In Vitro and In Vivo Synergistic Interactions Between the Runx2/Cbfa1 Transcription Factor and Bone Morphogenetic Protein-2 in Stimulating Osteoblast Differentiation (original) (raw)
. Author manuscript; available in PMC: 2013 Feb 6.
Published in final edited form as: J Bone Miner Res. 2003 Apr;18(4):705–715. doi: 10.1359/jbmr.2003.18.4.705
Abstract
Bone regeneration requires interactions between a number of factors including bone morphogenetic proteins (BMPs), growth factors, and transcriptional regulators such as Runx2/Cbfa1 (Runx2). Because each component may provide a unique contribution to the overall osteogenic response, we hypothesized that bone formation may be enhanced by using combinations of complimentary factors. As an initial test of this concept, interactions between BMP2 and Runx2 were examined using adenovirus-based expression vectors (AdCMV-Runx2, AdCMV-BMP2) in the pluripotent C3H10T1/2 cell line. Cells transduced with AdCMV-Runx2 strongly expressed osteoblast markers, such as alkaline phosphatase and osteocalcin, but formed only a weakly mineralized extracellular matrix in vitro, whereas cells transduced with AdCMV-BMP2 exhibited higher levels of mineralization, but only expressed low levels of Runx2 and osteocalcin mRNA. Significantly, when cells were transduced with optimal titers of both viruses, osteoblast differentiation was stimulated to levels that were 10-fold greater than those seen with either AdCMV-Runx2 or AdCMV-BMP2 alone. To measure in vivo osteogenic activity, virally transduced cells were subcutaneously implanted into immunodeficient mice. Cells transduced with control virus produced only fibrous tissue while those with AdCMV-Runx2 produced limited amounts of both cartilage and bone. In contrast, cells transduced with either AdCMV-BMP2 alone or AdCMV-BMP2 plus AdCMV-Cbfa1 generated large ossicles containing cartilage, bone, and a marrow cavity. However, ossification in the AdCMV-BMP2 plus AdCMV-Cbfa1 group was more extensive in that both mineral content and fractional bone area were greater than that seen in the AdCMV-BMP2 group. Thus, the increased osteoblast differentiation observed with combined adenovirus treatment in vitro is also manifested by increased bone formation in vivo. These results suggest that Runx2 and BMP2 have distinct, but complementary, roles in osteogenesis and that their combined actions may be necessary for optimal bone formation.
Keywords: Runx2, bone morphogenetic protein-2, gene therapy, osteoblast, adenovirus
INTRODUCTION
A number of soluble factors involved in normal bone development also participate in bone regeneration and fracture healing including the bone morphogenetic proteins (BMPs), TGF-_β_s, members of the hedgehog family of factors, fibroblast growth factors (FGFs), and various cytokines.(1,2) Of these, the BMPs have been most extensively studied, in part, because they can, by themselves, induce osteoblast differentiation and bone formation either when directly administered as recombinant proteins(3) or through gene therapy–mediated delivery using adenovirus vectors.(4,5)
BMP-initiated signals are mediated by the receptor-regulated Smad proteins 1, 5, and 8 (R-Smads) and the common partner protein, Smad4. R-Smad–Smad4 complexes interact with specific enhancer sequences (Smad binding elements [SBEs]) on target genes to stimulate gene expression.(6,7) Although few direct target genes for BMP signaling are known, an attractive candidate is Runx2, the bone-related product of the Cbfa1 gene.(8) This transcription factor, Runt, related to the Drosophila protein, is essential for osteoblast and hypertrophic chondrocyte differentiation, and for this reason, skeletons of _Cbfa1_−/− animals are totally devoid of mineral.(9,10) Because BMPs can upregu-late Runx2 in certain systems,(8,11) one possible explanation for how BMPs act in bone would be to assume that their effects are principally mediated by this transcription factor, which would subsequently activate downstream genes necessary for the osteoblast phenotype. However, some BMP activities are independent of Runx 2. For example, BMP2 can induce low level expression of osteoblast marker genes like osteocalcin (OCN) and alkaline phosphatase (ALP) in calvarial cells from _Cbfa1_−/− animals, although these cells are not able to form a mineralized matrix.(10) Furthermore, R-Smad–Smad4 complexes can directly interact with and activate certain target genes, thus providing a possible mechanism for BMPs to stimulate osteoblast gene expression through a Runx2-independent pathway.(12,13) Also un-resolved is the extent to which Runx2 is responsible for expression of the osteoblast phenotype. Overexpression of Runx2 has been reported to induce osteoblast-related genes in certain non-osteoblast mesenchymal cell lines.(8,14) However, no previous studies described effects of Runx2 on either formation of a mineralized extracellular matrix in vitro or bone formation in vivo. Thus, it remains an open question whether Runx2 regulates osteoblast differentiation in a linear manner (i.e., activates all downstream signals necessary for osteoblast differentiation) or whether it acts in concert with other non–Runx2-dependent factors that must also be present for transcriptional activation to occur.
To begin addressing these issues, the present study was designed to first determine the extent to which Runx2 can induce functional osteoblast differentiation in a mesenchymal cell line and then to examine functional interactions between Runx2 and BMP2 in stimulation of the osteoblast phenotype. As will be shown, adenovirus-mediated expression of Runx2 was found to induce osteoblast marker genes and weakly stimulate mineralization in the murine C3H10T1/2 mesenchymal cell line as well as stimulate cartilage and bone formation in vivo. However, osteoblast differentiation was synergistically enhanced if cells were cotransduced with an adenovirus expressing BMP2. These results have important implications for our understanding of the role of Runx2 in osteoblast formation and suggest a therapeutic potential for the combined use of complimentary factors for gene therapy–based bone regeneration.
MATERIALS AND METHODS
Construction of AdCMV-Runx2 and AdCMV-BMP2
Recombinant adenoviruses were constructed using Cre/lox recombination.(15) Briefly, mouse Runx2 cDNA or BMP2 cDNA was inserted into pAdlox to produce pAd-loxRunx2 and pAdloxBMP2 plasmids. Each plasmid was cotransfected with Ψ5 DNA into CRE8 cells using the calcium phosphate precipitation method. Primary lysates were collected after 5 days and used to reinfect CRE8 cells. A plaque assay method was used to purify virus from this secondary lysate. Each plaque lysate was screened for the presence of Runx2 or BMP2 insert using gene-specific primers and polymerase chain reaction (PCR) analysis. Positive plaques were amplified in 293 cells, and the virus was purified by CsCl gradient ultracentrifugation. Purified virus was stored in 20% glycerol-phosphate–buffered saline (PBS) and titered using a plaque assay. AdCMV-lacZ was purchased from the University of Michigan Vector Core Laboratory (Ann Arbor, MI, USA). Previous studies in our laboratory demonstrated that this vector does not stimulate bone formation regardless of whether virus was directly implanted in experimental animals or used to transduce dermal fibroblasts before implantation.(4,5)
Cell culture
The murine pluripotent mesenchymal cell line C3H10T1/2 was obtained from American Type Culture Collection (Gaithersburg, MD, USA). Cells were plated at a confluent density of 50,000 cells/cm2, based on previous studies with osteoblast-like cells indicating that cultures must be confluent before differentiation can commence.(16,17) Cells were grown in _α_-MEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/ streptomycin. For in vitro transduction of cells, adenovirus at the desired titer was added to cells in serum-free medium. After 4 h, serum was added to a final concentration of 2%, and cells were grown for an additional 24 h. Cells were then transferred to medium containing 50 _μ_g/ml ascorbic acid and 5 mM β-glycerol-phosphate and fed every 2 days unless indicated otherwise.
Western blot detection of Runx2 and BMP2 protein
For detection of Runx2, cells were lysed by incubation at 4°C for 30 minutes in lysis buffer (20 mM sodium phosphate [pH 7.0] 250 mM NaCl, 30 mM sodium pyrophosphate, 0.1% NP-40, 5 mM EDTA, 10 mM NaF, 0.1 mM Na3VO3, and 1 mM phenylmethyl sulfonyl fluoride (PMSF) supplemented with 1 μg/ml leupeptin, 1 _μ_g/ml pepstatin, and 1 _μ_g/ml aprotinin). For BMP2 detection, cells were transferred to serum-free medium for 24 h and both medium and cell layers were harvested, pooled, and lyophilized. Samples were fractionated by SDS-PAGE on 10% gels and electrophoretically transferred to a nitrocellulose membrane (Schleicher & Schuel, Keene, NH, USA). Anti-Runx2 antibody, a gift from Dr Gerard Karsenty (Baylor College of Medicine, Houston, TX, USA), was used at a dilution of 1:1000. BMP2 antibody (R&D Biosystems, Inc., Minneapolis, MN, USA) was used at a dilution of 1:500. Second antibodies (horseradish peroxidase–conjugated goat anti-rabbit IgG or goat anti-mouse IgG) were used at dilutions of 1:10,000 or 1:5000, respectively. Blocking and reaction with antibodies was conducted as described.(18) Immunore-activity was detected by enhanced chemiluminescence (ECL; Amersham, Piscataway, NJ, USA).
RNA analysis
Total RNA was isolated from cells using Trizol reagent (Life Technologies, Inc., Gaithersburg, MD, USA) as described by the manufacturer and quantified by ultraviolet spectroscopy. Ten-microgram aliquots of total RNA were fraction on 1% agarose-formaldehyde gels and blotted onto nitrocellulose membranes (Stratagene, La Jolla, CA, USA).(19) Membranes were hybridized with a cDNA probe labeled with [_α_-32P]deoxy-CTP (NENA Media Inc., Los Angeles, CA, USA) using a random primer labeling kit (Boehringer-Mannheim, Mannheim, Germany). Mouse cDNA probes used for hybridization were obtained from the following sources: Runx2(8) from Dr Gerard Karsenty (Baylor College of Medicine), OCN(20) from Dr John Wozney (Genetics Institute, Boston, MA, USA), bone sialoprotein (BSP)(21) from Dr Marion Young (NIDCR, Bethesda, MD, USA), tissue nonspecific ALP(22) from Dr Enrico Garattini (Instituto di Ricerche Farmacolgiche Mario Negri, Milan, Italy), and _α_2(I) collagen(23) from Dr B de Crombrugghe (UT/MD Anderson Cancer Center, Houston, TX, USA). All cDNA inserts were previously excised from plasmid DNA with the appropriate restriction enzymes and purified by agarose gel electrophoresis. Hybridizations were performed as previously described using a Bellco Autoblot hybridization oven (Bellco glass, Inc., Vineland, NJ, USA) and quantitatively scanned using a Packard A2024 Instant-Imager (Packard Bioscience, Meriden, CT, USA) and subsequently exposed to Kodak BIOMAX film (Eastman Kodak Co., Rochester, NY, USA) at –70°C. All blots were normalized for RNA loading by stripping and reprobing with cDNA to 18S rRNA.(24)
Biochemical assays
ALP activity was measured in cell layers as previously described using a _p_-nitrophenyl phosphate substrate and an incubation temperature of 37°C.(25) For measurement of calcium, cell layers were extracted overnight with 15% trichloroacetic acid (TCA). Insoluble material was removed by low-speed centrifugation. DNA in pellets was measured according to the method of Schneider.(26) Supernatants were then assayed for calcium with a commercially available kit (Sigma Diagnostics, Inc., St. Louis, MO, USA). Cultures were also stained for mineral by the method of von Kossa as previously described.(27)
Animal experiments
All procedures were approved by the University Committee on the Use and Care of Animals and were in compliance with State and Federal laws. A total of 6 × 106 cells were infected with the indicated adenovirus at a titer of 250 pfu/cell. After 24 h, cells were trypsinized, and 5 × 106 cells were absorbed to gelatin sponges prewetted with medium. Transplants were subcutaneously implanted into 4- to 6-week-old immunodeficient mice (N: National Institutes of Health-bg-_ν_-xid) as previously described.(28) Surgery was performed under anesthesia achieved by inhalation of isoflurane (Mallinckrodt Veterinary, Mundelein, IL, USA). Four longitudinal skin incisions of about 0.5–0.6 cm in length were made on the dorsal surface of each mouse, and subcutaneous pockets were formed by blunt dissection. A single transplant was placed into each pocket and incisions were closed with surgical staples.
Histology
Implants were removed from mice and fixed in 4% paraformaldehyde (PFA) overnight at 4°C, and then decalcified in 10% formic acid for 48 h. The solution was changed every 24 h. Samples were dehydrated through a series of graded ethanols, xylol, and finally xylene, after which they were embedded in paraffin and sectioned at 8 _μ_m. Slides were stained with hematoxylin and eosin. Morphometric analysis of bone areas in histological sections was conducted using ImagePro software (Media Cybernetics, Inc., Carlsbad, CA, USA).
Statistical analysis
All experiments were repeated a minimum of two times, and qualitatively identical results were obtained. Statistical analyses were performed using Instat 3.0 (GraphPAD Software, Inc., San Diego, CA, USA). Where indicated, experiment data are reported as means and SD of triplicate independent samples.
RESULTS
Induction of osteoblast differentiation by transduction of C3H10T1/2 cells with AdCMV-Runx2
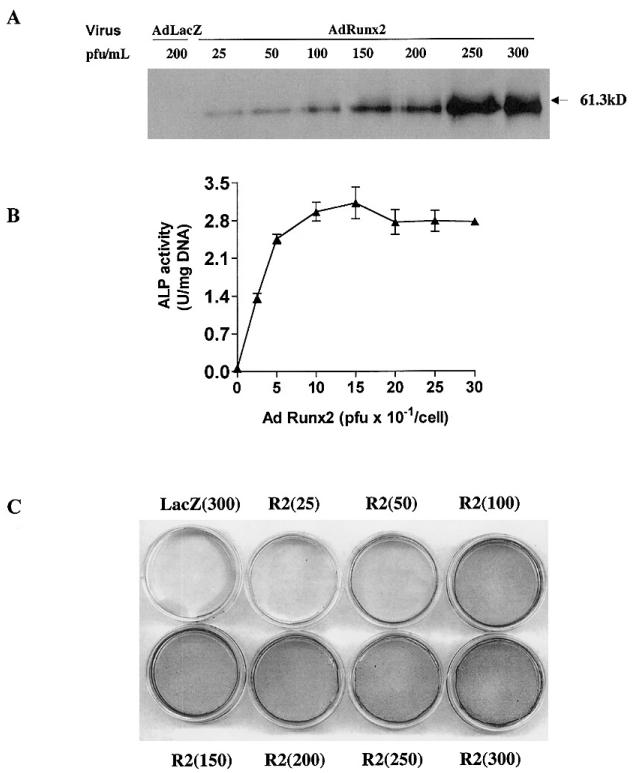
Initial studies were designed to examine whether Runx2 expression could induce an osteoblast phenotype in a pluripotent mesenchymal cell line. C3H10T1/2 cells were selected for study because of their known capacity to differentiate to the principle mesenchyme-derived cell types (i.e., myoblasts, adipocytes, chondrocytes, and osteoblasts) under the appropriate conditions(29) and because previous work had show that transient transfection with a Runx2 expression vector could induce osteoblast marker mRNAs in these cells.(8) To determine optimal conditions for virus treatment, C3H10T1/2 cells were transduced with increasing titers of AdCMV-lacZ or AdCMV-Runx2, and cells were assayed for Runx2 expression by Western blotting, ALP activity or mineralization (formation of a von Kossa–positive ECM). As shown in Fig. 1, adenovirus-mediated overexpression of Runx2 clearly induced Runx2 protein, ALP activity, and a modest degree of mineralization in C3H10T1/2 cells, whereas cells transduced with control lacZ virus contained no detectable Runx2 protein and failed to differentiate. Maximal induction of ALP activity and mineralization was observed at a viral titer of 100 pfu/cell. A somewhat higher viral titer of 250 pfu/cell was needed to maximally induce Runx2 protein expression. In separate experiments, cells transduced with increasing titers of AdCMV-lacZ were stained for _β_-galactosidase activity, and it was determined that 90–95% of C3H10T1/2 cells are transduced by a viral titer of 100 pfu/cell (result not shown).
FIG. 1.
Effect of adenovirus titer on Runx2 expression and induction of osteoblast differentiation in C3H10T1/2 cells. C3H10T1/2 cells were transduced with the indicated titer of AdCMV-LacZ (AdLacZ) or AdCMV-Runx2 (AdRunx2) as described in the Materials and Methods section. After 24 h, cells were fed with complete medium containing 10% FBS, 50 _μ_g/ml ascorbic acid, and 5 mM _β_-glycerol phosphate. (A) On day 3 after viral transduction, Runx2 protein levels were measured in whole cell extracts by Western blotting. (B) On day 6, cells were harvested for measurement of ALP activity. (C) A separate set of cultures was stained for mineralization by method of von Kossa after 12 days in culture.
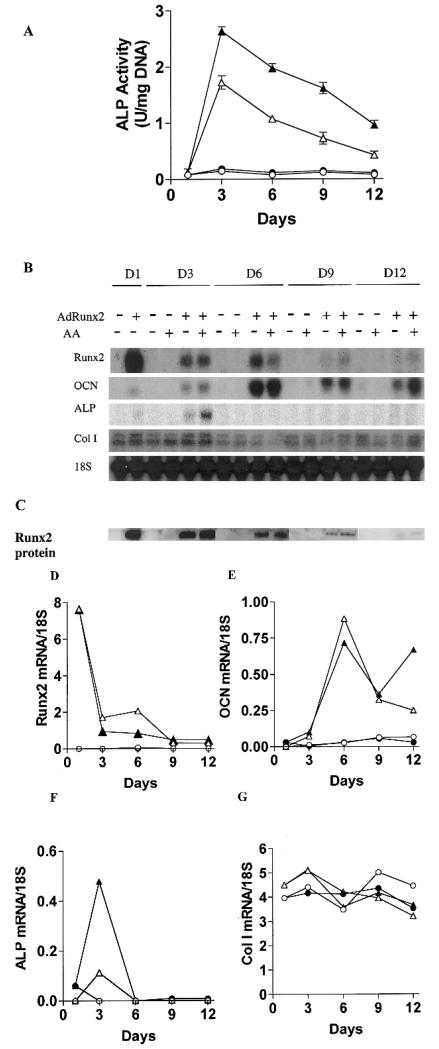
The experiments shown in Fig. 1 clearly indicate that Runx2 overexpression in C3H10T1/2 cells leads to the induction of features consistent with an osteoblast phenotype. To examine this phenomenon in greater detail, cells were transduced with either control virus (AdCMV-lacZ) or AdCMV-Runx2 and grown for increasing times in the presence or absence of ascorbic acid (Fig. 2). We previously showed that this cofactor induces in vitro osteoblast differ-entiation in preosteoblast-like cells by stimulating collagen matrix synthesis.(16) Unlike results with preosteoblast cell lines, C3H10T1/2 cells transduced with control virus had no detectable ALP activity (Fig. 2A) or osteoblast marker mRNA expression (Figs. 2B and 2D–2G), regardless of whether or not cells were treated with ascorbate. In contrast, cells transduced with AdCMV-Runx2 expressed maximal ALP activity after 3 days, with a gradual decline at later times. Furthermore, ALP activity was 1.6- to 2-fold higher when cells were also treated with ascorbate. Runx2 mRNA (Fig. 2B) or protein (Fig. 2C) was only detected in AdCMV-Runx2-transduced cells; it was present in the highest levels immediately after viral transduction (day 1) and declined to below the limits of detection by day 12. AdCMV-Runx2 also induced OCN and ALP mRNA beginning at day 1. ALP mRNA, like enzymatic activity, peaked at day 3, but was not detectable at later times. OCN mRNA was also detectable at day 1, continued to increase to day 6, and then gradually declined, but was still present in appreciable levels after 12 days. Thus, although Runx2 is known to be a direct regulator of the OCN gene, levels of osteoblast marker mRNA were not well correlated with either Runx2 mRNA or protein. Mineralization measured either as von Kossa–positive ECM or as cell layer–associated calcium was also modestly increased from day 6 onward (discussed below; Figs. 4B and 4G).
FIG. 2.
Time course of osteoblast differentiation in AdRunx2-transduced C3H10T1/2 cells. Cultures were transduced with AdLacZ (~) or AdRunx2 (r,p) at a titer of 250 pfu/cell. After 24 h, one-half the cells in each group were fed with basal medium (_α_-MEM, 10% FBS, open symbols) and one-half were fed with basal medium supplemented with 50 _μ_g/ml ascorbic acid (closed symbols). Cells were harvested at the times indicated and assayed for (A) ALP activity, (B) mRNA levels, or (C) Runx2 protein levels. (D–G) These panels show mRNA levels after imaging and normalization to 18S rRNA.
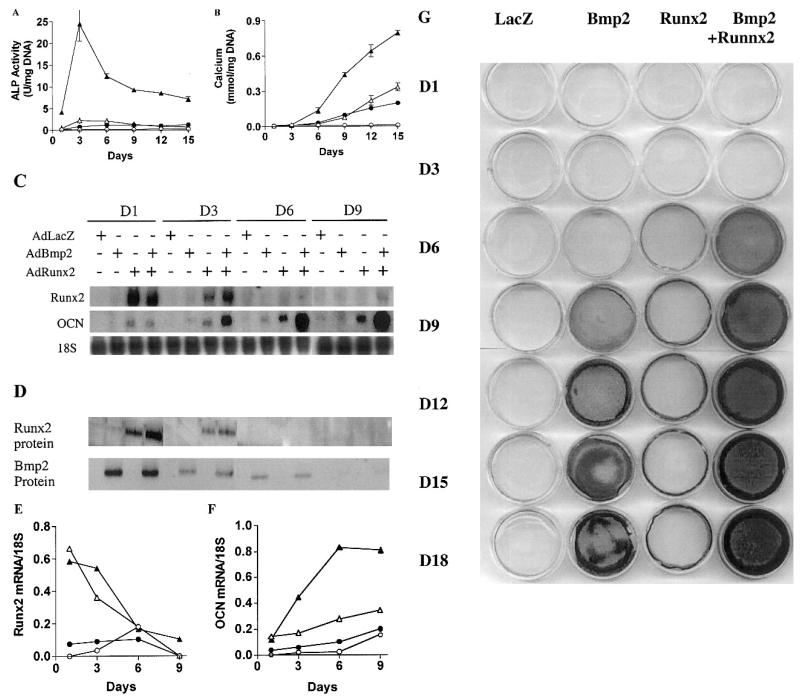
FIG. 4.
Time course of osteoblast differentiation in C3H10T1/2 cells transduced with AdBMP2 in the presence or absence of AdRunx2. Cells were transduced with the following combinations of adenovirus vectors: AdLacZ alone (200 pfu/cell), ●; AdLacZ (100 pfu/cell) plus AdBMP2 (100 pfu/cell), ●; AdLacZ (100 pfu/cell) plus AdRunx2 (100 pfu/cell), ▵; and AdBMP2 (100 pfu/cell) plus AdRunx2 (100 pfu/cell), ▴. Cells were harvested at the times indicated for measurement of (A) ALP activity, (B) calcium, (C) osteoblast marker mRNA expression, or (D) total Runx2 or BMP2 protein. Normalized mRNA levels are shown for (E) Runx2 mRNA and (F) OCN mRNA. (G) Replicate plates of cells were also stained for mineral by the method of von Kossa.
Although AdCMV-Runx2-transduced cells exhibited many characteristics of mature osteoblasts, certain differences were noted. Specifically, Col1A1 mRNA was not increased by Runx2 (Fig. 2B), although previous work indicated that this gene is, in part, regulated by this transcription factor.(30) Also notably absent was mRNA for bone sialoprotein and the parathyroid hormone (PTH)/parathyroid hormone–related protein (PTHrP) type I receptor (results not shown). However, AdCMV-Runx2–transduced cells did exhibit PTH-dependent increases in cyclic adenosine monophosphate (cAMP) activity (3-fold increase 3 days after virus treatment), which indicates that Runx2 expression likely did induce low levels of the PTH/PTHrP receptor (result not shown).
These results indicate that AdCMV-Runx2 transduction of C3H10T1/2 cells induced osteoblast marker mRNA and low levels of mineralization. Although virally expressed Runx2 mRNA and protein declined with time, detectable amounts of Runx2 were present for at least 9 days. Furthermore, OCN mRNA expression, which was seen exclusively in AdCMV-Runx2-transduced cells, persisted for at least 12 days after viral transduction. This suggests either that Runx2 levels were sufficient to activate this gene over this entire period or that Runx2 initiated additional downstream events that allowed OCN expression to continue. AdCMV-Runx2-transduced cells also resembled normal osteoblasts in other ways; they were partially dependent on ECM synthesis to fully express osteoblast markers and they exhibited a sequential pattern of gene expression with ALP being induced before OCN.
Cooperative interactions between Runx2 and BMP2 in inducing osteoblast differentiation in vitro
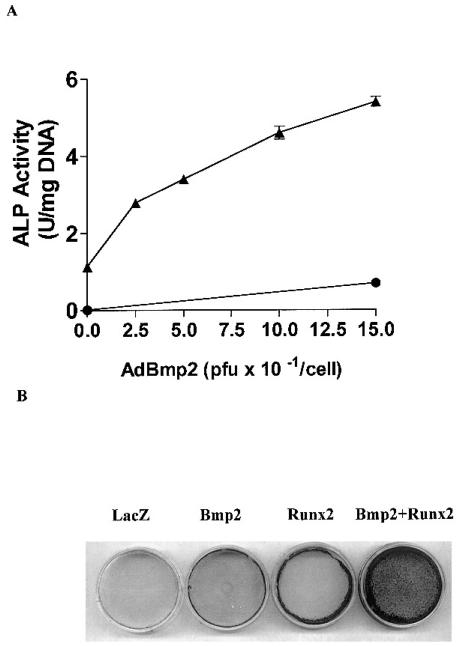
As noted in the Introduction, some osteogenic signals initiated by BMPs seem to be independent of Runx2 in that they are seen in _Cbfa1_−/− animals. In addition, BMP-dependent Smad proteins have been shown to cooperatively interact with Cbfa-family members to stimulate transcription in certain systems. In view of these findings, we hypothesized that BMP signals may be able to compliment Runx2-dependent gene expression to stimulate osteoblast differentiation. To test this concept, we examined the responsiveness of C3H10T1/2 cells to increasing titers of an adenovirus expressing BMP2 (AdCMV-BMP2) in the presence or absence of an optimal titer of AdCMV-Runx2 (100 pfu/cell). Analysis of secreted BMP2 protein by Western blotting demonstrated that maximal BMP2 production occurred in cells transduced with AdCMV-BMP2 at a titer of 150 pfu/cell (result not shown). Transduction of C3H10T1/2 cells with AdCMV-BMP2 alone induced only a modest increase in both ALP activity and von Kossa-positive mineral after 6 days in culture (Fig. 3). Comparable levels of differentiation were also seen with AdCMV-Runx2. However, when increasing titers of the BMP2 adenovirus were added in the presence of Runx2 virus, dramatic increases were seen in both ALP activity and mineralization. To avoid results being confounded by variations in total viral titer between groups, in this and all subsequent experiments, the total amount of virus was held constant at 250 pfu/cell by adding appropriate amounts of AdCMV-lacZ control virus.
FIG. 3.
Effect of Runx2 expression on AdBMP2-dependent induction of ALP and mineralization. Cells were transduced with the indicated titer of AdCMV-BMP2 in the presence (▴) or absence (●) of AdRunx2 at a titer of 100 pfu/cell. Total viral titer was held constant at 250 pfu/cell by addition of the appropriate titer of AdCMV-lac Z control virus. The cells were harvested (A) at day 6 for ALP assays or (B) at day 9 for measurement of mineralization by von Kossa staining.
Time course studies were next performed to examine cooperative interactions in greater detail (Fig. 4). C3H10T1/2 cells were divided into four groups and transduced with AdCMV-lacZ (250 pfu/cell), AdCMV-Runx2 (100 pfu/cell) plus AdCMV-lacZ (150 pfu/cell), AdCMV-BMP2 (150 pfu/cell) plus AdCMV-lacZ (100 pfu/cell), or AdCMV-Runx2 (100 pfu/cell) plus AdCMV-BMP2 (150 pfu/cell). Cells were analyzed for ALP activity (Fig. 4A), mineral (Figs. 4B and 4G), mRNA (Figs. 4C, 4E, and 4F), and transduced protein (Runx2 and BMP2) expression (Fig. 4D). As was seen in Fig. 3, cells transduced with individual viruses showed only modest amounts of ALP activity or mineral formation, whereas combined treatment gave a dramatic synergistic response that was observed at all time points examined. For example, at day 3, the ALP activity in cells treated with Runx2 plus BMP2 virus was approximately 10-fold greater than the sum of activities in cells individually treated with AdCMV-Runx2 or AdCMV-BMP2. ALP activity in all groups was highest at day 3 and declined at later times, whereas mineral measured either as cell layer–associated calcium or von Kossa–positive matrix continued to accumulate through the duration of the experiment. Synergistic interactions between AdCMV-Runx2 and AdCMV-BMP2 were also apparent when OCN mRNA levels were measured. The amount of synergy actually increased through day 9, although little Runx2 mRNA or protein was detected after days 3–6 (Figs. 4C-4F). BSP mRNA was not detected under any of the conditions tested (result not shown).
Surprisingly, transduction with AdCMV-BMP2 alone only slightly stimulated Runx2 mRNA or protein on days 1 and 3 and was without effect at later times. Nevertheless, this virus strongly induced expression of BMP2 protein (Fig. 4D) and modest amounts of OCN mRNA. BMP2 protein was highest at day 1 and gradually declined, being barely detectable at day 9, whereas OCN mRNA was detected on days 3–9. However, OCN mRNA levels were much lower than those seen with either AdCMV-Runx2 alone or combined virus treatment. Similarly, AdCMV-BMP2 had little effect on Runx2 protein or mRNA levels in cells cotransduced with Runx2 virus (Figs. 4C–4E). Under these conditions, AdCVM-BMP2 increased Runx2 mRNA or protein by only about 50% on days 1 and 3 (measured by scanning densitometry of blots). In contrast, during the same time interval, ALP activity increased 10-fold, whereas OCN mRNA increased 3-fold. Conversely, AdCMV-Runx2 did not induce BMP2 protein at any time examined and did not affect BMP2 levels in AdCMV-BMP2–transduced cells. A final point is that, although AdCMV-Runx2 was considerably more active than AdCMV-BMP2 in stimulating both ALP and OCN mRNA, it was a less potent inducer of mineralization (Figs. 4B and 4G).
In vivo osteogenic activity of virally transduced cells
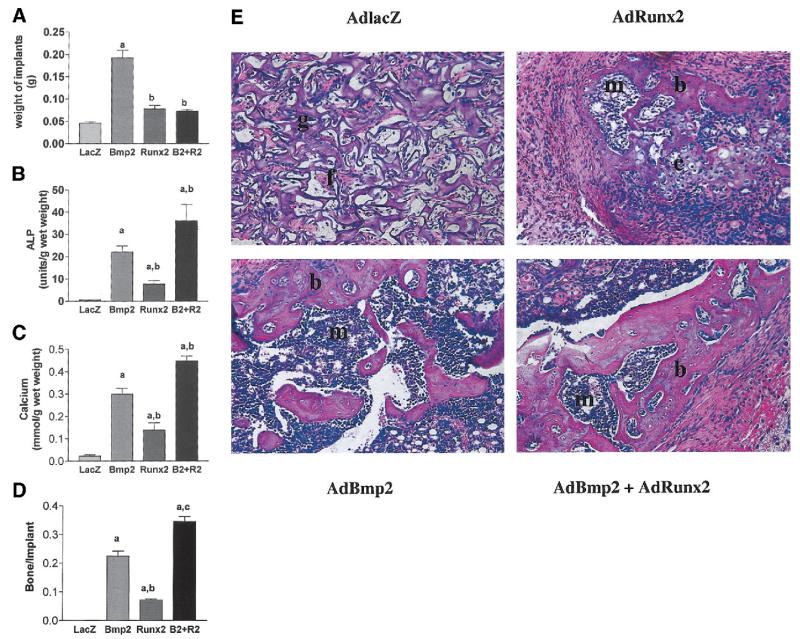
To provide in vivo confirmation of the phenotypic changes in C3H10T1/2 cells seen in vitro, virally transduced cells were adsorbed to collagen sponges and subcutaneously implanted into immunodeficient mice. After 4 weeks, implants were removed and either assayed for changes in wet weight (Fig. 5A), biochemical parameters of bone formation (ALP activity and calcium, Figs. 5B and 5C), or fixed and sectioned for histological analysis (Figs. 5D and 5E). Implants containing AdCMV-lacZ–transduced cells contained only fibrous tissue, residual carrier, and low levels of ALP and calcium. In contrast, clear evidence of bone formation was apparent in cell implants transduced with AdCMV-Runx2 and/or AdCMV-BMP2, although differences were noted between groups. First, the size of implants as reflected by wet weight was higher in the BMP2-only group (Fig. 5A). However, when biochemical parameters of bone for-mation normalized to implant weight were examined, both BMP2 and BMP2 plus Runx2 groups gave higher values than the Runx2-only group, with the highest values being seen in cells transduced with both viruses (Figs. 5B and 5C). Clear differences were also apparent when implant histology was examined (Figs. 5D and 5E). AdCMV-Runx2-implants contained zones of chondrogenesis adjacent to osteogenic regions as well as a small marrow space and fibrous tissue. However, all bone/cartilage formation was restricted to the space within the residual Gelfoam carrier. In contrast, in both AdCMV-BMP2–treated groups, bone formation extended beyond the carrier to form a clearly defined ossicle with a cortex that surrounded a zone of residual carrier, cartilage, and marrow. This cortical bone-like layer was thicker in the AdCMV-Runx2 plus AdCMV-BMP2–treated group, which may explain why the biochemical parameters of bone formation and fractional bone area as measured by histology were higher in these implants relative to the AdCMV-BMP2–only group.
FIG. 5.
In vivo bone formation by virally-transduced C3H10T1/2 cells. Cells were transduced with indicated adenoviruses and implanted into immunodeficient mice as described in Methods. After 4 weeks, transplants were harvested for determination of (A) total wet weigh, (B) ALP activity, or (C) calcium. (D) A morphometric analysis of histological sections from each treatment group. Results are expressed as the ratio of total bone area/total implant area. Statistical analysis: a, significantly different from AdLacZ (p < 0.001); b, significantly different from AdBMP2 (p < 0.05). (E) Histological sections of each treatment group. Implants of cells transduced with control virus contained residual Gelfoam carrier (g) and fibrous tissue (f), but no bone or cartilage. Cells transduced with AdRunx2 alone contained small areas of both bone (b) and cartilage (c) as well as a small marrow cavity (m). Both BMP2 alone and BMP2 plus Runx2 treated groups formed large ossicles with clearly defined cortical and trabecular bone as well as a marrow cavity.
DISCUSSION
This study examines whether bone formation can be enhanced by coexpressing factors that provide distinct, but complimentary, contributions to the osteogenic response. Using an adenovirus-based expression system in C3H10T1/2 mesenchymal cells, we were able to show that Runx2, a bone-related transcription factor, or BMP2, a bone morphogen, stimulate bone formation in cell culture and in vivo. Of particular significance, combined expression of both factors gave a greater osteogenic response than either factor alone. These studies provide further evidence that BMPs and Runx2 initiate separate, but complimentary, events during osteoblast differentiation, and that useful gene therapy–based strategies for bone regeneration may be achieved using the approach of overexpressing combinations of factors.
Osteogenic differentiation and Runx2
Although a large number of transcription factors have been associated with bone cells, only two molecules are known to be specifically required for osteoblast differentiation, Runx2 and, most recently, Osterix (Osx). Disruption of either of these genes by homologous recombination resulted in animals with severe defects in skeletal mineralization.(8,31) Skeletons of Runx2-deficient mice were essentially devoid of mineralization foci, whereas the Osx knockout had mineralized hypertrophic cartilage but not bone. These and related studies on the role of Runx2 in cartilage differentiation(32,33) indicate a generalized role for this molecule in both chondrocyte and osteoblast differentiation, with a more selective requirement for Osx in the formation of osteoblasts.
Consistent with these results, we find that AdCMV-Runx2-transduced C3H10T1/2 cells form both cartilage and bone when implanted into immunodeficient mice. Although we have not yet looked for induction of hypertrophic cartilage markers in vitro, Runx2-transduced cells clearly expressed several osteoblast markers (ALP, OCN, PTH/PTHrP receptor) and formed a weakly mineralized ECM. Runx2-expressing cells shared several properties with primary osteoblasts including a requirement for ascorbic acid–dependent ECM synthesis for optimal differentiation and a clear temporal sequence in the expression of differentiation markers (i.e., induction of ALP at early times followed by OCN and mineralization). Although other laboratories previously showed that overexpression of Runx2 in nonosteogenic cells induced osteoblast-related mRNA,(8,14) ours is the first demonstration that such cells are capable of forming a mineralized ECM in vitro and bone and cartilage in vivo.
Despite the obvious similarities between Runx2-transduced C3H10T1/2 cells and osteoblasts, certain differences were noted. We were unable to detect bone sialoprotein(34) in any C3H10T1/2 cultures regardless of treatment conditions (i.e., transduction with AdCMV-Runx2 or AdCMV-BMP2 individually or in combination). Also, Col1A1 mRNA was not affected by AdCMV-Runx2 treatment, although this gene is known to require Runx2 for optimal expression.(30) These results may indicate that C3H10T1/2 cells require additional factors to fully express an osteoblast phenotype and/or be an indication that transduced cells are expressing both chondrocyte and osteoblast markers as was suggested by the in vivo data. Because only a single cell line was examined in our studies, it is also not clear whether other pluripotent mesenchymal cells or lines will respond to Runx2 and BMP2 in the same way as C3H10T1/2 cells. Further studies will be required to resolve these points.
A second issue raised by our in vitro studies is related to the temporal relationship between the expression of Runx2 and other osteoblast markers. Because Runx2 is known to directly regulate the OCN gene by interacting with specific enhancer sequences in the proximal promoter,(8) we were surprised to find that OCN mRNA was only weakly inducedat day 1 after transduction when Runx2 protein levels were highest, peaked on day 6 when Runx2 had already decreased by more than 90%, and remained elevated to day 12 when Runx2 was no longer detectable (Fig. 2). There are several possible explanations for this result. The simplest is that OCN mRNA is relatively stable and continues to accumulate even after Runx2-dependent transcription of the OCN gene has begun to decrease. Although we have not directly examined this possibility, based on previous work with cultured osteoblasts, we consider it unlikely because (1) during osteoblast differentiation, OCN mRNA levels are generally closely correlated with rates of gene transcription(35,36); and (2) OCN mRNA half-life in differentiating osteoblasts is too short (estimated at 15 h(37)) to explain the persistently high mRNA levels observed in this study. A second possibility is that Runx2 can be inhibitory when present at the high concentrations found immediately after viral transduction and that the residual Runx2 seen at later times is sufficient to maintain OCN expression. In fact, certain in vitro studies support an inhibitory role for Runx2 in controlling its own expression and that of the bone sialoprotein gene.(38,39) However, viral titration studies did not support this view in that induction of Runx2 protein and differentiation markers increased with increasing AdCMV-Runx2 titer (Fig. 1). A final possibility is that Runx2 induces secondary factors that further enhance OCN transcription at later times. Such factors might function independently or in concert with Runx2. Further studies will be required to resolve which of these mechanisms best explains our results.
Relationship between BMP2 and Runx2 in the induction of osteoblast differentiation
BMP2 or BMP7 can upregulate Runx2 mRNA and induce osteoblastic marker genes in C2C12 or C3H10T1/2 cells,(8,11) which has led to the speculation that these BMP responses are mediated by Runx2. While this may be partially correct, BMPs clearly have other activities in bone that are independent of Runx2 in that they can induce osteoblast marker genes in calvarial cells from Cbfa1−/− animals.(10) Previous work by Lee et al.(11) also supports the hypothesis that BMPs must stimulate factors in addition to Runx2 for osteoblast differentiation to occur. Specifically, transfection with a Runx2 expression vector was unable to induce osteoblast differentiation in C2C12 cells unless BMP-related signals were also activated by either direct BMP treatment or transfection with a Smad5 expression vector. This view is also supported by our in vitro studies that detected marked differences in the behavior of AdCMV-BMP2– and AdCMV-Runx2–transduced C3H10T1/2 cells. BMP2-expressing cells exhibited weak induction of ALP and OCN mRNA and barely detectable increases in Runx2 mRNA or protein. Osteoblast markers were more strongly induced in the AdCMV-Runx2 transduced group, but levels were still considerably less than those seen with the combined Runx2 and BMP2 virus treatment. On the other hand, BMP2-expressing cells formed a more extensively mineralized ECM than the Runx2 expressing group (Fig. 4). These results cannot be explained by differences in BMP2 versus Runx2 expression because comparable signals were obtained for both proteins on Western blots. Rather, they suggest that BMP2 and Runx2 each provide unique osteogenic signals to C3H10T1/2 cultures.
To further pursue this concept, we examined the consequences of combined transduction of cells with optimal titers of Runx2 and BMP2-expressing adenoviruses and found a strongly synergistic interaction that was sustained throughout the 15-day time course examined (Figs. 3 and 4). By all criteria examined (ALP activity, OCN mRNA induction, mineral deposition), combined viral transduction stimulated osteoblast differentiation to levels that were 2- to 10-fold greater than the sum of individual levels found in cell transduced with either AdCMV-Runx2 or AdCMV-BMP2. The simplest interpretation of this result is to assume that Runx2 and BMP2 are contributing separate, but complimentary signals to synergistically stimulate osteoblast gene expression. Although we do not currently understand the basis for this synergy, it clearly cannot be explained by AdCMV-BMP2 upregulating endogenous Runx2 or by AdCMV-Runx2 inducing BMP2. Western blot analysis showed that BMP2 virus had a minimal effect on Runx2 levels in the presence or absence of AdCMV-Runx2. Similarly, total (cell layer plus medium) BMP2 protein was not detected in cells transduced only with AdCMV-Runx2, and Runx2 expression did not affect BMP2 levels in the presence of AdCMV-BMP2 (Fig. 4). Other possible explanations for our results include Runx2 stimulation of cellular responsiveness to BMP2 by regulation of BMP receptors or Smad signaling or modulation of Runx2/Smad transcriptional activity by interactions between these two factors on the promoters of target genes. Studies are in progress to clarify this issue.
These cell culture studies are to be contrasted with our in vivo results, which showed that the osteogenic activity of BMP2-expressing cells was clearly greater than the AdCMV-Runx2–transduced group and that the effects of combined virus treatment were additive rather than synergistic (Fig. 5). These differences are likely explained by the fact that the virally expressed BMP2 is a diffusable molecule while Runx2, a nuclear transcription factor, remains in the cell where it was first synthesized. Our previous in vivo work showed that fibroblasts transduced with BMP-expressing adenoviruses form bone that is composed of both host and donor cells.(5) Thus, BMP2 secreted from implanted C3H10T1/2 cells can act in an autocrine manner to stimulate their differentiation as well as recruit host cells to proceed down an osteogenic pathway to generate the prominent ossicle shown in Fig. 5E. In contrast, AdCMV-Runx2–transduced cells in vivo will behave much as they do in vitro and differentiate to osteoblasts as well as chondrocytes without having a major effect on cells from the host. The situation encountered when animals were implanted with cells expressing both BMP2 and Runx2 is considerably more complex but can still be understood by extrapolating from in vitro results. The size of the ossicles formed in this case (as reflected by the wet weight; Fig 5A) was approximately the same as with cells transduced with AdCMV-Runx2 alone and considerably less than the AdCMV-BMP2–transduced group. However, the fraction of each ossicle that contained bone, measured by ALP activity or calcium/g wet weight or by the fractional bone area of implants (Figs. 5B, 5C, and 5E), was clearly greater with combined virus treatment. Based on our in vitro results, C3H10T1/2 cells transduced with BMP2, and Runx2 adenoviruses would be predicted to differentiate into osteoblasts to a greater extent than cells transduced with either virus alone. However, this more rapid bone formation might be expected to restrict the release of BMP2 to host cells and thereby reduce the overall size of the resulting ossicle. Clearly, a definitive proof of this hypothesis will require separate examination of the contribution of implanted and host cells to the overall osteogenic response as might be obtained if cells were implanted in diffusion chambers to separate them from the host.
In summary, our results clearly show that the osteogenic activity of BMP-expressing cells can be modulated by coexpressing factors like Runx2 that are major regulators of the osteoprogenitor lineage. Our studies may have important clinical implications in that it is through manipulations of this type that it may be possible to control both the extent and type of bone formed in regenerating sites.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health Grants DE13386 (RTF), DE11723 (RTF), and DE13835 (PHK).
Footnotes
The authors have no conflict of interest.
REFERENCES
- 1.Yamaguchi A, Komori T, Suda T. Regulation of osteoblast differentiation mediated by bone morphogenetic proteins, hedgehogs, and Cbfa1. Endocr Rev. 2000;21:393–411. doi: 10.1210/edrv.21.4.0403. [DOI] [PubMed] [Google Scholar]
- 2.Lian JB, Stein GS. Osteoblast biology. In: Marcus R, Feldman D, Kelsey J, editors. Osteoporosis. 2nd ed. Vol. 1. Academic Press; San Diego, CA, USA: 2001. pp. 21–71. [Google Scholar]
- 3.Geesink RG, Hoefnagels NH, Bulstra SK. Osteogenic activity of OP-1 bone morphogenetic protein (BMP-7) in a human fibular defect. J Bone Joint Surg Br. 1999;81:710–718. doi: 10.1302/0301-620x.81b4.9311. [DOI] [PubMed] [Google Scholar]
- 4.Franceschi RT, Wang D, Krebsbach PH, Rutherford RB. Gene therapy for bone formation: In vitro and in vivo osteogenic activity of an adenovirus expressing BMP7. J Cell Biochem. 2000;78:476–486. doi: 10.1002/1097-4644(20000901)78:3<476::aid-jcb12>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 5.Krebsbach PH, Gu K, Franceschi RT, Rutherford RB. Gene therapy-directed osteogenesis: BMP-7-transduced human fibroblasts form bone in vivo. Hum Gene Ther. 2000;11:1201–1210. doi: 10.1089/10430340050015248. [DOI] [PubMed] [Google Scholar]
- 6.Baker JC, Harland RM. From receptor to nucleus: The Smad pathway. Curr Opin Genet Dev. 1997;7:467–473. doi: 10.1016/s0959-437x(97)80072-x. [DOI] [PubMed] [Google Scholar]
- 7.Whitman M. Smads and early developmental signaling by the TGFbeta superfamily. Genes Dev. 1998;12:2445–2462. doi: 10.1101/gad.12.16.2445. [DOI] [PubMed] [Google Scholar]
- 8.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 9.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 10.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 11.Lee KS, Kim HJ, Li QL, Chi XZ, Ueta C, Komori T, Wozney JM, Kim EG, Choi JY, Ryoo HM, Bae SC. Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol Cell Biol. 2000;20:8783–8792. doi: 10.1128/mcb.20.23.8783-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jonk LJ, Itoh S, Heldin CH, ten Dijke P, Kruijer W. Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-beta, activin, and bone morphogenetic protein-inducible enhancer. J Biol Chem. 1998;273:21145–21152. doi: 10.1074/jbc.273.33.21145. [DOI] [PubMed] [Google Scholar]
- 13.Hanai J, Chen LF, Kanno T, Ohtani-Fujita N, Kim WY, Guo WH, Imamura T, Ishidou Y, Fukuchi M, Shi MJ, Stavnezer J, Kawabata M, Miyazono K, Ito Y. Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Calpha promoter. J Biol Chem. 1999;274:31577–1582. doi: 10.1074/jbc.274.44.31577. [DOI] [PubMed] [Google Scholar]
- 14.Xiao ZS, Hinson TK, Quarles LD. Cbfa1 isoform overexpression upregulates osteocalcin gene expression in non-osteoblastic and pre-osteoblastic cells. J Cell Biochem. 1999;74:596–605. [PubMed] [Google Scholar]
- 15.Hardy S, Kitamura M, Harris-Stansil T, Dai Y, Phipps ML. Construction of adenovirus vectors through Cre-lox recombination. J Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franceschi RT, Iyer BS. Relationship between collagen synthesis and expression of the osteoblast phenotype in MC3T3–E1 cells. J Bone Miner Res. 1992;7:235–246. doi: 10.1002/jbmr.5650070216. [DOI] [PubMed] [Google Scholar]
- 17.Owen TA, Aronow M, Shalhoub V, Barone LM, Wilming L, Tassinari MS, Kennedy MB, Pockwinse S, Lian JB, Stein GS. Progressive development of the rat osteoblast phenotype in vitro: Reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. J Cell Physiol. 1990;143:420–430. doi: 10.1002/jcp.1041430304. [DOI] [PubMed] [Google Scholar]
- 18.Winnard RG, Gerstenfeld LC, Toma CD, Franceschi RT. Fibronectin gene expression, synthesis, and accumulation during in vitro differentiation of chicken osteoblasts. J Bone Miner Res. 1995;10:1969–1977. doi: 10.1002/jbmr.5650101217. [DOI] [PubMed] [Google Scholar]
- 19.Thomas PS. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci USA. 1980;77:5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Celeste AJ, Rosen V, Bueker JL, Kriz R, Wang EA, Wozney JM. Isolation of the human gene for bone gla protein utilizing mouse and rat cDNA clones. EMBO J. 1986;5:1885–1890. doi: 10.1002/j.1460-2075.1986.tb04440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young MF, Ibaraki K, Kerr JM, Lyu MS, Kozak CA. Murine bone sialoprotein (BSP): CDNA cloning, mRNA expression, and genetic mapping. Mamm Genome. 1994;5:108–111. doi: 10.1007/BF00292337. [DOI] [PubMed] [Google Scholar]
- 22.Terao M, Studer M, Gianni M, Garattini E. Isolation and characterization of the mouse liver/bone/kidney-type alkaline phosphatase gene. Biochem J. 1990;268:641–648. doi: 10.1042/bj2680641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liau G, Yamada Y, de Crombrugghe B. Coordinate regulation of the levels of type III and type I collagen mRNA in most but not all mouse fibroblasts. J Biol Chem. 1985;260:531–536. [PubMed] [Google Scholar]
- 24.Renkawitz R, Gerbi SA, Glatzer KH. Ribosomal DNA of fly Sciara coprophila has a very small and homogeneous repeat unit. Mol Gen Genet. 1979;173:1–13. doi: 10.1007/BF00267685. [DOI] [PubMed] [Google Scholar]
- 25.Manolagas SC, Burton DW, Deftos LJ. 1, 25-Dihydroxyvitamin D stimulates the alkaline phosphatase activity of osteoblast-like cells. J Biol Chem. 1981;256:7115–7117. [PubMed] [Google Scholar]
- 26.Schneider WC. Determination of nucleic acids in tissues by pentose analysis. Methods Enzymol. 1957;3:680–684. [Google Scholar]
- 27.McCauley LK, Koh AJ, Beecher CA, Cui Y, Decker JD, Franceschi RT. Effects of differentiation and transforming growth factor beta 1 on PTH/PTHrP receptor mRNA levels in MC3T3–E1 cells. J Bone Miner Res. 1995;10:1243–1255. doi: 10.1002/jbmr.5650100815. [DOI] [PubMed] [Google Scholar]
- 28.Krebsbach PH, Kuznetsov SA, Satomura K, Emmons RV, Rowe DW, Robey PG. Bone formation in vivo: Comparison of osteogenesis by transplanted mouse and human marrow stromal fibroblasts. Transplantation. 1997;63:1059–1069. doi: 10.1097/00007890-199704270-00003. [DOI] [PubMed] [Google Scholar]
- 29.Taylor SM, Jones PA. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17:771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- 30.Kern B, Shen J, Starbuck M, Karsenty G. Cbfa1 contributes to the osteoblast-specific expression of Type I collagen genes. J Biol Chem. 2001;276:7101–7107. doi: 10.1074/jbc.M006215200. [DOI] [PubMed] [Google Scholar]
- 31.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 32.Inada M, Yasui T, Nomura S, Miyake S, Deguchi K, Himeno M, Sato M, Yamagiwa H, Kimura T, Yasui N, Ochi T, Endo N, Kitamura Y, Kishimoto T, Komori T. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev Dyn. 1999;214:279–290. doi: 10.1002/(SICI)1097-0177(199904)214:4<279::AID-AJA1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 33.Kim IS, Otto F, Zabel B, Mundlos S. Regulation of chondrocyte differentiation by Cbfa1. Mech Dev. 1999;80:159–170. doi: 10.1016/s0925-4773(98)00210-x. [DOI] [PubMed] [Google Scholar]
- 34.Ganss B, Kim RH, Sodek J. Bone sialoprotein. Crit Rev Oral Biol Med. 1999;10:79–98. doi: 10.1177/10454411990100010401. [DOI] [PubMed] [Google Scholar]
- 35.Shalhoub V, Bortell R, Jackson ME, Marks SC, Jr, Stein JL, Lian JB, Stein GS. Transcriptionally active nuclei isolated from intact bone reflect modified levels of gene expression in skeletal development and pathology. J Cell Biochem. 1994;55:182–189. doi: 10.1002/jcb.240550205. [DOI] [PubMed] [Google Scholar]
- 36.Xiao G, Cui Y, Ducy P, Karsenty G, Franceschi RT. Ascorbic acid-dependent activation of the osteocalcin promoter in MC3T3–E1 preosteoblasts: Requirement for collagen matrix synthesis and the presence of an intact OSE2 sequence. Mol Endocrinol. 1997;11:1103–1113. doi: 10.1210/mend.11.8.9955. [DOI] [PubMed] [Google Scholar]
- 37.Franceschi RT, Iyer BS, Cui Y. Effects of ascorbic acid on collagen matrix formation and osteoblast differentiation in murine MC3T3–E1 cells. J Bone Miner Res. 1994;9:843–854. doi: 10.1002/jbmr.5650090610. [DOI] [PubMed] [Google Scholar]
- 38.Javed A, Barnes GL, Jasanya BO, Stein JL, Gerstenfeld L, Lian JB, Stein GS. Runt homology domain transcription factors (Runx Cbfa, and AML) mediate repression of the bone sialoprotein promoter: Evidence for promoter context-dependent activity of Cbfa proteins. Mol Cell Biol. 2001;21:2891–2905. doi: 10.1128/MCB.21.8.2891-2905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drissi H, Luc Q, Shakoori R, Chuva De, Sousa Lopes S, Choi JY, Terry A, Hu M, Jones S, Neil JC, Lian JB, Stein JL, Van Wijnen AJ, Stein GS. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J Cell Physiol. 2000;184:341–350. doi: 10.1002/1097-4652(200009)184:3<341::AID-JCP8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]