Mutations in Proline 82 of p53 Impair Its Activation by Pin1 and Chk2 in Response to DNA Damage (original) (raw)
Abstract
Tumor suppression by the p53 protein largely depends on the elimination of damaged cells by apoptosis. Mutations in the polyproline region (PPR) of p53 impair its apoptotic function. Deletion of the PPR renders p53 more sensitive to inhibition by Mdm2 via an unknown mechanism. We have explored the mechanism by which the PPR modulates the p53/Mdm2 loop. Proline 82 of p53 was identified to be essential for its interaction with the checkpoint kinase 2 (Chk2) and consequent phosphorylation of p53 on serine 20, following DNA damage. These physical and functional interactions are regulated by Pin1 through _cis_-trans isomerization of proline 82. Our study unravels the pathway by which Pin1 activates p53 in response to DNA damage and explains how Pin1 protects p53 from Mdm2. Further, we propose a role for Pin1-dependent induction of p53 conformational change as a mechanism responsible for the enhanced interaction between p53 and Chk2 following DNA damage. Importantly, our findings elucidate the selection for mutations in the Pin1 target Thr81/Pro82 motif within the PPR of p53 in human cancer.
The p53 tumor suppressor protein is essential for the proper control of cell growth and death. The p53 protein is activated when a cell encounters various stress conditions, such as DNA damage. p53 triggers cell growth arrest and/or apoptosis in order to prevent the replication of damaged cells. These functions of p53 are often lost during tumorigenesis. This loss is achieved either by direct mutations in the gene, which occur in about 50% of human cancer cells, or by modifications of p53 regulators (reviewed in reference 20). In nonstressed cells, p53 is maintained at low levels, primarily by the action of Mdm2 through an autoregulatory feedback loop. However, in response to stress, this loop is interrupted and p53 is activated in a proper temporal and spatial manner (reviewed in references 12 and 14). Multiple mechanisms have been identified to describe how this loop is interrupted, including posttranslational modifications of p53 and Mdm2, subcellular transportation, and interaction with specific modulators (reviewed in references 12 and 14). One important activating modification of p53 invoked by DNA damage involves the phosphorylation of p53 on serine 20 (Ser20) by checkpoint kinase 2 (Chk2) within the Mdm2 binding site (4, 13, 19). This phosphorylation contributes to the weakening of the p53/Mdm2 interaction, thereby protecting p53 from Mdm2 (5, 24). The physiological relevance of this pathway is demonstrated by the impaired activation of p53 in _Chk2_-null mice and by the lack of p53 mutations in cancers bearing Chk2 mutations (2). Interestingly, certain mutations in p53 and Chk2 abrogate their interaction and consequently prevent the phosphorylation of p53 on Ser20 by Chk2 (7, 10).
A fascinating insight into how the p53/Mdm2 loop is modulated came from recent studies describing a newly identified role for Pin1, a peptidyl-prolyl isomerase, that has been implicated in numerous aspects of cell cycle regulation (reviewed in references 15 and 16). Pertinently, Pin1 is also critical for the activation of p53 in response to genotoxic drugs, UV light, ionizing radiation (IR), or deregulated oncogene expression (28-30). DNA damage induces the interaction between Pin1 and p53, which is mediated through the WW domain of Pin1 and specific proline-directed phosphorylation of p53 on several Ser/Thr-Pro motifs at Ser33, Ser46, Ser315, and Thr81 (28-30). Thus, in response to DNA damage, Pin1 interacts with p53 and induces _cis_-trans isomerization, which protects p53 from Mdm2, leading to its accumulation and activation (28-30).
One of the Ser/Thr-Pro targets of Pin1 resides within the polyproline region (PPR) (amino acids 62 to 91) of p53, which contains five partially conserved PXXP motifs. A role for the PPR in the regulation of p53-dependent apoptosis, but not growth arrest (18, 27), has been suggested by the observation that human p53 lacking the PPR (p53ΔPro) exhibits a reduced specificity for its apoptotic target genes (25, 31). This impairment is only partially conserved in mouse p53 mutants bearing an equivalent deletion (9). Importantly, a closer examination of p53 target genes at the endogenous levels revealed that the altered specificity of p53ΔPro was not confined to apoptotic target genes (31). Therefore, this altered specificity is insufficient to explain the specific impairment of p53ΔPro to induce apoptosis. It is likely that additional mechanisms are responsible for this impaired apoptotic activity. In our search for a source of this impairment, we identified a new role for the PPR of p53 in its regulation by Mdm2. p53 lacking PPR has an increased binding affinity for Mdm2 and consequently becomes more sensitive to Mdm2-mediated ubiquitination, nuclear export, and degradation (3). This novel regulatory role for the PPR is consistent with the identification of a germ line mutation within the PPR: the replacement of proline 82 with leucine (Pro82Leu) in cancer patients with Li-Fraumeni syndrome (22) and ovarian carcinoma, and somatic mutations in prolines 85 and 89 (from proline to serine) in patients with bladder tumors (23).
In this study, we searched for a mechanism explaining how the PPR modulates the p53/Mdm2 loop (3). We found that the PPR, and more specifically Pro82, is essential for the p53/Chk2 interaction in response to DNA damage and the subsequent Ser20 phosphorylation. This physical and functional interaction is regulated by Pin1, which requires proline 82 of p53 for this action. Our results provide a mechanistic explanation for how Pin1 protects p53 from Mdm2, leading to its accumulation and activation. This offers an explanation for the selection for mutation in Pro82 of p53 in human cancer.
MATERIALS AND METHODS
Cells and transfection assays.
U2OS osteosarcoma cells, expressing wild-type (wt) p53 and 293 kidney epithelial cells, were grown at 37°C in Dulbecco's modified Eagle's medium, and H1299 lung adenocarcinoma and Saos-2 osteosarcoma cells, both lacking p53 expression, were grown in RPMI medium supplemented with 10% fetal calf serum. Mouse embryo fibroblasts (MEFs), both wt and Pin1 deficient, were grown in Dulbecco's modified Eagle's medium supplemented with 20% heat-inactivated fetal calf serum. Transfections were carried out as previously described (24). The amounts of expression plasmids used are indicated in the figure legends. To maintain a constant amount of plasmid DNA in each sample, a relevant empty vector was added. Western blot analysis and a luciferase assay were carried out as previously described (11).
Plasmids and antibodies.
The expression plasmids used were as follows: human wt p53 (pRC/CMV wtp53), human p53 lacking the PPR (pRC/CMV p53DproAE) (27); human mutant p53 with a substitution of isoleucine for proline 82 (pRC/CMV p53P82I); p53 triple mutant with three substitutions of alanine for serine 33, threonine 81, and serine 315 (pcDNA3 p53S33A,T81A,S315A); human Chk2 (pcDNA3 Flag-Chk2); human wt Pin1 (pcDNA3 Ha-Pin1); and human mutant Pin1 with a substitution of alanine for cysteine 109 (pcDNA3 Ha-Pin1C109A) (29). The reporter plasmid used was the p21 luciferase.
The antibodies used were as follows: anti-human p53 monoclonal antibodies PAb421, PAb1801, and DO1; anti-Chk2 monoclonal antibody DCS-273 (Sigma); antiactin monoclonal antibody AC-40 (Sigma); anti-influenza hemagglutinin epitope (anti-Ha) monoclonal antibody HA.11 (16B12; Covance); and anti-phospho-p53 Ser20 and anti-Pin1 polyclonal antibodies (Cell Signaling).
Binding assays.
For in vitro binding, glutathione _S_-transferase (GST) alone and GST-human Chk2 were purified from bacteria by using glutathione agarose beads (Pharmacia). Human wt p53, p53ΔPro, and p53P82I were transcribed and translated in vitro using the TNT T7-coupled wheat germ extract system (Promega). In vitro-translated p53 proteins were incubated with GST fusion proteins immobilized on beads for 2 h at 4°C in binding buffer (50 mM Tris-HCl [pH 8], 150 mM NaCl, 0.5% Nonidet P-40 [NP-40], 0.5 M EDTA, 100 mM NaF, 1 mM sodium orthovanadate, 10 μg/ml of aprotinin, 50 μg/ml of phenylmethylsulfonyl fluoride). The beads were then washed three times with washing buffer (50 mM Tris-HCl [pH 7.6], 500 mM NaCl, 0.5% NP-40, 0.5 M EDTA, and 5% sucrose) and subjected to Western blot analysis. For the in vivo GST binding assay, transfected H1299 cells were exposed to IR (where indicated) and then lysed in lysis buffer (same as binding buffer but with 300 mM NaCl). A GST pulldown assay with GST-Pin1 or GST control was carried out as described above.
A search for the identified p53 mutation was done with the IARC TP53 Mutation Database (http://www-p53.iarc.fr/index.html).
RESULTS
Proline 82 is important for the proper phosphorylation of p53 on Ser20 in response to DNA damage.
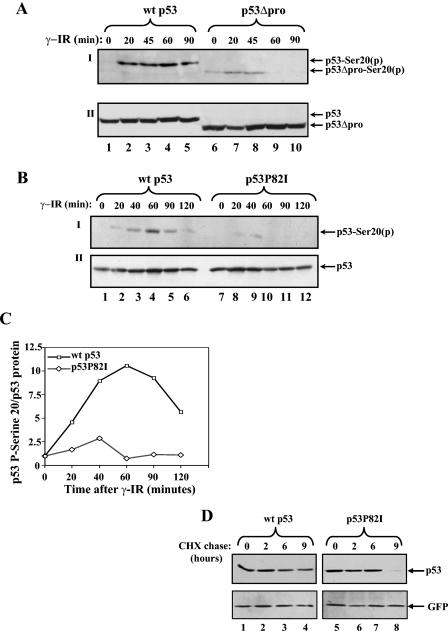
In order to dissect the mechanism by which the PPR of p53 modulates its regulation by Mdm2 (3), we searched for posttranslational modifications that may be involved. Since Ser20 phosphorylation of p53 impairs the physical and functional interaction between p53 and Mdm2 (5, 24), the effect of the PPR on this phosphorylation was tested. Saos-2 osteosarcoma cells were transfected with expression vectors for wt p53 or p53 lacking the PPR (p53ΔPro). Forty hours posttransfection, cells were either left untreated or exposed to IR (10 Gy). At selected time points, cells were harvested and the extent of p53 Ser20 phosphorylation was determined by Western blot analysis using anti-p53 phospho-Ser20 antibody (Fig. 1A). While the level of phosphorylation of wt p53 increased with time after IR and was maintained for 90 min (Fig. 1A, panel I, lanes 1 to 5), the level of phosphorylation of p53ΔPro was severely impaired for the duration of phosphorylation (reduced after 45 min) (Fig. 1A, panel I, lanes 6 to 10). This difference does not reflect changes in the expression levels of these p53 proteins (Fig. 1A, panel II).
FIG. 1.
Pro82 is important for the efficient and prolonged Ser20 phosphorylation of p53 in response to DNA damage. Saos-2 cells (A) or H1299 cells (B) were transiently transfected with expression plasmids for either wt p53 or p53ΔproAE (A) or for p53P82I (B) (1 μg each). Forty hours posttransfection, cells were exposed to γ-IR (10 Gy) before being harvested at the indicated times. By use of Western blot analysis, phosphorylation (p) of p53 Ser20 was determined with anti-p53-phospho-Ser20 antibodies (I), and the level of p53 expression was determined with a mixture of anti-p53 antibodies (PAb1801 and DO1) (II). (C) The extent of Ser20 phosphorylation (B, panel I**)** was calculated relative to the level of p53 expression (B, panel II**)** using densitometry. The ratios of wt p53 to the p53P82I mutant versus time following IR treatment were plotted on a graph. (D) H1299 cells were transfected and treated as described above, with the exception that cycloheximide (CHX) (10 μg/ml) was added 10 min before IR treatment. Cells were harvested at the indicated times, and the levels of p53 were monitored as described above. The transfection efficiencies were determined by introducing a GFP expression plasmid into each sample and monitoring the expression with anti-GFP antibody.
It can be argued that this difference reflects major structural changes incurred by the loss of the entire PPR of p53. We therefore examined whether a substitution of proline (Pro) 82 in p53, which is targeted for mutation in familial human breast cancer (22), results in the same impairment. The levels of Ser20 phosphorylation in response to IR for wt p53 and a substitution mutant of p53 (p53P82I, which substitutes isoleucine for proline 82) were compared; such an analysis has been described for p53ΔPro but in H1299 cells, lung carcinoma cells lacking p53 expression. A marked impairment in Ser20 phosphorylation was observed for p53P82I (Fig. 1B). Quantification of the phosphorylation signal, relative to the amount of p53, revealed a reduction in the extent of phosphorylation of p53P82I at early and late time points, with a maximal difference at 1 h following IR (Fig. 1C). This difference did not reflect changes in the levels of p53 expression (Fig. 1B, panel II). To examine whether the reduced phosphorylation of p53P82I is due to the reduced stability of this mutant, the stabilities of wt p53 and the p53P82I mutant were compared. H1299 cells were transfected with expression vectors for wt p53 or p53P82I as described above. Forty hours posttransfection, cells were treated with cycloheximide (10 μg/ml) and exposed to IR (10 Gy). Cells were harvested at 0, 2, 6, and 9 h after IR and cycloheximide treatment, and p53 levels were determined. As shown in Fig. 1D, the half-life of the p53P82I mutant was shorter than that of wt p53, consistent with our previous findings (3). However, there was no measurable difference in the stabilities of wt p53 and the p53P82I mutant within the first 6 h after IR and cycloheximide treatment. Since the phosphorylation assays were performed at earlier time points, it can be concluded that the observed difference in Ser20 phosphorylation is not due to differences in protein stability. It is important to note that the substitution of another proline residue, Pro89, a somatic mutation derived from a human bladder tumor (23), in this region did not result in this impairment (data not shown). These results support a positive role for Pro82 in the regulation of Ser20 phosphorylation in response to IR.
Pro82 is essential for DNA damage-induced p53/Chk2 interaction in vivo but not in vitro.
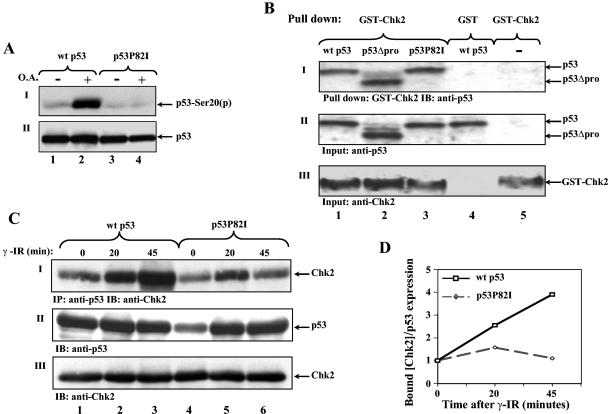
The observed impairment in Ser20 phosphorylation of p53P82I may result from an increased susceptibility to phosphatase and/or a reduced responsiveness to the kinase. To test the former, the effect of a phosphatase inhibitor (okadaic acid) on Ser20 phosphorylation was measured. H1299 cells were transfected with expression plasmids for either wt p53 or p53P82I. Twenty-four hours posttransfection, cells were treated with 300 nM okadaic acid for 2 h, or left untreated, before Ser20 phosphorylation was determined by Western blot analysis. The phosphorylation of wt p53 was markedly enhanced following okadaic acid treatment, whereas no effect was detected in the p53P82I mutant (Fig. 2A). This result suggests that the impaired Ser20 phosphorylation of p53P82I does not involve a phosphatase activity.
FIG. 2.
Pro82 facilitates DNA damage-induced p53/Chk2 interaction in vivo but not in vitro. (A) H1299 cells were transfected with expression plasmids for wt p53 or the p53P82I mutant (1 μg each). Forty hours posttransfection, cells were treated with a 300 nM concentration of the phosphatase inhibitor okadaic acid (O.A.) for 2 h (+) or left untreated (−) before being harvested. Serine 20 phosphorylation (p) (I) and p53 expression levels (II) were monitored as described in the legend to Fig. 1. (B) Equal amounts of in vitro-translated proteins of p53 (wt p53, p53P82I, and p53ΔPro) were incubated in vitro with bacterially expressed GST-Chk2 or GST alone followed by a pulldown assay. The amounts of bound p53 proteins were monitored by Western blot analysis using a mixture of anti-p53 antibodies (PAb1801 and DO1) (I). The amounts of input p53 forms and GST-Chk2 were determined by removing aliquots of each sample prior to the pulldown assay and subjecting these to Western blot analysis using anti-p53 antibodies (DO1 and PAb1801) (II) or anti-Chk2 antibody (DCS-273) (III). The binding results between wt p53 and GST and GST-Chk2 without p53 were used as the controls (lanes 4 and 5). (C) 293 cells were transfected with expression plasmids for Chk2 (4 μg) together with wt p53 or p53P82I (2 μg each). Forty hours posttransfection, cells were exposed to IR (10 Gy), and at the times indicated, cell extracts were subjected to an immunoprecipitation (IP) assay using anti-p53 antibody (PAb421), followed by immunoblotting (IB) with anti-Chk2 antibody (DCS-273) (I). The levels of p53 (II) and Chk2 (III) expression in each sample were monitored by removing aliquots of cell extracts prior to immunoprecipitation and subjecting these to Western blot analysis as described above. (D) The intensities of the bound Chk2 and the amounts of p53 shown in panel C were determined by densitometry, and the ratios of bound Chk2 to p53 expression were plotted. The ratio at time zero was taken as 1.
To test whether the impaired phosphorylation involves the Chk2, the p53/Chk2 interaction in response to DNA damage was evaluated (7, 10). Initially, the interaction was examined in vitro between in vitro-translated wt p53, p53ΔPro, or p53P82I protein and bacterially expressed GST-Chk2. Equal amounts of each p53 protein (Fig. 2B, panel II) were incubated with GST-Chk2 fusion protein linked to glutathione beads. Following a pulldown assay, the amounts of bound p53 were monitored by Western blot analysis using an anti-p53 antibody. As shown in Fig. 2B (panel I), all three forms of p53 bound equally to GST-Chk2.
Since the p53/Chk2 interaction was enhanced in response to DNA damage (7, 10), the interaction between p53 or p53P82I and Chk2 was determined in vivo in response to DNA damage. Kidney epithelial 293 cells were transfected with expression plasmids for wt p53 or p53P82I together with Chk2. Forty hours posttransfection, cells were either left untreated or exposed to γ-IR (10 Gy) for different periods. Cell extracts were subjected to coimmunoprecipitation using anti-p53 monoclonal antibody PAb421, followed by Western blot analysis with anti-Chk2 monoclonal antibody DCS-273. The amount of Chk2 bound to wt p53 increased significantly with time after irradiation, whereas the amount of p53P82I remained unchanged relative to the levels of p53 expression (Fig. 2C and D). These results clearly indicate that Pro82 plays a positive role in the p53/Chk2 interaction in response to DNA damage in vivo. This marked difference between the results obtained in vitro and those in vivo supports the notion that the p53/Chk2 interaction is facilitated by a modification involving Pro82. Such a modification occurs in vivo following DNA damage but not in vitro.
Pin1 is essential for the efficient phosphorylation of p53 on Ser20.
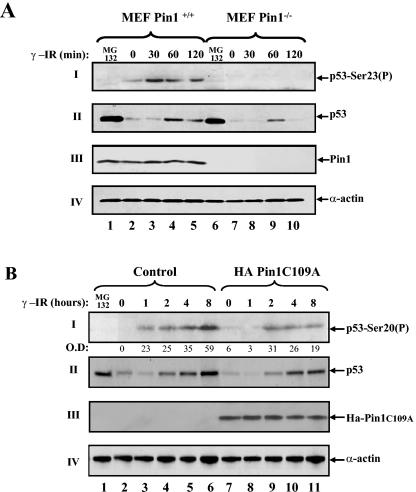
Among the p53-modifying enzymes that may be affected by the P82I substitution, Pin1 is an attractive candidate because the p53/Pin1 interaction requires phosphorylation on the adjacent residue, Thr81 (29, 30). This encouraged us to test whether the impaired phosphorylation of p53P82I results from a failure of this mutant to undergo isomerization by Pin1. The effects of Pin1 on Ser20 phosphorylation were compared between MEFs derived from Pin1 knockout mice and those of their normal counterparts. Cells were exposed to IR (10 Gy) and, at different time points, were harvested to determine the extent of Ser23 (corresponding to the Ser20 of human p53) phosphorylation. The level of Ser23 phosphorylation in the wt MEFs increased with time after IR exposure (Fig. 3A, panel I, lanes 2 to 5). By marked contrast, Ser23 phosphorylation was not detected in the _Pin1_-null MEFs (Fig. 3A, panel I, lanes 7 to 10). This strongly implicates Pin1 as an important regulator of p53 Ser20 phosphorylation in response to IR at physiological levels of p53 and Pin1. Importantly, the accumulation of p53 follows Ser23 phosphorylation (Fig. 3A, panel II, lanes 3 and 4), providing further support for the link between this phosphorylation and the role of Pin1 in the accumulation of p53 in response to DNA damage. In the absence of DNA damage, Ser23 phosphorylation was not detected even when a large amount of p53 accumulated from treatment with the proteasome inhibitor MG132 (Fig. 3A, panel II, lanes 1 and 6). As previously reported (29), the accumulation of p53 was impaired in the _Pin1_-null MEFs (Fig. 3A, panel II, lanes 7 to 10).
FIG. 3.
Pin1 is essential for Ser20 phosphorylation of p53 in response to DNA damage. (A) MEFs from wt or _Pin1_-null mice were exposed to IR (10 Gy), treated with MG132 (20 μM) for 4 h, or left untreated. At the indicated times, cells were harvested and the levels of Ser23 phosphorylation (P) and p53 expression were determined as described in the legend to Fig. 1. The expression levels of Pin1 were determined with anti-Pin1 polyclonal antibody, and the amount of extract loaded was determined by use of antiactin. (B) A pool of mock-transfected U2OS cells and a pool of stably transfected Ha-Pin1C109A mutants were either left untreated or exposed to IR (20 Gy). At the indicated times, the levels of Ser20 phosphorylation (I) and p53 expression (II) were monitored as described for panel A, while Ha-Pin1 was monitored by anti-Ha antibody (III). As a control, U2OS cells were exposed to the proteasome inhibitor MG132 (20 μM) for 4 h. The amount of extract loaded was determined by reprobing the same membrane with anti-α-actin (IV). O.D., optical density.
In addition to performing the MEF experiment, we examined the role of Pin1 in Ser20 phosphorylation in human cells at endogenous levels of p53. For this purpose, Pin1 activity was downregulated using a Pin1C109A mutant. This mutant binds, but cannot isomerize, p53 (29, 30) and hence is likely to act as a dominant negative mutant. U2OS osteosarcoma cells expressing wt p53 were stably transfected with the Pin1C109A mutant or with an empty vector. Cells were exposed to IR (20 Gy), and the amount of Ser20 phosphorylation of endogenous p53 was determined. Whereas phosphorylation appeared at 1 h after IR in the control cells, it was detected only after 2 h in the Pin1C109A transfectant pool. Likewise, the extent of phosphorylation was threefold higher with the control cells than with the Pin1C109A-expressing pool (Fig. 3B, panel I, lanes 6 and 11). It should be noted that, here too, Ser20 phosphorylation preceded p53 accumulation by 1 h (Fig. 3B, compare panels I and II). Further, Ser20 phosphorylation was dependent on DNA damage and was not detected in accumulated p53 by MG132 (Fig. 3B, panel II, lane 1). Together, these results indicate that Pin1 plays an important role in the regulation of Ser20 phosphorylation of p53.
Pin1 enhances p53/Chk2 interaction in response to DNA damage.
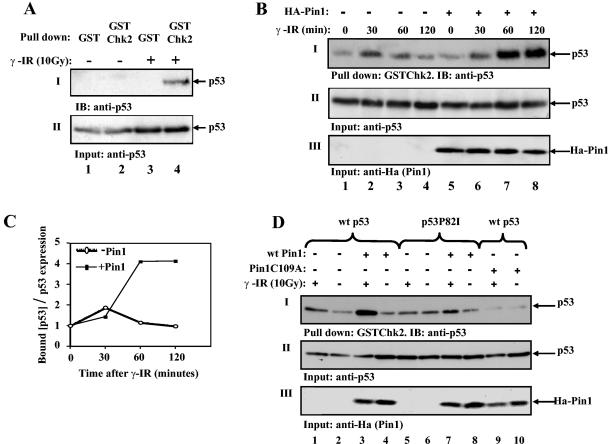
The finding that Pin1 plays an important role in the regulation of Ser20 phosphorylation of p53 begged the question as to whether Pin1 facilitates p53/Chk2 binding in response to DNA damage. To address this question, H1299 cells were transfected with wt p53 and Ha-Pin1. Twenty-four hours later, cells were exposed to IR (10 Gy) or were left untreated. Ninety minutes after IR, cell extracts were subjected to a pulldown assay using bacterially derived GST-Chk2 or GST alone. As shown in Fig. 4A, p53 interacted with GST-Chk2 but not with GST alone, and this interaction was dependent on DNA damage. The role of Pin1 in this interaction was determined by transfecting H1299 cells with p53 alone or together with Ha-Pin1 expression plasmids. Forty hours later, cells were irradiated (10 Gy), and at different times after IR, cell extracts were subjected to a GST pulldown assay using bacterially expressed GST-Chk2. The interaction between p53 and Chk2 increased within 30 min after irradiation, irrespective of the presence of exogenous Pin1 (Fig. 4B, lanes 2 and 6), suggesting that the endogenous levels of Pin1 are sufficient for the efficient initiation of this interaction. Thereafter, the extent of this interaction was reduced in the absence of exogenous Pin1 but was significantly increased and maintained for over 2 h in the presence of exogenous Pin1 (Fig. 4B and C). This result supports a role for Pin1 in modulating the extent and duration of p53/Chk2 interaction in response to DNA damage. Further, since Chk2 was bacterially expressed, it could not have been subjected to modification by Pin1, supporting the _cis_-trans isomerization of p53 by Pin1 as the relevant modification.
FIG. 4.
Pin1 enhances p53/Chk2 interaction in response to DNA damage. (A) H1299 cells were transfected with expression plasmids for p53 (3 μg) and Ha-Pin1 (5 μg). Twenty-four hours posttransfection, cells were exposed to IR (10 Gy) (+) or left untreated (−). Ninety minutes after IR, cell extracts were subjected to a pulldown assay using bacterially expressed GST-Chk2 or GST beads alone. The amounts of bound p53 and input p53 were monitored by immunoblotting (IB) as described in the legend to Fig. 1. (B) H1299 cells were transfected with expression plasmids for p53 alone (3 μg) (−) or together with Ha-Pin1 (5 μg) (+). Twenty-four hours posttransfection, cells were either left untreated or irradiated (10 Gy). At the indicated times, cell extracts were subjected to a pulldown assay using bacterially expressed GST-Chk2 beads. The amount of bound p53 was determined by Western blotting using anti-p53 antibodies (PAb1801 and DO1) (I). The amounts of input p53 (II) and Ha-Pin1 (III) were monitored with anti-p53 and anti-Ha antibodies, respectively. (C) The intensities of the bound p53 and amounts of p53 shown in panel B were determined by densitometry, and the ratios of bound p53 to p53 expression were plotted. The ratio at time zero was taken as 1. (D) H1299 cells were transfected with the indicated expression plasmids. Twenty-four hours posttransfection, cells were either left untreated or irradiated (10 Gy). Ninety minutes after irradiation, cells were subjected to a GST-Chk2 pulldown assay as described for panel A. The levels of input p53 (II) and Ha-Pin1 (III) are shown.
Next, we asked whether the impaired interaction between Chk2 and p53P82I results from a defective response of the latter to Pin1-mediated isomerization. To address this question, we compared the effects of Pin1 on the Chk2/p53 interaction between wt p53 and p53P82I following IR. H1299 cells were transfected with different combinations of expression vectors for wt p53 or p53P82I and wt Ha-Pin1 or Ha-Pin1C109A. Twenty-four hours posttransfection, cells were either left untreated or exposed to IR (10 Gy), and 90 min later, cells were harvested. Cell extracts were subjected to a pulldown assay using bacterially expressed GST-Chk2 as described above. The interaction between Chk2 and wt p53, but not between Chk2 and the p53P82I mutant, increased in response to IR (Fig. 4D, panel I, lanes 1, 2, 5, and 6), consistent with the results shown in Fig. 2C. Importantly, coexpression of wt Pin1 markedly increased the p53/Chk2 binding (Fig. 4D, panel I, lanes 3 and 4), while only a slight increase was observed with p53P82I (Fig. 4D, panel I, lanes 7 and 8). In contrast to wt Pin1, the Pin1C109A mutant abrogated p53/Chk2 binding, even in response to IR (Fig. 4D, panel I, lanes 9 and 10). These results suggest that Pin1 facilitates the interaction between p53 and Chk2 through _cis_-trans isomerization, which requires Pro82 in p53.
Pro82 is essential for binding and activation of p53 by Pin1.
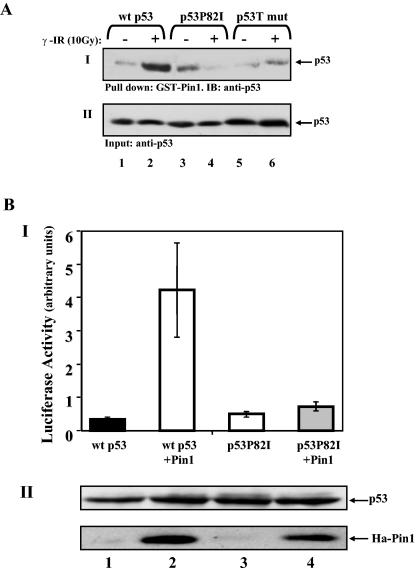
The demonstrated role for Pro82 in the interaction and phosphorylation of p53 by Chk2 raised the possibility that the substitution for Pro82 impairs interaction and _cis_-trans isomerization of p53 by Pin1. To test this prediction, we compared the interactions between Pin1 and wt p53 and between Pin1 and the p53P82I mutant in response to IR by a pulldown assay using GST-Pin1. As a control, a triple mutant of p53 (p53S33A,T81A,S315A [p53T]), which is deficient in Pin1 binding, was used (29, 30). H1299 cells were transfected with expression plasmids for wt p53, p53P82I, or p53T, and 24 h later, cells were either left untreated or exposed to IR (10 Gy). Forty-five minutes after IR, cell extracts were subjected to a pulldown assay using GST-Pin1 attached to beads. The amount of p53 bound to Pin1 was monitored by Western blot analysis using an anti-p53 antibody. While exposure of cells to IR significantly increased wt p53/Pin1 binding, no detectable increase was observed in the binding of Pin1 to the p53P82I or p53T mutants (Fig. 5A). This result clearly indicates that Pro82 is essential for the Pin1/p53 interaction in response to DNA damage. A prediction from this result was that the p53P82I mutant would not be activated by Pin1. This prediction was tested by a transcriptional assay using a p21 promoter-driven luciferase reporter gene. U2OS cells were transfected with the p21 luciferase reporter plasmid along with wt p53 or p53P82I expression plasmids alone or together with Ha-Pin1. Whereas the transcriptional activity of wt p53 was enhanced by Pin1, that of the p53P82I mutant was almost unaffected (Fig. 5B, panel I). This differential response did not reflect differences in the levels of protein expression between the two forms of p53, as determined by Western blot analysis in the same assay (Fig. 5B, panel II).
FIG. 5.
Pro82 is essential for binding and activation of p53 by Pin1. (A) H1299 cells were transfected with expression plasmids for either wt p53, the p53P82I mutant, or the p53S33A,S315A,T81A triple mutant (p53T mut). Twenty-four hours posttransfection, cells were either left untreated (−) or irradiated (10 Gy) (+), and 45 min later, cell extracts were subjected to a pulldown assay using GST-Pin1 beads. The amounts of bound p53 (I) and input p53 (II) were determined by Western blot analysis using anti-p53 antibodies (DO1 and PAb1801). (B) U2OS cells were transfected with the indicated expression plasmids (0.1 μg for p53 plasmids and 1 μg for Ha-Pin1 plasmids) together with a luciferase reporter plasmid (0.3 μg) driven by the p21 promoter. Twenty-four hours after transfection, the luciferase activity was determined for each sample (I). The luciferase activity is expressed in arbitrary units, and the averages and standard deviations for three independent experiments are shown. The amounts of p53 and Ha-Pin1 were monitored by Western blot analysis (II) using anti-p53 and anti-Ha antibodies, respectively.
DISCUSSION
The PPR of p53 is essential for p53-mediated apoptosis (18, 25, 27, 31), particularly in response to chemotherapeutic agents (1). This role is supported by the identification of germ line and somatic mutations within this region in human cancers (references 22 and 23 and the TP53 Mutation Database). Positioned between the N-terminal transactivation domain and the core DNA binding domain, the PPR can serve as a modulator for multiple functions of p53, predominantly transcriptional activities. Indeed, the lack of the PPR alters p53 specificity for certain target promoters (25, 31). This partially explains the impaired apoptotic activity of p53ΔPro. We have previously shown that the deletion of the PPR impairs p53 activities, including p53-mediated apoptosis, by increasing p53's sensitivity to inhibition and degradation by Mdm2 (3). This results from an enhanced binding of Mdm2 to p53 lacking PPR (3).
How the PPR modulates the p53/Mdm2 interaction is an important question that has been addressed in this study. The phosphorylation of p53 on Ser20 that abrogates the p53/Mdm2 interaction (5, 24) is defective in the p53ΔPro mutant and, more importantly, in the Pro82 mutant p53P82I (22). These PPR mutations severely impair both the extent and the duration of Ser20 phosphorylation in response to DNA damage (by IR). The defect in Ser20 phosphorylation of the p53P82I mutant results from a reduced ability of this mutant to bind Chk2 in vivo but not in vitro. This clearly links Pro82 with the regulation of p53 phosphorylation by Chk2. Further, the striking difference between the in vivo and in vitro binding results implies that Pro82-dependent regulation of Chk2/p53 binding requires a DNA damage-induced modification of p53 in vivo.
The location of Pro82 within key Ser/Thr-Pro target motifs of p53 for Pin1 (29, 30) spotlighted it as the prime modifying enzyme candidate. Using MEFs null for Pin1, we demonstrated that at endogenous expression levels, Pin1 plays a major role in the Ser20 phosphorylation of p53 in response to IR. It should be noted that this phosphorylation preceded p53 accumulation in response to DNA damage, supporting a link between Ser20 phosphorylation and the role of Pin1 in the accumulation of p53 in response to DNA damage. This effect of Pin1 is achieved by facilitating an IR-induced p53/Chk2 interaction, and it provides a novel insight into how the p53/Chk2 interaction is enhanced in response to DNA damage (10).
How the _cis_-trans isomerization of Pro82 of p53 affects Chk2 binding is not clear; however, it is likely to involve a conformational change that affects at least one of the two Chk2 docking sites located in conserved boxes II and V of p53 (7). Our results are consistent with previous studies demonstrating a critical role for Pin1 in the activation and accumulation of p53 in response to DNA damage (28-30). Importantly, our link between Pro82 and Chk2-dependent regulation of p53 clarifies how Pin1 protects p53 from Mdm2-mediated inhibition (29). We found that Pin1 modulates both the extent and the duration of Ser20 phosphorylation in response to DNA damage. This raises the possibility that Pin1 may affect the threshold of p53 signaling in response to DNA damage, which in turn may influence the outcome of the p53 response, for instance, by tipping the balance between growth arrest and apoptosis in a given cellular context (26). While our study focused on the Chk2 pathway, it did not exclude the contributions of parallel pathways activated by Pin1 that can affect p53 regulation directly or p53 functional outcome indirectly. The effects of Pin1 on β-catenin (17) and Cdc25 (8, 21) exemplify this notion. Recently, a role for the PPR in the transcriptional-independent apoptotic activity of p53 has been reported (6). It would be interesting to evaluate the role of Pin1 in this activity.
The substitution for Pro82, such as that in the p53P82I mutant, is sufficient to abrogate the p53/Pin1 interaction and, consequently, the Pin1-dependent activation of p53 through the Chk2 pathway. This suggests the first molecular explanation for why a missense mutation in Pro82 of p53 was selected in human cancer. Interestingly, the adjacent threonine (Thr81) within this Thr-Pro motif of Pin1 is also mutated in human cancer (TP53 Mutation Database). Moreover, mutations have also been identified in the other Pin1 target motifs in p53, Ser315/Pro316 and Ser46/Pro47, in a variety of human cancers (TP53 Mutation Database). This raises the intriguing possibility that these mutations may affect p53 regulation by impairing relevant posttranslational modifications of p53, as in the case of Pro82. Exploring the mechanisms by which these mutations affect p53 activities should provide explanations for their selection in cancer and unravel a new layer of complexity in p53 regulation.
Acknowledgments
We are grateful to Sue Haupt for critical comments. We thank Matthias Dobbelstein, Thanos Halazonetis, Moshe Oren, and Arnie Levine for the generous gifts of plasmids.
This work was supported by grants from the Israel Science Foundation, the Israel Cancer Research Fund, the Israel Cancer Association, the German-Israeli Foundation for Scientific Research and Development, the Association for International Cancer Research, and EC FP6 funding (contract 503576).
REFERENCES
- 1.Baptiste, N., P. Friedlander, X. Chen, and C. Prives. 2002. The proline-rich domain of p53 is required for cooperation with anti-neoplastic agents to promote apoptosis of tumor cells. Oncogene 21**:**9-21. [DOI] [PubMed] [Google Scholar]
- 2.Bartek, J., and J. Lukas. 2003. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3**:**421-429. [DOI] [PubMed] [Google Scholar]
- 3.Berger, M., R. Vogt Sionov, A. J. Levine, and Y. Haupt. 2001. A role for the polyproline domain of p53 in its regulation by Mdm2. J. Biol. Chem. 276**:**3785-3790. [DOI] [PubMed] [Google Scholar]
- 4.Chehab, N. H., A. Malikzay, M. Appel, and T. D. Halazonetis. 2000. Chk2/hCds1 functions as a DNA damage checkpoint in G1 by stabilizing p53. Genes Dev. 14**:**278-288. [PMC free article] [PubMed] [Google Scholar]
- 5.Chehab, N. H., A. Malikzay, E. S. Stavridi, and T. D. Halazonetis. 1999. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc. Natl. Acad. Sci. USA 96**:**13777-13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chipuk, J. E., T. Kuwana, L. Bouchier-Hayes, N. M. Droin, D. D. Newmeyer, M. Schuler, and D. R. Green. 2004. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303**:**1010-1014. [DOI] [PubMed] [Google Scholar]
- 7.Craig, A., M. Scott, L. Burch, G. Smith, K. Ball, and T. Hupp. 2003. Allosteric effects mediate CHK2 phosphorylation of the p53 transactivation domain. EMBO Rep. 4**:**787-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crenshaw, D. G., J. Yang, A. R. Means, and S. Kornbluth. 1998. The mitotic peptidyl-prolyl isomerase, Pin1, interacts with Cdc25 and Plx1. EMBO J. 17**:**1315-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards, S. J., L. Hananeia, M. R. Eccles, Y. F. Zhang, and A. W. Braithwaite. 2003. The proline-rich region of mouse p53 influences transactivation and apoptosis but is largely dispensable for these functions. Oncogene 22**:**4517-4523. [DOI] [PubMed] [Google Scholar]
- 10.Falck, J., C. Lukas, M. Protopopova, J. Lukas, G. Selivanova, and J. Bartek. 2001. Functional impact of concomitant versus alternative defects in the Chk2-p53 tumour suppressor pathway. Oncogene 20**:**5503-5510. [DOI] [PubMed] [Google Scholar]
- 11.Haupt, Y., Y. Barak, and M. Oren. 1996. Cell type-specific inhibition of p53-mediated apoptosis by mdm2. EMBO J. 15**:**1596-1606. [PMC free article] [PubMed] [Google Scholar]
- 12.Hayon, I. L., and Y. Haupt. 2002. p53: an internal investigation. Cell Cycle 1**:**111-116. [PubMed] [Google Scholar]
- 13.Hirao, A., Y. Y. Kong, S. Matsuoka, A. Wakeham, J. Ruland, H. Yoshida, D. Liu, S. J. Elledge, and T. W. Mak. 2000. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287**:**1824-1827. [DOI] [PubMed] [Google Scholar]
- 14.Iwakuma, T., and G. Lozano. 2003. MDM2, an introduction. Mol. Cancer Res. 1**:**993-1000. [PubMed] [Google Scholar]
- 15.Joseph, J. D., E. S. Yeh, K. I. Swenson, A. R. Means, and Winkler. 2003. The peptidyl-prolyl isomerase Pin1. Prog. Cell Cycle Res. 5:477-487. [PubMed] [Google Scholar]
- 16.Lu, K. P. 2003. Prolyl isomerase Pin1 as a molecular target for cancer diagnostics and therapeutics. Cancer Cell 4**:**175-180. [DOI] [PubMed] [Google Scholar]
- 17.Ryo, A., M. Nakamura, G. Wulf, Y. C. Liou, and K. P. Lu. 2001. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nat. Cell Biol. 3**:**793-801. [DOI] [PubMed] [Google Scholar]
- 18.Sakamuro, D., P. Sabbatini, E. White, and G. C. Prendergast. 1997. The polyproline region of p53 is required to activate apoptosis but not growth arrest. Oncogene 15**:**887-898. [DOI] [PubMed] [Google Scholar]
- 19.Shieh, S. Y., J. Ahn, K. Tamai, Y. Taya, and C. Prives. 2000. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 14**:**289-300. [PMC free article] [PubMed] [Google Scholar]
- 20.Sionov, R. V., and Y. Haupt. 1999. The cellular response to p53: the decision between life and death. Oncogene 18**:**6145-6157. [DOI] [PubMed] [Google Scholar]
- 21.Stukenberg, P. T., and M. W. Kirschner. 2001. Pin1 acts catalytically to promote a conformational change in Cdc25. Mol. Cell 7**:**1071-1083. [DOI] [PubMed] [Google Scholar]
- 22.Sun, X. F., O. Johannsson, S. Hakansson, G. Sellberg, B. Nordenskjold, H. Olsson, and A. Borg. 1996. A novel p53 germline alteration identified in a late onset breast cancer kindred. Oncogene 13**:**407-411. [PubMed] [Google Scholar]
- 23.Taylor, J. A., Y. Li, M. He, T. Mason, C. Mettlin, W. J. Vogler, S. Maygarden, and E. Liu. 1996. p53 mutations in bladder tumors from arylamine-exposed workers. Cancer Res. 56**:**294-298. [PubMed] [Google Scholar]
- 24.Unger, T., T. Juven-Gershon, E. Moallem, M. Berger, R. Vogt Sionov, G. Lozano, M. Oren, and Y. Haupt. 1999. Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J. 18**:**1805-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venot, C., M. Maratrat, C. Dureuil, E. Conseiller, L. Bracco, and L. Debussche. 1998. The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J. 17**:**4668-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogt Sionov, R., L. I. Hayon, and Y. Haupt. 2001. The regulation of p53 growth suppression, p. 106-125. In M. V. Blagosklonny (ed.), Cell cycle checkpoints and cancer. Landes Bioscience, Georgetown, Tex.
- 27.Walker, K. K., and A. J. Levine. 1996. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc. Natl. Acad. Sci. USA 93**:**15335-15340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wulf, G. M., Y. C. Liou, A. Ryo, S. W. Lee, and K. P. Lu. 2002. Role of Pin1 in the regulation of p53 stability and p21 transactivation, and cell cycle checkpoints in response to DNA damage. J. Biol. Chem. 277**:**47976-47979. [DOI] [PubMed] [Google Scholar]
- 29.Zacchi, P., M. Gostissa, T. Uchida, C. Salvagno, F. Avolio, S. Volinia, Z. Ronai, G. Blandino, C. Schneider, and G. Del Sal. 2002. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature 419**:**853-857. [DOI] [PubMed] [Google Scholar]
- 30.Zheng, H., H. You, X. Z. Zhou, S. A. Murray, T. Uchida, G. Wulf, L. Gu, X. Tang, K. P. Lu, and Z. X. Xiao. 2002. The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature 419**:**849-853. [DOI] [PubMed] [Google Scholar]
- 31.Zhu, J., J. Jiang, W. Zhou, K. Zhu, and X. Chen. 1999. Differential regulation of cellular target genes by p53 devoid of the PXXP motifs with impaired apoptotic activity. Oncogene 18**:**2149-2155. [DOI] [PubMed] [Google Scholar]