Blm3 is part of nascent proteasomes and is involved in a late stage of nuclear proteasome assembly (original) (raw)
Abstract
Proteasomes are multisubunit proteases that are responsible for regulated proteolysis. The degradation of the proteasomal maturation factor, named Ump1 in yeast, completes the autocatalytic processing of inactive precursor complexes into the proteolytically active core particle (CP) of the proteasome. We have identified Blm3, a conserved nuclear protein, as a new component of Ump1-associated precursor complexes. A lack of Blm3 resulted in an increased rate of precursor processing and an accelerated turnover of Ump1, which suggests that Blm3 prevents premature activation of proteasomal CPs. On the basis of biochemical fractionation experiments combined with in vivo localization studies, we propose that Blm3 joins nascent CPs inside the nucleus to coordinate late stages of proteasome assembly in yeast.
Introduction
Proteasomes, the main proteases in the nucleoplasm and cytoplasm of eukaryotic cells, are involved in a variety of biological processes. Two multisubunit complexes, the proteolytically active core particle (CP) and the regulatory particle (RP), constitute the proteasome. The RP recognizes protein substrates, gates the CP to translocate substrates into its catalytic cavity and confers ATP dependence on protein degradation (Glickman & Maytal, 2002).
The CP consists of two rings of seven β-subunits, which contains the active sites, flanked by two rings of seven non-catalytic α-subunits. The assembly of the CP is a multistep reaction, in which rings of α-subunits associated with pro-β subunits form early assembly intermediates (Frentzel et al., 1994). In yeast, early assembly intermediates are known as 15S precursor complexes or half-assembled core particles (h-CPs; Chen & Hochstrasser, 1996; Ramos et al., 1998). According to current models, the dimerization of two h-CPs into an as yet uncharacterized, short-lived late assembly intermediate has to take place before the release of active sites is triggered by the auto-catalytic processing of distinct pro-β subunits (Chen & Hochstrasser, 1996). These late assembly intermediates are known as unstable processing intermediates, pre-holo-proteasomes or nascent CPs (n-CPs). A conserved maturation factor, originally named Ump1 in yeast, is associated with the h-CP. Trapped inside the n-CP, Ump1 ensures efficient active-site formation and becomes the first substrate of the n-CP (Ramos et al., 1998;Heinemeyer, 2000; Krüger_et al_., 2001).
In yeast, ∼80% of proteasomes are localized in the nuclear periphery, which is the main subcellular location of proteasomal proteolysis (Enenkel et al., 1998; Wilkinson et al., 1998; Russell_et al_., 1999). Moreover, we found that Ump1-associated precursor complexes that resemble h-CPs are mainly nuclear in yeast. Our data suggested that h-CPs are imported into the nucleus before being assembled into the active CP and that proteasomal maturation takes place at the subcellular site of proteasomal proteolysis (Lehmann et al., 2002).
To gain further insight into the pathway of nuclear CP assembly, we screened Ump1-associated precursor complexes from nuclear extracts for new interacting proteins and discovered Blm3. In vivo localization studies verified that Blm3 is a nuclear protein. Using native gel electrophoresis and green fluorescent protein (GFP)-labelling techniques, Ump1-associated precursor complexes were fractionated into h-CPs and n-CPs. Blm3 was absent from the h-CP fraction but part of the n-CP fraction. We found that the kinetics of precursor processing are accelerated in blm3Δ null mutants compared with wild-type cells, which indicates that Blm3 delays the conversion of the n-CP into the mature CP. We propose that Blm3 is a new coordinator of nuclear CP maturation in yeast.
Results and Discussion
Recent studies from our laboratory carried out in yeast suggested that the nuclear CP originates from Ump1-associated precursor complexes that are localized in the nucleus. To identify new components involved in nuclear CP assembly and maturation, we analysed the protein composition of Ump1-associated precursor complexes from nuclear and endoplasmic reticulum (ER) extracts using IgG affinity-chromatography of protein-A-tagged Ump1 (Lehmann et al., 2002). In such precursor preparations, we consistently co-purified a 240-kDa protein (Fig. 1A). Peptide sequences derived from the 240-kDa protein were searched for in the yeast genome database and were assigned to Blm3, the gene product of BLM3-YFL006 (see supplementary information online). Blm3 was also found in preparations of proteasomal complexes that were affinity purified using protein-A-tagged versions of α-subunits (data not shown;Gavin et al., 2002).
Figure 1.
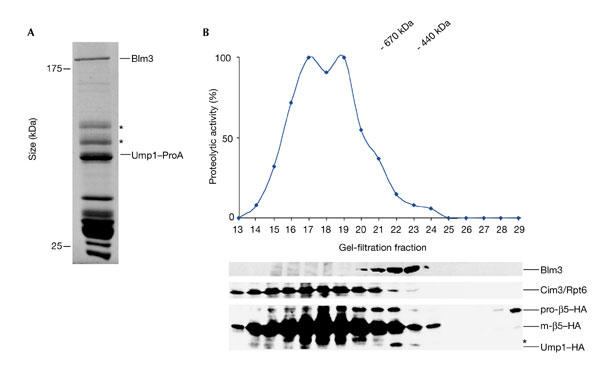
Identification of Blm3 as part of Ump1-associated precursor complexes. (A) Coomassie-blue-stained SDS–polyacrylamide gel of Ump1-associated precursor complexes that were isolated by IgG affinity-chromatography from nuclear/endoplasmic reticulum extracts from yeast wild-type cells expressing protein A (ProA)-tagged Ump1 instead of the endogenous protein (Lehmann et al., 2002). Blm3 and Ump1–ProA are indicated. Proteins indicated by asterisks are heat shock proteins. (B) Extracts from wild-type cells expressing haemagglutinin (HA)-tagged β5 and Ump1 instead of the endogenous proteins (wild-type strain JD133; Ramos et al., 1998) were fractionated on Superose 6 (Amersham). Fractions were assayed for peptide-cleavage activity against Cbz-Leu-Leu-Glu-β-naphthylamide (upper panel). Protein samples of each fraction were run on SDS–polyacrylamide gels, blotted and probed for Blm3, the regulatory particle subunit Cim3/Rpt6, the core particle subunit β5–HA and Ump1–HA (lower panel). Unprocessed and matured β5 subunits are abbreviated as pro-β5 and m-β5, respectively. A degradation band of m-β5 is indicated by an asterisk. The Superose 6 column was calibrated using the standards thyroglobulin (670 kDa) and ferritin (440 kDa).
The name of the BLM3 gene originated from a genetic study in yeast. BLM3 was cloned by complementing the bleomycin hypersensitivity that is conferred by the blm3-1 mutation, which suggests that Blm3 functions in protecting cells from DNA damage (Evans Febres_et al_., 2001). A structural homologue of Blm3 exists in mammalian cells, and has 17% identity with Blm3 and is named PA200 (Ustrell et al., 2002).
Before we continued our biochemical studies of the function of Blm3 in CP assembly, we used basic yeast genetics. First, we deleted the_BLM3-YFL006_ gene. The resulting blm3Δ null mutant is viable, but temperature sensitive (see supplementary information online), indicating that Blm3 is required under stress conditions. Second, we overexpressed Blm3 by chromosomal disruption of the BLM3 promoter using an inducible_GAL1_ promoter. The overexpression of Blm3 was found to inhibit cell growth (see supplementary information online).
Further evidence that Blm3 is a precursor-interacting protein was provided by gel-filtration experiments and density-gradient ultracentrifugations. Both fractionation methods showed that Blm3 does not coincide with the peak fractions of active proteasomes. In gel-filtration experiments, Blm3 co-eluted with Ump1-associated precursor complexes (Fig. 1B). In density-gradient ultracentrifugation experiments, Blm3 co-sedimented with fractions that comprised Ump1-associated precursor complexes (see supplementary information online).
If Blm3 functions in CP assembly and maturation, a deficiency for Blm3 may affect CP maturation, as seen in cells lacking Ump1 (Ramos et al., 1998). The lack of Ump1 causes the stabilization of precursor complexes, which is manifested in increased levels of pro-β5 subunits, the crucial determinants of CP maturation (Chen & Hochstrasser, 1996; Jäger_et al_., 1999). To test how a deletion of BLM3 affects CP maturation, steady-state levels of pro-β5 subunits were determined in_blm3Δ_ cells compared with isogenic wild-type cells that express haemagglutinin (HA)-tagged β5 instead of the endogenous protein (Fig. 2A). In parallel, ump1Δ cells were analysed as a control for precursor stabilization. Immunoblot analysis showed that pro-β5 subunits are present in wild-type cells (Fig. 2A, lane 1), hardly detectable in blm3Δ cells (Fig. 2A, lane 2) and accumulate in ump1Δ cells (Fig. 2A, lane 3). One possible explanation for this finding is that the half-life of pro-β5 subunits is decreased in blm3Δ cells compared with wild-type cells. Consequently, Blm3 would delay precursor processing.
Figure 2.
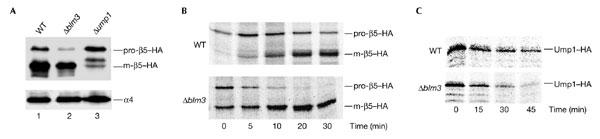
Blm3 delays core particle maturation. (A) Wild-type (WT),Δblm3 and Δump1 cells were grown to logarithmic phase (optical density of 1 at 600 nm), harvested, boiled immediately in sample buffer and subjected to SDS–polyacrylamide gel electrophoresis followed by immunoblotting. Proteins were probed for haemagglutinin (HA)-tagged β5 and α4 subunits, the latter of which was used as a loading control. Unprocessed (pro-) β5, matured (m-) β5 and α4 are indicated. (B) Pulse-chase analysis, comparing the rates of pro-β5 processing and Ump1 degradation in wild-type and blm3Δ cells. Cells were pulse-labelled with 35S-Met/Cys for 5 min and chased for the lengths of time indicated. HA-tagged β5 and Ump1 were precipitated with anti-HA antibodies. Pro-β5, m-β5 and Ump1 are indicated. Each pulse-chase analysis comparing blm3Δ and wild-type cells was performed three times in parallel.
If this conclusion is correct, the kinetics of pro-β5 processing and the turnover of Ump1 should be accelerated in blm3Δ cells compared with wild-type cells. To measure the kinetics of CP maturation, pulse-chase experiments were performed. A lack of Blm3 led to an increased processing rate of pro-β5 and an accelerated degradation of Ump1 (Fig. 2B), suggesting that Blm3 does delay the maturation process of the CP.
Is Blm3 an antagonist of Ump1? A blm3Δ ump1Δ double-knockout strain was created to test whether a lack of Blm3 can be compensated for by a lack of Ump1. However, blm3Δ ump1Δ cells showed an ump1Δ phenotype, which is manifested in the stabilization of precursor complexes (data not shown).
Next, we investigated whether Blm3 is associated with early or late Ump1-associated precursor complexes, namely the h-CP or the n-CP. For this purpose, we analysed precursor complexes from wild-type and blm3Δ cells by native polyacrylamide gel electrophoresis (PAGE). As precursor complexes are not detectable by chromogenic peptide-cleavage products because they are proteolytically inactive, we used GFP-streptactin (GFPS)–tagged Ump1, which allowed the affinity purification of precursor complexes under native conditions (Lehmann et al., 2002). GFPS-tagged precursor complexes from wild-type and blm3Δ cells were run on native PAGE gels. Phosphofluoroimaging of the native gels identified the species of Ump1-associated precursor complexes (Fig. 3A). Compared with the pattern in wild-type cells,blm3Δ cells lacked the more slowly migrating species (Fig. 3A, compare bands III and IV in lanes 1 and 2). Immunoblotting analysis showed that the more slowly migrating species in wild-type cells are Blm3-associated precursor complexes (Fig. 3B). To analyse the protein composition, the species of Ump1-associated precursor complexes from wild-type cells (Fig. 3A, bands I–III) were excised from the native gel, subjected to SDS–PAGE, and blotted and probed for Ump1 and β5. The predominant fraction of precursor complexes (Fig. 3A,B, band I) contained Ump1–GFPS and pro-β5 (Fig. 3C, lane 1). The fractions of precursor complexes (Fig. 3A,B, bands II and III) that migrated slower than the predominant fraction contained Ump1–GFPS, pro-β5 and mature β5 (m-β5; Fig. 3C, lanes 2 and 3).
Figure 3.
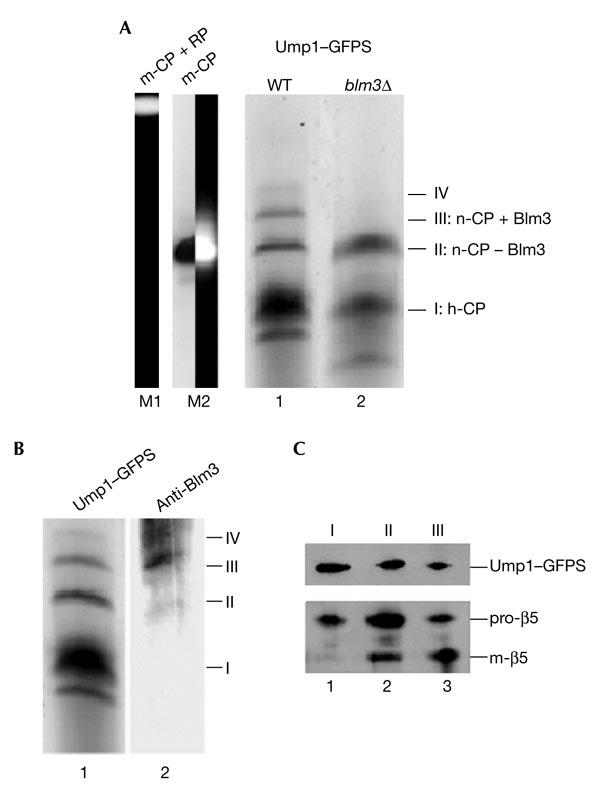
Blm3 is associated with the nascent core particle but not with the half-assembled core particle. (A) Green-fluorescent-protein–streptactin (GFPS)-tagged precursor complexes, which were isolated by streptactin affinity chromatography from wild-type (WT; lane 1) and blm3Δ (lane 2) cells expressing Ump1–GFPS instead of the endogenous protein, respectively, were separated by native polyacrylamide gel electrophoresis. The GFP-labelled precursor complexes were visualized by scanning the gel with a phosphofluoroimager. Bands were numbered (I–IV) and assigned to the half-assembled core particle (h-CP), or to the nascent CP (n-CP) with (+) or without (−) Blm3. Species I was assigned to the h-CP, as this species represents the main fraction of precursor complexes (Chen & Hochstrasser, 1996; Ramos et al., 1998; Lehmann et al., 2002). The n-CP was found to behave like the mature CP (m-CP) in native gels. Thus, the m-CP (Lehmann et al., 2002) was run as a size marker (lane M2; the left half shows phosphofluoroimaging of the GFPS-tagged m-CP; the right half shows the chromogenic peptide-cleavage activity of the GFPS-tagged m-CP. As another control, the migration of the m-CP in association with the regulatory particle (RP) is shown (lane M1). (B) Native gels of Ump1-associated precursor complexes were scanned for Ump1–GFPS (lane 1), blotted, and probed for Blm3 (lane 2). Bands were assigned as above. Band IV probably represents the n-CP capped by two Blm3 molecules, but could not be characterized further due to limiting amounts of protein. (C) The wild-type precursor complex species (bands I–III) were excised from the native gel, subjected to SDS–PAGE, blotted, and probed for Ump1 and β5 (lanes 1–3 are derived from bands I–III, respectively). Ump1–GFPS, pro-β5 and mature β5 (m-β5) are indicated. Subunits of the RP were not detected in any of our preparations of Ump1-associated precursor complexes (data not shown).
As the h-CP contains only unprocessed β5 subunits and represents the predominant fraction of precursor complexes in yeast (Chen & Hochstrasser, 1996; Ramos_et al_., 1998), we assigned the precursor species I to the h-CP. Blm3 was not found in the fraction of the h-CP (Fig. 3B), which suggets that Blm3 has no impact on early stages of CP assembly. The ongoing processing of pro-β5 involves the short-lived n-CP. Thus, we assigned the precursor species II and III to the n-CP. As shown inFig. 3B, Blm3 is part of the n-CP. These data show that Blm3 accompanies Ump1-associated precursor complexes only at a late stage of CP assembly.
We were now able to interpret our result that blm3Δ ump1Δ mutants have the same phenotype as ump1Δ mutants. A deficiency of Ump1 disrupts the formation of precursor complexes that are already at an early stage of assembly, which results in the accumulation of h-CPs (Ramos et al., 1998). As Blm3 follows Ump1 at a later stage of precursor assembly, a lack of Ump1 cannot be compensated for by a lack of Blm3.
As Blm3 plays a major role in CP maturation, Blm3 was expected to localize to the nucleus, similar to Ump1. To monitor the subcellular localization of Blm3 in living cells, BLM3 was replaced in the chromosome by a GFP fusion construct using yeast genetics (Fig. 4A). Blm3–GFP behaved like endogenous Blm3 in density-gradient ultracentrifugation (see supplementary information online). Direct fluorescence microscopy (Fig. 4B) and biochemical fractionation of wild-type cells that expressed Blm3–GFP (Fig. 4C) showed that Blm3 localizes to the nucleus to the same extent as Ump1 (Lehmann et al., 2002). The nuclear localization of Blm3 remained unchanged in ump1Δ cells (see supplementary information online), which indicates that Blm3 does not require Ump1 for efficient nuclear import. GFP-labelled Ump1 and α4 subunits of the CP localized mainly to the nuclear periphery of_blm3Δ_ cells, as seen in wild-type cells (seesupplementary information online), which shows that Ump1-associated precursor complexes are not stabilized in the yeast nucleus due to the presence of Blm3.
Figure 4.
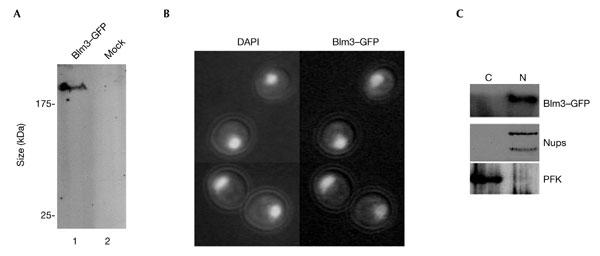
Blm3 is nuclear. (A) Western blot analysis of cells in which Blm3 is chromosomally replaced by a green fluorescent protein (GFP)-tagged version (lane 1) compared with mock-transfected (mock) cells (lane 2). Anti-GFP antibodies were used. (B) Direct fluorescence microscopy was used (Enenkel et al., 1998) to monitor GFP-tagged Blm3 in living cells (left panel, DAPI (4′,6-diamidino-2-phenylindole) staining of yeast nuclei, as visualized by the ultraviolet light filter superimposed with Nomarski optics; right panel, GFP filter superimposed with Nomarski optics). (C) Spheroplast lysates were fractionated into a cytosolic (C) and a nuclear (N) fraction, separated by SDS–polyacrylamide gel electrophoresis, blotted, and probed for Blm3, nuclear (Nups) and cytoplasmic (PFK) controls (Enenkel et al., 1998).
Conclusions
In this study, we describe Blm3 as a new component of nuclear CP assembly. Blm3 is part of the n-CP, a fraction of Ump1-associated precursor complexes. Our in vivo localization studies combined with biochemical fractionations show that Blm3 is associated with the n-CP in the nucleus. Thus, Blm3 is involved in the coordination of a late stage of nuclear CP maturation, and this complements our recently proposed model of nuclear CP assembly (Lehmann et al., 2002).
As a lack of Blm3 results in an increased rate of precursor processing and accelerated degradation of Ump1, we assume that Blm3 functions as a checkpoint protein in preventing premature activation of the nuclear CP. Under stress conditions, this function of Blm3 may be particularly required in the biological context of DNA repair, which may explain why Blm3 was originally identified as a protein that protects cells from DNA damage (Evans Febres et al., 2001).
With regard to PA200, the structural homologue of Blm3 in mammalian cells (Ustrell et al., 2002), it remains to be determined whether Blm3 and PA200 are functionally related. PA200 has a stimulatory effect on the peptide-cleavage activity of the nuclear CP in mammalian cells. In yeast, our data provide no evidence that Blm3 functions as an activator of the mature CP. By contrast, we saw an inhibitory effect of Blm3 on proteasomal proteolysis in vitro and in vivo (seesupplementary information online), consistent with our findings that Blm3 delays precursor processing as part of the n-CP. We assume that Blm3 joins the n-CP during the transition into the mature CP and that Blm3 interacts with the de novo synthesized m-CP.
As yeast cells that lack Blm3 are temperature sensitive and those that overproduce Blm3 are growth-inhibited, and because Blm3 controls a late stage of nuclear CP maturation, we propose that Blm3 is generally required for the regulation of nuclear CP maturation. In consequence, Blm3 may have an impact on nuclear CP activation.
Methods
Plasmids and strains.
Using homologous recombination techniques (Enenkel_et al_., 1998), endogenous BLM3 was replaced by_BLM3–GFP–HA_ by integrative transformation of a GFP–HA tag at the 3′ end of BLM3-YFL006 using_Spe_I/_Xho_I-digested pBS–YFL006–GFP–HA-URA3-HIS3. The blm3Δ::HIS3 allele contains a deletion of 5,415 nucleotides starting from position −53 (numbered with reference to the ATG).blm3Δ cells and isogenic wild-type cells that express tagged versions of β5 and Ump1 instead of the endogenous proteins were generated as described in Lehmann et al. (2002). The chromosomal replacement of β5 (Pre2) by an HA-tagged version was achieved by using _Sal_I/_Xho_I-digested pBS–Pre2HA–URA3–HIS3.
The strains used in this study are derived from the WCGa wild-type strain and are listed in Table 1.
Table 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| WCGa | MATa1, his3-11,15 leu2-3,112 ura3 can GAL | Enenkel et al., 1998 |
| UCE1 | MATa1, UMP1–ProA::YIp5 | Derivative of WCGa |
| UCE2 | MATa1, UMP1–GFPS::YIp5 | Derivative of WCGa |
| UCE3 | MATa1, UMP1–HA::YIp5 | Derivative of WCGa |
| UCE4 | MATa1, ump1Δ::LEU2 | Derivative of WCGa; Ramos et al., 1998 |
| UCE5 | MATa1, blm3Δ::HIS3,UMP1–GFPS::YIp5 | Derivative of BMF1 |
| BMF1 | MATa1, blm3Δ::HIS3 | Derivative of WCGa |
| BMF2 | MATa1, blm3Δ::HIS3,ump1Δ::LEU2 | Derivative of UCE4 |
| BMF3 | MATa1, blm3Δ::HIS3,UMP1–HA::YIp5 | Derivative of BMF1 |
| BMF4 | MATa1, blm3Δ::HIS3, PRE2–HA::URA3 HIS3 | Derivative of BMF1 |
| BBH1 | MATa1, PRE2-HA::URA3 HIS3 | Derivative of WCGa |
| BBH2 | MATa1, ump1Δ::LEU2, PRE2–HA::URA3 HIS3 | Derivative of UCE4 |
| BAL1 | MATa1, Blm3–GFP–HA::URA3 HIS3 | Derivative of WCGa |
Protein chemistry.
Protein-A- and GFPS-tagged protein complexes were isolated by affinity chromatography using IgG sepharose and streptactin matrices, respectively (Lehmann et al., 2002). Fractionation of yeast-cell extracts into soluble and nuclear/ER fractions was performed as described (Lehmann et al. 2002).
Gel filtration on Superose 6 (Amersham) was performed in 50 mM Tris-HCl, pH 7.5, 2 mM ATP, 5 mM MgCl2, 1 mM dithiothreitol (DTT), 15% glycerol, as described in Ramos et al. (1998).
Proteasomal activity was measured in 20 mM Tris-HCl, pH 7.8, 5 mM MgCl2, 10 mM KCl, 1 mM DTT, for 30 min at 37 °C using 100 μM chromogenic substrates (Ustrell et al., 2002).
Native gel electrophoresis.
Polyacrylamide gels (3.5–6.0%) were cast as described inFrentzel et al. (1994). Freshly prepared protein samples contained 10% glycerol and traces of xylene–cyanol blue. Gels were run at a cold temperature (8 °C) at 100 V for 30 min, and then at 200 V for ∼2 h in 0.09 M Tris base, 0.09 M H3BO4, 2 mM MgCl2, pH 8.1–8.4, 2 mM ATP, 2 mM DTT. GFP-tagged proteasomal complexes were visualized using Advanced Image Data Analysis (AIDA; Version 2.0; www.raytest.de) installed on a Phospho-Fluoro Imager FLA3000 (Fujifilm). For analysing the protein composition of the GFP-labelled complexes, the bands visualized by phosphofluoroimaging were excised from the native gel, boiled in sample buffer and subjected to SDS–PAGE.
Other methods.
Anti-Blm3 peptide antibodies were raised in guinea pig against amino-terminal residues 2–18 of Blm3 (J. Pineda). Pulse-chase experiments were carried out as described in Ramos et al. (1998). Direct fluorescence microscopy was performed as described inEnenkel et al. (1998).
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/4-embor938-s1.pdf).
Supplementary Material
supplementary information
Acknowledgments
We thank K. Janek for peptide-mass fingerprint analysis, R. Kraft for microsequence analysis, J. Dohmen, C. Mann and D. Wolf for strains and antibodies, B. Braun for pBS–Pre2HA–URA3–HIS3, B. Dahlmann for advice about proteasome enzymology and P.-M. Kloetzel and W. Heinemeyer for discussions and critical reading of the manuscript. This work was supported by a Deutsche Forschungsgemeinschaft grant (EN 301/4-2) to C.E.
References
- Chen P. & Hochstrasser M. ( 1996) Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell, 86, 961–972. [DOI] [PubMed] [Google Scholar]
- Enenkel C., Lehmann A. & Kloetzel P.M. ( 1998) Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope–ER network in yeast. EMBO J., 17, 6144–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans Febres D., Pramanik A., Caton M., Doherty K., McKoy J., Garcia E., Alejo W. & Moore C.W. ( 2001) The novel BLM3 gene encodes a protein that protects against lethal effects of oxidative damage. Cell. Mol. Biol., 47, 1149–1162. [PubMed] [Google Scholar]
- Frentzel S., Pesold-Hurt B., Seelig A. & Kloetzel P.M. ( 1994). 20S proteasomes are assembled via distinct precursor complexes. Processing of LMP2 and LMP7 proproteins takes place in 13–16S preproteasome complexes. J. Mol. Biol., 236, 975–981. [DOI] [PubMed] [Google Scholar]
- Gavin A.-C. et al. ( 2002). Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- Glickman M.H. & Maytal V. ( 2002) Regulating the 26S proteasome. Curr. Top. Microbiol. Immunol., 268, 43–72. [DOI] [PubMed] [Google Scholar]
- Heinemeyer W. ( 2000) in Proteasomes: the World of Regulatory Proteolysis (eds Hilt, W. & Wolf, D.H.), 48–70 (Eurekah.com/Landes Biosciences Publ. Co., Georgetown, Texas, USA). [Google Scholar]
- Jäger S., Groll M., Huber R., Wolf D.H. & Heinemeyer W. ( 1999) Proteasome β-type subunits: unequal roles of propeptides in core particle maturation and a hierarchy of active site function. J. Mol. Biol., 291, 997–1013. [DOI] [PubMed] [Google Scholar]
- Krüger E., Kloetzel P.-M. & Enenkel C. ( 2001) 20S proteasome biogenesis. Biochimie, 83, 1–5. [DOI] [PubMed] [Google Scholar]
- Lehmann A., Janek K., Braun B., Kloetzel P.-M. & Enenkel C. ( 2002). 20S proteasomes are imported as precursor complexes into the nucleus of yeast. J. Mol. Biol., 317, 401–413. [DOI] [PubMed] [Google Scholar]
- Ramos P.C., Hockendorff J., Johnson E.S., Varshavsky A. & Dohmen R.J. ( 1998) Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell, 92, 489–499. [DOI] [PubMed] [Google Scholar]
- Russell S.J., Steger K.A. & Johnston S.A. ( 1999) Subcellular localization, stoichiometry, and protein levels of 26S proteasome subunits in yeast. J. Biol. Chem., 274, 21943–21952. [DOI] [PubMed] [Google Scholar]
- Ustrell V., Hoffman L., Pratt G. & Rechsteiner M. ( 2002) PA200, a nuclear proteasome activator involved in DNA repair. EMBO J., 21, 3516–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson C.R., Wallace M., Morphew M., Perry P., Allshire R., Javerzat J.P., McIntosh J.R. & Gordon C. ( 1998). Localization of the 26S proteasome during mitosis and meiosis in fission yeast. EMBO J., 17, 6465–6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary information