Structural studies of the interaction of S-adenosylmethionine with the [4Fe-4S] clusters in biotin synthase and pyruvate formate-lyase activating enzyme (original) (raw)
Abstract
The diverse reactions catalyzed by the radical-SAM superfamily of enzymes are thought to proceed via a set of common mechanistic steps, key among which is the reductive cleavage of S-adenosyl-L-methionine (SAM) by a reduced [4Fe-4S] cluster to generate an intermediate deoxyadenosyl radical. A number of spectroscopic studies have provided evidence that SAM interacts directly with the [4Fe-4S] clusters in several of the radical-SAM enzymes; however, the molecular mechanism for the reductive cleavage has yet to be elucidated. Selenium X-ray absorption spectroscopy (Se-XAS) was used previously to provide evidence for a close interaction between the Se atom of selenomethionine (a cleavage product of Se-SAM) and an Fe atom of the [4Fe-4S] cluster of lysine-2,3-aminomutase (KAM). Here, we utilize the same approach to investigate the possibility of a similar interaction in pyruvate formate-lyase activating enzyme (PFL-AE) and biotin synthase (BioB), two additional members of the radical-SAM superfamily. The results show that the latter two enzymes do not exhibit the same Fe-Se interaction as was observed in KAM, indicating that the methionine product of reductive cleavage of SAM does not occupy a well-defined site close to the cluster in PFL-AE and BioB. These results are interpreted in terms of the differences among these enzymes in their use of SAM as either a cofactor or a substrate.
Keywords: Biotin synthase (BioB), iron-sulfur cluster, pyruvate formate-lyase activating enzyme (PFL-AE), radical SAM superfamily, X-ray absorption fine structure, X-ray absorption spectroscopy
Iron-sulfur (Fe-S) clusters constitute one of the most structurally and functionally diverse classes of biological prosthetic groups. The primary involvement of Fe-S clusters in biological systems is the mediation of electron transport. However, alternative functions have emerged in recent years, such as regulation of genes and structural roles (Beinert et al. 1997; Beinert 2000). Recently, an additional new role has been revealed involving the initiation of radical catalysis via the reductive cleavage of S-adenosylmethionine (SAM) by an Fe-S cluster (Cheek and Broderick 2001; Fontecave et al. 2001; Frey 2001). This class of enzymes, characterized by a C-X3-C-X2-C conserved binding motif and dependence on SAM, has been identified as the radical-SAM superfamily (Sofia et al. 2001). The diverse range of functions in this superfamily is remarkable, as represented by the most well-characterized enzymes, the activatases for pyruvate formate-lyase (PFL; Broderick et. al. 1997; Külzer et al. 1998) and anaerobic ribonucleotide reductase (ANAR; Ollagnier et al. 1997), which are responsible for creation of stable protein-based radicals; KAM, (Lieder et al. 1998), which catalyzes the interconversion of α-L-lysine and β-L-lysine; and BioB (Sanyal et al. 1994; Duin et al. 1997; Guianvarc’h et al. 1997) and lipoate synthase (LipA; Busby et. al. 1999; Ollagnier-de Choudens and Fontecave 1999), which are involved in the terminal step of the biosyntheis of the vitamins, biotin and lipoic acid, respectively.
PFL-AE generates a stable glycyl radical on PFL, which is responsible for the first step in bacterial anaerobic glucose metabolism, the conversion of pyruvate and coenzyme-A to formate and acetyl-CoA (Knappe et al. 1993; Wong and Kozarich 1994; Broderick et. al. 1997). Anaerobically isolated PFL-AE contains mostly cuboidal [3Fe-4S]+ clusters and minor quantities of [4Fe-4S]2+, [2Fe-2S]2+, and linear [3Fe-4S]+ clusters (Broderick et al. 2000; Krebs et. al. 2000); however, a homogeneous [4Fe-4S]2+,+ form is prepared under reducing conditions. The reduced [4Fe-4S]+ cluster in PFL-AE has been demonstrated to be catalytically essential for cleaving SAM and generating the protein-based radical on PFL (Henshaw et. al. 2000).
The final step of the biotin biosynthetic pathway is catalyzed by BioB and involves the insertion of S into dethiobiotin (DTB) at unactivated C-H bonds. Biotin synthases from Escherichia coli (Sanyal et al. 1994), Bacillus sphaericus (Méjean et al. 1995), and Arabidopsis thaliana (Baldet et al. 1997) have been purified to homogeneity from recombinant strains of E. coli or B. sphaericus. The E. coli enzyme is a 78-kD homodimer and, as isolated, contains one [2Fe-2S]2+ cluster per monomer (Sanyal et al. 1994; Ollagnier-De Choudens et al. 2000). The presence of [2Fe-2S]2+ clusters was also reported for the B. sphaericus (Méjean et al. 1995) and A. thaliana (Baldet et al. 1997) enzymes. Spectroscopic studies have demonstrated that anaerobic reconstitution of apo BioB with Fe and S results in the formation of one [4Fe-4S]2+ cluster per monomer (Ollagnier-de Choudens et al. 2000). Recently, Fontecave and colleagues (Ollagnier-de Choudens et al. 2002) provided direct evidence that the [4Fe-4S]+ cluster is responsible for reductive cleavage of SAM and is ligated by the cysteines in the C-X3-C-X2-C motif.
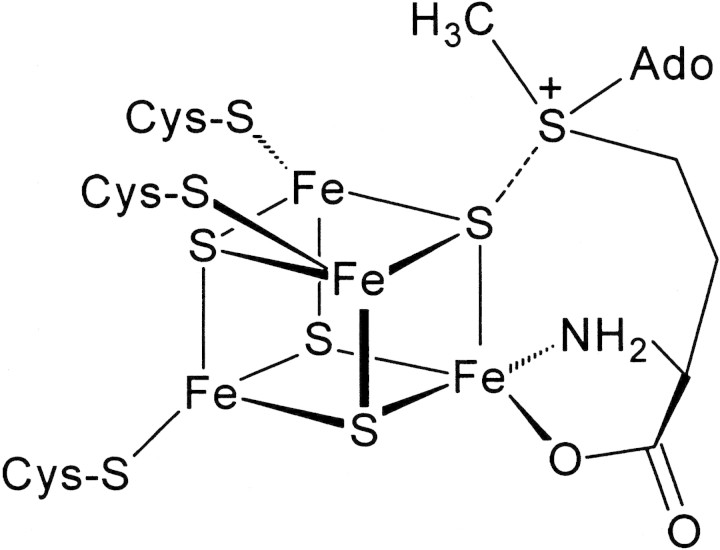
Mechanistic details of Fe-S cluster-mediated SAM cleavage are beginning to emerge through spectroscopic studies aimed at examining the nature of the interaction of SAM with the [4Fe-4S]2+,+ cluster (Cosper et al. 2000; Krebs et al. 2002; Walsby et al. 2002a,b). Despite the diversity in functions of the enzymes in the radical-SAM superfamily, cleavage of SAM by a reduced [4Fe-4S] cluster has been postulated to occur by a similar mechanism. Recent experiments have suggested that there is a close interaction of SAM with the [4Fe-4S]2+,+ clusters of KAM, PFL-AE, and BioB. A non-cysteinyl-ligated unique Fe site in the [4Fe-4S]2+ cluster of PFL-AE has been identified by Mössbauer spectroscopy (Krebs et al. 2002). Moreover, the Mössbauer parameters of this unique Fe can be dramatically affected by the addition of SAM to PFL-AE, suggesting that SAM must be interacting with the unique Fe (Krebs et al. 2002). Furthermore, electron-nuclear double-resonance (ENDOR) spectroscopy has shown that SAM coordinates the unique Fe in the [4Fe-4S]2+,+ cluster (Walsby et al. 2002a,b). These studies led to a model in which the amino acid moiety of SAM coordinates the unique Fe, whereas the sulfonium ion interacts with one of the μ3-bridging sulfides of the [4Fe-4S] cluster (Fig. 1▶). Similarly, a combination of EPR, resonance Raman, and Mössbauer spectroscopies has provided evidence for the interaction of SAM at a unique Fe site of the [4Fe-4S]2+ cluster in BioB (Cosper et. al. 2002).
Figure 1.
Proposed mode of interaction of SAM with the [4Fe-4S] cluster of PFL-AE. The model is based on electron-nuclear double resonance and Mössbauer studies (Krebs et al. 2002; Walsby et al. 2002a,b).
In contrast, Se XAS studies on KAM; however, have led to the suggestion that interaction between a cluster Fe and the sulfonium ion plays an important role in the reductive cleavage of SAM. Specifically, a 2.7 Å interaction between an Fe atom of the [4Fe-4S]2+ cluster and the Se in selenomethionine (SeMet), a cleavage product of Se-adenosyl-L-selenomethionine (Se-SAM), was observed when [4Fe-4S]2+ KAM was incubated with Se-SAM, dithionite, and the substrate analog, _trans_-3,4-dehydrolysine. The same interaction was observed upon incubation of KAM with SeMet, deoxyadenosine, and _trans_-3,4-dehydrolysine (Cosper et al. 2000). These observations led to the proposal of a mechanism by which an Fe atom of the [4Fe-4S] cluster interacts directly with the sulfonium ion of SAM, allowing for electron transfer to SAM and subsequent adenosyl radical formation (Cosper et al. 2000). In light of the different interpretations of the SAM-cluster interactions for PFL-AE and BioB relative to KAM, it was of interest to determine whether the phenomenon of the sulfonium-Fe interaction is unique to KAM or is a general feature of the radical-SAM family. We have therefore used Se XAS to probe the interaction between Se-SAM and the [4Fe-4S] clusters in BioB and PFL-AE.
Results and Discussion
Previous Se XAS results for KAM identified a ∼2.7 Å FT peak, which was interpreted as an interaction between a Fe atom from the [4Fe-4S] cluster and the Se atom of SeMet (Cosper et al. 2000). This interaction was observed only in the presence of both the cleavage products of Se-SAM and the substrate analog _trans_-4,5-dehydrolysine. Such samples could be prepared either by incubating [4Fe-4S]2+-KAM with Se-SAM, reductant and the substrate analog, the combination of which promoted reductive cleavage of Se-SAM to 5′-deoxyadenosine and Se-Met, or by incubating the enzyme with the products of Se-SAM cleavage (SeMet and 5′-deoxyadenosine) and substrate or substrate analog. Of these two methods, the latter provides the simplest assessment of the Se-Fe interaction, because it can be done in the presence of the native substrate rather than the substrate analog. Although there was no evidence for interaction of the Se of Se-SAM with a cluster Fe, the discovery that the Se of SeMet lies close to an Fe site of the cluster (Se-Fe distance of ∼2.7 Å), led to the hypothesis that reductive cleavage of SAM involves interaction between a cluster Fe and the sulfonium ion of SAM.
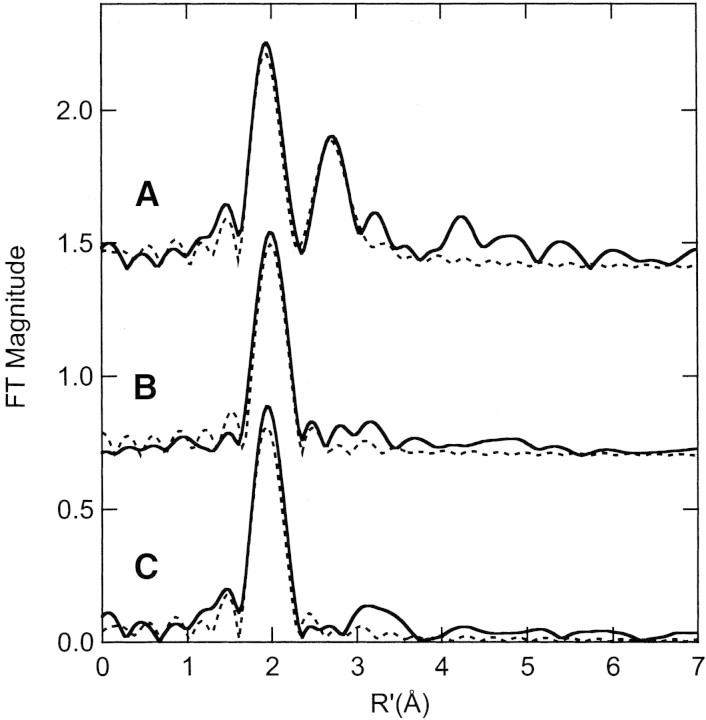
Se XAS data were collected for BioB and PFL-AE using both the turnover technique (in which Se-SAM and substrate are added to the active enzyme) and the addition of products technique. Incubation of BioB with SeMet, 5′-deoxyadenosine and biotin or of PFL-AE with SeMet, 5′-deoxyadenosine and PFL, results in XAS and FT spectra that show no evidence for a close Se-Fe interaction (Fig. 2▶). The Se EXAFS spectra for both BioB with SeMet, 5′-deoxyadenosine, and biotin and PFL-AE with SeMet, 5′-deoxyadenosine, and PFL can be best fit assuming a Se-C2 coordination environment with a Se-C distance of 1.93–1.96 Å and with reasonable Debye-Waller factor values (Fits 2, 6; Table 1▶). These fits are similar to those obtained for the model compound, SeMet (Cosper et al. 2000). Attempts to include a 2.7 Å Se-Fe interaction provided unsatisfactory fits. Additionally, no Se-Fe interaction was observed by use of the turnover technique, in which samples of BioB and PFL-AE were incubated with Se-SAM and substrate or when PFL-AE was incubated with Se-SAM alone (see supplemental material).
Figure 2.
Fourier transforms (over k = 2–12.5 Å−1) for (A) lysine 2,3-aminomutase incubated with SeMet, 5′deoxyadenosine, and didehydrolysine (solid line) and the calculated spectra for Se-C,Fe (Cosper et al. 2000); (B) PFL-AE [4Fe-4S]2+ incubated with SeMet and PFL (solid line) and the calculated spectra for Se-C2 (broken line; Fit 2, Table 1▶); and (C) BioB incubated with SeMet, d-biotin, and 5′deoxyadenosine (solid line) and the calculated spectra for Se-C2 (broken line; Fit 6, Table 1▶).
Table 1.
Curve fitting results for Se EXAFS of PFL-AE and BioB incubated with SeMeta
| Sample filename (k range) Δ_k_3χ | Fit | Shell | Ras (Å) | σas2 (Å2) | ΔE0 (eV) | f‘b |
|---|---|---|---|---|---|---|
| PFL-AE + SeMet, PFL, 5′deoxyAdo | 1 | Se-C | 1.96 | −0.0016 | 2.07 | 0.144 |
| EPRPA (2–12.5 Å−1) | 2 | Se-C2 | 1.96 | 0.0016 | 1.05 | 0.119 |
| Δ_k_3χ = 5.70 | 3 | Se-C3 | 1.95 | 0.0039 | −1.26 | 0.121 |
| 4 | Se-C4 | 1.94 | 0.0060 | −4.41 | 0.136 | |
| BioB + SeMet, d-biotin, 5′deoxyAdo | 5 | Se-C | 1.94 | −0.0018 | −1.97 | 0.130 |
| EBBDA (2–12.5 Å−1) | 6 | Se-C2 | 1.93 | 0.0012 | −4.75 | 0.106 |
| Δ_k_3χ = 5.34 | 7 | Se-C3 | 1.93 | 0.0035 | −6.95 | 0.111 |
| 8 | Se-C4 | 1.92 | 0.0054 | −8.64 | 0.129 |
The differences in the Se-XAS results of KAM (Cosper et al. 2000) compared with those reported herein for PFL-AE and BioB may reflect differences in the involvement of SAM in the mechanisms of these enzymes. One major difference between KAM and the other well-characterized radical-SAM enzymes is that KAM uses SAM catalytically. In KAM, SAM is cleaved reversibly by the [4Fe-4S]+ cluster to generate a 5′-deoxyadenosyl radical intermediate and methionine, and the SAM is regenerated after each interconversion of α-lysine to β-lysine; in other words, one SAM is involved in catalyzing numerous lysine rearrangements (Moss and Frey 1987). In the other well-characterized members of the radical-SAM family, such as PFL-AE and BioB, SAM is a reagent, being converted to methionine and 5′-deoxyadenosine with each substrate turnover (Knappe et. al. 1984; Guianvarc’h et al. 1997).
These differing roles of SAM in KAM compared with BioB and PFL-AE—as a catalytic cofactor in the former and as a substrate in the latter—may be reflected in differences in the mode of SAM binding as suggested previously (Walsby et al. 2002a) and/or differences in the binding affinity of the methionine product. For KAM, the products of SAM cleavage must remain in close proximity in the active site, as completion of catalysis is accompanied by reformation of SAM from methionine and the 5′-deoxyadenosyl radical. Hence, it is possible that interaction between an Fe atom of the [4Fe-4S] cluster in KAM and the methionine thioether serves to increase the methionine-binding affinity, thereby holding the methionine in place to rejoin the 5′-deoxyadenosyl radical. In contrast, the products of SAM cleavage must leave the active site of PFL-AE and BioB after each turnover, being replaced by a new SAM to be used in the next catalytic cycle. Because no interaction between an Fe atom from the [4Fe-4S] cluster and the Se atom of SeMet was observed for either PFL-AE or BioB, the SeMet is either absent from the active site, bound in a similar configuration, but with the Se atom more distant from a cluster Fe atom, or occupying several different orientations near the [4Fe-4S]2+ cluster. In the absence of information on methionine-binding affinity for KAM, PFL-AE, and BioB, it is not possible to discriminate between these possibilities or address the possibility of differences in interaction of SAM with the [4Fe-4S] cluster among members of the radical-SAM superfamily. However, what is clear from the results presented herein, is that a well-defined interaction between the product methionine and an Fe of the [4Fe-4S] cluster as seen in KAM (Cosper et al. 2000) is not a general property of radical-SAM family of Fe-S enzymes. Rather, it may be confined to members of this class, which use SAM in a catalytic role rather than as a substrate.
Materials and methods
E. coli [4Fe-4S]2+ BioB (Ollagnier-de Choudens et al. 2000; Cosper et. al. 2002) and Se-SAM (Park et al. 1996; Walsby et al. 2002a), were prepared as described previously. SeMet, 5′-deoxyadenosine, biotin, and DTB were purchased from Sigma-Aldrich. Four BioB samples were prepared in 50 mM Hepes (pH 7.5) as follows: (1) 1.3 mM [4Fe-4S]2+ BioB; (2) 1.3 mM [4Fe-4S]2+ BioB, 3.0 mM 5′-deoxyadenosine, and 1.3 mM SeMet; (3) 1.3 mM [4Fe-4S]2+ BioB, 3.2 mM 5′-deoxyadenosine, 1.3 mM SeMet, and 2.7 mM biotin; (4) 1.5 mM [4Fe-4S]2+ BioB, 1.6 mM Se-SAM, and 3.0 mM DTB. Four additional BioB samples were prepared in 50 mM Tris-HCl (pH 8.5) with 0.2 M NaCl, 1 mM DTT as follows: (1) 1.3 mM [4Fe-4S]2+ BioB; (2) 1.3 mM [4Fe-4S]2+ BioB with 1.0 mM Se-SAM; (3) 2.8 mM [2Fe-2S]2+/[4Fe-4S]2+ BioB; and (4) 2.8 mM [2Fe-2S]2+/[4Fe-4S]2+ BioB with 2.1 mM Se-SAM. After each sample was prepared and incubated at ambient temperature for 30 min, ethylene glycol was added to a final concentration of 20%. All preparation and manipulation of the eight BioB samples was performed inside a Vacuum Atmosphere glove box with O2 levels <5 ppm.
E. coli PFL-AE was prepared as described previously (Broderick et al. 2000; Walsby et al. 2002a). Five different PFL-AE samples were prepared in 50 mM HEPES (pH 7.5) as follows: (1) 1 mM PFL-AE, 25% (v/v) glycerol, 1 mM sodium dithionite, 200 μM 5-deazariboflavin, and 0.95 mM Se-SAM; (2) 1 mM PFL-AE, 25% (v/v) glycerol, and 0.95 mM Se-SAM; (3) 1 mM PFL-AE, 25% glycerol, 0.95 mM SeMet, and 4 mM 5′-deoxyadenosine; (4) 0.44 mM PFL-AE, 25% glycerol, 0.38 mM SeMet, 4 mM 5′-deoxyadenosine, and 1.1 mM PFL; and (5) 0.4 mM PFL-AE, 25% (v/v) glycerol, 0.8 mM sodium dithionite, 100 μM 5-deazariboflavin, 0.4 mM PFL, and 0.35 mM Se-SAM. Samples 1 and 5 were prepared by combining PFL-AE, glycerol, dithionite, and 5-deazariboflavin and illuminating for 30–60 min to generate the [4Fe-4S]+ state of PFL-AE. Following illumination, Se-SAM (1) or Se-SAM and PFL (5) were added. Sample 5 represents the turnover sample, whereas sample 4 represents the product addition method sample. All samples were prepared in an inert atmosphere box (Mbraun) operating at <3 ppm O2.
XAS data were collected at Stanford Synchrotron Radiation Laboratory (SSRL), beamline 7-3, with the SPEAR storage ring at 3.0 GeV and 60–100 mA. Fluorescence data were collected using a Ge solid-state array detector and a Si(220) double-crystal monochromator that was 50% detuned for harmonic rejection. Calibration was achieved using a powdered elemental Se standard (first inflection, 12658 eV). EXAFS analysis was performed with the EXAFSPAK software (www-ssrl.slac.stanford.edu/exafspak.html), according to standard procedures (Scott 1985). Fourier transform plots were generated with sulfur-based phase correction. Other XAS parameters were as described previously (Cosper et al. 2000, 2001).
Electronic supplemental material
Plots of XAS data for BioB and PFL-AE are available.
Acknowledgments
The XAS data were collected at SSRL, which is operated by the Department of Energy, Division of Chemical Sciences. The SSRL Biotechnology program is supported by the National Institutes of Health, Biomedical Resource Technology Program, Division of Research Resources. This investigation was supported in part by the National Institutes of Health (GM42025 to R.A.S., GM62542 to M.K.J., GM54608 to J.B.B., and NIH National Research Service Postdoctoral Fellowship, DK 59730 to M.M.C.). The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
- SAM, S-adenosyl-L-methionine
- Se-XAS, selenium X-ray absorption spectroscopy
- KAM, lysine-2,3-aminomutase
- PFL-AE, pyruvate formate-lyase activating enzyme
- BioB, biotin synthase
- Fe-S, iron-sulfur
- PFL, pyruvate formate-lyase
- ANAR, anaerobic ribonucleotide reductase
- LipA, lipoate synthase
- DTB, dethiobiotin
- ENDOR, electron-nuclear double resonance
- SeMet, selenomethionine
- Se-SAM, Se-adenosyl-L-selenomethionine
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0302203.
References
- Baldet, P., Alban, C., and Douce, R. 1997. Biotin synthesis in higher plants: Purification and characterization of bioB gene product equivalent from Arabidopsis thaliana overexpressed in Escherichia coli and its subcellular localization in pea leaf cells. FEBS Lett. 419 206–210. [DOI] [PubMed] [Google Scholar]
- Beinert, H. 2000. Iron-sulfur proteins: Ancient structures, still full of surprises. J. Biol. Inorg. Chem. 5 2–15. [DOI] [PubMed] [Google Scholar]
- Beinert, H., Holm, R.H., and Münck, E. 1997. Iron-sulfur clusters: Nature’s modular, multipurpose structures. Science 277 653–659. [DOI] [PubMed] [Google Scholar]
- Broderick, J.B., Duderstadt, R.E., Fernandez, D.C., Wojtuszewski, K., Henshaw, T.F., and Johnson, M.K. 1997. Pyruvate formate-lyase activating enzyme is an iron-sulfur protein. J. Am. Chem. Soc. 119 7396–7397. [Google Scholar]
- Broderick, J.B., Henshaw, T.F., Cheek, J., Wojtuszewski, K., Smith, S.R., Trojan, M.R., McGhan, R.M., Kopf, A., Kibbey, M., and Broderick, W.E. 2000. Pyruvate formate-lyase-activating enzyme: Strictly anaerobic isolation yields active enzyme containing a [3Fe-4S]+ cluster. Biochem. Biophys. Res. Commun. 269 451–456. [DOI] [PubMed] [Google Scholar]
- Busby, W.B., Schelvis, J.P.M., Yu, D.S., Babcock, G.T., and Marletta, M.A. 1999. Lipoic acid biosynthesis: LipA is an iron-sulfur protein. J. Am. Chem. Soc. 121 4706–4707. [Google Scholar]
- Cheek, J., and Broderick, J.B. 2001. Adenosylmethionine-dependent iron-sulfur enzymes: Versatile clusters in a radical new role. J. Biol. Inorg. Chem. 6 209–226. [DOI] [PubMed] [Google Scholar]
- Cosper, M.M., Jameson, G.N.L., Davydov, R., Eidsness, M.K., Hoffman, B.M., Huynh, B.H., and Johnson, M.K. 2002. The [4Fe-4S]2+ cluster in reconstituted biotin synthase binds _S_-adenosyl-L-methionine. J. Am. Chem. Soc. 124 14006–14007. [DOI] [PubMed] [Google Scholar]
- Cosper, N.J., Booker, S.J., Ruzicka, F., Frey, P.A., and Scott, R.A. 2000. Direct FeS cluster involvement in generation of a radical in lysine 2,3-aminomutase. Biochemistry 39 15668–15673. [DOI] [PubMed] [Google Scholar]
- Cosper, N.J., D’Souza, V.M., Scott, R.A., and Holz, R.C. 2001. Structural evidence that the methionyl aminopeptidase from Escherichia coli is a mononuclear metalloprotease. Biochemistry 40 13302–13309. [DOI] [PubMed] [Google Scholar]
- Duin, E.C., Lafferty, M.E., Crouse, B.R., Allen, R.M., Sanyal, I., Flint, D.H., and Johnson, M.K. 1997. [2Fe-2S] to [4Fe-4S] cluster conversion in Escherichia coli biotin synthase. Biochemistry 36 11811–11820. [DOI] [PubMed] [Google Scholar]
- Fontecave, M., Mulliez, E., and Ollagnier-de-Choudens, S. 2001. Adenosylmethionine as a source of 5′-deoxyadenosyl radicals. Curr. Opin. Chem. Biol. 5 506–511. [DOI] [PubMed] [Google Scholar]
- Frey, P.A. 2001. Radical mechanisms of enzymatic catalysis. Annu. Rev. Biochem. 70 121–148. [DOI] [PubMed] [Google Scholar]
- Guianvarc’h, D., Florentin, D., Tse Sum Bui, B., Nunzi, F., and Marquet, A. 1997. Biotin synthase, a new member of the family of enzymes which uses _S_-adenosylmethionine as a source of deoxyadenosyl radical. Biochem. Biophys. Res. Commun. 236 402–406. [DOI] [PubMed] [Google Scholar]
- Henshaw, T.F., Cheek, J., and Broderick, J.B. 2000. The [4Fe-4S]1+ cluster of pyruvate formate-lyase activating enzyme generates the glycyl radical on pyruvate formate-lyase: EPR-detected single turnover. J. Am. Chem. Soc. 122 8331–8332. [Google Scholar]
- Knappe, J., Neugubauer, F.A., Blaschkowski, H.P., and Gänzler, M. 1984. Post-translational activation introduces a free radical into pyruvate formate-lyase. Proc. Natl. Acad. Sci. 81 1332–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappe, J., Elbert, S., Frey, M., and Wagner, A.F. 1993. Pyruvate formate-lyase mechanism involving the protein-based glycyl radical. Biochem. Soc. Trans. 21 731–734. [DOI] [PubMed] [Google Scholar]
- Krebs, C., Henshaw, T.F., Cheek, J., Huynh, B.H., and Broderick, J.B. 2000. Conversion of 3Fe-4S to 4Fe-4S clusters in native pyruvate formate-lyase activating enzyme: Mössbauer characterization and implications for mechanism. J. Am. Chem. Soc. 122 12497–12506. [Google Scholar]
- Krebs, C., Broderick, W.E., Henshaw, T.F., Broderick, J.B., and Huynh, B.H. 2002. Coordination of adenosylmethionine to a unique iron site of the [4Fe-4S] of pyruvate formate-lyase activating enzyme: A Mössbauer spectroscopic study. J. Am. Chem. Soc. 124 912–913. [DOI] [PubMed] [Google Scholar]
- Külzer, R., Pils, T., Kappl, R., Hüttermann, J., and Knappe, J. 1998. Reconstitution and characterization of the polynuclear iron-sulfur cluster in pyruvate formate-lyase-activating enzyme. Molecular properties of the holoenzyme form. J. Biol. Chem. 273 4897–4903. [DOI] [PubMed] [Google Scholar]
- Lieder, K.W., Booker, S., Ruzicka, F.J., Beinert, H., Reed, G.H., and Frey, P.A. 1998. _S_-Adenosylmethionine-dependent reduction of lysine 2,3-aminomutase and observation of the catalytically functional iron-sulfur centers by electron paramagnetic resonance. Biochemistry 37 2578–2585. [DOI] [PubMed] [Google Scholar]
- Méjean, A., Bui, B.T.S., Florentin, D., Ploux, O., Izumi, Y., and Marquet, A. 1995. Highly purified biotin synthase can transform dethiobiotin into biotin in the absence of any other protein, in the presence of photoreduced deazaflavin. Biochem. Biophys. Res. Commun. 217 1231–1237. [DOI] [PubMed] [Google Scholar]
- Moss, M. and Frey, P.A. 1987. The role of _S_-adenosylmethionine in the lysine-2,3-aminomutase reaction. J. Biol. Chem. 262 14859–14862. [PubMed] [Google Scholar]
- Ollagnier, S., Mulliez, E., Schmidt, P.P., Eliasson, R., Gaillard, J., Deronzier, C., Bergman, T., Gräslund, A., Reichard, P., and Fontecave, M. 1997. Activation of the anaerobic ribonucleotide reductase from Escherichia coli. The essential role of the iron-sulfur center for _S_-adenosylmethionine reduction. J. Biol. Chem. 272 24216–24223. [DOI] [PubMed] [Google Scholar]
- Ollagnier-de Choudens, S. and Fontecave, M. 1999. The lipoate synthase from Escherichia coli is an iron-sulfur protein. FEBS Lett. 453 25–28. [DOI] [PubMed] [Google Scholar]
- Ollagnier-de Choudens, S., Sanakis, Y., Hewitson, K.S., Roach, P., Baldwin, J.E., Münck, E., and Fontecave, M. 2000. Iron-sulfur center of biotin synthase and lipoate synthase. Biochemistry 39 4165–4173. [DOI] [PubMed] [Google Scholar]
- Ollagnier-de Choudens, S., Sanakis, Y., Hewitson, K.S., Roach, P., Münck, E., and Fontecave, M. 2002. Reductive cleavage of _S_-adenosylmethionine by biotin synthase from Escherichia coli. J. Biol. Chem. 277 13449–13454. [DOI] [PubMed] [Google Scholar]
- Park, J., Tai, J., Roessner, C.A., and Scott, A.I. 1996. Enzymatic synthesis of _S_-adenosyl-L-methionine on the preparative scale. Bioorg. Med. Chem. 4 2179–2185. [DOI] [PubMed] [Google Scholar]
- Sanyal, I., Cohen, G., and Flint, D.H. 1994. Biotin synthase: Purification, characterization as a [2Fe-2S] cluster protein, and in vitro activity of the Escherichia coli bioB gene product. Biochemistry 33 3625–3631. [DOI] [PubMed] [Google Scholar]
- Scott, R.A. 1985. Measurement of metal-ligand distances by EXAFS. Methods Enzymol. 117 414–459. [Google Scholar]
- Sofia, H.J., Chen, G., Hetzler, B.G., Reyes-Spindola, J.F., and Miller, N.E. 2001. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: Functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsby, C.J., Hong, W., Broderick, W.E., Cheek, J., Ortillo, D., Broderick, J.B., and Hoffman, B.M. 2002a. Electron-nuclear double resonance spectroscopic evidence that _S_-adenosylmethionine binds in contact with the catalytically active [4Fe-4S]+ cluster of pyruvate formate-lyase activating enzyme. J. Am. Chem. Soc. 124 3143–3151. [DOI] [PubMed] [Google Scholar]
- Walsby, C.J., Ortillo, D., Broderick, W.E., Broderick, J.B., and Hoffman, B.M. 2002b. An anchoring role for FeS clusters: Chelation of the amino acid moiety of _S_-adenosylmethionine to the unique iron site of the [4Fe-4S] cluster of pyruvate formate-lyase activating enzyme. J. Am. Chem. Soc. 124 11270–11271. [DOI] [PubMed] [Google Scholar]
- Wong, K.K. and Kozarich, J.W. 1994. _S_-adenosylmethionine-dependent radical formation in anaerobic systems. Met. Ions Biol. Sys. 30 279–313. [Google Scholar]