Identification In Vivo of Different Rate-Limiting Steps Associated with Transcriptional Activators in the Presence and Absence of a GAGA Element (original) (raw)
Abstract
We analyzed the impact of a GAGA element on a transgenic promoter in Drosophila melanogaster that was activated by proteins composed of the Teton DNA binding domain and either the heat shock factor (HSF) activation domain or a potent subdomain of VP16. Permanganate footprinting was used to monitor polymerase II (Pol II) on the transgenic promoters in vivo. Activation by Teton-HSF but not by Teton-VP16A2 required the GAGA element; this correlated with the ability of the GAGA element to establish a paused Pol II. Although the GAGA element was not required for activation by Teton-VP16A2, the GAGA element greatly accelerated the rate of activation. The permanganate data also provided evidence that Pol II encountered different rate-limiting steps, following initiation in the presence of Teton-HSF and Teton-VP16A2. The rate-limiting step in the presence of Teton-HSF was release of Pol II paused about 20 to 40 nucleotides downstream from the start site. The rate-limiting step in the presence of Teton-VP16A2 occurred much closer to the transcription start site. Several biochemical studies have provided evidence for a structural transition shortly after Pol II initiates transcription. The behavior of Pol II in the presence of Teton-VP16A2 provides the first evidence that this transition occurs in vivo.
The GAGA element in Drosophila melanogaster is a paradigm for a class of DNA elements that can function to establish the transcriptional potential of genes without necessarily leading to gene expression. Another member of this class of elements could be the GC box recognized by Sp1, as Sp1 and its cognate binding site have been implicated in setting up polymerase II (Pol II) on the A20 promoter prior to activation by NF-κB (1). The existence of this class of regulatory elements is predicated on the identification of numerous genes that have Pol II associated with their promoters prior to transcriptional activation (1, 8, 14, 27, 28). Most of these elements remain to be identified, and their contributions to transcription are largely unknown.
The impact of the GAGA element on Pol II-promoter interactions is probably best characterized for the heat shock genes in Drosophila. hsp70 is rapidly induced during heat shock by heat shock factor (HSF) (19). Prior to heat shock induction, RNA Pol II initiates transcription but stably pauses 20 to 40 nucleotides further downstream. In vivo protein-DNA cross-linking reveals that the GAGA factor, which binds the GAGA element, associates with the hsp70 promoter prior to heat shock induction (25, 44). Mutation of a GAGA element in hsp70 diminishes the level of paused Pol II (16), but the decrease is likely to be caused by a defect in initiation rather than pausing (18, 32). Biochemical studies indicate that the GAGA factor functions with the chromatin remodeling factor, NURF, to form a nuclease sensitive region over the hsp70 promoter in reconstituted chromatin (35, 36). Although the contribution of the GAGA elements to the nuclease sensitivity of the hsp70 promoter has not been rigorously evaluated in vivo, analyses of the hsp26 heat shock gene in Drosophila tissues reveal that the GAGA elements in this promoter contribute to formation of a nuclease sensitive chromatin structure prior to heat shock induction (17, 20, 21). Collectively, these data indicate that the GAGA element functions prior to heat shock induction to establish an open chromatin structure and to allow transcription to occur, resulting in a paused Pol II.
In addition to the heat shock genes, GAGA factor has been implicated in regulation of numerous genes having a variety of cellular functions (39, 42). A genome-wide analysis of the distribution of GAGA factor in Drosophila cells identified approximately 250 potential target genes (38). The function of the GAGA factor at these genes is largely unknown.
To learn more about how the GAGA element contributes to transcription, we developed a transgenic system that allowed us to compare the function of different activators on promoters that contain or lack a GAGA element. For the present study, we compared an activator containing amino acids 610 to 691 from HSF to an activator containing three copies of a 13-amino-acid array derived from VP16. Amino acids 610 to 691 from Drosophila HSF fused to the Gal4 DNA binding domain (DBD) had previously been shown to activate transcription of a promoter located on extrachromosomal DNA (43). This information has been used to understand the mechanism of activation by HSF (23, 26, 45), yet it is not clear whether this information has relevance to activation in the context of the GAGA element or a paused Pol II. The three tandem repeats of the N-terminal portion of the VP16 activation domain fused to the tetracycline repressor of Escherichia coli had previously been shown to be a potent activator of a chromosomal copy of gene in human cells (2). We wanted to know whether the GAGA element would have any impact on such a potent activation domain.
Each activation domain was fused to a derivative of the DBD from the tetracycline repressor that could be induced to bind DNA upon addition of the tetracycline analog, doxycycline (37). This provided the opportunity to analyze the effect of the GAGA element on promoter activity under steady-state conditions and on the rate of transcriptional activation. The latter was of interest because one function of the GAGA element suggested by the heat shock genes is that it provides a means for rapidly inducing a gene.
MATERIALS AND METHODS
Construction of transgenic flies.
Detailed descriptions of plasmid constructs, including sequences, are available upon request. All P-element transformation vectors were based on pCaSpeR (34). Fly transformation was done by the method of Robertson et al. (30). The TRE5 transgenic flies were produced with a plasmid, pTRE5.70w, which was identical to the plasmid pG5-70w (33), with the exception that the region containing the five Gal4p binding sites was replaced with five tet operators derived from pTRE (Clonetech). The TRE5-GA transgenic flies were produced with a plasmid called pTRE5-GA.70w that was generated by inserting a GAGA element consisting of the sequence CGAGAGAGC between the TATA box and the tet operators in pTRE5.70w.
To generate transgenic flies that express Teton-VP16A2 or Teton-DBD, the appropriate regions were amplified from plasmid pUHrT62-1 (37) and inserted downstream of the hsp70 promoter in pCaSpeR-hs to produce the plasmids pCaSpeR-hs-Teton-VP16A2 and pCaSpeR-hs-Teton-DBD. To generate transgenic flies that express Teton-HSF protein, the sequence encoding amino acids 610 to 691 of HSF was PCR amplified from pHSF20 (4) with an additional simian virus 40 nuclear localization signal encoded at the N terminus. The PCR fragment was inserted into an XmaI site located 3′ to sequences encoding Teton-DBD in pCaSpeR-hs-Teton-DBD to yield the plasmid called pCaSpeR-hs-Teton-HSF.
Tissue staining for β-galactosidase activity.
Tissues were dissected from third-instar larvae in phosphate-buffered saline (PBS) and placed in 100 μl of PBS in a well slide. PBS was removed and replaced with 100 μl of 1% glutaraldehyde in PBS. Tissues were incubated at 22°C for 15 min and washed two times with 100 μl of PBS. Tissues were incubated in 100 μl of stain solution, consisting of 10 mM NaPO4 (pH 7.2), 150 mM NaCl, 1 mM MgCl2, 6 mM K4[FeII(CN)6], 6 mM K3[FeIII(CN)6], and 0.3% of X-Gal (5-bromo-4-chloro-3-indolyl-ā-d-galactopyranoside) for the times indicated in the legend to Fig. 1.
FIG. 1.
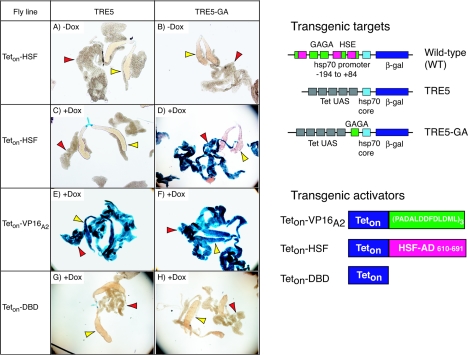
The HSF activation domain requires a GAGA element in the target promoter to achieve activation. Transgenic fly lines encoding Teton-HSF (left, A to D), Teton-VP16A2 (E and F), or Teton-DBD (G and H) were crossed with transgenic lines containing TRE5 (A, C, E, and G) or TRE5-GA (B, D, F, and H) promoters. The offspring were raised in the presence or absence of doxycycline (+ Dox, −Dox), heat shocked for 30 min to induce synthesis of Teton proteins, and allowed to recover overnight at 22°C. Salivary glands (yellow arrowheads) and fat body (red arrowheads) were isolated and incubated in X-Gal solution. Tissues expressing Teton-HSF or Teton-DBD were incubated with X-Gal for 5 h, and tissues expressing Teton-VP16A2 were incubated for 30 min. (Right) Schematics of the transgenic target genes and transgenic activators used in this study. The wild type (WT) is a transgene that has the hsp70 promoter from −194 to +84 positioned upstream from sequences encoding β-galactosidase. GAGA elements are green, heat-shock elements (HSE) are pink, the hsp70 core promoter region spanning −44 to +84 is light blue, and the β-galactosidase sequence is dark blue. TRE5 has five binding sites for Teton (grey boxes) located upstream from the hsp70 core promoter. TRE5-GA has a GAGA element inserted between the core promoter and the Tet upstream activation sequence (UAS). All three of the transgenic activators share the Teton DBD. Teton-VP16A2 has an activation domain consisting of three tandem copies of a 13-amino-acid peptide derived from VP16 (2). Teton-HSF has an activation domain consisting of the last 81 amino acids of Drosophila HSF. Teton-DBD lacks an activation domain.
Spectrophotometric assay for β-galactosidase activity.
Fat bodies from five larvae were homogenized in 200 μl of 50 mM potassium phosphate (pH 7.5), 1 mM MgCl2, and 0.2% NP-40. Homogenates were centrifuged at 13,000 × g for 5 min at 4°C. The supernatant was transferred to a new Eppendorf tube. The extract (50 μl for Teton-VP16A2 and 100 μl for Teton-HSF and Teton-DBD) was added to a 1-ml final volume of 50 mM potassium phosphate (pH 7.5), 1 mM MgCl2, 1 mM chlorophenol red-beta-d-galactopyranoside in a disposable cuvette and incubated at 37°C. The _A_574 was recorded for 3 h at 30-min intervals. The total amount of protein in the assay was determined by the Bio-Rad protein assay. β-Galactosidase activity was defined as the number of absorbance units per hour per milligram of protein. Results are shown as the mean value of three independent assays.
Genomic footprinting with permanganate.
Tissues were dissected from 10 larvae and placed in 100 μl of Schneider 2 medium (GibcoBRL catalogue no. 11590-056) with or without 1 μg of doxycycline/ml. For the kinetic analyses (see Fig. 5 and 6), fat bodies or salivary glands were placed in medium without doxycycline and then transferred to 100 μl of medium containing 5 μg of doxycycline/ml for various times. For permanganate treatment, tissues were transferred to a tube containing 200 μl of cold 20 mM potassium permanganate dissolved in 130 mM NaCl, 5 mM KCl, and 1.5 mM CaCl2 and incubated for 2 min on ice. The permanganate reaction was stopped with 200 μl of 20 mM Tris-HCl (pH 7.5), 20 mM NaCl, 40 mM Na2EDTA, 1% sodium dodecyl sulfate (SDS), and 400 mM 2-mercaptoethanol. Purification of genomic DNA and subsequent analysis by ligation-mediated PCR with primers TR-1, TR-2, and TR-3 was done as previously described (33, 41).
FIG. 5.
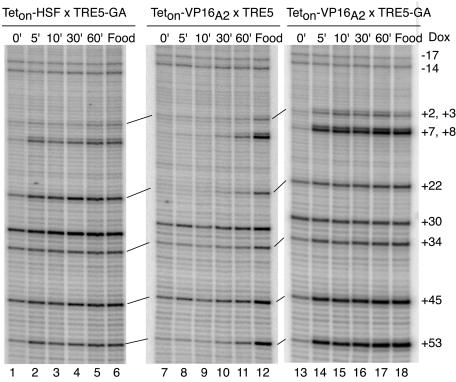
Kinetic analysis of activation in fat bodies. Fly lines containing transgenes for Teton-HSF or Teton-VP16A2 were crossed with fly lines containing the TRE5 or the TRE5-GA transgenes. Larvae were raised in the absence of doxycycline. Fat bodies were isolated from larvae, following production of the Teton proteins in larvae by heat shock and recovery. The fat bodies were incubated in medium containing 5 μg of doxycycline/ml for 0, 5, 10, 30, or 60 min, followed by permanganate treatment. Lanes 1 to 5, permanganate reactivity of TRE5-GA at various times following the induction of binding of Teton-HSF; lanes 7 to 11, permanganate reactivity of TRE5 at various times following the induction of binding of Teton-VP16A2; lanes 13 to 17, permanganate reactivity of TRE5-GA at various times following the induction of binding of Teton-VP16A2. Lanes 6, 12, and 18 show permanganate sensitivity of target genes in fat bodies when larvae were fed doxycycline.
FIG. 6.
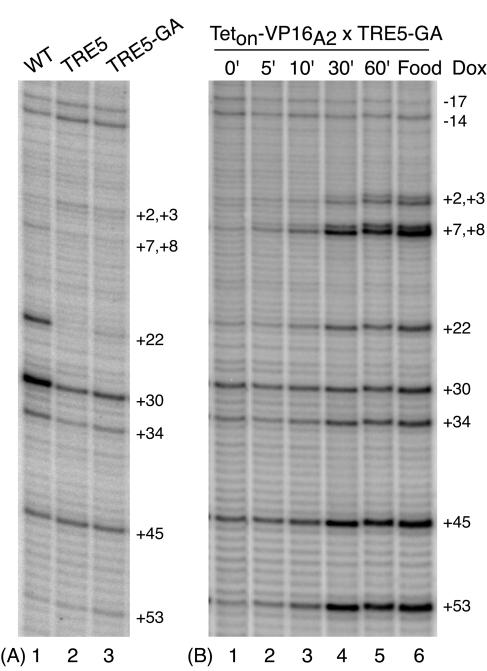
Kinetic analysis of TRE5-GA activation by Teton-VP16A2 in salivary glands. (A) Permanganate footprinting of transgenic targets in salivary glands. See the legend to Fig. 1 for a description of the target genes WT, TRE5, and TRE5-GA. (B) Fly lines expressing Teton-VP16A2 were crossed with fly lines containing TRE5-GA. Larvae were raised in the absence of doxycycline. Production of Teton-VP16A2 was induced by 30-min heat shock, followed by 1-h recovery at room temperature. Salivary glands were isolated from larvae and subjected to doxycycline induction and permanganate footprinting as described for fat bodies.
Preparation of antibodies against the Teton-DBD.
A sequence encoding Teton-DBD was amplified from pUHrT62-1 by PCR and introduced into the protein expression vector pET-28-(a) (Novagen). The recombinant protein was expressed in bacteria and purified with Ni-nitrilotriacetic acid resin (QIAGEN), following the manufacture's instructions. The purified protein was dialyzed against 20 mM HEPES (pH 7.6), 10% glycerol, 100 mM NaCl, 10 mM β-mercapethanol, 0.2 mM phenylmethylsulfonyl fluoride, and 0.1% NP-40 and used to raise antibodies in rabbits (Pocono Research Laboratory and Farm).
Western blot analyses of Teton-fusion proteins.
Larvae from transgenic flies were heat shocked at 37°C for 30 min, followed by 1 h of recovery to induce expression of Teton proteins. Fat bodies from 10 larvae were placed in 30 μl of SDS loading buffer. Fifteen microliters of sample was run on an SDS-12% polyacrylamide gel electrophoresis gel and transferred to a nitrocellulose filter. The ECL Western Blotting system (RPN2108; Amersham Biosciences) was used to detect antibodies bound to proteins on the nitrocellulose filter.
RESULTS
The GAGA element stimulates the steady-state level of activation by Teton-HSF but not by Teton-VP16A2.
To determine if the presence of the GAGA element affected the activity of the HSF and VP16A2 activation domains, two transgenic target genes were made that differed by the presence or absence of a GAGA element (Fig. 1). Both contained five binding sites for Teton located upstream from the hsp70 core promoter region. Both targets also had sequences encoding β-galactosidase so that expression could be assayed by staining tissues with X-Gal. A short GAGA element was inserted between the Teton binding sites and the TATA box in the construct designated TRE5-GA. DNase I footprinting analysis showed that the GAGA factor expressed in and purified from E. coli bound to this element (data not shown). The construct lacking the GAGA element was designated TRE5.
Flies containing either the TRE5 or TRE5-GA transgene were mated with flies containing transgenes encoding Teton-HSF, Teton-VP16A2, or Teton-DBD. Teton-DBD consisted of only the DBD. The progeny were raised on food in the presence or absence of doxycycline to control the interaction of the Teton protein with the target promoters. Expression of the Teton derivatives was placed under control of the heat shock gene promoter. Previously, we determined that a 30-min heat shock, followed by a 1-h recovery at room temperature resulted in production of enough Gal4p to see activation of a target transgene (33). Importantly, the 1-h recovery was sufficient to restore the non-heat-shocked state of the Drosophila larvae.
To measure expression from the target genes, larval tissues were assayed for the presence of β-galactosidase by staining with X-Gal. In the absence of doxycycline, neither transgenic target showed evidence of activation by Teton-HSF, Teton-VP16A2, or Teton-DBD (Fig. 1A and B and data not shown). This indicates that the GAGA element alone did not cause expression of the transgene. In the presence of doxycycline, Teton-HSF was observed to induce expression of TRE5-GA in fat bodies but not salivary glands (Fig. 1D), and no induction was observed for TRE5 in either tissue (Fig. 1C). In contrast, Teton-VP16A2 induced expression of both target promoters in salivary glands and in fat bodies (Fig. 1E and F). Teton-DBD failed to induce expression in any tissues, indicating that the DBD alone had no transcriptional activity (Fig. 1G and H). Similar results for the different Teton derivatives were observed for TRE5 transgenes inserted in two different chromosomal locations and for TRE5-GA transgenes inserted in three different locations (data not shown).
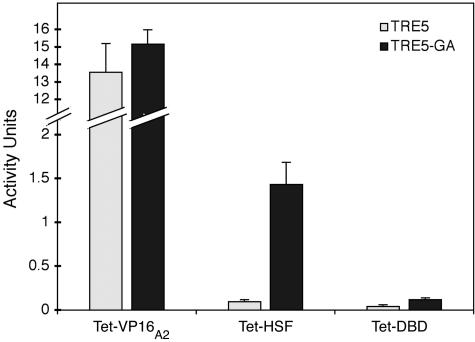
Based on the intensity of the blue pigment and the duration of the staining periods, we estimated that the level of expression of both targets by Teton-VP16A2 in fat bodies was 10 times higher than the level of expression of TRE5-GA by Teton-HSF. Quantification of β-galactosidase activity in fat bodies verified this estimate (Fig. 2). In addition, the level of β-galactosidase induced from TRE5 by Teton-HSF in fat body was <10% of the level induced from TRE5-GA and comparable to the background defined by Teton-DBD.
FIG. 2.
Measurement of β-galactosidase activity by various Teton-fusion proteins. Transgenic fly lines encoding Teton-VP16A2, Teton-HSF, and Teton-DBD were crossed with transgenic lines containing TRE5 or TRE5-GA promoters. The offspring were raised in the presence of doxycycline, heat shocked for 30 min to induce synthesis of Teton proteins, and allowed to recover for 3 h. Fat bodies from five larvae were collected for the chlorophenol red-beta-d-galactopyranoside assay. β-Galactosidase activity was defined as absorbance units per hour per milligram of protein.
Genomic footprinting with permanganate indicates that the capacity of Teton-HSF to activate transcription correlates with promoter proximal pausing mediated by the GAGA element.
To investigate the basis for the differences between Teton-HSF and Teton-VP16A2 in fat bodies, we analyzed the transgenic targets using permanganate footprinting. Permanganate footprinting has been widely used to monitor the presence of Pol II on DNA (8, 18, 31, 40, 41, 44). Permanganate reacts preferentially with thymines located in transcription bubbles associated with transcriptionally engaged Pol II. We chose this approach for three reasons. First, it allows detection of the paused Pol II on transgenes (41). An alternative method, chromatin immunoprecipitation, cannot distinguish between a paused Pol II and one that is in a preinitiated state (5). Second, it could be applied to small amounts of tissue. Third, there was the potential to use this method to carry out a kinetic analysis of changes in Pol II interactions at the promoter (7).
Larvae were raised in the presence or absence of doxycycline to control the association of the Teton derivatives with either TRE5-GA or TRE5. Synthesis of each Teton derivative was induced by a 30-min heat shock followed by a 1-h recovery. Fat bodies were isolated and subjected to permanganate footprinting as previously described for salivary glands (33, 41). Transcriptional activation by Teton-VP16A2 was readily apparent. The intensities of bands corresponding to most thymines between positions +2 and +53 were markedly higher in samples from larvae that were exposed to doxycycline than those that were not (Fig. 3A, lanes 1 to 4). This difference extends well downstream into the body of the gene (data not shown) and represents a snap shot of a train of Pol II molecules moving along the gene.
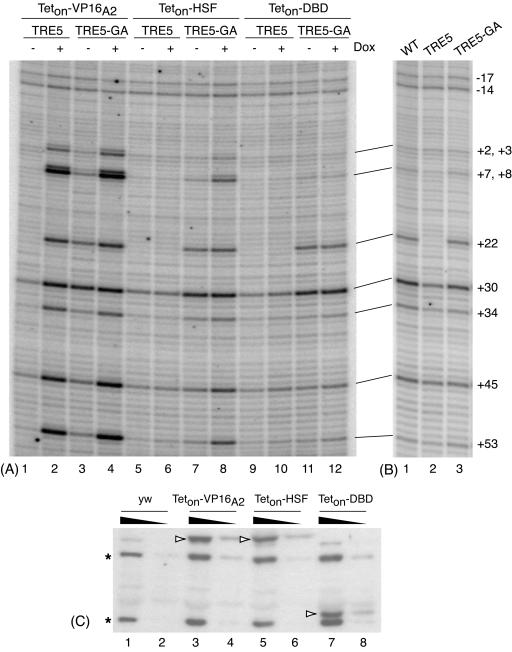
FIG. 3.
Permanganate footprinting analysis of TRE5 and TRE5-GA target genes subjected to the action of various Teton fusion proteins. (A) Fly lines containing transgenes for Teton-VP16A2 (lanes 1 through 4), Teton-HSF (lanes 5 through 8), or Teton-DBD (lanes 9 through 12) were crossed with fly lines containing the TRE5 (lanes 1, 2, 5, 6, 9, and 10) or TRE5-GA (lanes 3, 4, 7, 8, 11, and 12) targets. Offspring were raised either in the absence (odd-numbered lanes) or presence (even-numbered lanes) of doxycycline. Larvae were heat shocked for 30 min, followed by a 1-h recovery to induce synthesis of the Teton proteins. Fat bodies were then subjected to permanganate genomic footprinting. (B) Permanganate footprinting of transgenic targets in fat bodies. See the legend to Fig. 1 for a description of the target genes WT, TRE5, and TRE5-GA. No Teton derivatives were present in these tissues. (C) Western blot analysis of various Teton-fusion proteins produced in fat bodies. Fat bodies from five larvae (lanes 1, 3, 5, and 7) or one-third of this amount (lanes 2, 4, 6, and 8) were run on the gel. Teton fusion proteins (open arrowheads) were detected with antibody raised against the Teton-DBD. The two bands marked by asterisks represent background proteins detected with this antibody, and they serve as loading controls.
In contrast to Teton-VP16A2, Teton-HSF only enhanced the permanganate reactivity of TRE5-GA (Fig. 3A, lanes 5 to 8). Changes in permanganate reactivity caused by association of Teton-HSF were most apparent at positions +8, +45, and +53. In accord with differences in the level of expression (Fig. 2), the intensities of the bands at +8, +45, and +53 for TRE5-GA induced by Teton-HSF were less than the intensities of the bands seen for transgenes induced by Teton-VP16 A2 (Fig. 3A, lanes 4 and 8). None of the changes in permanganate reactivity occurred with Teton-DBD, indicating that the DBD alone had no intrinsic effect on either promoter (Fig. 3A, lanes 10 to 12).
Close inspection of Fig. 3A provided an explanation for why Teton-HSF was able to induce TRE5-GA but not TRE5 in fat bodies. In each case where doxycycline was absent, the intensities of the bands at positions +22 and +30 for TRE5-GA were greater than the corresponding bands for TRE5 (Fig. 3A, compare lanes 3, 7, and 11 to 1, 5, and 9). The enhanced reactivity at positions +22 and +30 for TRE5-GA suggested the presence of paused Pol II. This was verified by comparing the permanganate patterns detected on a transgenic version of the hsp70 promoter to the patterns detected on TRE5 and TRE5-GA (Fig. 3B). The transgenic hsp70 promoter spanned sequences from −194 to +84 and was previously shown to establish a paused Pol II prior to heat shock induction (41). Enhanced permanganate reactivity at +22 and +30 in non-heat-shocked cells is a hallmark of paused Pol II (8, 41). Both the wild-type promoter and TRE5-GA exhibited higher levels of permanganate reactivity at these positions than TRE5 (Fig. 3B, lanes 1 to 3). Hence, the ability of Teton-HSF to induce transcription appeared to depend on the presence of paused Pol II, which in turn depended on the GAGA element.
Western blot analyses were done to compare the levels of each Teton derivative produced in fat bodies. As shown in Fig. 3C, comparable levels of each of the derivatives were produced. Hence, the differences between the derivatives depended on the activation domain rather than the amount of activator. To strengthen this conclusion, we compared the activation of TRE5 in larvae that had one or three transgenes encoding Teton-HSF or one transgene encoding Teton-VP16A2. Western blotting analysis verified that the increase in the number of transgenes encoding Teton-HSF caused an increase in the amount of activator (Fig. 4B, lanes 1 and 2). Despite the increase in Teton-HSF, no activation was detected on TRE5 (Fig. 4A, lanes 2 and 4).
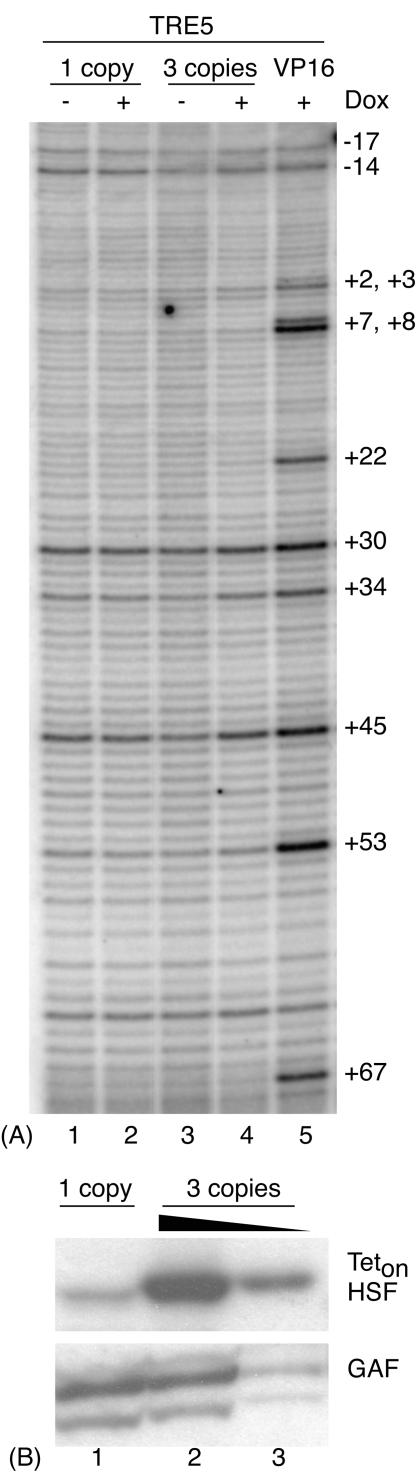
FIG. 4.
Activation of TRE5 cannot be achieved by increasing the amount of Teton-HSF in fat bodies. (A) Fat bodies were isolated from larvae containing one copy of the TRE5 transgene and either one or three copies of the transgene for Teton-HSF or one copy of the transgene for Teton-VP16A2. Larvae were raised either in the absence (lanes 1 and 3) or presence (lanes 2, 4, and 5) of doxycycline. Fat bodies were subjected to permanganate footprinting. (B) Western blot analysis of Teton-HSF proteins produced in fat bodies. Teton-HSF was detected in extracts from the transgenic flies that contain one copy (lane 1) or three copies (lane 2, one-third dilution in lane 3) of Teton-HSF transgene (with antibody against Teton-DBD). Samples were also analyzed for the presence of GAGA factor (GAF) as a control for sample loading.
The GAGA element provides for rapid induction by both Teton-HSF and Teton-VP16A2 in fat bodies.
A primary reason for fusing the activation domains to Teton was so we could determine if the GAGA element enhanced the rate of transcriptional activation. Since Teton-HSF failed to activate TRE5, we could only measure how quickly Teton-HSF activated TRE5-GA. The results shown in Fig. 5 indicated that Teton-HSF reached its maximal level of induction of TRE5-GA within 5 min of the addition of doxycycline to fat bodies isolated from larvae that had not been exposed to doxycycline. The levels of permanganate reactivity at thymines downstream from position +2, following the 5-min doxycycline treatment of the fat bodies, were comparable to the levels detected when larvae were exposed continuously to doxycycline (Fig. 5, compare lanes 2 and 6). Note that this pattern of permanganate reactivity represents a snap shot of a dynamic process in which Pol II molecules initiate transcription, clear the promoter region, and are replaced by newly initiated Pol II molecules. In other words, the position +8 permanganate reactivity 60 min after the addition of doxycycline was most likely not due to the same molecule of Pol II as that which was present at 5 min.
Next, we determined if the GAGA element affected the rate of activation by Teton-VP16A2. Recall that the steady-state levels of induction for TRE5 and TRE5-GA by Teton-VP16 A2 were comparable (Fig. 2 and 3A). In striking contrast, the presence of the GAGA element markedly increased the rate of induction by Teton-VP16A2. As was seen for Teton-HSF, the maximal level of induction of TRE5-GA by Teton-VP16A2 was achieved within 5 min of the addition of doxycycline (Fig. 5, lanes 14 to 18). In contrast, induction of TRE5 by Teton-VP16A2 did not become clearly evident until 60 min after the addition of doxycycline (Fig. 5, lane 11).
Interestingly, the relative intensities of the permanaganate reactivity at positions +8 and +22 were different for the target genes activated by Teton-HSF and Teton-VP16A2 (Fig. 5, lanes 2 to 18). In the case of Teton-HSF, the reactivity at +22 was greater than that at +8, whereas the opposite occurred for Teton-VP16A2. As will be discussed later, this suggests that Pol II encounters different rate-limiting steps in the presence of these activators.
Rapid induction correlates with the presence of paused Pol II.
The presence of the GAGA element greatly accelerated the rate of induction achieved by Teton-VP16A2 (Fig. 5). To determine if this was due to the mere presence of the GAGA element or the presence of paused Pol II, we analyzed the kinetics of induction by Teton-VP16A2 in salivary glands. In salivary glands, the level of paused Pol II detected on TRE5-GA in the absence of activator was significantly less than the level detected on a transgenic version of the hsp70 promoter (Fig. 6A, compare lanes 1 and 3). This differed from the situation in fat bodies, where comparable levels of paused Pol II were detected on the two transgenic promoters (Fig. 3B).
The kinetic analysis showed that Teton-VP16A2 induced TRE5-GA more slowly in salivary glands than in fat bodies. Figure 6B (lanes 1 to 6) shows that induction first became evident 10 min after the addition of doxycycline but required 30 min before the steady-state level of activation was reached. In contrast, full activation by Teton-VP16A2 in fat bodies was achieved within 5 min (Fig. 5, lanes 13 to 18). The slower rate of activation in salivary glands was not due to a delay in the association of doxycycline with Teton-VP16A2, since we could detect changes in transcriptional activity caused by Tetoff-VP16 within 5 min of adding doxycycline to salivary glands that had this activator bound to TRE5 (data not shown). Tetoff-VP16 was induced to dissociate from the DNA upon the addition of doxycycline (10). Hence, rapid induction correlated with the presence of paused Pol II rather than the mere presence of the GAGA element.
DISCUSSION
We developed a transgenic system to investigate the impact of a GAGA element on the activity of two transcriptional activation domains. This transgenic system is unique in that it allowed us to examine both steady-state and kinetic features of transcriptional activation in vivo. The use of permanganate footprinting allowed us to interrogate the behavior of the Pol II on the DNA at a level not readily available to other methods, such as chromatin immunoprecipitation or nuclear run-on. Certainly, neither of these latter methods would have detected the high density of Pol II that is evident immediately downstream from the transcription start site in the presence of Teton-VP16A2. We anticipate that further application of this overall experimental approach will provide significant insight into transcriptional control mechanisms in living cells.
This study was motivated by two main questions. First, would a small GAGA element be sufficient to establish a paused Pol II in the absence of an activator? This question is important because GAGA elements are found associated with a broad spectrum of promoters, making it important to learn if the presence of a GAGA element correlates with promoter proximal pausing. Second, what impact does the GAGA element have on activation by different activators? This information could help us understand what purpose the GAGA element serves in regulating gene expression. We examined the HSF activation domain, which had been defined by transient transfection analyses, to determine if this activation domain depended on the GAGA element. We examined the VP16 activation domain because we anticipated its strength might obviate the requirement for the GAGA element. In addressing these two questions, we have obtained convincing evidence in vivo that Pol II encounters different rate-limiting steps after initiating transcription in the presence of these two activators.
A GAGA element can establish paused Pol II but with tissue specificity.
The expression patterns of β-galactosidase produced by the TRE5 and TRE5-GA trangenes in the presence of Teton-HSF provided us with the first evidence that the GAGA element established paused Pol II on TRE5-GA in fat bodies. Subsequent permanganate data verified this possibility. Our results complement another study showing that mutation of a GAGA element in the hsp70 promoter caused a loss of paused Pol II (16). Our results indicate that it is not necessary to have extensive arrays of GAGA elements as is present in the hsp70 promoter to establish paused Pol II. This has immediate relevance to the β1-tubulin promoter in Drosophila. This promoter has paused Pol II (8), yet visual inspection of the promoter region only reveals two short GAGA elements separated by over 100 bp. Chromatin immunoprecipitation data show 12-fold-less GAGA factor cross-linking to β1-tubulin than to hsp70 (44a). The results reported here indicate that this low level of GAGA factor could be sufficient to account for the paused Pol II.
Our results have also uncovered a complexity not previously known. TRE5-GAGA failed to establish a paused Pol II in salivary glands. This is not due to the absence of factors required for pausing, since paused Pol II was evident on the endogenous hsp70 gene. We are lead to conclude that the small GAGA element is not sufficient for setting up the paused Pol II in all tissues. One obvious difference between the endogenous hsp70 promoter and TRE5-GA is the number of GAGA elements—the endogenous promoter has numerous elements distributed throughout a 150-bp region located upstream from the TATA element (9). Modest differences in the availability of the GAGA factor in different tissues could impact on the activity of the GAGA factor at a particular promoter because GAGA factor binds cooperatively to multiple GAGA elements (13, 41). Identifying conditions that allow establishment of the paused Pol II on the transgene in salivary glands provides a new avenue for investigating the mechanism of promoter proximal pausing.
The GAGA element significantly affects the rate of activation without itself causing activation.
Based on our assessment of levels of β-galactosidase accumulated over the course of many hours, our initial impression was that Teton-VP16A2 functioned independently of the GAGA element. However, the kinetic analysis made possible through the use of the Teton DNA binding domain results in a strikingly different conclusion. Based on the intensity of the permanganate bands at positions +45 and +53 and at additional sites further downstream (data not shown), we conclude that Teton-VP16A2 achieved maximal induction of the TRE5-GA promoter within 5 min of adding doxycycline. In contrast, a 60-min delay occurred before partial induction of the TRE5 promoter was clearly evident. Rapid induction correlated better with the presence of paused Pol II than the mere presence of the GAGA element, since slow induction occurred for TRE5-GA in salivary glands where paused Pol II was not evident despite the presence of the GAGA element.
We consider two ways the GAGA element might accelerate the rate of activation. By establishing a paused Pol II, activation could bypass processes involved in altering chromatin structure or recruiting TFIID. Alternatively, the GAGA element could render the adjacent region of DNA accessible to Teton-VP16A2. Recent results suggest that these two processes could be interdependent, as mutations in either the TATA element or the GAGA element diminished the DNase I hypersensitivity of the hsp26 promoter (17).
It is interesting that Teton-VP16 A2 and Teton-HSF induced TRE5-GA with rapid kinetics, yet the latter is clearly less potent, based on the weaker intensity of permanganate reactivity at positions +45 and +53 and the lower level of β-galactosidase that accumulates in the fat bodies. Thus, rapid induction does not necessarily require an activator that gives a high level of expression. Achieving rapid but correct levels of transcriptional activation is likely to be important for cells to respond appropriately to both external and internal signals.
Analysis in vivo of the functions of _cis_-acting elements and _trans_-acting factors from a kinetic standpoint has received relatively little attention. The facile way in which we were able to execute the kinetic analysis highlights a strength of the permanganate technique and renders this aspect of transcriptional regulation accessible to further investigation.
Permanganate footprinting reveals that Pol II molecules encounter different rate-limiting steps during transcription in the presence of Teton-HSF and Teton-VP16A2.
A significant advantage of the permanganate footprinting technique for monitoring the behavior of Pol II is that it provides a high-resolution view of the location of the Pol II on the DNA once Pol II has initiated transcription. This assertion is supported by both in vitro and in vivo experiments. Previously, we observed that Pol II stalled in vitro at position +1, +3, or +15 produced distinct patterns of permanganate reactivity that correlated well with the predicted location of the Pol II (18). Lis and colleagues were able to deduce from permanganate patterns that the location of paused Pol II on the hsp26 gene was shifted 10 nucleotides downstream relative to the location on the hsp70 gene (8), and this conclusion was subsequently verified by nuclear run-on experiments (29).
Teton-HSF and Teton-VP16A2 caused different patterns of permanganate reactivity to occur on TRE5-GA. In the case of Teton-HSF, permanganate reactivity at +22 and +30 exceeded the permanganate reactivity at +8, whereas the opposite occurred for Teton-VP16A2. These results indicate that the distribution of Pol II differs on TRE5-GA in the presence of these activators. In the case of Teton-HSF, the density of Pol II appears higher in the region 20 to 30 nucleotides downstream from the start than in the immediate vicinity of the transcription start. This suggests that each Pol II molecule continues to pause in the promoter proximal region, albeit transiently, even in the presence of Teton-HSF. O'Brien et al. (24) concluded on the basis of in vivo cross-linking data that promoter proximal pausing persisted as a rate-limiting step on the endogenous hsp70 gene during heat shock and suggested that release of the paused Pol II was the step targeted by HSF. Our results agree with this hypothesis and extend the conclusion by demonstrating that amino acids 610 to 691 of HSF are sufficient to release paused Pol II. This region of HSF has been shown to interact with TBP, mediator, and NURF, but the mechanism by which HSF releases a paused Pol II remains a mystery (23, 26, 45). In subsequent work, we found that the region encompassing amino acids 610 to 664 functions as well as amino acids 610 to 691 (Y. V. Wang and D. S. Gilmour, unpublished data). A deeper analysis of the HSF activation domain based on the approach described here could help pinpoint which protein-protein interaction correlates with releasing paused Pol II.
In the presence of Teton-VP16 A2, there is a higher density of Pol II in the immediate vicinity of the transcription start site than in the region 20 to 30 nucleotides further downstream. By considering the permanganate patterns that were detected previously when Pol II was stalled at different positions on hsp70 in vitro (18), we conclude that the majority of the Pol II detected in the promoter proximal region of TRE5-GA has initiated transcription but not reached position +15. The permanganate pattern observed on TRE5-GA in vivo best matched the pattern when Pol II was stalled at +3. When Pol II was stalled at +3 in vitro, strong permanganate reactivity at +8 was flanked by weaker reactivity at +3 and +22. In contrast, Pol II held at +1 by the presence of alpha-amanitin caused strong permanganate reactivity at −12 and +3 and weaker reactivity at +8, whereas Pol II stalled at +15 caused strong permanganate reactivity at +22 and +30 but weaker reactivity at +8.
Interestingly, the pattern of permanganate reactivity associated with Teton-VP16A2 was strikingly similar to the pattern we observed on the hsp70 core promoter when the promoter was induced in vivo by Gal4p (33). We propose that a Pol II molecule rapidly initiates transcription but encounters a slow phase during elongation immediately downstream from the start site. The relatively weak intensity of permanganate reactivity at +22 and +30 indicates that the density of Pol II in this region was less than at the region closer to the promoter. This is consistent with there being a high density of Pol II near the transcription start, since these Pol II molecules would occlude Pol II molecules from being in the +22 region.
Several biochemical studies have provided evidence for changes in the enzymatic and structural properties of Pol II several nucleotides downstream from the transcription start (11, 12, 15, 22). In vitro, a single molecule of Pol II can execute multiple initiation events without leaving the promoter (6). The Pol II appears to elongate a few nucleotides, releases the short nascent transcript, and starts over without dissociating from the promoter. This is known as abortive initiation and has only been observed in vitro, where it has been possible to isolate the short abortive transcripts. Recent X-ray structure and chemical mapping data indicate that TFIIB has a finger-like domain that extends into the active site of the Pol II, and it has been proposed that growth of the nascent RNA chain shortly after initiation results in steric clash between TFIIB and the RNA (3, 15). The capacity of the Pol II to continue elongation could depend on displacement of TFIIB from Pol II. The high-resolution information obtained through our permanganate footprinting analysis provides evidence that this could be a slow step during transcription in vivo when transcription is mediated by a potent activator.
Acknowledgments
We thank Hermann Bujard for plasmid pUHrT62-1. We thank Maria Horvat Gordon for help producing transgenic fly lines, Jody Washinsky for construction of the TRE5-GA transgene, and all members of the Gilmour lab for comments on the manuscript.
This work was supported by grant GM47477 from the National Institutes of Health and grant MCB-9723537 from the National Science Foundation.
REFERENCES
- 1.Ainbinder, E., M. Revach, O. Wolstein, S. Moshonov, N. Diamant, and R. Dikstein. 2002. Mechanism of rapid transcriptional induction of tumor necrosis factor alpha-responsive genes by NF-κB. Mol. Cell. Biol. 22**:**6354-6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron, U., M. Gossen, and H. Bujard. 1997. Tetracycline-controlled transcription in eukaryotes: novel transactivators with graded transactivation potential. Nucleic Acids Res. 25**:**2723-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, H. T., and S. Hahn. 2004. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell 119**:**169-180. [DOI] [PubMed] [Google Scholar]
- 4.Clos, J., J. T. Westwood, P. B. Becker, S. Wilson, K. Lambert, and C. Wu. 1990. Molecular cloning and expression of a hexameric Drosophila heat shock factor subject to negative regulation. Cell 63**:**1085-1097. [DOI] [PubMed] [Google Scholar]
- 5.Dellino, G. I., Y. B. Schwartz, G. Farkas, D. McCabe, S. C. Elgin, and V. Pirrotta. 2004. Polycomb silencing blocks transcription initiation. Mol. Cell 13**:**887-893. [DOI] [PubMed] [Google Scholar]
- 6.Dvir, A. 2002. Promoter escape by RNA polymerase II. Biochim Biophys Acta 1577**:**208-223. [DOI] [PubMed] [Google Scholar]
- 7.Giardina, C., and J. T. Lis. 1995. Dynamic protein-DNA architecture of a yeast heat shock promoter. Mol. Cell. Biol. 15**:**2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giardina, C., M. Perez-Riba, and J. T. Lis. 1992. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 6**:**2190-2200. [DOI] [PubMed] [Google Scholar]
- 9.Gilmour, D. S., G. H. Thomas, and S. C. R. Elgin. 1989. Drosophila nuclear proteins bind to regions of alternating C and T residues in gene promoters. Science 245**:**1487-1490. [DOI] [PubMed] [Google Scholar]
- 10.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89**:**5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holstege, F. C., U. Fiedler, and H. T. Timmers. 1997. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 16**:**7468-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadesch, T. R., and M. J. Chamberlin. 1982. Studies of in vitro transcription by calf thymus RNA polymerase II using a novel duplex DNA template. J. Biol. Chem. 257**:**5286-5295. [PubMed] [Google Scholar]
- 13.Katsani, K. R., M. A. Hajibagheri, and C. P. Verrijzer. 1999. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 18**:**698-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krumm, A., T. Meulia, M. Brunvand, and M. Groudine. 1992. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 6**:**2201-2213. [DOI] [PubMed] [Google Scholar]
- 15.Kugel, J. F., and J. A. Goodrich. 2002. Translocation after synthesis of a four-nucleotide RNA commits RNA polymerase II to promoter escape. Mol. Cell. Biol. 22**:**762-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, H., K. W. Kraus, M. F. Wolfner, and J. T. Lis. 1992. DNA sequence requirements for generating paused polymerase at the start of hsp70. Genes Dev. 6**:**284-295. [DOI] [PubMed] [Google Scholar]
- 17.Leibovitch, B. A., Q. Lu, L. R. Benjamin, Y. Liu, D. S. Gilmour, and S. C. R. Elgin. 2002. GAGA factor and the TFIID complex collaborate in generating an open chromatin structure at the Drosophila melanogaster hsp26 promoter. Mol. Cell. Biol. 22**:**6148-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, B., J. A. Weber, Y. Chen, A. L. Greenleaf, and D. S. Gilmour. 1996. Analyses of promoter-proximal pausing by RNA polymerase II on the hsp70 heat shock gene promoter in a Drosophila nuclear extract. Mol. Cell. Biol. 16**:**5433-5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lis, J. 1998. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 63**:**347-356. [DOI] [PubMed] [Google Scholar]
- 20.Lu, Q., L. L. Wallrath, B. D. Allan, R. L. Glaser, J. T. Lis, and S. C. R. Elgin. 1992. Promoter sequence containing (CT)n.(GA)n repeats is critical for the formation of the DNase 1 hypersensitive sites in the Drosophila hsp26 gene. J. Mol. Biol. 225**:**985-998. [DOI] [PubMed] [Google Scholar]
- 21.Lu, Q., L. L. Wallrath, H. Granok, and S. C. R. Elgin. 1993. (CT)n · (GA)n repeats and heat shock elements have distinct roles in chromatin structure and transcriptional activation of the Drosophila hsp26 gene. Mol. Cell. Biol. 13**:**2808-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luse, D. S., and G. A. Jacob. 1987. Abortive initiation by RNA polymerase II in vitro at the adenovirus 2 major late promoter. J. Biol. Chem. 262**:**14990-14997. [PubMed] [Google Scholar]
- 23.Mason, P. B., Jr., and J. T. Lis. 1997. Cooperative and competitive protein interactions at the hsp70 promoter. J. Biol. Chem. 272**:**33227-33233. [DOI] [PubMed] [Google Scholar]
- 24.O'Brien, T., and J. T. Lis. 1991. RNA polymerase II pauses at the 5′ end of the transcriptionally induced Drosophila hsp70 gene. Mol. Cell Biol. 11**:**5285-5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Brien, T., R. C. Wilkins, C. Giardina, and J. T. Lis. 1995. Distribution of GAGA protein on Drosophila genes in vivo. Genes Dev. 9**:**1098-1110. [DOI] [PubMed] [Google Scholar]
- 26.Park, J. M., J. Werner, J. M. Kim, J. T. Lis, and Y. J. Kim. 2001. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell. 8**:**9-19. [DOI] [PubMed] [Google Scholar]
- 27.Pinaud, S., and J. Mirkovitch. 1998. Regulation of c-fos expression by RNA polymerase elongation competence. J. Mol. Biol. 280**:**785-798. [DOI] [PubMed] [Google Scholar]
- 28.Plet, A., D. Eick, and J. M. Blanchard. 1995. Elongation and premature termination of transcripts initiated from c-fos and c-myc promoters show dissimilar patterns. Oncogene 10**:**319-328. [PubMed] [Google Scholar]
- 29.Rasmussen, E. B., and J. T. Lis. 1993. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl. Acad. Sci. USA 90**:**7923-7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robertson, H. M., C. R. Preston, R. W. Phillis, D. M. Johnson-Schlitz, W. K. Benz, and W. R. Engels. 1988. A stable source of P-element transposase in Drosophila melanogaster. Genetics 118**:**6341-6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasse-Dwight, S., and J. D. Gralla. 1989. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J. Biol. Chem. 264**:**8074-8081. [PubMed] [Google Scholar]
- 32.Shopland, L. S., K. Hirayoshi, M. Fernandes, and J. T. Lis. 1995. HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes Dev. 9**:**2756-2769. [DOI] [PubMed] [Google Scholar]
- 33.Tang, H., Y. Liu, L. Madabusi, and D. S. Gilmour. 2000. Promoter-proximal pausing on the hsp70 promoter in Drosophila melanogaster depends on the upstream regulator. Mol. Cell. Biol. 20**:**2569-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thummel, C. S., A. M. Boulet, and H. D. Lipshitz. 1988. Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene 74**:**445-456. [DOI] [PubMed] [Google Scholar]
- 35.Tsukiyama, T., P. B. Becker, and C. Wu. 1994. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature 367**:**525-532. [DOI] [PubMed] [Google Scholar]
- 36.Tsukiyama, T., and C. Wu. 1995. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell 83**:**1011-1020. [DOI] [PubMed] [Google Scholar]
- 37.Urlinger, S., U. Baron, M. Thellmann, M. T. Hasan, H. Bujard, and W. Hillen. 2000. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl. Acad. Sci. USA 97**:**7963-7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Steensel, B., J. Delrow, and H. J. Bussemaker. 2003. Genomewide analysis of Drosophila GAGA factor target genes reveals context-dependent DNA binding. Proc. Natl. Acad. Sci. USA 100**:**2580-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wada, T., G. Orphanides, J. Hasegawa, D. K. Kim, D. Shima, Y. Yamaguchi, A. Fukuda, K. Hisatake, S. Oh, D. Reinberg, and H. Handa. 2000. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH.Mol. Cell 5**:**1067-1072. [DOI] [PubMed] [Google Scholar]
- 40.Wang, W., M. Carey, and J. D. Gralla. 1992. Polymerase II promoter activation: closed complex formation and ATP-driven start site opening. Science 255**:**450-453. [DOI] [PubMed] [Google Scholar]
- 41.Weber, J. A., D. J. Taxman, Q. Lu, and D. S. Gilmour. 1997. Molecular architecture of the hsp70 promoter after deleting the TATA box or the upstream regulatory region. Mol. Cell. Biol. 17**:**3799-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilkins, R. C., and J. T. Lis. 1997. Dynamics of potentiation and activation: GAGA factor and its role in heat shock gene regulation. Nucleic Acids Res. 25**:**3963-3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wisniewski, J., A. Orosz, R. Allada, and C. Wu. 1996. The C-terminal region of Drosophila heat shock factor (HSF) contains a constitutively functional transactivation domain. Nucleic Acids Res. 24**:**367-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, C. H., Y. Yamaguchi, L. R. Benjamin, M. Horvat-Gordon, J. Washinsky, E. Enerly, J. Larsson, A. Lambertsson, H. Handa, and D. Gilmour. 2003. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 17**:**1402-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Wu, C. H., C. Lee, R. Fan, M. J. Smith, Y. Yamaguchi, H. Handa, and D. S. Gilmour. 2005. Molecular characterization of Drosophila NELF. Nucleic Acids Res. 33**:**1269-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao, H., R. Sandaltzopoulos, H. M. Wang, A. Hamiche, R. Ranallo, K. M. Lee, D. Fu, and C. Wu. 2001. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell 8**:**531-543. [DOI] [PubMed] [Google Scholar]