters in S. typhimurium at low concentrations; similar effects have been obtained with E. coli and Pseudomonas aeruginosa (results not shown). As examples, erythromycin (an inhibitor of translation) and rifampicin (an inhibitor of transcription) modulate (activate or repress) transcription of a significant proportion of genes (≈5%) in S. typhimurium and E. coli. Studies with other inhibitors, including trimethoprim (targeting dihydrofolate reductase) and fosfomycin (which blocks cell-wall synthesis), confirm that compounds with different structures and modes of action exert effects on bacterial transcription at subinhibitory concentrations (results not shown). Thus, it appears that many antibiotic inhibitors, when used at low concentrations, have in common the ability to activate or repress gene transcription, which is distinct from their inhibitory effects. Interestingly, the two contrasting responses occur with the binding of the antibiotics to their “normal” targets, because resistant mutants affecting binding to the cell targets (ribosomes and RNA polymerase, respectively, in the case of erythromycin and rifampicin) showed significantly reduced responses.
What biochemical mechanisms are responsible for these effects? We suggest that all macromolecular processes are coupled to the transcription machinery such that even minor (non-growth-retarding) effects of the binding of SMs to a macromolecular target lead to alteration of the rate of mRNA production. Because this result occurs with a variety of SMs active at different sites on the ribosome (chloramphenicol, aminoglycosides, macrolides, and tetracycline), or in cell-wall synthesis (some _β_-lactams and fosfomycin), the transcription machinery must have the means to sense these subtle conformational or stoichiometric changes and respond by specific up- or down-regulation. In the case of rifampicin, which acts on the RNA polymerase _β_-subunit, a direct interaction must operate. We cannot eliminate the possibility that some antibiotics have occult binding sites whose effects are detected only by the sensitive assay system being used, or that the antibiotics activate one or more metabolic networks (stress responses) through functional interactions with their “normal” targets.
We have shown that stress responses such as SOS and the universal response have no significant effect on SM activation, but it is conceivable that hitherto-unidentified stress responses, bacterial regulons, or signal transduction processes are responsible for the effects observed. The isolation and genetic analysis of mutants that do not respond to antibiotics should throw some light on this matter. In addition, it is known that bacterial metabolism is a complex network of interacting pathways, and negative effects on one pathway often lead to compensatory adjustments in other pathways (a form of homeostasis), as shown by expression profiling studies (23, 24). This result may occur through coordinate changes in transcription rates, which would be reflected as an apparent activation or repression of promoter activity. In prokaryotes, regulation of transcription in response to external signals is rapid and efficient. At present, such explanations for the global transcriptional changes observed in the presence of low antibiotic concentrations cannot be excluded.
In summary, antibiotics of different structure and known inhibitory activity show extensive stimulation or inhibition of a large number of promoters when target bacteria are exposed to subinhibitory concentrations of the drugs. A variety of chromosomal gene promoters are activated, including those involved in virulence, metabolic, and adaptive functions; others remain to be identified. The extent and magnitude of the effects observed suggest that the transcriptional modulation by commonly used antibiotics could lead to negative consequences during the treatment of bacterial infections in human hosts. The up-regulation of quorum-sensing systems by low concentrations of antibiotics would lead to the precocious activation of bacterial regulons, including the production of virulence factors in pathogens (10). Such untoward activity against host cells and tissues may contribute to the deleterious side reactions that accompany antibiotic therapy. Low concentrations of antibiotics remaining in the host after the treatment of infection could contribute to the physiological state normally described as the postantibiotic effect. In addition, subinhibitory concentrations of antibiotics may disrupt the ecology of the normal flora by transcriptional modulation, as described here.
Concerning the role(s) of SMs in the chemical ecology of the environment, our studies indicate that SMs may be significant elements in the dynamics of bacterial communities in nature, contributing both competitive and interactive responses. Inhibition occurs when high concentrations are attained, transcriptional changes occur at low concentrations. Many of the promoters identified in our studies regulate genes of unknown function; it is possible that a number of these are associated with processes that would not be important under laboratory conditions.
The SMs identified as antibiotics may have evolved to play two distinct roles in natural microbial communities (2). Antibiotics are a complex class of molecules, differing from other small molecule effectors such as amino acid derivatives. They do not have any enzyme-substrate activity (apart from their resistance enzymes, which may regulate their effects on transcription); in fact, antibiotic resistance genes are frequently present in strains that do not make antibiotics (25). Numerous examples of metabolic interdependence in microbial consortia are known (5), and within communities SMs may influence population structure and dynamics, and interspecies stimulation because of SMs appears to be common between streptomycetes (26). The modulatory effects of SMs suggest many possibilities as modalities of microbial communication. Even those SMs that are ineffective inhibitors (e.g., erythromycin and rifampicin) can enter cells of Gram-negative bacteria and bind to macromolecular targets in concentrations sufficient to exert the transcriptional responses demonstrated here. The same is likely to be true for a wide range of other SMs and could include both prokaryotic and eukaryotic cells.
At first sight, the differences in response seen when bacteria are exposed to SMs in liquid or on solid media may appear unusual, but it is well known that colonies growing on agar plates have distinct community structures containing bacteria in different physiological states; the generation times of bacteria are different in sessile compared with planktonic growth. The most striking differences are seen when bacteria form community structures known as biofilms on solid supports (5). Differences in SM-induced changes may be considered a further manifestation of the two states.
Finally, the effects of SMs at low concentrations on transcription may provide the basis for novel approaches to the identification of biologically active SMs from natural sources for use as pharmaceutical agents. Employing transcriptional responses to test microbial or plant extracts for their abilities to interact with intracellular targets would be a very sensitive measure of biological activity that is readily adaptable to high-throughput methods. Stress-response promoter–reporters have been described for this purpose (27, 28), but greater discrimination could be attained with a broader range of promoters, as described here. Because active small molecules exert their transcriptional effects by binding to specific intracellular targets, testing compounds against defined panels of promoter–reporter constructs should provide the means for the identification of primary cellular targets important to mechanism of action studies.
Acknowledgments
C. Gross, B. Ahmer, M. Cashel, A. Dahlberg, R. Gourse, K. Sanderson, and R. Hengge-Aronis provided bacterial strains and advice. M. Zaharik and B. Finlay were helpful in carrying out luminescence studies. A. Bölmstrom generously provided Etest strips. T. Beatty, R. Hancock, D. Mazel, and C. J. Thompson made useful comments on the manuscript, which was diligently prepared by D. Davies. We thank the Canadian Bacterial Diseases Network, the National Science and Engineering Council of Canada, and the Alberta Heritage Foundation for financial support. M.G.S. is an Alberta Heritage Foundation for Medical Research (AHFMR) Senior Scholar and a Canada Research Chair in Microbial Gene Expression.
Abbreviations
- SM, small molecule
- MICs, minimal inhibitory concentrations
References
- 1.Waksman S. A. (1961) Perspect. Biol. Med. 4**,** 271-286. [Google Scholar]
- 2.Kell D. B., Kaprelyants, A. S. & Grafen, A. (1995) Trends Ecol. Evol. 10**,** 126-129. [DOI] [PubMed] [Google Scholar]
- 3.Demain A. L. & Fang, A. (2000) in History of Modern Biotechnology, ed. Fiechter, A. (Springer, Berlin), Vol. 1, pp. 2–39. [Google Scholar]
- 4.Piepersberg W. (2001) in Molecular Medical Microbiology, ed. Sussman, M. (Academic, New York), Vol. 1, pp. 561–585. [Google Scholar]
- 5.Dunny G. M. & Winans, S. C., (1999) Cell–Cell Signaling in Bacteria (Am. Soc. Microbiol., Washington, DC).
- 6.Strohl W. R., (1997) Biotechnology of Antibiotics (Dekker, New York).
- 7.Hastings J. W. & Greenberg, E. P. (1999) J. Bacteriol. 181**,** 2667-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chadwick D. & Whelan, J., (1992) Secondary Metabolites: Their Function and Evolution, CIBA Foundation Symposium 171 (Wiley, Chichester, U.K.). [PubMed]
- 9.Parsek M. R. & Greenberg, E. P. (2000) Proc. Natl. Acad. Sci. USA 97**,** 8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams P., Camara, M., Hardman, A., Swift, S., Milton, D., Hope, V. J., Winzer, K., Middleton, B., Pritchard, D. I. & Bycroft, B. W. (2000) Philos. Trans. R. Soc. London B 355**,** 667-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregory S. T. & Dahlberg, A. E. (1995) Nucleic Acids Res. 23**,** 4234-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brickman E., Söll, L. & Beckwith, J. (1973) J. Bacteriol. 116**,** 582-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin D. J. & Gross, C. A. (1988) J. Mol. Biol. 202**,** 45-58. [DOI] [PubMed] [Google Scholar]
- 14.Xiao H., Kalman, M., Ikehara, K., Zemel, S., Glaser, G. & Cashel, M. (1991) J. Mol. Biol. 266**,** 5980-5990. [PubMed] [Google Scholar]
- 15.Lange R. & Hengge-Aronis, R. (1991) Mol. Microbiol. 5**,** 49-59. [DOI] [PubMed] [Google Scholar]
- 16.Winson M. K., Swift, S., Fish, L., Throup, J. P., Jørgensen, F., Chhabra, S. R., Bycroft, B. W., Williams, P. A. & Stewart, G. S. A. B. (1998) FEMS Microbiol. Lett. 163**,** 185-192. [DOI] [PubMed] [Google Scholar]
- 17.Swift S., Karlyshev, A. B., Fish, L., Durant, E. L., Winson, M. K., Chhabra, S. R., Williams, P., Macintyre, S. & Stewart, G. S. A. B. (1997) J. Bacteriol. 179**,** 5271-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michael B., Smith, J. N., Swift, S., Heffron, F. & Ahmer, B. M. M. (2001) J. Bacteriol. 183**,** 5733-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beeston A. L. & Surette, M. G. (2002) J. Bacteriol. 184**,** 3450-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker C. N., Stocker, S. A., Culver, D. H. & Thornsberry, C. (1991) J. Clin. Microbiol. 29**,** 533-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storz G. & Hengge-Aronis, R., (2000) Bacterial Stress Responses (Am. Soc. Microbiol., Washington, DC).
- 22.Hengge-Aronis R. (2000) in Bacterial Stress Responses, eds. Storz, G. & Hengge-Aronis, R. (Am. Soc. Microbiol., Washington, DC), pp. 161–178.
- 23.Hommais F., Krin, E., Laurent-Winter, C., Soutourina, O., Malpertuy, A., Le Caer, J.-P., Danchin, A. & Bertin, P. (2001) Mol. Microbiol. 40**,** 20-36. [DOI] [PubMed] [Google Scholar]
- 24.Schuster S., Fell, D. A. & Dandekar, T. (2000) Nat. Biotechnol. 18**,** 326-332. [DOI] [PubMed] [Google Scholar]
- 25.Yamashita F., Hotta, K., Kurasawa, S., Okami, Y. & Umezawa, H. (1985) J. Antibiot. 38**,** 58-63. [DOI] [PubMed] [Google Scholar]
- 26.Ueda K., Kawai, S., Ogawa, H., Kiyama, A., Kubota, T., Kawanobe, H. & Beppu, T. (2000) J. Antibiot. 53**,** 979-982. [DOI] [PubMed] [Google Scholar]
- 27.Ben-Israel O., Ben-Israel, H. & Ulitzur, S. (1998) Appl. Environ. Microbiol. 64**,** 4346-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bianchi A. A. & Baneyx, F. (1999) Appl. Environ. Microbiol. 65**,** 5023-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]