How Many Mutant p53 Molecules Are Needed To Inactivate a Tetramer? (original) (raw)
Abstract
The tumor suppressor p53 is transcription factor composed of four identical subunits. The majority of the mutations in p53 are missense mutations that impair DNA binding. On the other hand, the _p53_-related p63 and p73 genes are rarely mutated, but many cell types express natural variants lacking the N-terminal transactivation domain (NΔ). Compelling evidence indicates that both the DNA binding-defective and NΔ mutants can impair the function of wild-type p53 in a dominant-negative manner. Interestingly, it is uncertain how many mutant subunit(s) a p53 tetramer can tolerate. In this study, we first made theoretical predictions based on the number of mutant p53 monomers needed to inactivate a tetramer and then tested how well the experimental data fit the predicted values. Surprisingly, these experiments reveal that DNA binding-defective p53 mutants (R249S and R273H) are very ineffective in impairing the transcriptional activity of p53: at least three mutants are required to inactivate a tetramer. In marked contrast, p53NΔ is a very potent inhibitor of p53: one NΔ subunit per tetramer is sufficient to abolish the transcriptional activity. DNA binding is not necessary for the NΔ proteins to inactivate p53. Similarly, NΔ variants of p63 and p73 are also powerful inhibitors of members of the p53 family. These results have important implications for our thinking about the mechanism of tumorigenesis involving missense p53 mutants or the N-terminally truncated isoforms.
Mutation of the p53 gene is one of the most common steps in tumorigenesis and is found in more than half of all cancer cases. Germ line mutations of p53 are found in cancer-prone families with Li-Fraumeni syndrome (37). Somatic mutations are frequently associated with exposure to carcinogenic agents. For example, dietary aflatoxin B1 exposure is strongly correlated with the R249S (herein designated RS) mutation in hepatocellular carcinoma (19), and cigarette smoke component benzo(a)pyrene undergoes metabolic activation and can cause mutations of residues 175, 248, and 273 in cultured cells, the same mutational hotspots in lung cancer (7).
The p53 gene encodes a protein with a central DNA binding domain, flanked by an N-terminal transactivation domain, and a C-terminal tetramerization domain (25). The majority of the mutations in p53 are missense point mutations clustered in the DNA binding domain (17). The structure of the DNA binding domain consists of a large β-sandwich that acts as a scaffold for three loop-based elements that contact the DNA (4). Importantly, the residues most frequently mutated in cancers are all at or near the protein-DNA interface, and over two-thirds of the missense mutations are within the DNA binding loops (40).
The active form of p53 is a tetramer of four identical subunits, consisting of a dimer of a dimer (22). The tetramerization domain contains a β-strand and an α-helix, which associates with another monomer across an antiparallel β-sheet and an antiparallel helix-helix interface. The two dimers are held together by a large hydrophobic surface of each helix pair. Consistent with its tetrameric state, p53 binds DNA sites that contain four repeats of the pentamer sequence motif 5′-Pu-Pu-Pu-C-A/T-3′ (Pu is purine).
The functions of p53 are primarily mediated through the regulation of cell cycle checkpoints, apoptosis, and genome stability (41). Stresses including DNA damage and aberrant growth signals activate p53. Among other downstream targets, activated p53 enhances the transcription of the cyclin-dependent kinase inhibitor p21_CIP1/WAF1_, facilitating the cell cycle arrest. The transcriptional activity and stability of p53 are highly regulated by posttranslational mechanisms involving protein-protein interactions, phosphorylation, acetylation, ubiquitination, and sumoylation. MDM2, also one of the transcriptional targets of p53, binds to the transactivation domain, inhibits p53-mediated transcription, shuttles p53 out of the nucleus, and targets p53 for ubiquitin-mediated proteolysis (41). After DNA damage or other stresses, the inactivation of p53 by MDM2 is disrupted by phosphorylation of the N-terminal region of p53.
The various functional domains in p53 are conserved in the two p53-related proteins p63 and p73 (20). In contrast to p53, the p63 and p73 genes produce multiple transcripts as a result of alternative splicing and alternative promoter utilization. Importantly, several of these isoforms lack the N-terminal transactivation domain (ΔNp63 and ΔNp73). Indeed, the ΔN variants of p63 and p73 are the most abundant isoforms expressed in several cell types (32, 34, 49, 51). Unlike p53, p63 and p73 are rarely mutated in cancers. Instead, they are implicated in stem cell identity, neurogenesis, natural immunity, and homeostatic control (48).
Several mechanisms have been postulated to inactivate p53 (39, 41). Deletion of one or both p53 alleles reduces the expression of the tetramers. Nonsense or splice site mutations that result in the deletion of the tetramerization domain also reduce the abundance of tetramers. Amplification of the MDM2 gene, deletion of the ARF gene, or expression of some viral oncogenes stimulates p53 degradation. Mislocalization of p53 to the cytoplasm also appears to be a mechanism in several types of cancers. Finally, missense mutations of the DNA binding domain are probably the most common mechanism for p53 inactivation. These mutations disrupt the DNA binding capability of p53. But whether the DNA binding-defective mutants can also act in a dominant-negative manner to disrupt normal p53 function, and further reduce the functional active tetramers, remains an important question.
Many attempts have been made to address the issue of whether p53 mutants can impair the function of the wild-type protein in a dominant-negative manner. Mutated p53 present within a tetramer is thought to abolish the DNA binding capacity of the entire complex. This has the important implication that a heterozygous mutation in p53 could result in the functional inactivation of cellular p53. Early experiments revealed that complexes between wild-type and mutant p53 take on a mutant protein conformation (9, 10, 27, 33). Extensive data from studies in cell culture suggest that many missense mutant p53 proteins can inhibit the transactivation of target genes (for example, see references 16, 23, and 35). Since the relative expression of wild-type and mutant p53 is usually not known, it is uncertain how many mutants per tetramer are needed to inactivate transactivation. Conflicting data that show a lack of dominant-negative effect were obtained when similar levels of wild-type and mutant p53 were expressed (1, 12). Moreover, despite the possible dominant-negative function of the missense mutants, the remaining wild-type allele is generally mutated or lost in cancers, suggesting that complete loss of normal p53 can further promote tumorigenesis (18). Hence, the fundamental question of the efficacy of mutant p53 on wild-type function remains unanswered.
In the present study, we approach the problem by first predicting how the transactivation of promoters would decrease if a different number of p53 mutants is needed to inactivate a tetramer. We then tested how well the experimental data fit with the predicted values by careful comparison of the relative levels of wild type and mutant. One of the key findings is that at least three DNA binding-defective mutants are required to inactivate the p53 tetramer. In marked contrast, one N-terminally deleted mutant per tetramer is sufficient to turn off the transcriptional activity.
MATERIALS AND METHODS
Materials.
All reagents were obtained from Sigma-Aldrich (St. Louis, Mo.) unless stated otherwise.
DNA constructs.
Human p53 in pRcCMV (43), p63α in pRc/CMV (42), hemagglutinin (HA)-tagged simian p73 in pcDNA3 (30), p73(R293H) (30), MDM2 promoter-luciferase reporter construct (43), p21_CIP1/WAF1_ promoter-luciferase reporter construct (42), and β-galactosidase construct (30) were obtained from sources as previously described. The isoforms used in this study were p63α and p73β. Constructs for FLAG-p53NΔ (NΔ90) and FLAG-p53(L22Q+W23S) were as previously described (24). The NcoI fragment of p53 was ligated into this construct to create FLAG-p53 in pUHD-P1. The resulting construct was cut with NcoI (partial)-BamHI and ligated into pUHD-P2 (47) to create HA-p53 in pUHD-P2. The NcoI-EcoRI fragment of FLAG-p53NΔ in pUHD-P1 was ligated into pUHD-P1 (45) to produce FLAG-p53NΔ159 in pUHD-P1. The NheI-SspI fragment of FLAG-p53 in pUHD-P1 or FLAG-p53NΔ in pUHD-P1 was ligated into NheI-EcoRV pcDNA3.1− (Invitrogen, Carlsbad, Calif.) to create FLAG-p53CΔ (CΔ326) in pcDNA3.1− or FLAG-p53NΔCΔ in pcDNA3.1−, respectively. FLAG-p63 in pUHD-P1 was as previously described (42). The construct was cut with NheI-SphI, treated with Klenow and dNTP, and religated to create p63NΔ (NΔ58) in pUHD-P1. A three-HA-tagged p63 was created by subcloning p63 into pCAGGS (a gift from Katsumi Yamashita, Kanazawa University, Kanazawa, Japan). HA-p73 in pcDNA3 was amplified by PCR with 5′GTCTAGAATGGCCCAGTCCACCGCCAC3′ and a Sp6 primer; the PCR product was cut with XbaI and ligated into pUHD-P1 to create FLAG-p73 in pUHD-P1. The NcoI fragment of p73 was put into pUHD-P1 to produce FLAG-p73NΔ (containing residues 71 to 445) in pUHD-P1. The resulting construct was amplified by PCR with a plasmid forward primer and 5′TGGGATCCTTACGTGTCCTCGTCTCC3′; the PCR product was cut with NcoI-BamHI and ligated into pUHD-P1 to produce FLAG-p73NΔCΔ (containing residues 71 to 354) in pUHD-P1. Site-directed mutagenesis was carried out with a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) by using the following primers and their antisense: 5′TGAACCGGAGTCCCATCCTCAC3′ (RS) and 5′TTTGAGGTGCATGTTTGTGC3′ (R273H; hereafter designated RH).
Cell culture.
H1299 (non-small cell lung carcinoma) (3) cells were obtained from the American Type Culture Collection (Rockville, Md.). Cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) fetal bovine serum (Invitrogen) in a humidified incubator at 37°C in 5% CO2. Cells were transfected by the calcium phosphate precipitation method (2). The amount of total DNA transfected was adjusted to the same level with blank vectors. Cell extracts were prepared as described previously (31). The protein concentration of cell lysates was measured with the bicinchoninic acid protein assay system (Pierce, Rockford, Ill.) with bovine serum albumin as a standard.
Chromatin fractionation.
The protocol was modified from that described in Fujita et al. (13). Cells from 10-cm plates were harvested, washed in phosphate-buffered saline, and lysed with 1 ml of CSK buffer (10 mM PIPES [piperazine-N,_N_′-bis(2-ethanesulfonic acid), pH 6.8], 100 mM NaCl, 300 mM sucrose, 1 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 5 mM β-glycerophosphate, 5 mM NaF, 0.5% Triton X-100, 50 mM Na3VO4, 2 mM ATP, 1 mM phenylmethylsulfonyl fluoride) at 4°C for 20 min. After 10% of the lysates were taken for loading total cell lysate, the rest was spun in a microcentrifuge at 70 × g for 3 min. The supernatant was clarified by centrifugation at 7,600 × g for 5 min and mixed with an equal volume of sodium dodecyl sulfate sample buffer. The pellet was washed once more with 1 ml of CSK buffer, resuspended in 100 μl of CSK buffer, and mixed with an equal volume of sodium dodecyl sulfate sample buffer.
Luciferase and β-galactosidase assays.
Luciferase assays and β-galactosidase assays were performed exactly as described previously (24).
Antibodies and immunological methods.
Immunoblotting and immunoprecipitation were performed as described (31). Signals on immunoblots were analyzed with the National Institutes of Health Image program (Bethesda, Md.) by using the appropriate serial dilutions of the samples for calibration. Indirect immunofluorescence microscopy was performed as described previously (46). Rabbit polyclonal antibodies against FLAG tag (47), monoclonal antibody 12CA5 against HA tag (47), monoclonal antibody M2 against FLAG tag (14), and monoclonal antibody A17 against CDC2 (5) were obtained from sources as described previously. Monoclonal antibody 4A4 against p63 (sc-8431), monoclonal antibody DO1 against p53 (sc-126), and polyclonal antibodies against p21_CIP1/WAF1_ (sc-397) were from Santa Cruz Biotechnology (Santa Cruz, Calif.). Monoclonal antibody 421 against p53 (a gift from Tim Hunt, Cancer Research UK) was used in experiments with p53(LW).
RESULTS
Theoretical considerations.
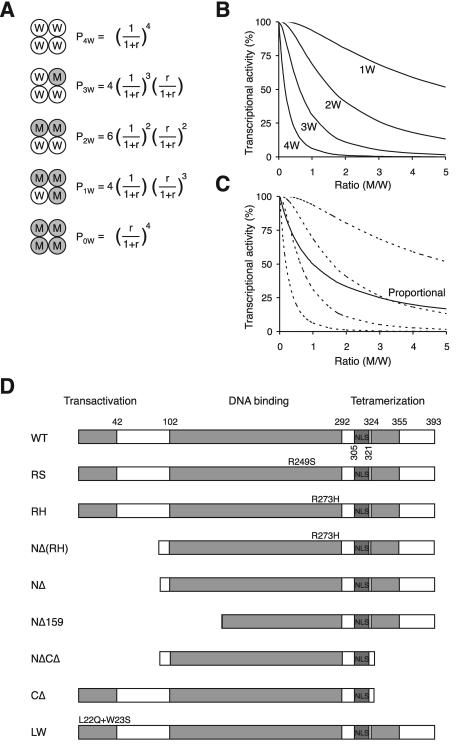
It is expected that if both wild-type and mutant p53 are expressed in a cell, a mixture of tetramers composed of different numbers of wild-type (W) and mutant (M) subunits will be generated. These range from tetramers compose entirely of wild type to those with mutants only (Fig. 1A). If we assume equal expression of W and M (as in the case when one copy of the p53 gene is mutated), then the probability of forming a tetramer containing a mixture of M and W is obviously greater than that with only M or W. The probabilities P are given by the following: _P_4W = 1/16; _P_3W = 4/16; _P_2W = 6/16; _P_1W = 4/16; and _P_0W = 1/16. These probabilities will be altered when the relative expression between W and M is shifted. In general, the values of P are given in Fig. 1A, where the ratio (r) is defined as M/W.
FIG. 1.
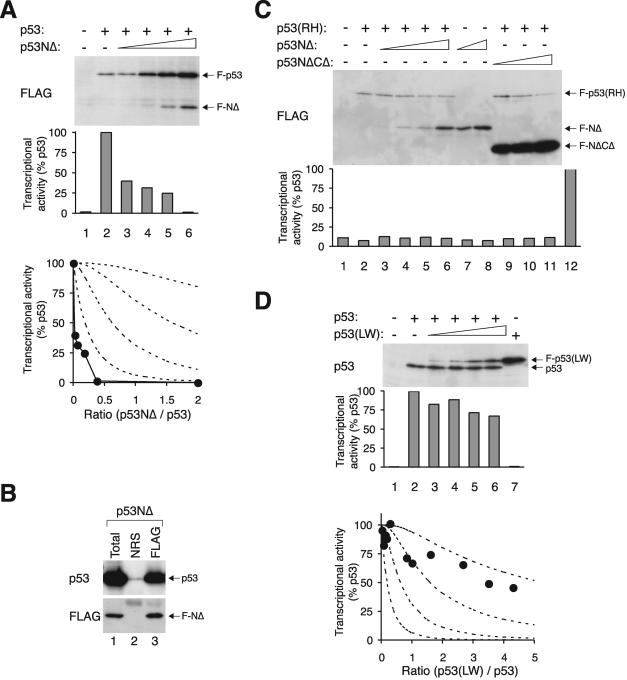
Theoretical prediction of the activity of p53 tetramer. (A) The probabilities of the formation of tetramers with different compositions. W denotes wild-type and M denotes mutant p53. The probabilities (P) of forming different tetramers are given, where the ratio r is the concentration of M/concentration of W. (B) Theoretical prediction of p53 activity with different relative levels of mutant p53. The activity against the ratio of M/W is calculated from the probability of forming the different tetramers shown in panel A. The various curves are based on the assumption that tetramers are only active with the indicated number of wild-type p53 subunits as follows: 4W, 4; 3W, ≥3; 2W, ≥2; or 1W, ≥1. (C) Theoretical prediction of p53 activity for proportional inhibition by a p53 mutant. The activity is calculated according to the assumption that it is directly proportional to the number of wild-type molecules in the tetramer. The four dotted lines are identical to those from panel B for reference. (D) Schematic diagram of the p53 constructs used in this study. The positions of the various structural elements are shown to scale. WT, wild type; LW, point mutation L22Q+W23S.
If we hypothesize that only a tetramer containing 4W is active as a transcription factor, then the final activity A equals _P_4W. On the other hand, if tetramers containing 4W and 3W are both active, then the equation A(≥3W) = _P_4W + _P_3W holds. Similarly, the equations A(≥2W) = _P_4W + _P_3W + _P_2W and A(≥1W) = _P_4W + _P_3W + _P_2W + _P_1W hold. For each scenario, the activity can be calculated against the M/W ratio (Fig. 1B). Therefore, each scenario has a characteristic inhibition curve when the concentration of mutant p53 is increased relative to the wild type, testable by experimental approaches.
Tetramers are postulated to be either fully active or inactive in the above theoretical predictions. An alternative analysis is that the activity of the tetramer is proportional to the number of wild-type subunits present. If this is the case, the activity of the tetramer is given by the formula A = _P_4w + _P_3w(3/4) + _P_2w(1/2) + _P_1w(1/4). Figure 1C shows that the inhibition curve of the proportional decrease hypothesis lies between the curves for the “all-or-none” hypothesis of ≥3W and ≥2W. Implications will be discussed later.
At least three molecules of DNA binding-defective p53 mutants per tetramer are required to inactivate the transactivation of MDM2 and p21_CIP1/WAF1_ promoters.
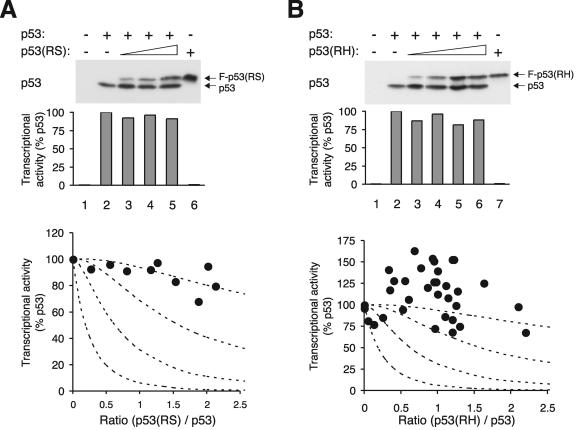
We next tested the hypotheses described above for two hotspot DNA binding-defective p53 mutants, RS and RH. The various constructs of wild-type and mutant p53 used in this study are shown in Fig. 1D. Typically, a constant amount of wild-type p53 and an increasing amount of mutant p53 were coexpressed in H1299 cells (a p53 null cell line). The transcriptional activity of p53 was quantified by cotransfection of a p53-responsive promoter luciferase reporter. The luciferase activities were normalized with the β-galactosidase activity from a cotransfected plasmid to correct for transfection efficiency. Expression of the recombinant p53 was detected by immunoblotting. In cases when the mutant p53 protein was the same size as the wild-type protein, one of them was tagged with a FLAG epitope so that the two proteins could be detected together with a monoclonal antibody against p53 and distinguished by their different gel mobility (Fig. 2A). In cases where the mutant is smaller than the wild type (truncation mutants), both proteins were tagged with FLAG and detected with a monoclonal antibody against the FLAG epitope (see Fig. 4A). Since the wild-type and mutant p53 were detected together with the same antibody on the same blot, their relative levels could be quantified by densitometry with the appropriate serial dilution standards. Given that this approach depended only on the relative expression between wild-type and mutant p53, data from several independent experiments could be pooled.
FIG. 2.
DNA binding-defective p53 mutants inhibit p53 tetramer inefficiently. (A) Inhibition curve of p53(RS) on p53 activity. H1299 cells were transfected with plasmids expressing an MDM2-promoter luciferase reporter and β-galactosidase. Plasmids expressing wild-type p53 and increasing amounts of FLAG-p53(RS) were cotransfected as indicated. Cell extracts were prepared at 24 h after transfection. Both p53 and FLAG-p53(RS) were detected together by immunoblotting with a monoclonal antibody against p53. The luciferase activity was measured, normalized with the β-galactosidase activity to correct for variations in transfection efficiency between samples, and plotted as a percentage of p53 alone (middle panel). The relative expression level of p53 and FLAG-p53(RS) was quantified as described in Materials and Methods and plotted against p53 activity (filled circles, bottom panel). Data from several experiments were pooled. The dotted lines represent the predicted inhibition curves as shown in Fig. 1B. (B) RH mutant. Experiments were performed as described for panel A except that FLAG-p53(RH) was used instead of p53(RS).
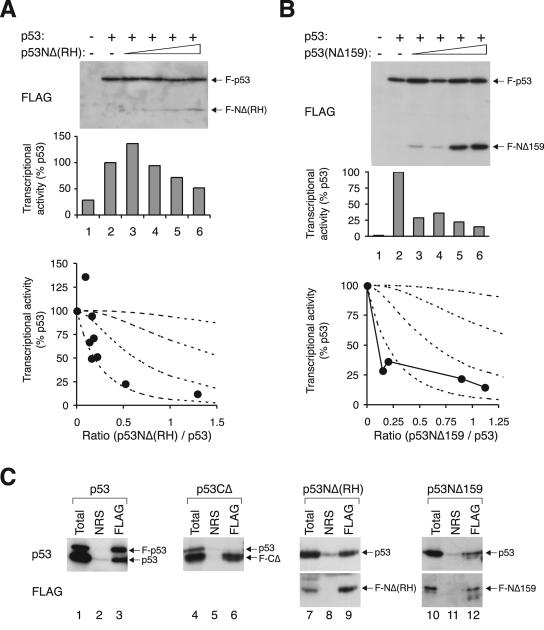
FIG. 4.
Inhibition of p53 tetramer by transcriptional-incompetent mutants. (A) Inhibition curve of p53NΔ(RH). Transcriptional assays were performed with FLAG-p53 and various levels of FLAG-p53NΔ(RH), similar to the method described in the legend of Fig. 2. The recombinant proteins were detected by immunoblotting with a monoclonal antibody against the FLAG epitope. (B) Inhibition curve of p53NΔ159. Transcriptional assays were performed with FLAG-p53 and various levels of FLAG-p53NΔ159 according to the method described in the legend of Fig. 2. (C) Binding of p53 to different truncation mutants. Untagged p53 was coexpressed with FLAG-tagged p53, p53CΔ, p53NΔ(RH), or p53NΔ159 as indicated. Cell extracts were prepared and subjected to immunoprecipitation followed by immunoblotting for p53 and FLAG according to the method described in the legend of Fig. 3E. NRS, normal rabbit serum.
In order to compare the theoretical predictions to the actual effects of mutant p53, it is critical that both the transcriptional activities and protein expression be quantifiable. We assumed that both wild-type and mutant p53 were equally sensitive to the antibodies on immunoblots. This is probably a fair assumption since only monoclonal antibodies were used. Although the signals from immunoblotting were far from linear, the relative expression of the wild type and mutants was readily quantified by comparing the serial dilutions of the samples. For the transcriptional activities, we found that the luciferase reporter assays were approximately linear over more than a 10-fold dilution of p53 (our unpublished data). Hence, we believe that our measurements of the transcriptional activity with different levels of protein expression were within the linear range.
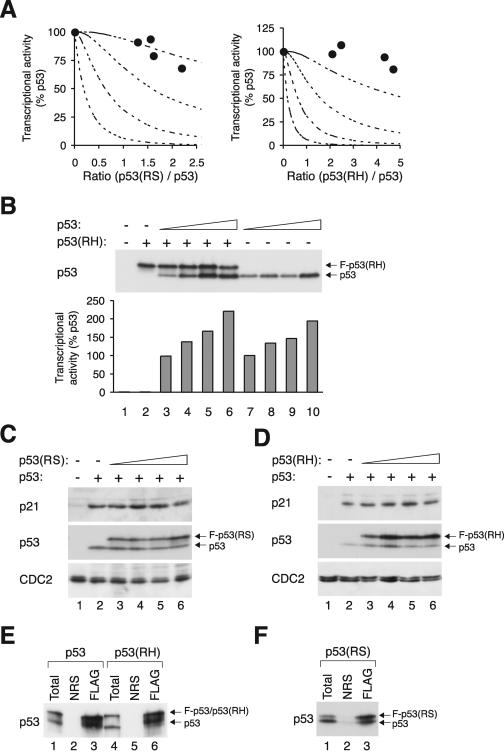
Figure 2A summarizes the experiments that examined the effects of p53(RS). Examples of the expression of the two proteins and the corresponding transcriptional activities are shown. As expected, p53 robustly transactivated the MDM2 promoter (lane 2). In contrast, p53(RS) alone did not display any activity (lane 6). Unexpectedly, we found that p53 was very resilient to inhibition by p53(RS). When compared to p53 alone (when r = 0), the presence of p53(RS) only slightly decreased the transactivation. Over 80% of the transcriptional activity was retained even when there was twice as much mutant as wild type. Notably, the profile of the inhibition curve was most similar to the profile predicted for the scenario when three mutants per tetramer are needed for inactivation.
To exclude the possibility that the large number of mutants needed to inactivate the tetramer is peculiar for p53(RS), the effects of p53(RH) on p53 were also examined. Similarly to p53(RS), p53(RH) did not significantly inhibit p53 when the proteins were expressed at comparable levels (Fig. 2B). We do not fully understand why the transcription activities were frequently enhanced in the presence of p53(RH) (see Discussion).
We next examined the effects of mutant p53 on the transactivation of another p53-responsive promoter. Figure 3A shows that, similar to the transactivation of the MDM2 promoter, transactivation of the p21_CIP1/WAF1_ promoter by p53 decreased only slightly in the presence of p53(RS) or p53(RH). These data show that the ineffectiveness of mutant p53 on the tetramer is common to both the MDM2 promoter and p21_CIP1/WAF1_ promoter.
FIG. 3.
RS and RH mutants form complexes with p53 without significant effect on its transcriptional activities. (A) At least three p53(RS) or p53(RH) mutants per tetramer are needed to impair the transactivation of p21_CIP1/WAF1_ promoter. Wild-type p53 was coexpressed with various levels of FLAG-tagged p53(RS) or p53(RH) as described in the legend of Fig. 2, except that the p21_CIP1/WAF1_ promoter-luciferase reporter was used instead of the MDM2 promoter. The dotted lines represent the predicted inhibition curves as shown in Fig. 1B. (B) p53(RH) does not inhibit p53 transcriptional activity. Cells were transfected with plasmids expressing an MDM2-promoter luciferase reporter and β-galactosidase. A constant amount of FLAG-p53(RH) and various amounts of wild-type p53 were expressed as indicated. Cell extracts were prepared, and the luciferase and β-galactosidase activities were determined (lower panel). The transcriptional activity was expressed as a percentage of the lowest concentration of p53 used (lane 7). The expression of p53 and FLAG-p53(RH) was detected by immunoblotting. (C) Expression of endogenous p21_CIP1/WAF1_ is only weakly impaired by p53(RS). Cells were transfected with plasmids expressing wild-type p53 and FLAG-p53(RS) as indicated. Cell extracts were prepared at 24 h after transfection, and the expression of p21_CIP1/WAF1_ was detected by immunoblotting. Recombinant p53 and FLAG-p53(RS) were detected together by immunoblotting with a monoclonal antibody against p53. Equal loading of lysates was confirmed by immunoblotting for CDC2. (D) Expression of endogenous p21_CIP1/WAF1_ is only weakly impaired by p53(RH). The experiment was performed as described for panel C except that p53(RH) was used instead ofp53(RS). (E) p53-p53(RH) complexes are formed as efficiently as p53-p53. Untagged p53 was coexpressed with either FLAG-p53 (lanes 1 to 3) or FLAG-p53(RH) (lanes 4 to 6). Cell extracts were prepared and 100 μg was subjected to immunoprecipitation with either control normal rabbit serum (NRS) or FLAG antiserum as indicated. The immunoprecipitates were immunoblotted for p53. Total cell lysates (10 μg) were applied to indicate the inputs. (F) p53(RS) binds efficiently to wild-type p53. Untagged p53 was coexpressed with FLAG-p53(RS). Cell extracts were prepared, immunoprecipitated with NRS or anti-FLAG antiserum, and immunoblotted for p53 as described for panel C.
We next performed the converse experiment in which p53(RH) was expressed at a constant level and increasing amounts of wild-type p53 were introduced (Fig. 3B). As expected, p53(RH) alone did not display any transcriptional activity (lane 2). Titrating in p53 progressively elevated the transcriptional activity. Consistent with the above results, the transcriptional activity was not significantly affected by the presence of p53(RH).
To see whether the expression of endogenous proteins was affected by p53 mutants, the level of endogenous p21_CIP1/WAF1_ was examined after transfection of the recombinant p53. As expected, expression of p21_CIP1/WAF1_ was induced by p53 (Fig. 3C and D). The level of expression was not significantly reduced in the presence of p53(RS) (Fig. 3C) or p53(RH) (Fig. 3D). The relatively small influence of mutant p53 on the induction of p21_CIP1/WAF1_ by wild-type p53 is in agreement with the results from the above transactivation assays.
To confirm that the RS and RH mutants could form complexes with wild-type p53, their physical interaction was examined by coimmunoprecipitation. The results in Fig. 3E show that p53 was coimmunoprecipitated with a FLAG-tagged p53 (lane 3) but not when the immunoprecipitation was performed with control serum (lane 2). Similarly, p53 was coimmunoprecipitated with FLAG-p53(RH) (Fig. 3E) or FLAG-p53(RS) (Fig. 3F). These data validate the finding that the two non-DNA binding mutants could bind to p53 under our experimental conditions.
Taken together, these results indicate that although non-DNA binding mutants can form tetramers with p53, they have a weak influence on the transactivation of the MDM2 promoter or the p21_CIP1/WAF1_ promoter. Our data suggest that one or two molecules of RS or RH mutants are unable to inactivate a tetramer.
One molecule of p53 lacking the transactivation domain is sufficient to inactivate a tetramer.
Given that the DNA binding-defective mutants have only a slight influence on the tetramer, we next investigated whether the transcriptional activity of the mutant is important for the activity of the tetramer as a whole. To this end, the N-terminal region (including the transactivation domain) was removed from p53(RH), creating a version [p53NΔ(RH)] that was defective in both DNA binding and transactivation but retained the ability to form a tetramer. We found that in contrast to p53(RH), p53NΔ(RH) was very potent in impeding the transcriptional activity of p53 (Fig. 4A). The transcriptional activity decreased sharply as the relative level of p53NΔ(RH) increased, dropping to less than 25% of that of the control when the wild type and mutant were expressed at the same level. The inhibition curve followed the profile predicted for the scenario when only tetramers with four wild-type subunits are active.
This idea is further corroborated by experiments with p53NΔ159, in which both the transactivation domain and part of the DNA binding domain were eliminated (Fig. 1D). Similar to p53NΔ(RH), p53NΔ159 was a powerful inhibitor of p53 (Fig. 4B). The results shown in Fig. 4C confirm that both p53NΔ(RH) and p53NΔ159 could form complexes with full-length p53. Interaction between p53 and FLAG-p53 was used as a positive control (lanes 1 to 3), and the lack of binding between p53 and p53CΔ (with the tetramerization domain deleted) served as a negative control (lanes 4 to 6).
To see whether the inhibition of the tetramer by p53NΔ(RH) was mainly due to the lack of the transactivation domain or the lack of DNA binding, a p53NΔ construct (without a transactivation domain but competent in DNA binding) was generated. The results in Fig. 5A show that p53NΔ was a very potent inhibitor of p53. The physical association between p53 and p53NΔ was confirmed by immunoprecipitation (Fig. 5B).
FIG. 5.
p53 tetramer is strongly inhibited by a mutant lacking the transactivation domain. (A) Inhibition curve of p53NΔ. Transcriptional assays were performed with FLAG-p53 and various levels of FLAG-p53NΔ according to the method described in the legend of Fig. 2. The recombinant proteins were detected by immunoblotting with a monoclonal antibody against the FLAG epitope. (B) Association between p53 and p53NΔ. Untagged p53 was coexpressed with FLAG-p53NΔ. Cell extracts were prepared and subjected to immunoprecipitation followed by immunoblotting for p53 and FLAG according to the method described in the legend of Fig. 3E. (C) p53NΔ does not trigger the activation of non-DNA binding p53. Cells were transfected with plasmids expressing an MDM2-promoter luciferase reporter and β-galactosidase. FLAG-tagged p53(RH) was coexpressed with p53NΔ or p53NΔCΔ as indicated. Cell extracts were prepared, and the expression of the recombinant proteins was detected by immunoblotting. The luciferase activity was measured and normalized with the β-galactosidase activity. Lane 12 shows the luciferase activity of wild-type p53 as control. (D) Inhibition curve of p53(LW). Transcriptional assays were performed with p53 and various levels of FLAG-p53(LW) according to the method described in the legend of Fig. 2. The recombinant proteins were detected by immunoblotting with a monoclonal antibody 421 against p53. NRS, normal rabbit serum.
Collectively, these data indicate that the transactivation domain of p53 is critical for the transcriptional activity of the tetramer. It is likely that all four subunits of the tetramer must contain the transactivation domain to generate an active transcription factor. This idea is further supported by the experiment in which p53NΔ was titrated into p53(RH), with the expectation that the p53NΔ would carry the p53(RH) to the DNA. Consistent with the above results, no transactivation of the MDM2 promoter was observed when p53(RH) was coexpressed with p53NΔ (Fig. 5C).
Interestingly, the mutant p53(LW) (with point mutations L22Q+W23S that disrupted transactivation) did not inhibit the tetramer as effectively as p53(NΔ) (Fig. 5D). These observations indicate that the inhibition of p53 function by mutants lacking the entire N-terminal region was not simply due to their lack of intrinsic transcriptional activity.
p53 lacking both the N-terminal and C-terminal regions is mislocalized and cannot inhibit p53 tetramer.
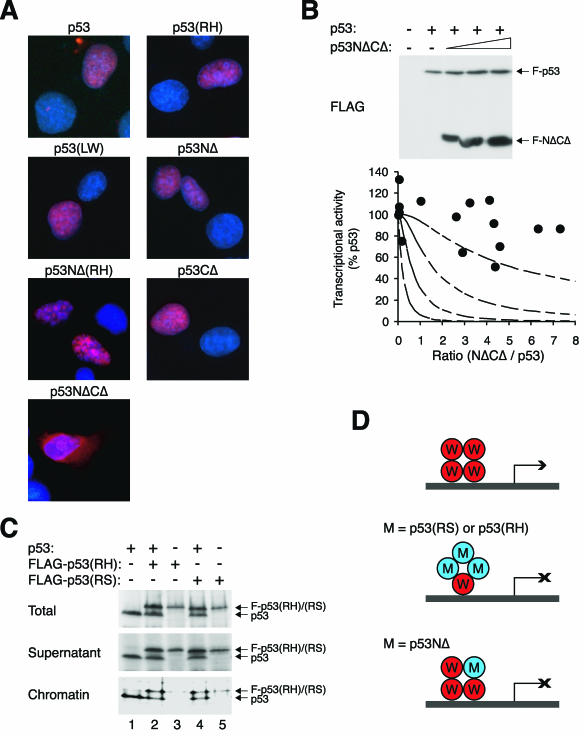
The various p53 constructs used in this study were generated with the consideration of retaining the major nuclear localization sequence (NLS) (Fig. 1D). To confirm that p53 and the various mutants were localized correctly to the nucleus, their subcellular localization was detected by indirect immunofluorescence microscopy. The results shown in Fig. 6A indicate that p53, p53(RH), p53NΔ(RH), p53NΔ, and p53(LW) as well as p53CΔ were localized exclusively to the nucleus. However, p53NΔCΔ was predominantly cytoplasmic. This is intriguing because p53NΔCΔ retained the NLS, and both p53NΔ and p53CΔ were localized to the nucleus. Since p53NΔCΔ was mislocalized and did not bind p53 (data not shown), it was used as a negative control for our analyses. As expected, p53NΔCΔ did not affect the transcriptional activity of p53 even when its expression was about eightfold over that of p53 (Fig. 6B).
FIG. 6.
Nuclear localization and chromatin binding of p53 mutants. (A) Localization of p53 mutants. FLAG-tagged p53 or the indicated mutants were transfected into H1299 cells. The cells were fixed, and the localization of the recombinant proteins was analyzed by indirect immunofluorescence microscopy with a monoclonal antibody against FLAG (red). Hoechst 33258 dye was used for nuclear staining (blue). (B) p53NΔCΔ mutant does not inhibit p53 tetramer. Transcriptional assays were performed with FLAG-p53 and various levels of FLAG-p53NΔCΔ according to the method described in the legend of Fig. 2. The recombinant proteins were detected by immunoblotting with a monoclonal antibody against the FLAG epitope. (C) Wild-type p53 triggers binding of RS and RH to chromatin. H1299 cells were transfected with plasmids expressing p53, FLAG-p53(RS), and FLAG-p53(RH) as indicated. The cells were harvested and subjected to chromatin fractionation as described in Materials and Methods. Total cell lysates, supernatant, and chromatin-bound fractions were subjected to immunoblotting for p53. The fractionation was validated by immunoblotting for histone H3 (chromatin bound) and prohibitin (supernatant) (data not shown). (D) Model of inhibition of tetramer by p53 mutants. For DNA binding-defective mutants, at least three mutant molecules per tetramer are required to inactivate transcription. For NΔ mutants, one molecule per tetramer is sufficient to abolish transcription.
To confirm that p53(RS) and p53(RH) did not bind DNA even though they were localized to the nucleus, cells were fractionated into chromatin-bound and soluble fractions. Figure 6C shows that while a fraction of p53 could be detected to bind DNA, little p53(RS) or p53(RH) was associated with DNA when these proteins were expressed on their own. Chromatin binding of p53(RS) and p53(RH) increased when they were coexpressed with p53, indicating that DNA binding tetramers containing a mixture of wild-type and mutant p53 were formed.
Collectively, these data are consistent with the idea that different modes of inhibition of p53 tetramers are used by DNA binding-defective mutants and NΔ mutants. A tetramer can probably tolerate at least three DNA binding-defective mutants, but cannot contain even one NΔ subunit (Fig. 6D).
Inhibition of the p53 family by p73NΔ.
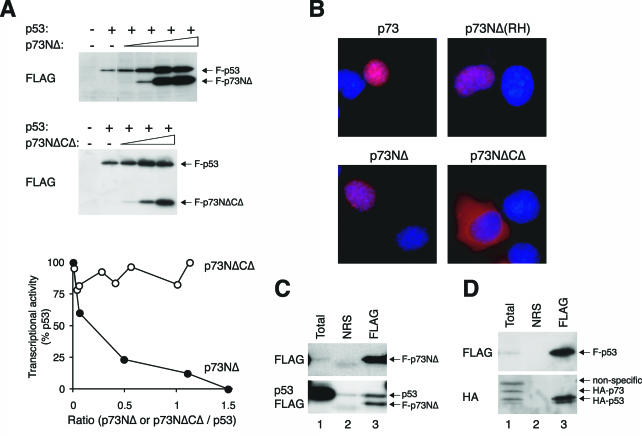
Given that N-terminally truncated isoforms of p63 and p73 are naturally expressed in cells (see introduction), we next investigated the efficacy of p53 inhibition by p73NΔ. The results shown in Fig. 7A indicate that p73NΔ efficiently diminished the transcriptional activity of p53. As a negative control, p73NΔCΔ, which was retained in the cytoplasm (Fig. 7B) and did not bind p53 (data not shown), failed to inactivate p53 (Fig. 7A). Complex formation between p53 and p73NΔ could be detected by coimmunoprecipitation (Fig. 7C). However, it is notable that the affinity between p53-p53 was considerably stronger than between p53-p73. When HA-tagged p53 and p73 were coexpressed to the same level, only HA-p53 was coimmunoprecipitated with FLAG-p53 (Fig. 7D). These data suggest that although p73NΔ has relatively weak affinity for p53, it is still a potent inhibitor of p53. This may partly reflect the requirement that all four subunits of the p53 tetramer contain the transactivation domain.
FIG. 7.
N-terminally truncated mutants of p73 can inactivate p53. (A) p73NΔ but not p73NΔCΔ inhibits p53. FLAG-tagged p53 was coexpressed with p73NΔ or p73NΔCΔ as indicated. Transcriptional assays were performed according to the method described in the legend of Fig. 2. A summary of several experiments is shown in the bottom panel. (B) Localization of p73 mutants. FLAG-tagged p73 or the indicated mutants were transfected into H1299 cells. The cells were fixed, and the localization of the recombinant proteins was analyzed by indirect immunofluorescence microscopy with a monoclonal antibody against FLAG (red). Hoechst 33258 dye was used for nuclear staining (blue). (C) p73NΔ can form a complex with p53. Untagged p53 was coexpressed with FLAG-p73NΔ. Cell extracts were prepared and subjected to immunoprecipitation followed by immunoblotting for p53 and FLAG according to the method described in the legend of Fig. 3E. (D) The affinity between p53 and p73 is significantly weaker than between p53 and p53. FLAG-p53, HA-p53, and HA-p73 were coexpressed in H1299 cells. Cell extracts were prepared and subjected to immunoprecipitation with normal rabbit serum (NRS) or FLAG antiserum. The immunoprecipitates were immunoblotted for FLAG and HA as indicated. Total cell lysates were applied in lane 1.
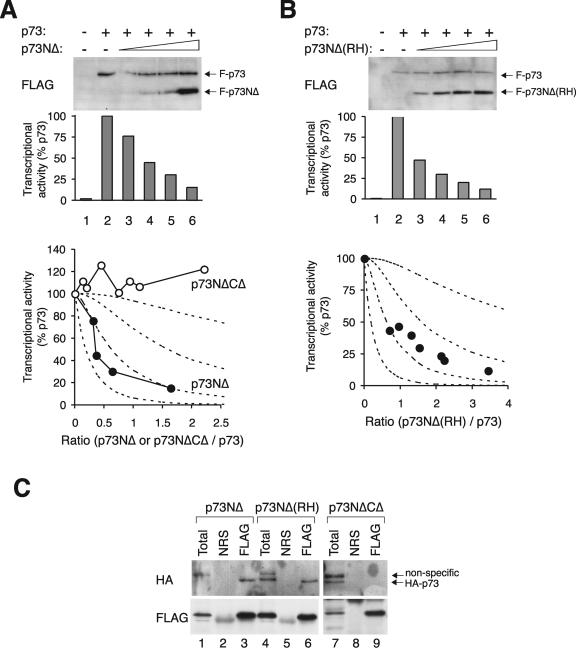
Not surprisingly, full-length p73 could also be inactivated by p73NΔ in a concentration-dependent manner (Fig. 8A). The p73NΔCΔ mutant acted as a negative control and had no effect on the transcriptional activity of p73. Inactivation of p73 by p73NΔ did not require DNA binding, as the transcriptional activity was also reduced by p73NΔ containing the R293H mutation (equivalent to the RH of p53) (Fig. 8B). Figure 8C confirms that p73NΔ and p73NΔ(RH) could form complexes with full-length p73. In contrast, p73NΔCΔ did not associate with p73 under similar conditions.
FIG. 8.
Inactivation of p73 by p73 lacking the transactivation domain. (A) p73NΔ but not p73NΔCΔ inhibits p73. FLAG-p73 was coexpressed with FLAG-p73NΔ. Transcriptional assays were performed according to the method described in the legend of Fig. 2. A summary of several experiments with p73NΔ as well as p73NΔCΔ is shown in the bottom panel. (B) Inhibition of p73 by p73NΔ(R293H). Transcriptional assays were performed according to the method described in the legend of Fig. 2 to assess the effects of FLAG-p73(RH) on FLAG-p73. (C) p73 binds to p73NΔ but not to p73 lacking the putative tetramerization domain. HA-p73 was coexpressed with FLAG-tagged p73NΔ, p73NΔ(RH), or p73NΔCΔ as indicated. Cell extracts were prepared, subjected to immunoprecipitation with FLAG antiserum, and followed by immunoblotting for HA and FLAG. NRS, normal rabbit serum.
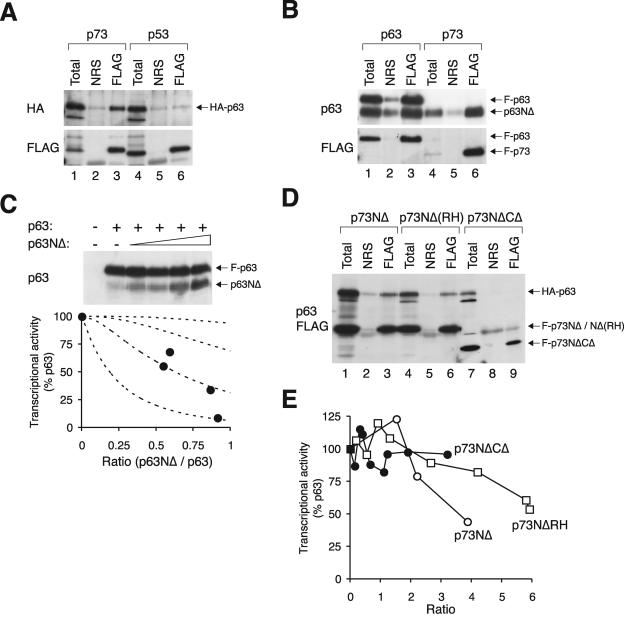
To complete our analysis, we found that p63 could bind to p73 (Fig. 9A). In contrast, binding between p63 and p53 was undetectable under similar conditions. Furthermore, p63NΔ could bind to full-length p63 and p73 (Fig. 9B) and inhibit the transactivation of the MDM2 promoter by p63 (Fig. 9C). Finally, p63 could form complexes with p73NΔ and p73NΔ(RH) but not with the p73NΔCΔ control (Fig. 7D). The transcriptional activity of p63 could be suppressed by p73NΔ and p73NΔ(RH), albeit a relatively high concentration of p73 is required (Fig. 9E). Taken together, these data indicate that the NΔ isoforms of p63 and p73 are potentially powerful inhibitors of all the members of the p53 family.
FIG. 9.
A high ratio of p63NΔ or p73NΔ is required for inhibition of p63. (A) p63 binds to p73 but not to p53. HA-p63 was coexpressed with either FLAG-p73 or FLAG-p53 as indicated. Cell extracts were prepared, subjected to immunoprecipitation with normal rabbit serum or FLAG antiserum, and followed by immunoblotting for HA and FLAG. (B) p63NΔ form complexes with p63 and p73. Untagged p63NΔ was coexpressed with either FLAG-p63 or FLAG-p73 as indicated. Cell extracts were prepared, subjected to immunoprecipitation with FLAG antiserum, and followed by immunoblotting for p63 and FLAG. (C) Inhibition of p63 by p63NΔ. Transcriptional assays were performed according to the method described in the legend of Fig. 2 to assess the effects of p63NΔ on FLAG-p63. The recombinant proteins were detected with a monoclonal antibody against p63. (D) p73NΔ can bind to p63. HA-p63 was coexpressed with FLAG-tagged p73NΔ, p73NΔ(RH), or p73NΔCΔ as indicated. Cell extracts were prepared, subjected to immunoprecipitation with normal rabbit serum or FLAG antiserum, and followed by immunoblotting for p63 and FLAG. (E) Inhibition curves of various p73 mutants on p63. Transcriptional assays were performed according to the method described in the legend of Fig. 2 to assess the effects of FLAG-tagged p73NΔ, p73NΔ(RH), or p73NΔCΔ on p63. NRS, normal rabbit serum.
DISCUSSION
Several assumptions were made in the prediction of the inhibition potential of mutant p53. We postulated that all the wild-type and mutant p53 proteins were present in tetramers, and monomers did not contribute to transcriptional activity. This idea is supported by data from a number of studies (26, 44). We found that p53 lacking the tetramerization domain (p53CΔ) only weakly transactivated the p53-responsive promoters (our unpublished data), indicating that the activity contributed by monomeric p53 (if any) was insignificant compared to that by tetrameric p53. We assumed that tetramers were formed first before binding to DNA, instead of each subunit being progressively assembled onto the DNA. The later scenario would involve a different calculation because the first subunit to assemble could only be a wild type. We also postulated that wild-type and mutant p53 had equal affinity for each other, and the formation of the tetramer was random, depending only on the relative concentration of the two proteins. Although it is clear that wild-type p53 can bind to the mutant form (Fig. 3E and F), we did not formally rule out the existence of some selectivity in their binding. Furthermore, it has been reported that dimerization (but not tetramerization) of p53 involves a cotranslational process (29).
Only a very simplistic view of the p53 tetramer is considered in this study. One can envision a more complicated model of the p53 tetramer. For example, rather than all four subunits of the tetramer being equal, it is possible that each position in the tetramer in relation to the DNA is different. This probably does not matter to our analyses, provided the assembly of the tetramer is random. The relative rough experimental readouts do not justify the examination of more complicated models.
In the model used here, tetramers are assumed to be either fully active or inactive (Fig. 1B). An alternative model assumed that the activity of the tetramer is proportional to the number of wild-type subunits present (Fig. 1C). It would be difficult to distinguish the two models if two to three wild-type p53 subunits could activate a tetramer. To add to the complication, the two models are not necessarily mutually exclusive. Fortunately, the effects of the mutant p53 examined here are clear-cut: they are either very potent (for NΔ) or very ineffective (for RS and RH). However, the inhibition profiles of p63NΔ/p73NΔ on p63/p73 lie in middle ground, and a definite distinction between the two models cannot be made without further experimentation.
Another prerequisite of our analysis is that both wild-type and mutant p53 were localized correctly. Indirect immunofluorescence microscopy revealed that apart from p53NΔCΔ, all the p53 constructs used in this study were localized to the nucleus (Fig. 6A). Similarly, p73NΔCΔ was also mislocalized to the cytoplasm (Fig. 7B). Since the major NLS was intact in p53NΔCΔ (Fig. 1D), it is unclear why the protein was mislocalized. Intriguingly, both p53NΔ and p53CΔ were localized correctly, suggesting that multiple redundant nuclear localization elements are present in both the N- and C-terminal regions. In this connection, another putative NLS at around residues 92 to 112 has been reported (15). It is possible that p53NΔCΔ disrupted both this NLS as well as the C-terminal NLS. Another possibility is that while the cytoplasmic p53CΔ was rapidly degraded, the degradation of p53NΔCΔ (lacking the MDM2 binding sites) was defective, thereby allowing the protein to accumulate in the cytoplasm.
Missense DNA binding-defective mutants.
We found that the two missense mutants RS and RH have surprisingly weak effects on the transcriptional activity of wild-type p53 (Fig. 2 and 3). On their own, these mutants can form oligomers (our unpublished data) but have no transcriptional activity (Fig. 2). Our data are consistent with the idea that at least three molecules of p53(RS) or p53(RH) per tetramer are required to inactivate transactivation (Fig. 6D). These results have vast implications for our thinking about the mechanisms of tumorigenesis involving missense p53 mutants. The cell expresses approximately equal amounts of wild-type and mutant p53 when missense mutation occurs in one copy of the p53 gene. Our data indicate that the overall p53 activity will not be severely compromised in these situations (over 80% activity retained in our in vitro assays).
We found that the transcription activities were frequently enhanced in the presence of p53(RH) (Fig. 2B), which was not observed with p53(RS) (Fig. 2A and 3A). These data are consistent with some previously published reports on the stimulation of p53 activity by RH (11, 53). One possibility is that the DNA binding ability of p53(RH) is not as defective as other missense mutants like p53(RS). Interaction with one or more wild-type p53 proteins may change the conformation and unleash the DNA binding potential of p53(RH).
Despite the inefficiency of mutant p53 to act in a dominant-negative manner in our assays, the missense mutation of one copy of p53 evidently plays a critical role in tumorigenesis. Germ line mutation of one copy of the p53 gene in Li-Fraumeni syndrome strongly predisposes the individual to a variety of cancers (37). Furthermore, the physiological expression of mutant p53 in mice can strongly limit the overall cellular p53 function (8). How can we reconcile these apparent discrepancies? Mutant p53 frequently are more stable than the wild type, and this may contribute to a higher relative concentration of mutant p53. However, this is unlikely to be a major factor because the transcriptional activity remained high even when there was twice as much mutant as wild type (Fig. 2 and 3). Another important consideration for in vivo function is that the promoter, rather than the p53 tetramer, is likely to be limiting. To measure the transcriptional activity of p53, excessive MDM2 or p21_CIP1/WAF1_ promoters were used in our assays. About fivefold less promoter could be transfected without reducing the luciferase activity (our unpublished data). It is possible that when the promoter is limiting in vivo, mutant p53 may have a more potent dominant-negative effect on the wild-type function. It is conceivable that when an inactive tetramer (containing three mutants) occupies a promoter, it completely precludes the transcription of the gene by active tetramers. This depends on the exchange rate of tetramers from the promoter. We do not favor this hypothesis because p53(RS) and p53(RH) were equally ineffective in inhibiting p53 when the experiments were performed with limiting promoter (our unpublished data). Moreover, the induction of endogenous p21_CIP1/WAF1_ by p53 was only weakly inhibited by mutant p53 (Fig. 3C and D). A more likely explanation is that even a slight reduction of p53 transcription activity is sufficient to promote tumorigenesis in the long run.
N-terminal truncation mutants.
Compelling evidence indicates that NΔ variants of p63 and p73 are antagonists of p53, p63, and p73 (50). In complete agreement with previously published data, we show that, in contrast to the DNA binding-defective mutants, N-terminally truncated mutants of p53, p63, or p73 are potent inhibitors of the respective full-length proteins. Moreover, cross-inhibition of other members of the family can occur, albeit a higher concentration of NΔ mutant was typically required (Fig. 7A and 9E and our unpublished data).
Two mechanisms have been proposed to explain the dominant-negative suppression of p53/p63/p73 by the NΔ proteins. In the first, NΔ proteins may simply compete with wild-type proteins for binding to target gene promoters. In the second, NΔ proteins may form tetramers with the wild-type proteins and decrease the transcriptional activity of the tetramer. As this study concerns the second mechanism, we tried to minimize the potential effects of the first by expressing excessive promoters in our assays. However, competition for promoter binding may be partly responsible for the effects of p53NΔ, as it inactivated p53 more readily than suggested by the predicted profile for one molecule per tetramer (Fig. 5A). Consistent with this idea, p53NΔ(RH) was slightly less effective than p53NΔ in inactivating p53 (Fig. 4A). Similarly, p73NΔ(RH) inhibited p73 slightly less efficiently than p73NΔ (Fig. 8A and B).
Interestingly, the transcriptional inactive mutant p53(LW) did not inhibit the tetramer as effectively as p53(NΔ) (Fig. 5D), suggesting that the inhibition of p53 function by p53NΔ was not simply due to a lack of intrinsic transcriptional activity. It is likely that p53NΔ disrupted structures of p53 and interaction with other proteins that were not affected in p53(LW). It is notable that two independent transactivation domains have been identified at the N-terminal region of p53 (residues 1 to 40 and 43 to 63) (54). It is possible that the point mutation (L22Q+W23S) disrupted the function of only the first transactivation domain. In this connection, we found that although the transactivation of MDM2 promoter by p53(LW) was much lower than by wild-type p53 (Fig. 5D), it was nevertheless higher than that of p53(NΔ) when the two were compared side by side (our unpublished data).
The tetramerization domain of p73 is similar to that in p53, and p73NΔ can form complexes with p73 (Fig. 8C). Several studies reported the absence of binding of p73 to p53 (6, 21, 36, 38). On the other hand, conflicting results were reported that demonstrated interaction between p73 and p53 (28, 32). Our data may provide a synthesis of these different results. We found that p53 could bind to p73NΔ (Fig. 7C), but the binding between p53 and p73 was considerably weaker than p53-p53 binding (Fig. 7D). Despite the relative weak affinity of p73NΔ for p53, p73NΔ was still a potent inhibitor of p53 (Fig. 7A). On face value, this argues that inhibition may in part be due to the competition for promoter binding. But another explanation is that since only one subunit of p73NΔ is probably sufficient to inactivate the p53 tetramer, inhibition could be very effective despite the relatively low affinity.
Another potential mechanism in which NΔ variants act may involve the binding to MDM2. In addition to the transactivation domain, the N-terminal region of p53 also contains the MDM2 binding site. Hence, it is expected that tetramers containing one or more NΔ subunit(s) are more stable and accumulate in the nucleus. This may further enhance the dominant-negative effect of NΔ mutants. By exerting their effects on transcriptional activity, promoter competition, and protein stability, the NΔ variants are exceedingly effective inhibitors of p53.
The effects of NΔ variants on p63 were considerably weaker than on p53 and p73 (Fig. 9). We think this may reflect the fact that the MDM2 and p21_CIP1/WAF1_ promoters may not be optimum targets for p63. Although p63 could transactivate both promoters, the activity was considerably lower than p53 and p73 when similar concentrations of proteins were expressed (our unpublished data).
Collectively, our data suggest that a single NΔ subunit within a tetramer is sufficient to turn off its transcriptional activity. NΔ variants are the major p73 proteins expressed in several cell types including sympathetic neurons (see above). Evidence from p73-deficient mice indicates that p73NΔ may suppress the p53-dependent apoptosis in sympathetic neurons, underscoring the importance of the dominant-negative functions of p73NΔ in vivo (32). Is there any physiological relevance with the dominant-negative effects of p53NΔ? Unlike p63 and p73, no NΔ forms of p53 are expressed from alternative promoters or splicing. However, translation from an alternative initiation site is induced by MDM2, producing a N-terminally truncated p53 (NΔ40 or NΔ44) without the transactivation domain (52). This truncated p53 is defective in transactivation of p21_CIP1/WAF1_ promoter (but is competent in transactivation of the MDM2, BAX, and GADD45 promoters) (52, 54). This truncated p53 can form tetramers with full-length p53 and is potentially a powerful inhibitor of p53 for some promoters.
Acknowledgments
We thank the members of the Poon laboratory for critical comments on the manuscript.
This work was supported in part by grants from the Research Grants Council grant HKUST6129/02 M and University Grants Council grant HIA03/04.SC01 to R.Y.C.P. R.Y.C.P. is a Croucher Foundation Senior Fellow.
REFERENCES
- 1.Aurelio, O. N., X. T. Kong, S. Gupta, and E. J. Stanbridge. 2000. p53 mutants have selective dominant-negative effects on apoptosis but not growth arrest in human cancer cell lines. Mol. Cell. Biol. 20**:**770-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl. 1991. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 3.Bodner, S. M., J. D. Minna, S. M. Jensen, D. D'Amico, D. Carbone, T. Mitsudomi, J. Fedorko, D. L. Buchhagen, M. M. Nau, A. F. Gazdar, et al. 1992. Expression of mutant p53 proteins in lung cancer correlates with the class of p53 gene mutation. Oncogene 7**:**743-749. [PubMed] [Google Scholar]
- 4.Cho, Y., S. Gorina, P. D. Jeffrey, and N. P. Pavletich. 1994. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science 265**:**346-355. [DOI] [PubMed] [Google Scholar]
- 5.Chow, J. P. H., W. I. Siu, T. K. Fung, W. M. Chan, A. Lau, T. Arooz, C.-P. Ng, K. Yamashita, and R. Y. C. Poon. 2003. DNA damage during the spindle-assembly checkpoint degrades CDC25A, inhibits cyclin-CDC2 complexes, and reverses cells to interphase. Mol. Biol. Cell 14**:**3189-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davison, T. S., C. Vagner, M. Kaghad, A. Ayed, D. Caput, and C. H. Arrowsmith. 1999. p73 and p63 are homotetramers capable of weak heterotypic interactions with each other but not with p53. J. Biol. Chem. 274**:**18709-18714. [DOI] [PubMed] [Google Scholar]
- 7.Denissenko, M. F., A. Pao, M. Tang, and G. P. Pfeifer. 1996. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science 274**:**430-432. [DOI] [PubMed] [Google Scholar]
- 8.de Vries, A., E. R. Flores, B. Miranda, H. M. Hsieh, C. T. van Oostrom, J. Sage, and T. Jacks. 2002. Targeted point mutations of p53 lead to dominant-negative inhibition of wild-type p53 function. Proc. Natl. Acad. Sci. USA 99**:**2948-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eliyahu, D., N. Goldfinger, O. Pinhasi-Kimhi, G. Shaulsky, Y. Skurnik, N. Arai, V. Rotter, and M. Oren. 1988. Meth A fibrosarcoma cells express two transforming mutant p53 species. Oncogene 3**:**313-321. [PubMed] [Google Scholar]
- 10.Finlay, C. A., P. W. Hinds, and A. J. Levine. 1989. The p53 proto-oncogene can act as a suppressor of transformation. Cell 57**:**1083-1093. [DOI] [PubMed] [Google Scholar]
- 11.Forrester, K., S. E. Lupold, V. L. Ott, C. H. Chay, V. Band, X. W. Wang, and C. C. Harris. 1995. Effects of p53 mutants on wild-type p53-mediated transactivation are cell type dependent. Oncogene 10**:**2103-2111. [PubMed] [Google Scholar]
- 12.Frebourg, T., M. Sadelain, Y. S. Ng, J. Kassel, and S. H. Friend. 1994. Equal transcription of wild-type and mutant p53 using bicistronic vectors results in the wild-type phenotype. Cancer Res. 54**:**878-881. [PubMed] [Google Scholar]
- 13.Fujita, M., T. Kiyono, Y. Hayashi, and M. Ishibashi. 1997. In vivo interaction of human MCM heterohexameric complexes with chromatin. Possible involvement of ATP. J. Biol. Chem. 272**:**10928-10935. [DOI] [PubMed] [Google Scholar]
- 14.Fung, T. K., W. Y. Siu, C. H. Yam, A. Lau, and R. Y. C. Poon. 2002. Cyclin F is degraded during G2-M by mechanisms fundamentally different from other cyclins. J. Biol. Chem. 277**:**35140-35149. [DOI] [PubMed] [Google Scholar]
- 15.Gu, J., H. Kawai, D. Wiederschain, and Z. M. Yuan. 2001. Mechanism of functional inactivation of a Li-Fraumeni syndrome p53 that has a mutation outside of the DNA-binding domain. Cancer Res. 61**:**1741-1746. [PubMed] [Google Scholar]
- 16.Hachiya, M., A. Chumakov, C. W. Miller, M. Akashi, J. Said, and H. P. Koeffler. 1994. Mutant p53 proteins behave in a dominant, negative fashion in vivo. Anticancer Res. 14**:**1853-1859. [PubMed] [Google Scholar]
- 17.Harris, C. C. 1993. p53: at the crossroads of molecular carcinogenesis and risk assessment. Science 262**:**1980-1981. [DOI] [PubMed] [Google Scholar]
- 18.Hollstein, M., D. Sidransky, B. Vogelstein, and C. C. Harris. 1991. p53 mutations in human cancers. Science 253**:**49-53. [DOI] [PubMed] [Google Scholar]
- 19.Hsu, I. C., R. A. Metcalf, T. Sun, J. A. Welsh, N. J. Wang, and C. C. Harris. 1991. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature 350**:**427-428. [DOI] [PubMed] [Google Scholar]
- 20.Irwin, M. S., and W. G. Kaelin. 2001. p53 family update: p73 and p63 develop their own identities. Cell Growth Differ. 12**:**337-349. [PubMed] [Google Scholar]
- 21.Ishimoto, O., C. Kawahara, K. Enjo, M. Obinata, T. Nukiwa, and S. Ikawa. 2002. Possible oncogenic potential of DeltaNp73: a newly identified isoform of human p73. Cancer Res. 62**:**636-641. [PubMed] [Google Scholar]
- 22.Jeffrey, P. D., S. Gorina, and N. P. Pavletich. 1995. Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science 267**:**1498-1502. [DOI] [PubMed] [Google Scholar]
- 23.Kern, S. E., J. A. Pietenpol, S. Thiagalingam, A. Seymour, K. W. Kinzler, and B. Vogelstein. 1992. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science 256**:**827-830. [DOI] [PubMed] [Google Scholar]
- 24.Leung, K. M., L. S. Po, F. C. Tsang, W. Y. Siu, A. Lau, H. T. Ho, and R. Y. C. Poon. 2002. The candidate tumor suppressor ING1b can stabilize p53 by disrupting the regulation of p53 by MDM2. Cancer Res. 62**:**4890-4893. [PubMed] [Google Scholar]
- 25.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88**:**323-331. [DOI] [PubMed] [Google Scholar]
- 26.McLure, K. G., and P. W. Lee. 1998. How p53 binds DNA as a tetramer. EMBO J. 17**:**3342-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milner, J., and E. A. Medcalf. 1991. Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation. Cell 65**:**765-774. [DOI] [PubMed] [Google Scholar]
- 28.Nakagawa, T., M. Takahashi, T. Ozaki, K. Watanabe Ki, S. Todo, H. Mizuguchi, T. Hayakawa, and A. Nakagawara. 2002. Autoinhibitory regulation of p73 by ΔNp73 to modulate cell survival and death through a p73-specific target element within the Δ_Np73_ promoter. Mol. Cell. Biol. 22**:**2575-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicholls, C. D., K. G. McLure, M. A. Shields, and P. W. Lee. 2002. Biogenesis of p53 involves cotranslational dimerization of monomers and posttranslational dimerization of dimers. Implications on the dominant negative effect. J. Biol. Chem. 277**:**12937-12945. [DOI] [PubMed] [Google Scholar]
- 30.Ongkeko, W. M., X. Q. Wang, W. Y. Siu, A. W. S. Lau, K. Yamashita, A. L. Harris, L. S. Cox, and R. Y. C. Poon. 1999. MDM2 and MDMX bind and stabilize the tumor suppressor p53-related protein p73. Curr. Biol. 9**:**829-832. [DOI] [PubMed] [Google Scholar]
- 31.Poon, R. Y. C., H. Toyoshima, and T. Hunter. 1995. Redistribution of the CDK inhibitor p27 between different cyclin. CDK complexes in the mouse fibroblast cell cycle and in cells arrested with lovastatin or ultraviolet irradiation. Mol. Biol. Cell 6**:**1197-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pozniak, C. D., S. Radinovic, A. Yang, F. McKeon, D. R. Kaplan, and F. D. Miller. 2000. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science 289**:**304-306. [DOI] [PubMed] [Google Scholar]
- 33.Rovinski, B., and S. Benchimol. 1988. Immortalization of rat embryo fibroblasts by the cellular p53 oncogene. Oncogene 2**:**445-452. [PubMed] [Google Scholar]
- 34.Sayan, A. E., B. S. Sayan, N. Findikli, and M. Ozturk. 2001. Acquired expression of transcriptionally active p73 in hepatocellular carcinoma cells. Oncogene 20**:**5111-5117. [DOI] [PubMed] [Google Scholar]
- 35.Srivastava, S., S. Wang, Y. A. Tong, K. Pirollo, and E. H. Chang. 1993. Several mutant p53 proteins detected in cancer-prone families with Li-Fraumeni syndrome exhibit transdominant effects on the biochemical properties of the wild-type p53. Oncogene 8**:**2449-2456. [PubMed] [Google Scholar]
- 36.Stiewe, T., C. C. Theseling, and B. M. Putzer. 2002. Transactivation-deficient Delta TA-p73 inhibits p53 by direct competition for DNA binding: implications for tumorigenesis. J. Biol. Chem. 277**:**14177-14185. [DOI] [PubMed] [Google Scholar]
- 37.Varley, J. M. 2003. Germline TP53 mutations and Li-Fraumeni syndrome. Hum. Mutat. 21**:**313-320. [DOI] [PubMed] [Google Scholar]
- 38.Vikhanskaya, F., M. D'Incalci, and M. Broggini. 2000. p73 competes with p53 and attenuates its response in a human ovarian cancer cell line. Nucleic Acids Res. 28**:**513-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogelstein, B., and K. W. Kinzler. 1992. p53 function and dysfunction. Cell 70**:**523-526. [DOI] [PubMed] [Google Scholar]
- 40.Vogelstein, B., and K. W. Kinzler. 1994. Tumour-suppressor genes. X-rays strike p53 again. Nature 370**:**174-175. [DOI] [PubMed] [Google Scholar]
- 41.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408**:**307-310. [DOI] [PubMed] [Google Scholar]
- 42.Wang, X., T. Arooz, W. Y. Siu, C. H. Chiu, A. Lau, K. Yamashita, and R. Y. C. Poon. 2001. MDM2 and MDMX can interact differently with ARF and members of the p53 family. FEBS Lett. 490**:**202-208. [DOI] [PubMed] [Google Scholar]
- 43.Wang, X. Q., W. M. Ongkeko, A. W. Lau, K. M. Leung, and R. Y. C. Poon. 2001. A possible role of p73 on the modulation of p53 level through MDM2. Cancer Res. 61**:**1598-1603. [PubMed] [Google Scholar]
- 44.Wang, Y., J. F. Schwedes, D. Parks, K. Mann, and P. Tegtmeyer. 1995. Interaction of p53 with its consensus DNA-binding site. Mol. Cell. Biol. 15**:**2157-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yam, C. H., R. W. Ng, W. Y. Siu, A. W. Lau, and R. Y. C. Poon. 1999. Regulation of cyclin A-Cdk2 by SCF component Skp1 and F-box protein Skp2. Mol. Cell. Biol. 19**:**635-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yam, C. H., W. Y. Siu, T. Arooz, C. H. Chiu, A. Lau, X. Q. Wang, and R. Y. C. Poon. 1999. MDM2 and MDMX inhibit the transcriptional activity of ectopically expressed SMAD proteins. Cancer Res. 59**:**5075-5078. [PubMed] [Google Scholar]
- 47.Yam, C. H., W. Y. Siu, A. Lau, and R. Y. C. Poon. 2000. Degradation of cyclin A does not require its phosphorylation by CDC2 and cyclin-dependent kinase 2. J. Biol. Chem. 275**:**3158-3167. [DOI] [PubMed] [Google Scholar]
- 48.Yang, A., M. Kaghad, D. Caput, and F. McKeon. 2002. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet. 18**:**90-95. [DOI] [PubMed] [Google Scholar]
- 49.Yang, A., M. Kaghad, Y. Wang, E. Gillett, M. D. Fleming, V. Dotsch, N. C. Andrews, D. Caput, and F. McKeon. 1998. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2**:**305-316. [DOI] [PubMed] [Google Scholar]
- 50.Yang, A., and F. McKeon. 2000. P63 and P73: P53 mimics, menaces and more. Nat. Rev. Mol. Cell Biol. 1**:**199-207. [DOI] [PubMed] [Google Scholar]
- 51.Yang, A., N. Walker, R. Bronson, M. Kaghad, M. Oosterwegel, J. Bonnin, C. Vagner, H. Bonnet, P. Dikkes, A. Sharpe, F. McKeon, and D. Caput. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404**:**99-103. [DOI] [PubMed] [Google Scholar]
- 52.Yin, Y., C. W. Stephen, M. G. Luciani, and R. Fahraeus. 2002. p53 stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat. Cell Biol. 4**:**462-467. [DOI] [PubMed] [Google Scholar]
- 53.Zacharatos, P. V., V. G. Gorgoulis, A. Kotsinas, E. N. Manolis, T. Liloglou, A. N. Rassidakis, P. Kanavaros, J. D. Field, T. Halazonetis, and C. Kittas. 1999. Modulation of wild-type p53 activity by mutant p53 R273H depends on the p53 responsive element (p53RE). A comparative study between the p53REs of the MDM2, WAFI/Cip1 and Bax genes in the lung cancer environment. WAFI/Cip1 = WAF1/Cip1. Anticancer Res. 19**:**579-587. [PubMed] [Google Scholar]
- 54.Zhu, J., W. Zhou, J. Jiang, and X. Chen. 1998. Identification of a novel p53 functional domain that is necessary for mediating apoptosis. J. Biol. Chem. 273**:**13030-13036. [DOI] [PubMed] [Google Scholar]