Microarray Analysis of Pneumococcal Gene Expression during Invasive Disease (original) (raw)
Abstract
Streptococcus pneumoniae is a leading cause of invasive bacterial disease. This is the first study to examine the expression of S. pneumoniae genes in vivo by using whole-genome microarrays available from The Institute for Genomic Research. Total RNA was collected from pneumococci isolated from infected blood, infected cerebrospinal fluid, and bacteria attached to a pharyngeal epithelial cell line in vitro. Microarray analysis of pneumococcal genes expressed in these models identified body site-specific patterns of expression for virulence factors, transporters, transcription factors, translation-associated proteins, metabolism, and genes with unknown function. Contributions to virulence predicted for several unknown genes with enhanced expression in vivo were confirmed by insertion duplication mutagenesis and challenge of mice with the mutants. Finally, we cross-referenced our results with previous studies that used signature-tagged mutagenesis and differential fluorescence induction to identify genes that are potentially required by a broad range of pneumococcal strains for invasive disease.
Streptococcus pneumoniae is the primary cause of community-acquired pneumonia and a major cause of invasive bacterial disease (28). Each year in the United States, pneumococci are responsible for 100,000 to 135,000 hospitalizations for pneumonia, 50,000 cases of bacteremia, and 3,000 cases of meningitis (18). Worldwide these diseases account for more than a million deaths a year. Currently, a 7-valent polysaccharide-protein conjugate vaccine is effective; however, it protects against only a small subset of the serotypes known to cause invasive disease (9). Invasive disease follows colonization of the nasopharynx and is the result of spread of the bacteria to the lungs and blood. Once in the bloodstream, infection may result in septicemia or meningitis, both of which have high mortality despite accepted antibiotic therapy (28).
Since Pasteur and Sternberg first described the pneumococcus, a number of virulence factors required for invasive disease have been identified. Major virulence determinants such as capsular polysaccharide, pneumolysin, and choline-binding proteins have clearly established roles in pathogenesis (18). More recently, large-scale identification of S. pneumoniae virulence determinants has been attempted. Studies, such as signature-tagged mutagenesis (STM), have used transposons and suicide vectors to pepper the chromosome of the bacteria with mutations and identify genes required for virulence (13, 25, 39). STM screens rely on negative selection of mutants through passage in animal models and, to date, STM has been used three times for the pneumococcus, each time in a different strain.
Ideally, characterization of pneumococcal virulence determinants should include analysis of gene expression during invasive disease. Confirmation of their expression in vivo would not only verify the contribution of these genes to pathogenesis but would also elucidate their contribution to discrete forms of disease (e.g., genes expressed during pneumonia versus those in the blood during bacteremia). Unfortunately, analysis of in vivo bacterial gene expression, much less at discrete sites, has been limited by the difficulties of isolating sufficient quantities of pure and intact bacterial RNA from infected host tissues. To circumvent the requirement for RNA, investigators have used differential fluorescence induction (DFI) to identify S. pneumoniae promoters that are induced during disease (29). In contrast to STM, DFI relies on the promoter activity of random fragments of DNA cloned upstream of an episomal, promoterless green fluorescent protein. Fluorescence is imparted by the promoter activity of the randomly integrated DNA fragment under the environmental conditions tested (in vivo). Fluorescent bacteria can then be sorted by fluorescence-activated cell sorting analysis, leaving only bacteria containing plasmids with active promoters. Using DFI, Marra et al. identified operons that are enhanced during bacterial growth in a chinchilla model of otitis media, lower respiratory tract infection in a mouse, and growth in an intraperitoneal chamber implant model (29).
Large-scale analysis of gene expression during invasive disease provides not only transcriptional data with regard to certain genes, such as virulence determinants but, after interpretation, also provides information as to the status of the bacteria during infection and the response of the bacteria to the host environment. In vivo gene expression therefore potentially identifies unknown or unappreciated targets for pharmaceutical or vaccine intervention since, presumably, the genes whose transcription is altered during invasive disease are those that are required by the bacteria for survival in the host (14).
We describe here a protocol for the collection of RNA from bacteria within infected blood and cerebrospinal fluid (CSF) in vivo and bacteria adherent to epithelial cells in vitro. Using genome-wide cDNA microarrays available from the Pathogen Functional Genomic Research Center (PFGRC) at The Institute for Genomic Research (TIGR; Rockville, Md.), we have examined pneumococcal gene expression during bacteremia, meningitis, and epithelial cell contact (ECC). Analysis of pneumococcal genes expressed in these models identified global patterns of expression unique to each condition tested. Patterns of expression were identified with regard to virulence factors, transporters, transcription factors, translation-associated proteins, metabolism, and genes with unknown function. These findings were then cross referenced to previous STM and DFI studies. Finally, we examined several candidate virulence genes with unknown function by insertion duplication mutagenesis and challenge of mice.
MATERIALS AND METHODS
Media and growth conditions.
S. pneumoniae was grown on tryptic soy agar (Difco, Detroit, Mich.) supplemented with 3% defibrinated sheep blood or in defined semisynthetic casein liquid medium (24) supplemented with 0.5% yeast extract (C+Y). Erythromycin (1 μg/ml) and kanamycin (400 μg/ml) (Sigma, St. Louis, Mo.) were added to the growth medium as appropriate. S. pneumoniae cultures were inoculated from frozen stock and incubated at 37°C in 5% CO2. Escherichia coli strains were grown in Luria-Bertani medium (Difco) at 37°C in an orbital shaker; erythromycin (1 mg/ml) was added to the E. coli cultures to maintain any plasmids.
Bacterial strains and construction of mutants.
D39 Xen7 (D39X), a stable bioluminescent isolate of S. pneumoniae, serotype 2, strain D39 (2), was created as previously described (11). TIGR4 Xen 35 (T4X) was created by transformation of TIGR4 with genomic DNA from D39X by using CSP-2 (35). Bioluminescent mutants deficient in genes identified as enhanced during invasive disease were created by insertion duplication mutagenesis (37). PCR primers with nested EcoRI and BamHI restriction sites were used to amplify 200- to 500-bp fragments from the N termini of the genes. Amplified PCR fragments were purified and digested with EcoRI and BamHI (New England BioLabs, Beverly, Mass.). Digested fragments were ligated into the vector pJDC9 and transformed into chemically competent XL1-Blue E. coli (Stratagene, La Jolla, Calif.). Single transformants containing the insert were identified and plasmid DNA from these clones was used to transform D39X by using CSP-1 or, alternatively, T4X with CSP-2 (7). Chromosomal integration of the vector at the correct locus was verified by PCR analysis and sequencing by using a primer contained within the integrated vector and a primer upstream of the gene of interest.
Isolation of bacterial RNA from infected mouse blood.
Female BALB/cJ mice (4 to 5 weeks old; The Jackson Laboratory, Bar Harbor, Maine) were maintained in a biosafety level 2 facility at St. Jude Children's Research Hospital. All experimental procedures were done with mice anesthetized with either inhaled isoflurane (Baxter Healthcare Corp., Deerfield, Ill.) at 2.5% or intraperitoneal MKX (1 ml of ketamine [Fort Dodge Laboratories, Fort Dodge, Iowa] at 100 mg/ml; 5 ml of xylazine [Miles Laboratories, Shawnee Mission, KA] at 100 mg/ml; 21 ml of phosphate-buffered saline [PBS]) at 5 μl/g of body weight. Bacteria in the blood were collected by using a modified version of the protocol described by Ogunniyi et al. (33). Mice were infected intratracheally with 105 CFU of D39X and imaged by using a charge-coupled device camera at 48 h postchallenge. Mice with severe sepsis, as determined by bioluminescent imaging (>50,000 relative light units/mouse) (11), were exsanguinated, and the blood was immediately transferred to a tube containing RNAprotect (Qiagen, Valencia, Calif.) at a ratio of 35:1 (vol/vol; RNAprotect/expected total mouse blood collected) and vortexed for 5 s. Bacteria were harvested by centrifugation of the suspension at 825 × g for 10 min to remove debris, followed by centrifugation at 15,000 × g for 15 min to pellet the bacteria. RNA isolation was performed by using a Qiagen RNeasy minikit (Qiagen) with the following modifications. Bacteria were lysed in the presence of 400 mg of 0.1-mm zirconia-silica beads (BioSpec Products, Inc., Bartlesville, Okla.) by using the Mini-Beadbeater 3110BX (BioSpec Products, Inc.) and then incubated at 70° C for 10 min. Bacterial lysate was spun through a Qiashredder column (Qiagen) to remove the beads, and the remaining solution was subsequently processed according to the manufacturer's protocol with an on-column DNase digestion step. Quantitation of RNA was performed by using a UV spectrophotometer (UV-1601; Shimadzu Corp., Kyoto, Japan) at an optical density at 260 nm (OD260), whereas degradation was assessed by visualization of the RNA on a 1% agarose Tris-borate-EDTA (TBE) gel.
Isolation of bacterial RNA from infected rabbit CSF.
Male New Zealand White rabbits (5 kg; Myrtles, Thompson Station, Tenn.) were anesthetized with 35 ml of 25% urethane administered subcutaneously and pentobarbital sodium (15 mg/kg) given intravenously. Anesthetized rabbits were immobilized on a stereotaxic frame and challenged by direct intracisternal injection with 108 CFU of T4X by using a 25-gauge spinal needle (27). At 4 h after challenge, CSF was collected and transferred immediately to a sterile tube containing RNAprotect at a ratio of 3:1 (RNAprotect/rabbit CSF). RNA was then collected as described above.
Isolation of bacterial RNA from pneumococci attached to Detroit cells.
A T-175 flask of confluent Detroit pharyngeal epithelial cells (38) was activated with tumor necrosis factor alpha (Sigma) at 10 ng/ml for 2 h. Cells were challenged with 25 ml of a 4 × 106-CFU/ml suspension of T4R (12), an unencapsulated derivative of TIGR4, and incubated at 37°C for 3 h to allow for adherence. After the incubation, cells were washed three times with PBS (BioWhittaker, Walkersville, Md.) to remove nonadherent bacteria and covered with 10 ml of RNAprotect. Cells were scraped off the surface of the flask, and the suspension was transferred to a new T-175 flask containing 3-mm glass beads. Cells were sonicated for 5 min by using a tabletop ultrasonic cleaner (FS20; Fisher Scientific, Pittsburgh, Pa.) to lyse the epithelial cells but leave the bacteria intact. The suspension was then centrifuged at 800 × g for 5 min to remove cellular debris and then centrifuged at 4,600 × g for 10 min to pellet the bacteria. RNA was collected from the bacterial/eukaryotic pellet by using a Qiagen RNeasy minikit. To remove the remaining eukaryotic RNA, bacterial RNA isolated from the bacterial/eukaryotic pellet was then enriched by using MICROBEnrich (Ambion, Austin, Tex.). Quantitation of RNA was performed by using a UV spectrophotometer at OD260, whereas degradation was assessed by visualization of the RNA on a 1% agarose TBE gel. As a control, RNA was collected from bacteria grown in tissue culture medium in parallel and was processed by using MICROBEnrich.
Microarray analysis of bacterial RNA.
Microarray experiments were performed by using whole-genome S. pneumoniae cDNA microarrays obtained from the PFGRC at TIGR (http://pfgrc.tigr.org.). The S. pneumoniae genome microarray consisted of PCR products representing segments of 2,131 open reading frames from S. pneumoniae strain TIGR4 (44) and 118 unique open reading frames from strains R6 (17) and G54 (39). Microarray experiments, including RNA quality control, Cy3 and Cy5 dye labeling, hybridization, washing, and scanning, were performed at the Functional Genomics lab, Hartwell Center for Bioinformatics and Biotechnology, St. Jude Children's Research Hospital, by using protocols from the PFGRC (http://pfgrc.tigr.org/protocols.shtml). The hybridization probe was constituted by mixture of differentially labeled cDNA derived from (i) total RNA isolated from S. pneumoniae obtained from either infected mouse blood or rabbit CSF or (ii) RNA from bacteria grown in C+Y. Alternatively, probe for ECC analysis was constituted with (i) RNA isolated from bacteria adhered to Detroit cells or (ii) RNA from parallel cultures grown in tissue culture media. RNA samples from both conditions were labeled with monofunctional Cy3 and Cy5 dyes by using an indirect amino-allyl labeling method, combined, and hybridized overnight to the printed slides. Slides were washed and scanned by using an Axon 4000B dual channel scanner (Axon Corp., Union City, Calif.) to generate a multi-TIFF image of each slide. Images were analyzed by using Axon GenePix 4.1 image analysis software, and the resulting text-data files were imported into Spotfire DecisionSite for Functional Genomics (version 7.2; Spotfire, Sommerville, Mass.). A series of filtration algorithms were applied to remove spots that consistently generated bad data (based on the frequency with which a particular spot failed to reach a minimum required signal-to-noise ratio [SNR] and the frequency with which a particular spot was flagged bad by the image analysis software GenePix Pro 4.1). Genes that were flagged ≥66% of the time were removed from analysis. Similarly, spots that failed to meet the SNR criteria 75% of the time (9 of 12 times for blood and ECC microarrays and 6 of 8 for CSF microarrays) were removed from consideration. Intensity-based global normalization was then performed to remove dye-specific bias, and background correction was performed by subtracting the normalized median pixel intensity of the background value from the normalized median pixel intensity of the spot itself. Cy5/Cy3 ratios (fold changes) were then calculated for every spot. Since each gene was spotted four times per glass microarray, only genes whose corresponding spots were not flagged at least 75% of the times were considered.
Statistical analysis of microarray data.
Microarray analysis examining RNA from bacteria adhered to Detroit cells and blood experiments was performed on total RNA isolated from three independent biological replicate experiments. Microarray analysis examining RNA from infected rabbit CSF was performed on total RNA isolated from two independent biological replicate experiments. Accuracy and statistical significance of the gene expression differential over the course of the replicate experiments was calculated by using a Student t test-analysis of variance algorithm available in Spotfire DecisionSite (19). Genes with high levels of significance (P > 0.001) and a minimum fold change of 2.0 were considered up- or downregulated.
Virulence assessment of mutants.
To assess the virulence potential of mutants deficient in genes with altered expression in vivo, mice were challenged intratracheally and monitored for 4 days or challenged intranasally and monitored for 7 days. Exponential cultures (OD620 = 0.5) of D39X, T4X, or their derivatives were centrifuged, and the bacteria were washed with and suspended in PBS. Mice were anesthetized with isoflurane and challenged intranasally with 107 CFU in 25 μl or intratracheally with 105 CFU in 100 μl of PBS. At 2 days postchallenge, all mice were sampled by intranasal lavage, blood collection from the tail vein, and bioluminescent imaging with a Xenogen IVIS camera. Bioluminescent imaging provided a noninvasive method for assessing the bacterial burden in the lungs of all of the mice (data not shown). After imaging, 6 to 10 mice were randomly selected from each cohort and sacrificed, and bacterial titers in lungs were determined. The number of CFU present per gram of homogenized lungs was used to assess bacterial burden in the lungs. Remaining mice (6 to 10 per group) were then observed to determine the percent survival over time. In all instances after challenge, the infectious dose was confirmed by serial dilution and plating of the bacterial suspension on blood agar.
RESULTS AND DISCUSSION
Microarray analysis is a powerful tool for transcriptional analysis of pneumococcal gene expression during invasive disease. Provided RNA is obtainable, microarrays directly measure transcription for each gene on the chromosome at once. To date, however, pneumococcal microarrays have only been used to examine bacteria in vitro, more specifically, to examine the pneumococcal response to a particular environment or to examine isogenic mutants. In the present study, we describe for the first time microarray analysis of in vivo gene expression during bacteremia and meningitis. We also examine gene expression in response to intimate contact of the pneumococcus with epithelial cells in vitro.
Collection of RNA samples from infected blood and CSF.
Collection of bacterial RNA from infected tissues has been limited by the inherent difficulties of separating large quantities of host cells and debris away from bacteria in a manner sufficiently timely to prevent degradation of RNA or to prevent novel gene expression in response to conditions presented during the separation process (14, 36). To avoid these problems, we harvested infected mouse blood and rabbit CSF directly into RNAprotect (Qiagen). RNAprotect served to stabilize bacterial RNA, allowed purification of the RNA without degradation (Fig. 1), and lysed contaminating host cells. Preliminary experiments determined that high titers of bacteria (>108 CFU/ml) in body fluids were required to obtain sufficient RNA (data not shown). Although strain T4X was used for meningitis, preliminary experiments determined its yield in blood was too low (data not shown). Therefore, for bacteremia we used strain D39, which attains titers in blood of 109 CFU/ml by 48 h after challenge (35). Bioluminescence of T4X and D39X permitted visualization of the bacteria in living mice and allowed us to identify animals with sufficiently high bacterial titers for RNA isolation. Bioluminescent imaging was necessary, since only half of the mice infected intratracheally had sufficient bacteria in the bloodstream 48 h postchallenge. The remaining mice eventually progressed to similar levels of bacteremia (∼109 CFU/ml of blood within 12 to 24 h); however, this occurred in a manner too staggered for satisfactory bacteria collection. Exsanguination of the mice collected at 48 h yielded ∼0.75 ml of blood from each infected animal, which in turn allowed collection of ∼25 μg of pure bacterial RNA from the pooled blood of three to four mice.
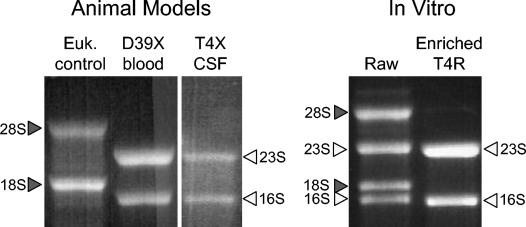
FIG. 1.
RNA collected from S. pneumoniae in vivo and in vitro. After isolation of pneumococcal RNA from infected blood, CSF, and bacteria adherent to the Detroit pharyngeal epithelial cell line, RNA was visualized on a 1% TBE gel to confirm purity and assess degradation. Bacterial RNA (open arrowheads) and eukaryotic RNA (shaded arrowheads) are indicated. Bacterial RNA isolated from the ECC model was enriched after collection of the initial pellet (raw) to remove contaminating eukaryotic RNA.
Sufficient bacteria could be harvested from the CSF of infected rabbits only upon challenge with 108 CFU of T4X (35); strain D39X did not grow to a sufficiently high titer. T4X is a derivative of TIGR4 (44), the fully sequenced clinical isolate obtained from a child with meningitis. At 4 h after intracisternal challenge, ∼1.0 ml of infected CSF was collected and yielded ∼5 μg of pure bacterial RNA from each rabbit (Fig. 1).
Collection of RNA samples from pneumococci attached to epithelial cells.
To examine bacterial gene expression in response to ECC, we infected confluent monolayers of Detroit cells (38), a pharyngeal epithelial cell line, with pneumococcus T4R. T4R is an unencapsulated derivative of T4 (12). As a control, RNA was collected from bacteria grown in the same tissue culture media without cells. These analyses therefore reflect only changes in gene expression that are a result of host cell exposure. Stimuli resulting in alterations in gene expression should therefore be limited to (i) intimate contact with the host cells, (ii) soluble factors released by the host cell during normal processes, and (iii) soluble factors released by the host cell in response to the bacterial infection. Collection of RNA from bacteria attached to Detroit cells resulted in the collection of ∼25 μg of mixed eukaryotic and prokaryotic RNA from the bacteria and host cell pellet. Using MICROBencrich, we were able to purify the prokaryotic RNA (Fig. 1), with a final yield of ∼10 μg per T-175 flask.
Microarray analysis of gene expression relies on the comparison of two RNA species. Levels of expression reported in the test condition are reported in relation to levels of transcription observed in the control condition. A direct comparison of test versus control can be set up in vitro with tissue culture (medium to medium plus Detroit cells). With regard to in vivo gene expression, an appropriate in vitro control condition does not exist. However, reference to the same arbitrary control media (blood versus C+Y or CSF versus C+Y) allows for indirect comparisons between the different test conditions (blood versus CSF). As a baseline, >99% of genes showed similar expression levels in D39 and T4 grown in C+Y (data not shown).
Microarray analysis of bacterial physiology in different body sites.
Filtration algorithms and SNR analysis of the microarrays by GenePix 4.1 removed genes whose corresponding spots on the microarray were not valid ≥75% of the time. As such, the results reported in the present study correspond to 69% of the genome during growth in the bloodstream, 53% of the genome during meningitis, and 68% of the genome after ECC. A list of the genes that did not meet the above criteria and therefore were not analyzed for alternate gene expression in vivo can be found at http://www.stjuderesearch.org/vivogene. Stringent requirements imposed by the filtration algorithms ensured that any alterations in gene expression that are reported here are rigorously supported by the data collected from the microarrays.
For genes that met the established criteria for transcription analysis, it was determined that the majority (92% in the blood, 85% in CSF, and 90% after ECC) were expressed in a fashion similar to growth in C+Y. Table 1 lists the genes with altered transcription sorted based on their putative cellular role and their location on the bacterial chromosome. Inclusion criteria were a >2-fold change in levels of expression and a P value of <0.01. Of the genes with altered expression in vivo, 55% had enhanced expression in blood, while 81% were enhanced during ECC. In contrast, only 35% were enhanced during growth in CSF. Overall, the patterns of gene expression were determined to be distinct for bacteria in each anatomic site. Figure 2 is a Venn diagram illustrating the differences and similarities between the expression profiles of the pneumococci in blood, in CSF, and during ECC. Highlighting the disparity between these conditions, only eight genes had similar alterations in gene expression during bacterial growth in blood, in CSF, or during ECC: two that encode the virulence determinants PspA (46) and PrtA (4), three that encode genes in the psa operon (manganese acquisition and transport) (30), two that are involved in energy metabolism (an enolase [_eno_] and a glycosyl hydrolase [SP0265]), and a transporter (SP1587). Their common expression warrants further investigation as potential targets for intervention, since they may reflect a core set of genes required for virulence.
TABLE 1.
Differential expression of S. pneumoniae genes as determined by microarray analysis
| Gene name and/or description | TIGR annotationb | Fold increase or decrease and P valuea in: | |||||
|---|---|---|---|---|---|---|---|
| Blood | CSF | ECC | |||||
| Fold change | P | Fold change | P | Fold change | P | ||
| Virulence determinants | |||||||
| blpU; bacteriocin | SP0041 | −3.1 | 3E-07 | ||||
| cps4A; capsular polysaccharide biosynthesis | SP0346 | 2.2 | 2E-03 | ||||
| cps4C; capsular polysaccharide biosynthesis | SP0348 | 3.9 | 2E-12 | ||||
| pspA; pneumococcal surface protein A | SP0117A | 3.4 | 2E-04 | 50.3 | 1E-08 | 4.0 | 1E-04 |
| cbpJ; choline-binding protein J | SP0378 | 3.6 | 9E-07 | ||||
| cbpG; choline-binding protein G | SP0390 | 1.7 | 5E-03 | ||||
| cbpF; choline-binding protein F | SP0391 | 2.3 | 2E-03 | ||||
| blpK; bacteriocin associated protein | SP0533 | −2.5 | 3E-06 | ||||
| blpY; Immunity protein | SP0545 | NA | NA | −16.8 | 8E-13 | NA | NA |
| prtA; protective antigen A | SP0641A,D | 15.5 | 7E-06 | 9.8 | 8E-13 | 3.3 | 1E-02 |
| spxB; pyruvate oxidase | SP0730 | −2.6 | 6E-06 | ||||
| lmb; adhesion lipoprotein | SP1002 | 3.4 | 2E-05 | −5.2 | 1E-09 | ||
| Conserved hypothetical protein | SP1003A | 8.8 | 3E-05 | −4.6 | 7E-09 | ||
| xseA; exodeoxyribonuclease VII, large subunit | SP1207 | −2.2 | 3E-07 | ||||
| pln; pneumolysin | SP1923A | −5.6 | 5E-06 | −9.8 | 3E-03 | ||
| lytA; autolysin | SP1937 | −7.6 | 9E-13 | ||||
| chpA; choline-binding protein A | SP2190A | 2.3 | 2E-02 | ||||
| chpD; choline-binding protein D | SP2201A | NA | NA | 12.0 | 2E-12 | ||
| htrA; serine protease | SP2239A | NA | NA | 8.2 | 4E-05 | ||
| spoJ; homologous to sporulation protein | SP2240D | 3.9 | 9E-05 | ||||
| Cell wall synthesis | |||||||
| bacA; bacitracin resistance protein | SP0457B | 2.5 | 4E-04 | ||||
| fibA; beta-lactam resistance factor | SP0615 | 4.0 | 1E-03 | ||||
| lytB; endo-β-_N_-acetylglucosaminldase | SP0965 | 3.5 | 4E-03 | ||||
| licC; phosphocholine cytidytyltransferase | SP1267 | 4.7 | 1E-09 | 14.4 | 7E-08 | ||
| licB; choline transport | SP1268 | 5.1 | 2E-09 | 15.4 | 1E-09 | ||
| pck; choline kinase | SP1269 | 4.4 | 6E-09 | 18.2 | 2E-07 | ||
| Alcohol dehydrogenase, zinc containing | SP1270 | 5.5 | 4E-09 | 17.2 | 9E-06 | ||
| Cytidine diphosphocholine pyrophosphorylase, putative | SP1271 | 5.7 | 5E-10 | 14.0 | 3E-08 | ||
| pgdA; _N_-acetylglucosamine deacetylase A | SP1479D | −3.2 | 2E-05 | ||||
| penA; penicillin-binding protein 2B | SP1673 | 2.4 | 2E-02 | ||||
| dltD; d-alanyl-lipotechoic acid biosynthesis | SP2173 | 2.8 | 7E-04 | 6.6 | 1E-03 | ||
| dltA; d-alanine activating protein | SP2176A | 3.6 | 1E-03 | ||||
| Cell membrane | |||||||
| Lipoprotein | SP0149 | 3.7 | 2E-09 | ||||
| Glycosyl transferase, group 1 | SP1366 | 2.8 | 6E-04 | ||||
| pgm; phosphoglucomutase | SP1498 | −2.1 | 8E-03 | ||||
| Competence/DNA transformation | |||||||
| comX1; transcriptional regulator | SP0014 | −12.1 | 6E-11 | ||||
| Hypothetical protein | SP0029 | NA | NA | −8.1 | 5E-09 | NA | NA |
| ccs16; competence-induced protein Ccs16 | SP0030 | NA | NA | −16.8 | 4E-08 | NA | NA |
| comA; competence factor transport protein | SP0042D | −32.9 | 8E-12 | ||||
| comB; competence factor transport protein | SP0043D,B | −48.8 | 2E-12 | ||||
| ccs 4; competence-induced protein | SP0200 | −5.7 | 2E-07 | 3.0 | 2E-02 | ||
| Hypothetical protein | SP0201 | −2.9 | 2E-05 | ||||
| dprA; DNA processing protein | SP1266 | −49.7 | 2E-11 | 6.2 | 9E-04 | ||
| cspC-related protein | SP1913 | 5.2 | 8E-04 | ||||
| cbf1; cmp-binding-factor 1 | SP1980 | −4.7 | 2E-09 | ||||
| ccs50; competence-induced protein | SP1981 | −4.5 | 2E-06 | ||||
| recA; DNA replication, recombination, and repair | SP1940 | −12.9 | 1E-12 | ||||
| cinA; competence/damage-inducible protein | SP1941A | −39.0 | 1E-11 | 2.1 | 8E-03 | ||
| Conserved hypothetical protein | SP1944 | −2.4 | 2E-05 | ||||
| Hypothetical protein | SP1945 | −12.1 | 5E-06 | ||||
| comE; response regulator ComE | SP2235 | −15.0 | 2E-14 | NA | NA | ||
| comD; sensor histidine kinase | SP2236A | −6.7 | 2E-08 | ||||
| Stress related | |||||||
| hcrA; heat-inducible transcription repressor | SP0515 | −4.5 | 1E-07 | −4.4 | 8E-04 | ||
| grpE; heat shock protein | SP0516 | −3.7 | 2E-08 | −3.7 | 2E-03 | ||
| dnaK; protein folding and stabilization | SP0517 | −3.1 | 6E-06 | −3.9 | 2E-03 | ||
| sodA; superoxide dismutase | SP0766A | −3.5 | 3E-09 | ||||
| Cadmium resistance transporter, putative | SP1625 | −3.9 | 1E-10 | ||||
| Conserved hypothetical protein | SP1644 | 2.3 | 3E-04 | ||||
| relA; GTP pyrophosphokinase | SP1645A | 2.2 | 1E-03 | 2.8 | 1E-03 | ||
| Conserved hypothetical protein | SP1801 | NA | NA | 3.2 | 4E-03 | ||
| Hypothetical protein | SP1802 | 5.5 | 1E-03 | ||||
| Conserved hypothetical protein | SP1803 | NA | NA | 4.1 | 6E-05 | ||
| General stress protein 24, putative | SP1804 | 2.7 | 1E-07 | 6.7 | 2E-03 | ||
| Hypothetical protein | SP1805 | 3.5 | 8E-10 | 3.4 | 2E-03 | ||
| groEL; chaperonin, 60 kDa | SP1906 | −6.1 | 4E-11 | ||||
| groES; chaperonin, 10 kDa | SP1907 | −6.9 | 2E-08 | ||||
| DNA repair, recombination, and modification | |||||||
| Conserved hypothetical protein | SP0022 | NA | NA | −3.5 | 4E-06 | NA | NA |
| radA; DNA repair protein | SP0023AA,C | NA | NA | −6.3 | 2E-07 | NA | NA |
| Hypothetical protein | SP0025D | −2.8 | 8E-07 | ||||
| hexB; DNA mismatch repair protein | SP0173 | 14.0 | 5E-14 | ||||
| Hypothetical protein | SP0792 | 2.3 | 4E-08 | ||||
| MutT/nudix family protein | SP0794 | 2.0 | 1E-09 | ||||
| radC; DNA repair protein | SP1088 | −63.2 | 9E-11 | ||||
| Type II restriction endonuclease, putative | SP1221 | 3.0 | 6E-03 | ||||
| recR; recombination protein | SP1672 | 2.1 | 3E-03 | ||||
| recG; ATP-dependent DNA helicase | SP1697 | 2.6 | 6E-04 | ||||
| Cell division | |||||||
| ftsW; cell division protein | SP1067 | 3.8 | 5E-03 | ||||
| ppc; phosphoenolpyruvate carboxylase | SP1068B | 5.0 | 6E-04 | ||||
| DivlVA; cell division protein | SP1661 | 1.9 | 3E-03 | ||||
| y/mH | SP1662 | 2.2 | 2E-02 | ||||
| y/mF | SP1664 | 1.8 | 4E-02 | ||||
| y/mE | SP1665 | 2.0 | 3E-02 | ||||
| spollLJ family protein | SP1975 | −2.2 | 1E-05 | ||||
| Energy metabolism | |||||||
| Glycosyl hydrolase, family 1 | SP0265A | 3.5 | 1E-03 | 2.2 | 1E-06 | 5.5 | 5E-03 |
| Glycosamine-fructose-6-phosphate | SP0266 | 2.9 | 1E-11 | ||||
| manN; PTS system, IIC component | SP0283 | −2.2 | 7E-03 | ||||
| Alcohol dehydrogenase, zinc-containing | SP0285 | −1.7 | 6E-03 | ||||
| pfl; formate acetyltransferase | SP0459 | 2.6 | 7E-04 | ||||
| fba; fructose-bisphosphate aldolase | SP0605 | −2.1 | 1E-07 | −2.2 | 2E-04 | ||
| Thioredoxin family protein | SP0659B | NA | NA | NA | NA | −3.7 | 2E-04 |
| manA; mannose-6-phosphate isomerase | SP0736 | 2.7 | 1E-11 | 4.2 | 8E-04 | ||
| lacR; lactose system repressor | SP0875 | −4.8 | 9E-04 | ||||
| 1-phosphofructokinase, putative | SP0876 | −4.0 | 3E-04 | ||||
| aid; alanine dehydrogenase, authentic frameshift | SP0952 | NA | NA | 3.7 | 2E-03 | ||
| Phosphoglycerate mutase family protein | SP0984 | NA | NA | 2.1 | 4E-03 | ||
| glgC; glucose-1-phosphate edenylyltransferase | SP1122 | 3.5 | 3E-08 | ||||
| eno; enolase | SP1128 | −4.2 | 3E-03 | −2.1 | 1E-05 | −3.3 | 4E-05 |
| Acetoin dehydrogenase, E1 component | SP1164 | 2.3 | 8E-03 | ||||
| lacG; 6-phospho-β-galaciosidase | SP1184 | NA | NA | 6.5 | 4E-03 | ||
| lacB; galactose-6-phosphate isomerase | SP1192 | NA | NA | 5.9 | 2E-06 | ||
| lacA; galactose-6-phosphate isomerase | SP1193A | NA | NA | 4.2 | 6E-03 | ||
| ldh; L-lactate dehydrogenase | SP1220 | −2.3 | 3E-04 | ||||
| fhs; formate-tetrahydrofolate ligase | SP1229 | 3.9 | 3E-04 | ||||
| nagB; glucosamine-6-phosphate isomerase | SP1415 | −7.5 | 3E-12 | ||||
| alpC; ATP synthase F1, epsilon subunit | SP1507 | 4.8 | 3E-06 | ||||
| atpD; ATP synthase F1, beta subunit | SP1508 | 4.0 | 6E-06 | ||||
| alpG; ATP synthase F1, gamma subunit | SP1509 | 3.4 | 5E-04 | ||||
| atpA; ATP synthase F1, alpha subunit | SP1510 | 4.1 | 1E-08 | ||||
| atpH; ATP synthase F1, delta subunit | SP1511 | 3.8 | 7E-03 | ||||
| atpF; ATP synthase F0, B subunit | SP1512 | NA | NA | NA | NA | 2.7 | 4E-04 |
| atpB; ATP synthase F0, A subunit | SP1513 | 5.3 | 4E-06 | ||||
| Glutathione _S_-transferase family protein | SP1550 | 5.1 | 2E-05 | ||||
| Cation-transporting ATPase, E1-E2 family | SP1551 | 3.7 | 9E-09 | ||||
| tpi; triosephosphate isomerase | SP1574 | 2.2 | 7E-03 | ||||
| Oxalate formate antiporter | SP1587 | 3.5 | 5E-05 | 4.3 | 2E-11 | 3.9 | 7E-04 |
| gpmA; phosphoglycerate mutase | SP1655 | −5.6 | 3E-03 | ||||
| ROK family protein | SP1675 | 6.0 | 2E-09 | ||||
| scrR; sucrose operon repressor | SP1725 | 3.8 | 4E-03 | 5.3 | 9E-10 | ||
| galK; galactokinase | SP1853 | 2.7 | 3E-03 | ||||
| gap; glyceraldehyde-3-phosphate dehydrogenase | SP2012 | −6.5 | 5E-04 | ||||
| Glycosyl hydrolase, family 1 | SP2021 | 5.8 | 6E-06 | 14.5 | 3E-03 | ||
| malA; maltose metabolism | SP2111 | 2.0 | 8E-04 | NA | NA | NA | NA |
| malR; maltose operon transcriptional repressor | SP2112 | 3.1 | 2E-04 | ||||
| Transketolase, N-terminal subunit | SP2128A | NA | NA | 5.2 | 3E-05 | ||
| PTS system, IIC component, putative | SP2129 | NA | NA | 6.8 | 4E-06 | ||
| argF; ornithine carbamoyltransferase | SP2150 | 7.6 | 8E-04 | ||||
| arcC; carbamate kinase | SP2151 | 9.2 | 2E-03 | ||||
| Fatty acid metabolism | |||||||
| cls; cardiolipin synthetase | SP0199A | 3.6 | 3E-02 | ||||
| enoyl-CoAc hydratase/isomerase family protein | SP0415 | −17.3 | 9E-09 | −3.0 | 6E-04 | ||
| acp; acyl carrier protein | SP0417 | −7.1 | 7E-04 | ||||
| fabH; 3-oxoacyl-(acyl carrier protein) synthase III | SP0418 | −4.9 | 3E-07 | ||||
| fabK; enoyl-(acyl carrier protein) reductase | SP0419 | −12.8 | 2E-09 | ||||
| fabD | SP0420 | −9.8 | 6E-07 | ||||
| fabG; 3-oxoacyl- (acyl carrier protein) reductase | SP0421 | −13.0 | 8E-10 | −2.1 | 4E-03 | ||
| fabF; 3-oxoacyl-(acyl carrier protein) synthase II | SP0422 | −10.6 | 4E-09 | ||||
| accB; acetyl-CoA carboxylase | SP0423 | −10.3 | 9E-11 | −2.1 | 1E-03 | ||
| fabZ | SP0424 | −6.3 | 9E-09 | −2.1 | 2E-03 | ||
| accC; acetyl-CoA carboxylase | SP0425 | −10.7 | 4E-11 | −2.3 | 6E-03 | ||
| accD; acetyl-CoA carboxylase, subunit beta | SP0426 | −8.8 | 5E-11 | ||||
| accA; acetyl-CoA carboxylase, subunit alpha | SP0427 | −7.7 | 1E-10 | ||||
| Amino acid biosynthesis and acquisition | |||||||
| Amino acid ABC transporter, ATP-binding protein | SP0111 | 2.3 | 1E-09 | ||||
| aliA; oligopeptide ABC transporter | SP0366 | 3.9 | 2E-03 | ||||
| ilvB; acetolactate synthase, large subunit | SP0445A | 7.4 | 4E-03 | ||||
| ilvN; acetolactate synthase, small subunit | SP0446 | 2.0 | 1E-10 | ||||
| ilvC; ketol-acid reductoisomerase | SP0447 | 5.5 | 4E-04 | ||||
| ilvA; threonine dehydratase | SP0450 | 4.5 | 5E-04 | ||||
| Transcriptional regulator, MerR family | SP0501 | −5.7 | 1E-10 | ||||
| Glutamine synthetase, type I | SP0502 | −4.4 | 1E-09 | ||||
| livH; branched-chain amino acid ABC transporter | SP0750 | 2.5 | 2E-06 | ||||
| livM; branched-chain amino acid ABC transporter | SP0751 | 2.3 | 3E-07 | ||||
| livG; branched-chain amino acid ABC transporter | SP0752 | 2.3 | 5E-09 | ||||
| livF; branched-chain amino acid ABC transporter | SP0753 | 2.3 | 3E-07 | ||||
| Conserved hypothetical protein | SP0783 | −4.7 | 1E-11 | ||||
| Glutathione reductase | SP0784 | −2.2 | 2E-06 | ||||
| lcEL; branched-chain amino acid aminotransferase | SP0856A | 3.5 | 3E-03 | ||||
| Oligopeptide-binding protein, internal deletion | SP0857 | 3.8 | 2E-04 | ||||
| proA; gamma-glutamyl phosphate reductase | SP0932B | 4.0 | 3E-03 | ||||
| dapA; dihydrodipicolinate synthase | SP1014 | 2.6 | 2E-10 | 3.6 | 4E-04 | ||
| hemK; conserved protein | SP1021D | 14.3 | 1E-04 | ||||
| amiE; ATP-binding protein | SP1888 | 2.4 | 6E-05 | 2.9 | 1E-08 | ||
| amiD; permease protein | SP1889A | 2.1 | 1E-04 | 2.8 | 9E-09 | ||
| AmiC; permease protein | SP1890A | 2.9 | 2E-11 | ||||
| Nucleoside/nucleotride metabolism | |||||||
| purA; adenylosuccinate synthetase | SP0019 | −3.9 | 5E-09 | ||||
| Phosphoribosytformylglycinamidine synthase | SP0045A,C,B | 6.1 | 5E-05 | NA | NA | ||
| purF; amidophosphoribosyltransferase | SP0046 | 11.6 | 1E-06 | NA | NA | ||
| purM; purine ribonucleotide biosynthesis | SP0047B | 8.4 | 1E-05 | NA | NA | ||
| purN; purine ribonucleotide biosynthesis | SP0048B | 9.7 | 1E-05 | NA | NA | ||
| purH; purine ribonucleotide biosynthesis | SP0050AA | 10.3 | 4E-06 | NA | NA | ||
| purE; purine ribonucleotide biosynthesis | SP0053C | 3.0 | 1E-04 | NA | NA | ||
| purK; purine ribonucleotide biosynthesis | SP0054C | 6.9 | 4E-05 | ||||
| Hypothetical protein | SP0055 | 2.4 | 4E-04 | NA | NA | ||
| purB; adenylosuccinate lyase | SP0056B | 2.7 | 5E-05 | ||||
| nrdD; anaerobic ribonucleoside triphosphate | SP0202 | 2.7 | 5E-05 | ||||
| Acetyltransferase. GNAT family | SP0204 | 3.2 | 2E-04 | ||||
| nrdG; anaerobic ribonucleoside-triphosphate | SP0205 | 3.5 | 3E-05 | 5.6 | 4E-07 | ||
| Hypothetical protein | SP0206 | 2.3 | 2E-04 | ||||
| Conserved domain protein | SP0207 | 2.9 | 5E-05 | ||||
| adk; adenylate kinase | SP0231 | −3.8 | 2E-03 | ||||
| Amino acid biosynthesis and acquisition | |||||||
| pyrF; orotidine 5-phosphate decarboxylase | SP0701D | −6.7 | 8E-04 | ||||
| pyrE; orotate phosphoribosyltransferase | SP0702D | −11.0 | 9E-05 | NA | NA | ||
| upp; uracil phosphoribosyltransferase | SP0745 | −3.9 | 4E-03 | ||||
| Hypothetical protein | SP0830 | −2.5 | 3E-03 | ||||
| deoD; purine nucleoside phosphorylase, family 2 | SP0831 | −3.3 | 2E-03 | ||||
| pyrK; dihydroorotate dehydrogenase, electron | SP0963 | −8.9 | 7E-05 | 3.4 | IE-03 | ||
| pyrDb; dihydroorotate dehydrogenase B | SP0964D | −5.7 | 2E-03 | 4.7 | 3E-04 | ||
| carA; carbamoyl-phosphate synthase, small subunit | SP1276 | −4.9 | 9E-04 | 2.2 | 2E-06 | ||
| pyrB; aspartate carbamoyltransferase | SP1277 | −6.9 | 1E-03 | 2.0 | 3E-06 | ||
| pyrR; pyrimidine operon regulatory protein | SP1278AA | −8.0 | 8E-05 | ||||
| uraA; uracil permease | SP1286A | −7.7 | 1E-04 | ||||
| Oxidoreductase, pyridine nucleotide-disulfide | SP1588 | −4.5 | 8E-04 | ||||
| Co factor metabolism | |||||||
| ribAB; riboflavin, FMN, and FAD biosynthesis | SP0176D,B | −4.2 | 8E-04 | ||||
| ribD; riboflavin, FMN, and FAD biosynthesis | SP0178D | −5.4 | 4E-04 | ||||
| folC; dihydrofolate synthetase | SP0290 | 2.1 | 8E-05 | 3.1 | 4E-03 | ||
| folE; GTP cyclohydrolase I | SP0291 | 2.3 | 8E-05 | ||||
| folE; GTP cyclohydrolase I | SP0291 | 2.3 | 8E-05 | ||||
| Bifunctional lolate synthesis protein | SP0292 | 2.6 | 1E-04 | 2.9 | 2E-03 | ||
| coaA; pantothenale kinase | SP0839 | 5.1 | 4E-03 | ||||
| Macrolide-efflux protein | SP1110 | −4.9 | 3E-07 | ||||
| Pyridoxine biosynthesis protein | SP1468 | 2.8 | 2E-10 | ||||
| coaD; phosphopantetheine adenylyltransferase | SP1968 | 2.3 | 2E-06 | ||||
| nadC; pyridine biosynthesis | SP2016 | −7.2 | 1E-07 | ||||
| Membrane protein | SP2017A,D | NA | NA | −15.7 | 5E-12 | ||
| 5-Formyltetrahydrofolate cyclo-ligase | SP2095A | 2.5 | 4E-10 | ||||
| Anion/cation aquisition | |||||||
| Iron | |||||||
| Non-heme Iron-containing ferritin | SP1572 | −8.4 | 8E-05 | −2.4 | 4E-06 | ||
| Alcohol dehydrogenase, iron-containing | SP2026 | −3.6 | 2E-03 | NA | NA | ||
| Potassium | |||||||
| Potassium uptake protein, Trk family | SP0480 | 2.6 | 4E-04 | ||||
| Phosphate | |||||||
| pstA; putative membrane protein | SP1393 | 2.8 | 2E-03 | ||||
| Magnesium | |||||||
| Magnesium transporter, CorA family | SP0185 | −2.1 | 5E-03 | ||||
| Manganese | |||||||
| psaB; ATP-binding protein | SP1648 | 7.3 | 1E-05 | 5.0 | 2E-09 | 3.7 | 3E-05 |
| psaC; permease, authentic frameshift | SP1649 | 9.3 | 2E-06 | 4.8 | 2E-11 | 7.6 | 3E-04 |
| psaA; manganese-binding adhesion liprotein | SP1650 | 9.6 | 3E-07 | 5.6 | 1E-08 | 8.0 | 3E-04 |
| Zinc | |||||||
| adcA; zinc-binding adhesion liprotein | SP2169 | 9.1 | 3E-06 | NA | NA | NA | NA |
| adcB; permease protein | SP2170A | 7.7 | 1E-05 | ||||
| adcC; ATP-binding protein | SP2171 | 6.8 | 3E-05 | ||||
| adcR; repressor | SP2172 | 4.2 | 8E-06 | ||||
| Transcription | |||||||
| mutR; transcriptional regulator | SP0141AA | 3.0 | 2E-04 | ||||
| Conserved hypothetical protein | SP0385A,D | −5.0 | 6E-10 | ||||
| Sensor histidine kinase, putative | SP0386D | −3.9 | 2E-07 | ||||
| DNA-binding response regulator | SP0387D | −4.3 | 1E-09 | ||||
| Hypothetical protein | SP0389 | −3.3 | 2E-06 | ||||
| Transcriptional regulator, MarR family | SP0416 | −5.3 | 1E-10 | ||||
| vncS; sensor histidine kinase | SP0604 | 2.2 | 5E-04 | 2.5 | 1E-02 | ||
| ciaR; DNA-binding response regulator | SP0798D | 3.1 | 4E-03 | ||||
| ciaH; sensor histidine kinase ClaH | SP0799D | 3.1 | 1E-03 | ||||
| Transcriptional regulator, putative | SP0908 | 2.1 | 7E-04 | ||||
| Conserved hypothetical protein | SP0909 | 2.2 | 2E-03 | NA | NA | ||
| prsA; ribose-phosphate pyrophosphokinase | SP1095D | −2.5 | 1E-06 | ||||
| dnaG; DNA primase | SP1072 | −5.2 | 3E-09 | ||||
| rpoD; RNA polymerase sigma-70 factor | SP1073 | −5.0 | 4E-06 | ||||
| Conserved hypothetical protein | SP1074 | −4.4 | 3E-10 | ||||
| vicX protein | SP1225 | 2.5 | 3E-05 | ||||
| Sensory box sensor histidine kinase | SP1226 | 2.4 | 3E-04 | ||||
| ATP-dependent RNA helicase, putative | SP1586 | 3.1 | 1E-05 | ||||
| Transcriptional regulator, Gn1R family | SP1714 | −6.6 | 2E-11 | ||||
| ABC transporter, ATP-binding protein | SP1715A,B | −6.7 | 1E-08 | ||||
| Conserved hypothetical protein | SP1716 | −12.0 | 3E-09 | ||||
| ABC transporter, ATP-binding protein | SP1717A | −13.3 | 1E-10 | ||||
| marR; transcriptional regulator | SP1863 | −2.3 | 7E-06 | ||||
| ccpA; catabolite control protein A | SP1999 | 4.2 | 3E-03 | ||||
| Translation | |||||||
| Ribosomal protein S10 | SP0208 | −4.8 | 3E-04 | ||||
| Ribosomal protein L3 | SP0209 | −4.2 | 1E-03 | 2.9 | 8E-04 | ||
| Ribosomal protein L23 | SP0211 | −4.0 | 4E-03 | 2.8 | 2E-05 | ||
| Ribosomal protein L2 | SP0212 | −4.1 | 1E-03 | ||||
| Ribosomal protein S19 | SP0213 | −3.9 | 3E-03 | ||||
| Ribosomal protein L14 | SP0219 | −3.6 | 4E-03 | ||||
| Translation initiation factor IF | SP0232 | −5.0 | 9E-04 | ||||
| Ribosomal protein L36 | SP0233 | −4.5 | 4E-03 | ||||
| Ribosomal protein S13 | SP0234 | −4.7 | 5E-04 | ||||
| Ribosomal protein S11 | SP0235 | −4.4 | 2E-03 | ||||
| Ribosomal protein L17 | SP0237 | −6.1 | 5E-04 | ||||
| serS; seryl-tRNA synthetase | SP0411 | 4.5 | 6E-03 | ||||
| Ribosomal protein L11 | SP0630 | −4.7 | 1E-03 | ||||
| Ribosomal protein L1 | SP0631 | −5.0 | 5E-04 | ||||
| Ribosomal protein S16 | SP0775 | −5.2 | 2E-04 | ||||
| rpsA; ribosomal protein S1 | SP0862 | 3.7 | 2E-04 | ||||
| Translation initiation factor IF-3 | SP0959 | −6.4 | 6E-05 | ||||
| Ribosomal protein L35 | SP0960 | −5.3 | 3E-03 | ||||
| Ribosomal protein L20 | SP0961 | −6.0 | 9E-04 | ||||
| Conserved hypothetical protein | SP1097 | −3.5 | 4E-05 | ||||
| Conserved hypothetical protein | SP1098 | 2.5 | 3E-02 | ||||
| Ribosomal large subunit pseudouridine synthase | SP1099 | −3.4 | 5E-05 | 2.1 | 2E-02 | ||
| Ribosomal protein L7/L12 | SP1354 | −7.8 | 8E-06 | ||||
| Ribosomal protein L10 | SP1355 | −7.3 | 3E-05 | ||||
| Ribosomal protein S21 | SP1414 | −3.8 | 4E-03 | ||||
| glyQ; glyl-tRNA synthetase, alpha subunit | SP1475 | −3.0 | 4E-06 | ||||
| Ribosomal protein S18 | SP1539 | −4.2 | 9E-04 | ||||
| Single-strand binding protein | SP1540 | −4.6 | 9E-04 | ||||
| Ribosomal protein S6 | SP1541 | −4.2 | 5E-03 | ||||
| Ribosomal protein S15 | SP1626 | −3.5 | 5E-03 | ||||
| fmt; methionyl-tRNA formyltransferase | SP1735 | 2.4 | 1E-07 | ||||
| oligoendopeptidase F. putative | SP1780A | 2.9 | 5E-05 | ||||
| Conserved hypothetical protein | SP1781 | 3.2 | 1E-04 | ||||
| prmA: ribosomal protein L11 methyltransferase | SP1782C | 2.4 | 1E-04 | ||||
| tgt: queuine tRNA-ribosyltransferase | SP2058 | −3.1 | 1E-05 | ||||
| Ribosomal subunit interface protein | SP2206 | −3.9 | 2E-07 | ||||
| Post translational protein alternations | |||||||
| ABC transporter, ATP-binding protein | SP0151 | 2.1 | 1E-06 | ||||
| ATP-dependent Clp protease | SP0338A | NA | NA | −5.8 | 2E-08 | NA | NA |
| Lipoate-protein ligase, putative | SP1160 | 3.1 | 9E-05 | 2.6 | 4E-07 | ||
| Peptidase, U32 family | SP1429 | −5.6 | 2E-03 | ||||
| secA: preprotein translocase | SP1702 | 2.2 | 4E-03 | ||||
| Serine/threonine protein kinase | SP1732 | 2.0 | 2E-03 | ||||
| Peptidase, M20/M25/M40 family | SP2096 | 2.9 | 3E-03 | ||||
| Conserved hypothetical protein | SP2143A | NA | NA | NA | NA | 11.9 | 8E-07 |
| Unknown function | |||||||
| Transporters | |||||||
| ABC transporter, ATP-binding protein | SP0636 | −4.9 | 1E-07 | ||||
| Conserved hypothetical protein | SP0638 | −4.8 | 1E-08 | ||||
| Hypothetical protein | SP0639 | −4.0 | 2E-06 | ||||
| ABC transporter, ATP-binding protein | SP0786 | 2.1 | 1E-04 | ||||
| Conserved hypothetical protein | SP0787 | 2.7 | 2E-04 | ||||
| ABC transporter, ATP-binding protein | SP1114 | NA | NA | −2.9 | 4E-04 | ||
| Atz/Trz family protein | SP1356D | 8.1 | 9E-06 | ||||
| ABC transporter, ATP-binding/permease protein | SP1357D | 3.9 | 4E-03 | ||||
| ABC transporter, ATP-binding/permease protein | SP1358D | 3.9 | 2E-04 | ||||
| Glycerol uptake facilitator protein, putative | SP1491 | 3.4 | 4E-04 | ||||
| Sodium/dicarboxylate symporter family protein | SP1753 | 10.2 | 2E-04 | ||||
| MATE efflux family protein | SP2065 | NA | NA | 2.4 | 5E-03 | ||
| ABC transporter, ATP-binding/permease protein | SP2073 | 2.2 | 2E-03 | NA | NA | NA | NA |
| ABC transporter, ATP-binding/permease protein | SP2075D | 3.0 | 5E-03 | ||||
| Cation-Transporting ATPase, E1-E2 family | SP2101A | 2.2 | 1E-07 | ||||
| ABC transporter, ATP-binding protein | SP2196 | 2.3 | 1E-08 | ||||
| ABC transporter, substrate-binding protein | SP2197 | −4.0 | 1E-03 | −2.8 | 4E-05 | ||
| Unknown | |||||||
| Hypothetical protein | SP0088 | 2.9 | 1E-07 | ||||
| LysM domain protein | SP0107 | 2.7 | 1E-04 | ||||
| Hypothetical protein | SP0115 | 18.0 | 9E-06 | NA | NA | ||
| Conserved hypothetical protein | SP0122 | −7.8 | 1E-06 | ||||
| Hypothetical protein | SP0124 | −7.9 | 1E-06 | ||||
| Hypothetical protein | SP0125 | −3.1 | 1E-05 | ||||
| Hypothetical protein | SP0142D | 13.2 | 6E-06 | ||||
| Conserved domain protein | SP0143D,C | 18.8 | 2E-05 | ||||
| Hypothetical protein | SP0144D | 13.6 | 1E-05 | ||||
| Conserved hypothetical protein | SP0145A,D | 13.0 | 7E-05 | ||||
| Conserved hypothetical protein | SP0159 | 4.4 | 2E-04 | ||||
| Hypothetical protein | SP0198A | 2.1 | 1E-03 | ||||
| Conserved hypothetical protein | SP0239 | 3.1 | 3E-03 | ||||
| Conserved hypothetical protein | SP0288 | 2.1 | 8E-04 | 6.7 | 3E-04 | ||
| IS_66_ family element: Orf3, degenerate | SP0362 | −2.6 | 4E-03 | ||||
| IS_3_-Spn 1: transposase, degenerate | SP0392 | −4.8 | 8E-04 | ||||
| Conserved hypothetical protein | SP0481 | 2.3 | 7E-07 | ||||
| ABC transporter, ATP-binding protein | SP0483B | 2.2 | 5E-12 | ||||
| Conserved hypothetical protein | SP0488 | −4.7 | 1E-03 | −2.9 | 3E-07 | ||
| Hypothetical protein | SP0582 | −161.9 | 3E-15 | ||||
| Conserved domain protein | SP0617 | 3.5 | 1E-03 | ||||
| Conserved hypothetical protein | SP0629B | 2.4 | 6E-07 | ||||
| Hypothetical protein | SP0703 | −4.0 | 2E-03 | NA | NA | ||
| Conserved hypothetical protein | SP0742C | −4.3 | 1E-09 | ||||
| KH domain protein | SP0776 | −5.0 | 7E-04 | ||||
| Conserved hypothetical protein | SP0787 | 3.7 | 6E-03 | ||||
| Hypothetical protein | SP0800 | 4.5 | 2E-05 | 2.1 | 4E-06 | ||
| Hypothetical protein | SP0816 | −3.5 | 2E-03 | ||||
| Hypothetical protein | SP0833 | −2.6 | 6E-04 | ||||
| Hemolysin-related protein | SP0834 | −3.7 | 3E-03 | ||||
| Conserved hypothetical protein | SP0841 | 3.3 | 1E-08 | ||||
| Conserved hypothetical protein | SP0868 | −3.9 | 5E-07 | ||||
| NifU family protein | SP0870 | −3.5 | 7E-09 | ||||
| Conserved hypothetical protein | SP0871 | 3.2 | 6E-03 | ||||
| Conserved hypothetical protein | SP0921 | −2.2 | 1E-03 | ||||
| Hypothetical protein | SP0958 | −6.7 | 1E-11 | ||||
| _O_-methyltransferase | SP0980D | −3.3 | 6E-06 | ||||
| Conserved hypothetical protein | SP1012 | −7.9 | 6E-04 | NA | |||
| Conserved hypothetical protein | SP1090 | −3.9 | 7E-09 | ||||
| Conserved domain protein | SP1174A | 6.6 | 1E-08 | −7.8 | 2E-14 | ||
| Conserved domain protein | SP1175A | 16.3 | 1E-07 | 3.4 | 1E-02 | ||
| Conserved hypothetical protein | SP1240 | 2.7 | 6E-04 | ||||
| Conserved hypothetical protein, strain TIGR4 | SP1249 | 3.6 | 1E-07 | 6.9 | 4E-03 | ||
| lemA | SP1284 | −2.1 | 4E-07 | ||||
| Transposase family protein, authentic frameshift | SP1484 | NA | NA | 3.6 | 6E-04 | ||
| Methyltransferase, putative | SP1578 | 3.4 | 2E-05 | ||||
| Conserved domain protein | SP1641 | −15.5 | 3E-11 | ||||
| Hypothetical protein | SP1852A | 2.1 | 3E-04 | NA | NA | ||
| Nitroreductase family protein | SP1710 | NA | NA | 2.2 | 2E-08 | NA | NA |
| Conserved hypothetical protein | SP1716 | 3.2 | 8E-03 | ||||
| Hypothetical protein | SP1779A | 2.8 | 8E-05 | ||||
| Hypothetical protein | SP1787 | −5.1 | 8E-09 | ||||
| Conserved hypothetical protein | SP1875 | 2.2 | 9E-05 | ||||
| Conserved hypothetical protein | SP1876 | 2.2 | 1E-04 | ||||
| Integrase/recombinase, phage integrase family | SP1877 | 2.1 | 7E-04 | ||||
| CBS domain protein | SP1878 | 2.1 | 7E-04 | ||||
| Oxidoreductase, short chain dehydrogenase/reductase | SP1909 | 5.9 | 1E-10 | ||||
| Conserved hypothetical protein | SP1922 | −5.8 | 2E-13 | ||||
| Hypothetical protein | SP1924 | −9.2 | 2E-09 | ||||
| Hypothetical protein | SP1925 | −4.1 | 3E-07 | ||||
| Hypothetical protein | SP1926 | −3.7 | 3E-06 | ||||
| Conserved hypothetical protein | SP2027D | NA | NA | 2.6 | 3E-03 | ||
| Conserved hypothetical protein | SP2045 | −3.0 | 9E-09 | ||||
| LysM domain protein, authentic frameshift | SP2063 | 4.9 | 5E-06 | NA | NA | ||
| Conserved hypothetical protein | SP2132 | 2.8 | 1E-03 | NA | NA | NA | NA |
| Conserved hypothetical protein | SP2152 | 18.2 | 4E-03 | ||||
| Transporter, truncation | NTL02SP0105 | 4.2 | 4E-05 | ||||
| Hypothetical protein | NTL02SP0107 | 5.0 | 6E-05 | ||||
| Conserved hypothetical protein | NTL02SP0108 | 13.7 | 2E-10 | ||||
| Hypothetical protein | NTL02SP1212 | −5.9 | 2E-04 | ||||
| Transposon | |||||||
| IS_66_ family element; Orf3, degenerate | SP0362 | −11.6 | 4E-15 | ||||
| IS_66_ family element; Orf3, degenerate | SP0644 | −12.9 | 2E-13 | ||||
| IS_66_ family element; Orf2, Interruption | SP0812 | 2.5 | 1E-07 | ||||
| IS_1381_; transposase OrfA | SP1310 | 5.3 | 2E-11 | ||||
| IS_66_ family element; Orf3, degenerate | SP1311 | −27.4 | 3E-07 | ||||
| IS_1167_; transposase | SP1692 | −2.4 | 3E-06 | ||||
| IS_1381_; transposase OrfA, internal deletion | SP2137 | 4.9 | 4E-08 |
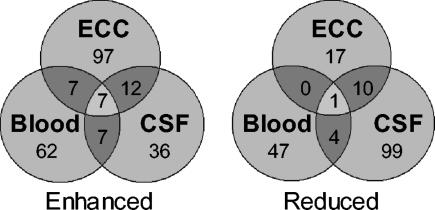
FIG. 2.
Venn diagrams highlighting disparity between infectious models for genes with altered expression. Numbers indicate the amount of genes determined to have altered expression that are either shared or exclusive to growth of the pneumococci in blood, CSF, or ECC.
Analysis of gene transcription indicated several features of bacterial physiology unique to each body compartment. Analysis of blood-dependent transcription revealed that cell wall and membrane synthesis, cell division, and competence were unchanged from in vitro. Nonetheless, the pneumococcus appears to be under stress. Supporting this view, 24 genes encoding ribosomal proteins had reduced transcription in the blood. We interpret this as indicating that the pneumococcus is reducing its translational capacity. Likewise, relA, the gene encoding the GTP pyrophosphokinase was enhanced during growth in blood (26). RelA is expressed in response to lack of nutrients and is responsible for ppGpp synthesis and entry of the bacteria into the stringent response. Finally, the genes encoding manganese and zinc transporters and the genes responsible for purine and folic acid biosynthesis were also increased.
In the CSF, the pneumococcus decreased transcription of at least 20 genes involved with competence (15), including comX1, comA, comB, comE, and comD. Likewise, 11 genes involved in the biosynthesis of fatty acids were also markedly reduced. Decreased expression of the fab operon (31) may indicate that sufficient fatty acids are available in the CSF. Other differences observed in CSF cultures included enhanced expression of the lic operon (50), which indicates that the bacteria are decorating their cell wall with phosphorylcholine. SP1804, a gene encoding a general stress protein, is enhanced in the CSF, perhaps indicating that the bacteria are under stress in adapting from the rich medium of blood to the poor medium of CSF.
Pneumococci adherent to epithelial cells also had unique alterations in their expression profiles. We observed enhanced expression of the lic operon (50) and the dlt operon (49) during ECC. These changes indicate enhanced addition of phosphorylcholine and d-alanine to teichoic acids. Enhanced expression of VncS and CiaR/H, genes encoding two-component systems (42), indicate global changes in the synthesis of cell wall polymers, peptide pheromones, bacteriocins, and htrA expression. Manganese acquisition also appears to be particularly important since the psa operon showed enhanced expression in all three of the conditions tested and has recently been shown to undergo enhanced transcription in vivo (30).
With regard to energy metabolism, during growth in blood the pneumococcus reduces the expression of the genes encoding 1-phosphofructokinase and fructose-bisphosphate aldolase. Phosphorylation of 1-phosphofructose by 1-phophofructokinase is the rate-limiting step of glycolysis and targets the molecule for glycolysis (22). Reducing expression levels of 1-phosphofructokinase suggests that the pneumococcus has a readily available carbon source in the blood. At the same time, concurrent enhanced expression of SP0265 and SP2021, glycosyl hydrolases, and enhanced expression of scrR and malR, the repressors for sucrose and maltose operons, suggest that the pneumococcus is also utilizing alternative sources of energy.
Phase variation.
S. pneumoniae undergoes spontaneous phase variation between a transparent phenotype and an opaque phenotype (21); the transparent phenotype has an enhanced capacity to adhere and colonize the nasopharynx, whereas the more phagocytosis-resistant opaque phenotype predominates in blood. An increased capacity to adhere by the transparent phenotype corresponds to higher levels of CbpA, phosphorylcholine, teichoic acid, and autolysin than do opaque variants (21, 41, 48). In contrast, the opaque phenotype produces more capsular polysaccharide and hydrogen peroxide and requires up to 30-fold more human immune serum to achieve 50% opsonophagocytic killing than related transparent strains (20, 43).
Analysis of the genes enhanced during ECC confirmed a selection for the transparent phenotype. We observed enhanced expression of cbpA and the lic and dlt operons. Similar results for bacteria in CSF taken together with downregulation of spxB in CSF indicate that the transparent phenotype may reappear after bacteria leave the bloodstream. Interestingly, we did not observe phenotype-enhanced expression of spxB or of the genes involved in capsule production in the blood.
Microarray analysis of known virulence determinants.
Microarray analysis of 20 known pneumococcal virulence determinants indicated site-specific expression for 17 of them (Table 1). Only two virulence determinants with altered gene expression, pspA and prtA, were expressed in a similar fashion in blood, CSF, or ECC (see above). pspA, encodes a choline-binding protein that inhibits complement deposition on the surface of the bacteria and is upregulated in blood by Northern analysis (46). prtA encodes a conserved serine protease (4).
Expression profiles were consistent with much of the current understanding of pneumococcal pathogenesis. During attachment to pharyngeal epithelial cells, pneumococci enhanced expression of the genes encoding choline-binding protein A (CbpA) and HtrA, two proteins shown to contribute to nasopharyngeal colonization (41, 42). Expression of ply was decreased, suggesting attenuation of the ability to injure host cells with the toxin pneumolysin. We observed enhanced expression in the blood of the operon encoding choline-binding protein G (CbpG) and choline-binding protein F (CbpF) (12). CbpG has been demonstrated in our laboratory to be required for the development of high-grade bacteremia (preliminary data). Lastly, we observed decreased expression of the genes encoding pneumolysin (ply) (8), autolysin (lytA) (45), and pyruvate oxidase (spxB) (43) in the CSF. Pyruvate oxidase, SpxB, is responsible for the production of hydrogen peroxide by the pneumococcus. This trio was striking since pneumolysin, hydrogen peroxide, and inflammatory cell wall components released by autolysin are the principle agents by which the pneumococcus induces neuronal damage (6, 47). Decreased expression of these genes in the CSF suggests that the pneumococcus adapts so as to decrease damage to neurons.
We also observed alterations in the gene expression of less-well-characterized virulence determinants. In the blood, we observed enhanced expression of lmb, a gene homologous to a laminin-binding protein in Streptococcus anginosus (1). lmb transcription was subsequently reduced in the CSF. Likewise, we observed a decrease in the expression of genes involved in bacteriocin synthesis and immunity (10). These expression profiles suggest a potentially underappreciated role for these genes during pathogenesis.
Cross-reference to STM studies and DFI analyses.
Investigators have used STM and DFI to identify pneumococcal genes required for invasive disease (13, 25, 29, 39). In Table 1, we highlight genes with altered expression that have been identified by STM or DFI. Comparison of the three STM studies determined that the majority of loci identified were hit by only one study. Although this may reflect differences in methodology, it is likely that the use of three different strains of _S. pneumoniae_—G54, a serotype 19F (39); strain 0100993, a serotype 3 (25); and T4, a serotype 4 (13)—contributed to the discrepancy observed between the three STM studies. This disparity suggests that strain-dependent variations may occur with regard to virulence. As such, genes identified by more than one method are therefore particularly interesting for the current analysis since they may represent a set of core virulence elements required by all pneumococci for the disease process. Moreover, core genes need not have altered transcription in vivo. The genes encoding immunoglobulin A1 protease (40), ZmpB (a metalloprotease that elicits inflammation in the lower respiratory tract) (5), and PavA (a fibronectin-binding protein) (16) all maintained unchanged levels of expression in vivo and yet were all identified by more than one STM study (Table 2). Table 2 is a comprehensive list of genes with unaltered expression that have been identified by more than one STM study. This approach is not comprehensive, however, since none of the major virulence determinants, such as pneumolysin, capsular polysaccharide, or autolysin, was identified by more than one STM study. This may indicate a more site-specific contribution to pathogenesis (34). Finally, the majority of genes hit by more than one STM analysis encoded proteins whose function is unknown (Table 1). Those whose expression was altered in vivo and that are characterized include the pur operon (32) and rib operon (23) for purine and riboflavin biosynthesis, respectively, as well as radA, a DNA repair enzyme (3).
TABLE 2.
Loci or genes within the same operon identified by multiple STM analyses with constitutive gene expression as determined by in vivo microarray analysis
| Gene name | TIGR annotationa | Cellular role |
|---|---|---|
| iga; immunoglobulin A1 protease | SP0071A,B | Pathogenesis: degradation of slgA |
| Formate acetyltransferase, putative | SP0251A,C | Energy metabolism: fermentation |
| Transcriptional regulator, putative | SP0306A,C | Regulatory functions: other |
| zmpB; zinc metalloprotease ZmpB | SP664A,B | Pathogenesis: protein and peptide secretion degradation of proteins |
| Transcriptional regulator, LysR family | SP927A,C | Regulatory functions: DNA interactions |
| pavA; adherence and virulence protein A | SP966A,C | Pathogenesis: cell adhesion |
| Cof family protein | SP1291A | Unknown |
| SAP domain protein | SP1292B | Unknown |
| Oxidoreductase, Gfo/Idh/MocA family | SP1482C | Unknown substrate |
| ATP-dependent RNA helicase | SP1483A | Transcription |
| msmK; sugar ABC transporter, ATP-binding protein | SP1580A,B | Carbohydrate transport |
| ROK family protein | SP2142A,C | Small molecule interactions |
| Antigen, cell wall surface anchor protein | SP2145A,B | Unknown |
Virulence assessment of mutants.
It is likely that bacterial genes with enhanced expression during invasive disease contribute to the survival of the bacteria in vivo and to the progression of disease. To test this hypothesis, we created mutants in a number of genes of unknown function that were enhanced during ECC or in the bloodstream. Some of these mutants corresponded to genes (SP0090, SP0092, SP1434, and SP2163) that did not meet the criteria listed here. These genes were examined nonetheless due to large changes in gene expression determined in vivo after less-stringent screening (data not shown). Mutants in genes enhanced during ECC were created in T4X and assessed in an intranasal model of infection. Mutants lacking genes enhanced during bacteremia were created in a D39X background and assessed in an intratracheal challenge model of bacteremia. T4X mutants that were determined to be attenuated were then subsequently transferred into a D39X background to identify strain differences (Table 3).
TABLE 3.
Virulence assessment of mutants deficient in genes with enhanced expression during ECC or growth in blood
| Challenge type and strain | Function | Log10 median titer 2 days postchallengera in: | % Survivalc | |||
|---|---|---|---|---|---|---|
| Nasal lavage or lungsb | Blood | |||||
| Titer | P | Titer | P | |||
| Intranasal challenge | ||||||
| T4X | 6.1 | NA | 5.6 | NA | 10 | |
| T4X SP0090- | ABC transporter, permease | 4.3 | 0.003 | <3.0 | <0.001 | 100 |
| T4X SP0092- | ABC transporter, substrate binding protein | 5.7 | 0.278 | 4.6 | 0.107 | 10 |
| T4X SP0498- | Endo-β-_N_-acetylglucosaminidase, putative | 5.5 | 0.026 | 5.7 | 0.377 | 10 |
| T4X SP1021- | hemK; conserved protein | 5.7 | 0.088 | 4.7 | 0.163 | 20 |
| T4X SP1753- | Sodium/dicarboxylate symporter family protein | 4.5 | 0.011 | 4.3 | 0.077 | 10 |
| T4X SP2143- | Conserved hypothetical protein | 5.0 | 0.029 | <3.0 | <0.001 | 100 |
| T4X SP2163- | PTS system, IIB component | 5.1 | 0.011 | <3.0 | <0.001 | 70 |
| D39X | 4.9 | NA | 7.1 | NA | 27 | |
| D39X SP0090- | ABC transporter, permease protein | 4.3 | 0.119 | 6.6 | 0.527 | 40 |
| D39X SP2143- | Conserved hypothetical protein | 4.2 | 0.111 | 6.9 | 0.748 | 27 |
| D39X SP2163- | PTS system, IIB component | 6.2 | 0.235 | 7.0 | 0.852 | 13 |
| Intratracheal challenge | ||||||
| D39X | 8.4 | NA | 9.0 | NA | 0 | |
| D39X SP0115- | Hypothetical protein | 8.4 | 0.902 | 9.1 | 0.535 | 0 |
| D39X SP0142- | Hypothetical protein | 8.6 | 0.805 | 9.1 | 0.710 | 0 |
| D39X SP0641- | Serine protease | 8.3 | 1.000 | 9.0 | 0.620 | 0 |
| D39X SP1002- | lmb; adhesion lipoprotein | 7.2 | 0.128 | 9.0 | 0.620 | 0 |
| D39X SP1175- | Conserved domain protein | 7.5 | 0.234 | 8.6 | 0.181 | 0 |
| D39X SP1434- | ABC transporter | 8.5 | 0.383 | 9.2 | 0.165 | 0 |
Analysis of mutants made in the T4X background determined that five of seven mutants had a significantly reduced capacity to colonize the nasopharynx. Three of these were completely attenuated and did not cross into the bloodstream, indicating that the genes deleted contributed to the pathogenic potential of the bacteria. Surprisingly, when these three genes were mutagenized in a D39X background, no differences in virulence were observed. This occurred despite the observation that several of the genes mutated in D39X had previously been identified by STM as required for in vivo passage. Bioluminescent imaging of the mice with the Xenogen IVIS camera agreed with bacterial titers collected from lungs of infected mice. These images are thus not shown since they are redundant with the values listed in Table 3. Overall, these results suggest that D39X is can overcome deficiencies in virulence traits that T4X cannot.
Considerations.
Although the present study is the first to directly measure pneumococcal gene expression during invasive disease, it must be acknowledged that to obtain sufficient quantities of bacterial RNA from the disease models, certain aspects of the experimental design were not optimal. Foremost, it was necessary to use a different strain for each disease model (D39X for blood, T4X for CSF, and T4R for ECC). Although preliminary data from in vitro experiments (data not shown) demonstrates that D39 and T4 respond in a similar manner to growth in vitro (>99% of the genes examined when grown in C+Y), how each strain responds to different anatomical sites remains unknown. It was also necessary to use a different host species for each disease model (mice for bloodstream infection, rabbits for CSF infection, and humans for ECC), as well as a different RNA extraction protocol for ECC. The relationship between different host species and the pneumococcus may also result in alterations of the pneumococcal response. As a result of these caveats, care must be taken when gene expression levels for different anatomical sites are compared.
A second consideration is the requirement for very high titers of pneumococci in the biological samples and the various durations of infection. For blood, bacterial titers were ca. 109 CFU/ml and were collected 2 days after intratracheal challenge. As such, the expression profile observed does not represent the initial phases of bloodstream infection. Likewise, rabbits were infected intracisternally with 108 CFU, and the bacteria were collected 4 h later. Since the bacteria were collected after only 4 h, our model represents bacterial gene expression during early meningitis. During a natural infection, only a few animals would have a bacterial titer as high as 108 CFU/ml in the CSF, and leukocytes and other host factors would be present in the CSF.
A final consideration is that not all of the genes in the genome were examined. Although our analysis of in vivo gene expression is the most comprehensive to date, we were unable to report on the expression of 31% of the genome during growth in blood, 47% in the CSF, and 32% after ECC. As such, the present study is not comprehensive and further studies, including confirmation by Northern analysis of specific genes, are warranted.
Conclusions.
Microarray analysis of pneumococcal gene expression during invasive disease sheds light on the complex mechanisms of pneumococcal pathogenesis. Despite limitations imposed by technical challenges associated with in vivo RNA collection, we observed dramatic changes in a variety of genes, thus providing important information as to the physiology of the pneumococcus during invasive disease. Most genes (∼90%) do not change expression from in vitro growth medium to in vivo. However, strikingly, the pneumococcus seems to adapt to the blood, CSF, and ECC in a site-specific manner. These changes may be a result of the pneumococcus adapting to nutrient availability and the stresses placed on the bacteria. In blood, cell wall and membrane synthesis and competence were unchanged, whereas the bacteria strongly decreased ribosomal transcription, reduced glycolysis, and increased CbpG and CbpF. In CSF, pneumococci drastically curtailed gene expression for competence, autolysis, and production of toxins, while increasing amino acid biosynthesis and choline on the cell surface.
Cross-referencing of microarray analyses with that of STM and DFI and the virulence assessment of our own mutants strongly implies that pneumococcal virulence is dependent on a network of genes, the majority of which are not fully understood. Although an understanding of this network may seem daunting, it seems that the continued use of global analytical techniques will shed light on the core genes that are required for virulence. Such an attempt will require synthesis of data from multiple strains to derive a general picture of virulence above the noise of strain variation.
Acknowledgments
We thank Robert Fleischmann and Scott Peterson at the PFGRC at TIGR for providing the pneumococcal microarrays necessary for this project. We also thank Alessandra Polissi, Sauli Haataja, and Jeremy Brown for providing data from their STM studies.
This study was supported by NIH grant R01 AI27913 and The American Lebanese Syrian Associated Charities.
REFERENCES
- 1.Allen, B. L., and M. Hook. 2002. Isolation of a putative laminin binding protein from Streptococcus anginosus. Microb. Pathog. 33**:**23-31. [DOI] [PubMed] [Google Scholar]
- 2.Avery, O. T., C. M. MacLeod, and M. McCarty. 1944. Studies on the chemical nature of the substance inducing transformation of the pneumococcal types: induction of transformation by the deoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 79**:**137-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beam, C. E., C. J. Saveson, and S. T. Lovett. 2002. Role for radA/sms in recombination intermediate processing in Escherichia coli. J. Bacteriol. 184**:**6836-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bethe, G., R. Nau, A. Wellmer, R. Hakenbeck, R. R. Reinert, H. P. Heinz, and G. Zysk. 2001. The cell wall-associated serine protease PrtA: a highly conserved virulence factor of Streptococcus pneumoniae. FEMS Microbiol. Lett. 205**:**99-104. [DOI] [PubMed] [Google Scholar]
- 5.Blue, C. E., G. K. Paterson, A. R. Kerr, M. Berge, J. P. Claverys, and T. J. Mitchell. 2003. ZmpB, a novel virulence factor of Streptococcus pneumoniae that induces tumor necrosis factor alpha production in the respiratory tract. Infect. Immun. 71**:**4925-4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, J. S., J. E. Sublett, D. Freyer, T. J. Mitchell, J. L. Cleveland, E. I. Tuomanen, and J. R. Weber. 2002. Pneumococcal pneumolysin and H2O2 mediate brain cell apoptosis during meningitis. J. Clin. Investig. 109**:**19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bricker, A. L., and A. Camilli. 1999. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol. Lett. 172**:**131-135. [DOI] [PubMed] [Google Scholar]
- 8.Cockeran, R., R. Anderson, and C. Feldman. 2002. The role of pneumolysin in the pathogenesis of Streptococcus pneumoniae infection. Curr. Opin. Infect. Dis. 15**:**235-239. [DOI] [PubMed] [Google Scholar]
- 9.Darkes, M. J., and G. L. Plosker. 2002. Pneumococcal conjugate vaccine (Prevnar; PNCRM7): a review of its use in the prevention of Streptococcus pneumoniae infection. Paediatr. Drugs 4**:**609-630. [DOI] [PubMed] [Google Scholar]
- 10.de Saizieu, A., C. Gardes, N. Flint, C. Wagner, M. Kamber, T. J. Mitchell, W. Keck, K. E. Amrein, and R. Lange. 2000. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 182**:**4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francis, K. P., J. Yu, C. Bellinger-Kawahara, D. Joh, M. J. Hawkinson, G. Xiao, T. F. Purchio, M. G. Caparon, M. Lipsitch, and P. R. Contag. 2001. Visualizing pneumococcal infections in the lungs of live mice using bioluminescent Streptococcus pneumoniae transformed with a novel gram-positive lux transposon. Infect. Immun. 69**:**3350-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gosink, K. K., E. R. Mann, C. Guglielmo, E. I. Tuomanen, and H. R. Masure. 2000. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect. Immun. 68**:**5690-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45**:**1389-1406. [PMC free article] [PubMed] [Google Scholar]
- 14.Hava, D. L., J. LeMieux, and A. Camilli. 2003. From nose to lung: the regulation behind Streptococcus pneumoniae virulence factors. Mol. Microbiol. 50**:**1103-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Havarstein, L. S. 1998. Identification of a competence regulon in Streptococcus pneumoniae by genomic analysis. Trends Microbiol. 6**:**297-300. [DOI] [PubMed] [Google Scholar]
- 16.Holmes, A. R., R. McNab, K. W. Millsap, M. Rohde, S. Hammerschmidt, J. L. Mawdsley, and H. F. Jenkinson. 2001. The pavA gene of Streptococcus pneumoniae encodes a fibronectin-binding protein that is essential for virulence. Mol. Microbiol. 41**:**1395-1408. [DOI] [PubMed] [Google Scholar]
- 17.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183**:**5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jedrzejas, M. J. 2001. Pneumococcal virulence factors: structure and function. Microbiol. Mol. Biol. Rev. 65**:**187-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerr, M. K., and G. A. Churchill. 2001. Bootstrapping cluster analysis: assessing the reliability of conclusions from microarray experiments. Proc. Natl. Acad. Sci. USA 98**:**8961-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, J. O., S. Romero-Steiner, U. B. Sorensen, J. Blom, M. Carvalho, S. Barnard, G. Carlone, and J. N. Weiser. 1999. Relationship between cell surface carbohydrates and intrastrain variation on opsonophagocytosis of Streptococcus pneumoniae. Infect. Immun. 67**:**2327-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, J. O., and J. N. Weiser. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J. Infect. Dis. 177**:**368-377. [DOI] [PubMed] [Google Scholar]
- 22.Kornberg, H. L. 2001. Routes for fructose utilization by Escherichia coli. J. Mol. Microbiol. Biotechnol. 3**:**355-359. [PubMed] [Google Scholar]
- 23.Kreger, A. S., and R. H. Olsen. 1968. Purification and properties of mutant and wild-type diaphorases from Diplococcus pneumoniae. J. Bacteriol. 96**:**1029-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lacks, S., and R. D. Hotchkiss. 1960. A study of the genetic material determining an enzyme activity in the pneumococcus. Biochim. Biophys. Acta 39**:**508-517. [DOI] [PubMed] [Google Scholar]
- 25.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40**:**555-571. [DOI] [PubMed] [Google Scholar]
- 26.Lemos, J. A., T. A. Brown, Jr., and R. A. Burne. 2004. Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect. Immun. 72**:**1431-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madu, A., C. Cioffe, U. Mian, M. Burroughs, E. Tuomanen, M. Mayers, E. Schwartz, and M. Miller. 1994. Pharmacokinetics of fluconazole in cerebrospinal fluid and serum of rabbits: validation of an animal model used to measure drug concentrations in cerebrospinal fluid. Antimicrob. Agents Chemother. 38**:**2111-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandell, L. A. 1995. Community-acquired pneumonia: etiology, epidemiology, and treatment. Chest 108**:**35S-42S. [DOI] [PubMed] [Google Scholar]
- 29.Marra, A., J. Asundi, M. Bartilson, S. Lawson, F. Fang, J. Christine, C. Wiesner, D. Brigham, W. P. Schneider, and A. E. Hromockyj. 2002. Differential fluorescence induction analysis of Streptococcus pneumoniae identifies genes involved in pathogenesis. Infect. Immun. 70**:**1422-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marra, A., S. Lawson, J. S. Asundi, D. Brigham, and A. E. Hromockyj. 2002. In vivo characterization of the psa genes from Streptococcus pneumoniae in multiple models of infection. Microbiology 148**:**1483-1491. [DOI] [PubMed] [Google Scholar]
- 31.Marrakchi, H., K. H. Choi, and C. O. Rock. 2002. A new mechanism for anaerobic unsaturated fatty acid formation in Streptococcus pneumoniae. J. Biol. Chem. 277**:**44809-44816. [DOI] [PubMed] [Google Scholar]
- 32.Ng, W. L., K. M. Kazmierczak, G. T. Robertson, R. Gilmour, and M. E. Winkler. 2003. Transcriptional regulation and signature patterns revealed by microarray analyses of Streptococcus pneumoniae R6 challenged with sublethal concentrations of translation inhibitors. J. Bacteriol. 185**:**359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogunniyi, A. D., P. Giammarinaro, and J. C. Paton. 2002. The genes encoding virulence-associated proteins and the capsule of Streptococcus pneumoniae are upregulated and differentially expressed in vivo. Microbiology 148**:**2045-2053. [DOI] [PubMed] [Google Scholar]
- 34.Orihuela, C. J., G. Gao, K. P. Francis, J. Yu, and E. Tuomanen. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J. Infect. Dis., in press. [DOI] [PubMed]
- 35.Orihuela, C. J., G. Gao, M. McGee, J. Yu, K. P. Francis, and E. I. Tuomanen. 2003. Organ-specific models of Streptococcus pneumoniae disease. Scand. J. Infect. Dis. 35**:**647-652. [DOI] [PubMed] [Google Scholar]
- 36.Orihuela, C. J., R. Janssen, C. W. Robb, D. A. Watson, and D. W. Niesel. 2000. Peritoneal culture alters Streptococcus pneumoniae protein profiles and virulence properties. Infect. Immun. 68**:**6082-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearce, B. J., Y. B. Yin, and H. R. Masure. 1993. Genetic identification of exported proteins in Streptococcus pneumoniae. Mol. Microbiol. 9**:**1037-1050. [DOI] [PubMed] [Google Scholar]
- 38.Peterson, W. D., Jr., C. S. Stulberg, and W. F. Simpson. 1971. A permanent heteroploid human cell line with type B glucose-6-phosphate dehydrogenase. Proc. Soc. Exp. Biol. Med. 136**:**1187-1191. [DOI] [PubMed] [Google Scholar]
- 39.Polissi, A., A. Pontiggia, G. Feger, M. Altieri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66**:**5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poulsen, K., J. Reinholdt, and M. Kilian. 1996. Characterization of the Streptococcus pneumoniae immunoglobulin A1 protease gene (iga) and its translation product. Infect. Immun. 64**:**3957-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenow, C., P. Ryan, J. N. Weiser, S. Johnson, P. Fontan, A. Ortqvist, and H. R. Masure. 1997. Contribution of novel choline-binding proteins to adherence, colonization, and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 25**:**819-829. [DOI] [PubMed] [Google Scholar]
- 42.Sebert, M. E., L. M. Palmer, M. Rosenberg, and J. N. Weiser. 2002. Microarray-based identification of htrA, a Streptococcus pneumoniae gene that is regulated by the CiaRH two-component system and contributes to nasopharyngeal colonization. Infect. Immun. 70**:**4059-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spellerberg, B., D. R. Cundell, J. Sandros, B. J. Pearce, I. Idanpaan-Heikkila, C. Rosenow, and H. R. Masure. 1996. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol. Microbiol. 19**:**803-813. [DOI] [PubMed] [Google Scholar]
- 44.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293**:**498-506. [DOI] [PubMed] [Google Scholar]
- 45.Tomasz, A., and S. Waks. 1975. Mechanism of action of penicillin: triggering of the pneumococcal autolytic enzyme by inhibitors of cell wall synthesis. Proc. Natl. Acad. Sci. USA 72**:**4162-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tu, A. H., R. L. Fulgham, M. A. McCrory, D. E. Briles, and A. J. Szalai. 1999. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect. Immun. 67**:**4720-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tuomanen, E., B. Hengstler, O. Zak, and A. Tomasz. 1986. Induction of meningeal inflammation by diverse bacterial cell walls. Eur. J. Clin. Microbiol. 5**:**682-684. [DOI] [PubMed] [Google Scholar]
- 48.Weiser, J. N., R. Austrian, P. K. Sreenivasan, and H. R. Masure. 1994. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect. Immun. 62**:**2582-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whiting, G. C., and S. H. Gillespie. 1996. Investigation of a choline phosphate synthesis pathway in Streptococcus pneumoniae: evidence for choline phosphate cytidylyltransferase activity. FEMS Microbiol. Lett. 143**:**279-284. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, J. R., I. Idanpaan-Heikkila, W. Fischer, and E. I. Tuomanen. 1999. Pneumococcal licD2 gene is involved in phosphorylcholine metabolism. Mol. Microbiol. 31**:**1477-1488. [DOI] [PubMed] [Google Scholar]