Sensitive Sequencing Method for KRAS Mutation Detection by Pyrosequencing (original) (raw)
Abstract
Both benign and malignant tumors represent heterogenous tissue containing tumor cells and non-neoplastic mesenchymal and inflammatory cells. To detect a minority of mutant KRAS alleles among abundant wild-type alleles, we developed a sensitive DNA sequencing assay using Pyrosequencing, ie, nucleotide extension sequencing with an allele quantification capability. We designed our Pyrosequencing assay for use with whole-genome-amplified DNA from paraffin-embedded tissue. Assessing various mixtures of DNA from mutant KRAS cell lines and DNA from a wild-type KRAS cell line, we found that mutation detection rates for Pyrosequencing were superior to dideoxy sequencing. In addition, Pyrosequencing proved superior to dideoxy sequencing in the detection of KRAS mutations from DNA mixtures of paraffin-embedded colon cancer and normal tissue as well as from paraffin-embedded pancreatic cancers. Quantification of mutant alleles by Pyrosequencing was precise and useful for assay validation, monitoring, and quality assurance. Our Pyrosequencing method is simple, robust, and sensitive, with a detection limit of approximately 5% mutant alleles. It is particularly useful for tumors containing abundant non-neoplastic cells. In addition, the applicability of this assay for DNA amplified by whole-genome amplification technique provides an expanded source of DNA for large-scale studies.
Screening and identification of gene mutations in benign and malignant tumors is commonly performed by polymerase chain reaction (PCR) amplification followed by dideoxy DNA sequencing or conformation-based separation such as single-strand conformation polymorphism (SSCP), denaturing gradient gel electrophoresis (DGGE), or high performance liquid chromatography (HPLC). Although SSCP, DGGE, and HPLC are useful screening methods, sequencing technologies remain the gold standard method to confirm and determine the identity of a specific mutation. Dideoxy DNA sequencing is the most commonly used sequencing method; however, it may not detect a minority of mutant sequences present in a background of abundant wild-type DNA sequence. Solid tumors are heterogenous tissue that typically contains tumor cells admixed with non-neoplastic cells, including inflammatory cells, vascular endothelial cells, and mesenchymal cells. Detection of gain-of-function mutations in oncogenes such as KRAS poses a particular challenge. Not only does each non-neoplastic cell in tumor tissue contribute two wild-type alleles, but also each tumor cell may carry one copy of its own wild-type allele. This problem is particularly evident in tumors with abundant desmoplastic stroma surrounding a minority of tumor cells, abundant inflammatory cells, or scanty tumor cells after chemotherapy or radiation therapy. A sensitive and cost-effective DNA sequencing assay that can be applied to a large number of samples is clearly needed.
Pyrosequencing (Biotage AB and Biosystems, Uppsala, Sweden) is real-time, nonelectrophoretic, nucleotide extension sequencing for various applications.1,2,3 In Pyrosequencing, pyrophosphate is generated only when a dispensed nucleotide anneals to the template and a primer extends by DNA polymerase. Subsequently, pyrophosphate is converted to fluorescence emission, the intensity of which is proportional to the amounts of annealed and extended nucleotide molecules. The Pyrosequencing assay for single-nucleotide polymorphisms (SNPs) can quantify the relative amount of each allele very accurately.1 It can also be applied to quantitative CpG island methylation analysis.4,5,6
In this study, we developed a sensitive sequencing assay for the detection of KRAS mutations using Pyrosequencing technology. Although Pyrosequencing typically reads at most 40 to 50 nucleotides, we demonstrate that it has higher analytical sensitivity for the detection of mutant DNA mixed in wild-type DNA when compared with dideoxy (eg, BigDye Terminator) sequencing. The detection limit of our KRAS Pyrosequencing was approximately 3 to 5% of mutant KRAS allele. Our Pyrosequencing method shows good precision, and its ability to quantify the amount of a mutant allele is very useful for assay validation and quality assurance. Moreover, our method effectively works on paraffin-embedded archival tumor tissue and on DNA samples after whole-genome amplification (WGA). High analytical sensitivity is particularly important to avoid bias caused by different features of tumors, such as tumors with abundant inflammatory cells seen in colon cancer with microsatellite instability, desmoplastic pancreatic cancer with cellular and abundant stroma, or previously treated tumors with only scanty remaining neoplastic cells. Our Pyrosequencing assay is a simple, robust, and high-throughput mutation detection assay that can be applied to large epidemiological studies and clinical trials.
Materials and Methods
Cell Lines, Paraffin-Embedded Tissue Blocks, Genomic DNA Extraction, and DNA Mixing
This study has been approved by the Dana-Farber/Harvard Cancer Center and Brigham and Women’s Hospital Institutional Review Boards. DNA was extracted using QIAmp DNA mini kit (Qiagen, Valencia, CA) and quantified from three cell lines; SW480 with the KRAS mutation c.35G>T (codon 12 GGT>GTT; p.Gly12Val), LoVo with the c.38G>A mutation (codon 13 GGC>GAC; p.Gly13Asp), and OVCAR with wild-type KRAS. DNA samples from each of the two _KRAS_-mutant cell lines were mixed with DNA samples from the KRAS wild-type cell line in various fractions of mutant DNA (50, 30, 20, 10, 5, 3, and 2%). All cell line DNA mixtures were prepared in duplicate. Paraffin-embedded tissue blocks of colon cancer with five different KRAS mutations [c.35G>T (codon 12 GGT>GTT; p.Gly12Val), c.35G>A (codon 12 GGT>GAT; p.Gly12Asp), c.34G>T (codon 12 GGT>TGT; p.Gly12Cys), c.34G>A (codon 12 GGT>AGT; p.Gly12Ser), and c.38G>A (codon 13 GGC>GAC; p.Gly13Asp)] were collected and anonymized from participants in the Nurses’ Health Study and Health Professionals Follow-Up Study who developed incident colorectal cancer.7 Tumor tissue on glass slides was manually dissected, and genomic DNA was extracted using QIAmp DNA mini kit (Qiagen). Each of these five colon cancer DNA samples with different KRAS mutations was mixed with DNA from normal paraffin tissue in various fractions of cancer DNA (50, 30, 20, 10, and 5%). Paraffin blocks of pancreatic cancers (nine cases) were randomly selected and anonymized from the archival files of the Department of Pathology at Brigham and Women’s Hospital and Massachusetts General Hospital. These pancreatic cancers had not been analyzed for KRAS mutations. Pancreatic cancer DNA was analyzed without mixing.
WGA, PCR, and Dideoxy (BigDye Terminator) Sequencing
Whole genome amplification (WGA) is a useful technique to preserve original study material for many different assays and for future studies. In WGA, genomic DNA is amplified by PCR using primers consisting of random sequence of 15 nucleotides.8 We have extensively used this technique and have shown that WGA does not significantly affect results of microsatellite analysis or sequencing of KRAS or BRAF by dideoxy sequencing (unpublished data). Each PCR mix contained 20 pmol of the random primers, 1.69 nmol each of dNTP, 2.5 mmol/L MgCl2, 1× PCR buffer (Applied Biosystems, Foster City, CA), 0.5 U of AmpliTaq Gold (Applied Biosystems), and 2 μl of template DNA solution in a total volume of 27 μl. PCR conditions consisted of initial denaturing at 94°C for 1 minute; 50 cycles of 94°C for 30 seconds, 37°C for 2 minutes, 41.5°C for 30 seconds, 46°C for 30 seconds, 50.5°C for 30 seconds, 55°C for 2 minutes, and 68°C for 30 seconds; and final extension at 72°C for 1 minute. PCR Primers for KRAS dideoxy sequencing were as follows: KRAS-F14, forward, 5′-tgt aaa acg acg gcc agt tgt gtg aca tgt tct aat ata gtc ac-3′; and KRAS-R7, reverse, 5′-aga atg gtc ctg cac cag taa-3′. Each PCR mix contained the forward and reverse primers (each 10 pmol), 1.69 nmol each of dNTP, 3 mmol/L MgCl2, 1× PCR buffer, 0.75 U of AmpliTaq Gold, and 2 μl of template WGA product in a total volume of 27 μl. PCR conditions consisted of initial denaturing at 94°C for 1 minute; 50 cycles of 95°C for 20 seconds, 50°C for 20 seconds, and 72°C for 40 seconds; and final extension at 72°C for 1 minute. The PCR products were purified using QIAquik PCR Purification kit (Qiagen). Cycle sequencing was performed using BigDye Terminator kit (Applied Biosystems), and analyzed by ABI 3730 (Applied Biosystems).
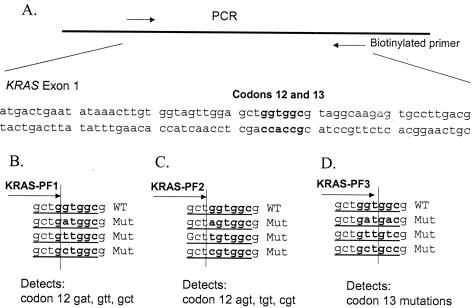
Development of KRAS Pyrosequencing Assay (Figure 1)
Figure 1.
KRAS Pyrosequencing assay design. A: PCR amplifies a segment of KRAS exon 1 containing codons 12 and 13. The reverse PCR primer is biotinylated. B: KRAS-PF1 Pyrosequencing primer can detect common codon 12 mutations. C: KRAS-PF2 Pyrosequencing primer can detect the rest of the codon 12 mutations. D: KRAS-PF3 Pyrosequencing primer can detect codon 13 mutations. WT, wild type; Mut, mutant.
PCR amplification primers for Pyrosequencing were as follows: KRAS-F, forward, 5′-nnn ggc ctg ctg aaa atg act gaa-3′; and KRAS-R, reverse biotinylated primer, 5′-tta gct gta tcg tca agg cac tct-3′. Each PCR mix contained the forward and reverse primers (each 20 pmol), 2.81 nmol each of dNTP, 3 mmol/L MgCl2, 1× PCR buffer, 1.25 U of AmpliTaq Gold, and 5 μl of template WGA product in a total volume of 50 μl. PCR conditions consisted of initial denaturing at 94°C for 1 minute; 50 cycles of 95°C for 20 seconds, 58°C for 20 seconds, and 72°C for 40 seconds; and final extension at 72°C for 1 minute. The PCR products were electrophoresed in an agarose gel to confirm successful amplification of the 82-bp PCR product. The PCR products (each 10 μl) were sequenced by Pyrosequencing PSQ96 HS System (Biotage AB) following the manufacturer’s instructions, using all three Pyrosequencing primers (see below). Nucleotide dispensation order was cyclic (CTAG from 5′ to 3′).
To increase sensitivity for the detection of all mutations in KRAS codons 12 and 13, we designed three slightly different Pyrosequencing primers, using software ADSW (Biotage AB) (Figure 1, B to D). One can use KRAS-PF2 primer to sequence all nucleotides of KRAS codons 12/13 and beyond; however, we found that the use of KRAS-PF1 and KRAS-PF3 primers increases the sensitivity of detection for specific KRAS mutations (data not shown). Moreover, the use of three different primers serves as a quality control measure because all codon 12/13 mutations, except for those in the first nucleotide of codon 12, can be detected by more than one primer. Pyrograms of wild-type KRAS codons 12 and 13 sequence by these primers are shown in Figure 2, A, D, and G. The primer KRAS-PF1 (5′-tgt ggt agt tgg agc tg-3′) ends at the first nucleotide of codon 12 to detect mutations in the second nucleotide of codon 12 [c.35G>A (codon 12 GAT), c.35G>T (codon 12 GTT), and c.35G>C (codon 12 GCT)] (Figure 1B). Because of cyclic CTAG nucleotide dispensation, all three mutations in the second nucleotide of codon 12 can be detected by dispensation of the first three nucleotides CTA with the highest signal-to-noise ratio (Figure 2, B and C). Pyrosequencing using KRAS-PF1 for the c.35G>T (codon 12 GTT) mutation is particularly sensitive, because the c.35G>T mutation (codon 12 GTT) creates a T dinucleotide run so that the first T peak on a Pyrogram by KRAS-PF1 primer is twice as high as a peak for one nucleotide change such as c.35G>A.
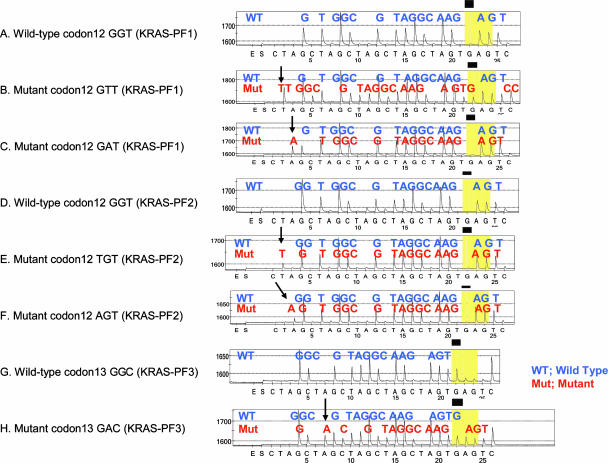
Figure 2.
Pyrograms of wild-type and mutant KRAS. A: Wild-type codon 12 by the KRAS-PF1 primer. B: c.35G>T (codon 12 GTT) mutation by the KRAS-PF1 primer. C: c.35G>A (codon 12 GAT) mutation by the KRAS-PF1 primer. D: Wild-type codon 12 by the KRAS-PF2 primer. E: c.34G>T (codon 12 TGT) mutation by the KRAS-PF2 primer. F: c.34G>A (codon 12 AGT) mutation by the KRAS-PF2 primer. G: Wild-type codon 13 by the KRAS-PF3 primer. H: c.38G>A (codon 13 GAC) mutation by the KRAS-PF3 primer. Arrows indicate the presence of mutant alleles. Note that B, C, E, F, and H show the presence of wild-type sequence, which is derived from non-neoplastic cells in tumor (and a normal allele in tumor cells).
The primer KRAS-PF2 (5′-tgt ggt agt tgg agc t-3′) ends at the third nucleotide of codon 11 to detect mutations in the first nucleotide of codon 12 [c.34G>A (codon 12 AGT), c.34G>T (codon 12 TGT), and c.34G>C (codon 12 CGT)] (Figure 1C). Because of cyclic CTAG nucleotide dispensation, all three mutations in the first nucleotide of codon 12 can be detected by the first three nucleotides CTA with the highest signal-to-noise ratio (Figure 2, E and F). The primer KRAS-PF3 (5′-tgg tag ttg gag ctg gt-3′) ends at the third nucleotide of codon 12 to detect all mutations in codon 13 [particularly c.38G>A (codon 13 GAC)] with high sensitivity (Figures 1D,2H).
Comparison of Analytical Sensitivities of Pyrosequencing and BigDye Terminator Sequencing and Statistical Analysis
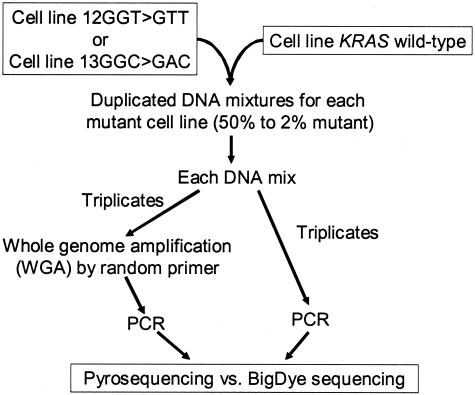
For cell line DNA mixing study (Figure 3), each mutant cell line DNA sample (SW480 or LoVo) was mixed with a wild-type DNA sample (OVCAR) in dilutions of 50, 30, 20, 10, 5, 3, and 2%. All cell line DNA mixtures were prepared in duplicate. All cell line DNA mixtures were amplified by WGA in triplicate. Subsequently, three aliquots from each triplicate WGA were independently amplified by PCR for KRAS and sequenced by Pyrosequencing and BigDye Terminator methods. In addition, three aliquots from each original DNA mixture without WGA were independently amplified by PCR for KRAS and sequenced by Pyrosequencing and BigDye Terminator methods. Thus, there were a total of 24 [= 2 cell lines × 2 duplicate × (3 + 3) aliquots] independent results by both sequencing methods for each dilution.
Figure 3.
Cell line DNA mixing study.
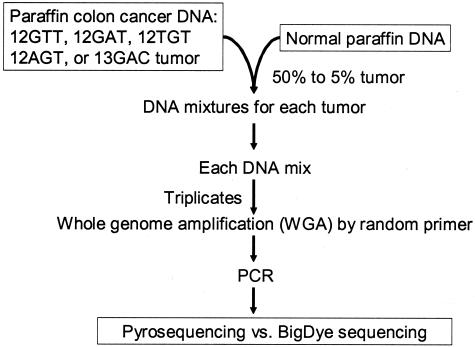
For paraffin colon cancer mixing study (Figure 4), each mutant DNA was mixed with normal DNA in dilutions of 50, 30, 20, 10, and 5%. From each DNA mixture and from each pancreatic cancer sample with unknown KRAS status, DNA was amplified by WGA in triplicate, and three aliquots (each from triplicate WGA) were subsequently amplified by PCR for KRAS, followed by Pyrosequencing and BigDye sequencing. Thus, for paraffin colon cancer mixing study, there were 15 (= 5 samples with different mutations × 3 aliquots) independent results by both sequencing methods for each dilution. For pancreatic cancers, there were 27 (= 9 tumors × 3 aliquots) independent results. Results of both sequencing methods were compared. Statistical analysis was performed using Fisher’s exact test on categorical data by SAS program (version 8.2; SAS Institute, Cary, NC).
Figure 4.
Paraffin colon cancer DNA mixing study.
Results
Precision and Reproducibility of Allele Quantification by KRAS Pyrosequencing
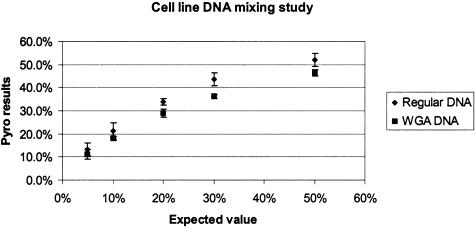
One of the advantages of Pyrosequencing is its ability to easily quantify amounts of a mutant allele or a SNP variant. To demonstrate assay precision and reproducibility, we quantified KRAS mutant alleles by Pyrosequencing in the various mixtures of DNA extracted from cell lines that contained mutated and wild-type KRAS, respectively (50, 30, 20, 10, and 5% mutant mixtures). Results are shown in Figure 5. SDs of the mutant allele quantifications in six repeated runs for each DNA mixing ratio ranged from 0.8 to 3.1%. WGA appeared to decrease the amounts of the mutant alleles detected by Pyrosequencing compared with KRAS Pyrosequencing without WGA, indicating subtle, but consistent bias introduced by WGA. Nonetheless, allele quantifications by Pyrosequencing assay were very precise and reproducible, even when whole-genome-amplified DNA samples were used. Allele quantification by Pyrosequencing allows us to perform quality control and quality assurance easily and objectively.
Figure 5.
Allele quantifications by KRAS Pyrosequencing. The horizontal axis shows expected value of the fraction of mutant KRAS based on various cell line DNA mixtures. The vertical axis shows observed quantification results by Pyrosequencing. “Regular DNA” indicates results without WGA. The vertical bar from each dot indicates ±1 SD.
Comparison of KRAS Pyrosequencing and BigDye Sequencing Using Mixed DNA Samples (Figure 6)
Figure 6.
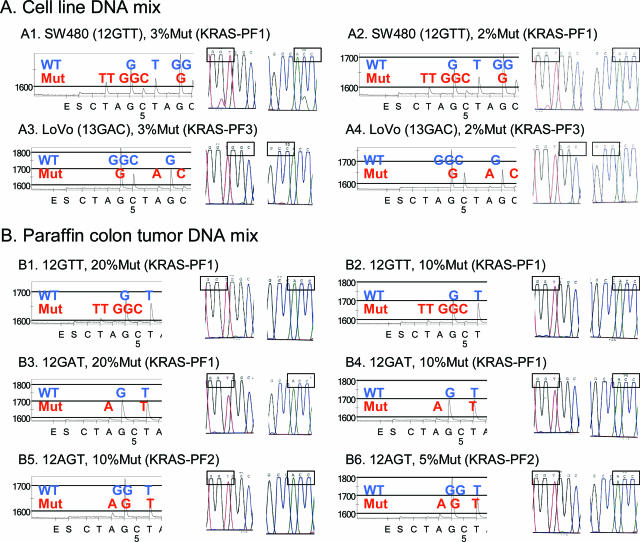
Comparison of Pyrosequencing and BigDye Terminator sequencing. A: Cell line DNA mixing study. A1: The c.35G>T (codon 12 GTT) mutation can be observed by both methods. A2: The c.35G>T (codon 12 GTT) mutation in the 2% mix can easily be observed by Pyrosequencing (left) but is subtle in BigDye sequencing (right). A3 and A4: The c.38G>A (codon 13 GAC) mutation can barely be observed by Pyrosequencing (left) but cannot be detected by BigDye sequencing (right). B: Paraffin colon tumor DNA mixing study. B1 and B2: The c.35G>T (codon 12 GTT) mutation can be barely observed by Pyrosequencing (left) but cannot be detected by BigDye sequencing (right). B3 and B4: The c.35G>A (codon 12 GAT) mutation can barely be observed by Pyrosequencing (left) but cannot be detected by BigDye sequencing (right). B5 and B6: The c.34G>A (codon 12 AGT) mutation can readily be observed by Pyrosequencing (left) but cannot be detected by BigDye sequencing (right).
To test analytical sensitivity of our KRAS Pyrosequencing method, we performed both Pyrosequencing and BigDye Terminator sequencing for KRAS on the same set of DNA mix samples containing various amounts of known KRAS mutation. Figure 6A shows representative results of cell line mixing study on the 3 and 2% mixes. In the cell line mixing study, there were no or insignificant differences between BigDye sequencing and Pyrosequencing when examining mixtures in which the proportion of mutant DNA was either high or very low (ie, mixtures of 50, 30, and 20% as well as 3 and 2%). However, there were significant differences in detection rates in the 24 runs for the 10 and 5% mutant mixtures (Table 1). Particularly, in the 5% mix, BigDye sequencing could detect mutations only in 8 of 24 runs, whereas Pyrosequencing could detect mutations in 23 of 24 runs (P = 0.000008).
Table 1.
Results of Cell Line DNA Mixing Study
| Fraction of mutant DNA | Diagnosis | Dideoxy (BigDye) sequencing | Pyrosequencing |
|---|---|---|---|
| 50% | Correct | 23/24 (96%) | 24/24 (100%) |
| Wild type (incorrect) | 0/24 (0%) | 0/24 (0%) | |
| Indeterminate | 1/24 (4%) | 0/24 (0%) | |
| 30% | Correct | 23/24 (96%) | 24/24 (100%) |
| Wild type (incorrect) | 0/24 (0%) | 0/24 (0%) | |
| Indeterminate | 1/24 (4%) | 0/24 (0%) | |
| 20% | Correct | 21/24 (88%) | 24/24 (100%) |
| Wild type (incorrect) | 0/24 (0%) | 0/24 (0%) | |
| Indeterminate | 3/24 (12%) | 0/24 (0%) | |
| 10% | Correct | 16/24 (67%)* | 24/24 (100%)* |
| Wild type (incorrect) | 0/24 (0%) | 0/24 (0%) | |
| Indeterminate | 8/24 (33%) | 0/24 (0%) | |
| 5% | Correct | 8/24 (33%)† | 23/24 (96%)† |
| Wild type (incorrect) | 4/24 (17%) | 0/24 (0%) | |
| Indeterminate | 12/24 (50%) | 1/24 (4%) | |
| 3% | Correct | 5/24 (21%) | 11/24 (46%) |
| Wild type (incorrect) | 14/24 (58%) | 9/24 (38%) | |
| Indeterminate | 5/24 (21%) | 4/24 (17%) | |
| 2% | Correct | 1/24 (4%) | 4/24 (17%) |
| Wild type (incorrect) | 21/24 (88%) | 16/24 (67%) | |
| Indeterminate | 2/24 (8%) | 4/24 (17%) |
When we repeated our analyses using whole-genome-amplified DNA, we found virtually identical results to those we obtained using the DNA mixtures without WGA. For the 5% mutant mixtures, Pyrosequencing detected mutations in 11 (92%) of 12 runs with whole-genome-amplified DNA and in 12 (100%) of 12 runs without WGA. In contrast, BigDye sequencing detected mutations in 7 (58%) of 12 runs with whole-genome-amplified DNA and in 9 (75%) of 12 runs without WGA. For the 3% mutant mixtures, Pyrosequencing detected mutations in 6 (50%) of 12 runs with whole-genome-amplified DNA and in 5 (42%) of 12 runs without WGA. In contrast, BigDye sequencing detected mutations in only 2 (17%) of 12 runs with whole-genome-amplified DNA and in 3 (25%) of 12 runs without WGA. Thus, the use of WGA did not significantly affect sensitivity of detection by Pyrosequencing or BigDye sequencing.
In the paraffin colon cancer mixing study, we repeated WGA reactions in triplicate using each DNA mixture as a template, followed by specific PCR for KRAS. There were no significant differences between BigDye sequencing results and Pyrosequencing results for the 50 and 30% mutant mixtures. However, for both the 20 and 10% mutant mixtures, significant differences were evident (Table 2). Figure 6B shows representative Pyrograms and corresponding BigDye sequencing results on the 20, 10, and 5% mutant mixtures. Pyrosequencing detected mutations in all (100%) of the 15 runs for the 20 and 10% mixtures, whereas BigDye sequencing detected mutations in only 8 (53%) of the 15 runs for the 20% mixture (P = 0.006 for the comparison between Pyrosequencing and BigDye sequencing) and in only 6 (40%) of the 15 runs for the 10% mixture (P = 0.0007). For the 5% mixture, Pyrosequencing could still detect mutations in 10 (67%) of the 15 runs, whereas BigDye sequencing could detect mutations in only 7 (47%) of the 15 runs; however, this difference did not reach the level of statistical significance.
Table 2.
Results of Paraffin Colon Cancer Tissue DNA Mixing Study
| Fraction of cancer DNA | Diagnosis | Dideoxy (BigDye) sequencing | Pyrosequencing |
|---|---|---|---|
| 50% | Correct | 15/15 (100%) | 15/15 (100%) |
| Wild type (incorrect) | 0/15 (0%) | 0/15 (0%) | |
| Indeterminate | 0/15 (0%) | 0/15 (0%) | |
| 30% | Correct | 14/15 (93%) | 15/15 (100%) |
| Wild type (incorrect) | 0/15 (0%) | 0/15 (0%) | |
| Indeterminate | 1/15 (7%) | 0/15 (0%) | |
| 20% | Correct | 8/15 (53%)* | 15/15 (100%)* |
| Wild type (incorrect) | 5/15 (33%) | 0/15 (0%) | |
| Indeterminate | 2/15 (13%) | 0/15 (0%) | |
| 10% | Correct | 6/15 (40%)† | 15/15 (100%)† |
| Wild type (incorrect) | 6/15 (40%) | 0/15 (0%) | |
| Indeterminate | 3/15 (20%) | 0/15 (0%) | |
| 5% | Correct | 7/15 (47%) | 10/15 (67%) |
| Wild type (incorrect) | 6/15 (40%) | 5/15 (33%) | |
| Indeterminate | 2/15 (3%) | 0/15 (0%) |
Detection of KRAS Mutations in Paraffin-Embedded Pancreatic Cancer Samples
For pancreatic cancer samples, we repeated WGA reactions in triplicate using template DNA extracted from each pancreatic tumor, followed by specific PCR for KRAS. Among all nine cases, Pyrosequencing demonstrated concordant results for each set of triplicates, whereas BigDye sequencing showed such triplicate concordant results in only five of nine cases. Of the nine cases, seven cases had KRAS mutations. By Pyrosequencing, KRAS mutations were identified in 21 of 21 runs for the seven mutant tumors, whereas for BigDye sequencing, KRAS mutations were identified in only 14 of 21 runs (P = 0.01 for the comparison of Pyrosequencing with BigDye sequencing). These results suggest that for pancreatic carcinoma, in which tumor cells tend to be sparsely admixed in desmoplastic stroma, Pyrosequencing appeared more sensitive for the detection of KRAS mutations than BigDye sequencing.
Discussion
Pyrosequencing is nonelectrophoretic nucleotide extension sequencing for various applications,1,9 including SNP genotyping,2 bacterial strain typing,3 mutation detection in tumor,10,11 and quantitative CpG island methylation analyses.4,5,6 In this study, we developed a sensitive sequencing assay for the detection of KRAS mutations using Pyrosequencing technology. Pyrosequencing reads a relatively shorter length of nucleotides (up to 40 to 50) than dideoxy sequencing. However, the relatively shorter reading length of Pyrosequencing as compared with dideoxy sequencing does not represent any real disadvantage for KRAS sequencing, because KRAS mutations are concentrated in codons 12 and 13. We demonstrate that Pyrosequencing has a higher analytical sensitivity (a lower detection limit) than dideoxy (BigDye) sequencing for the detection of mutant KRAS mixed with wild-type KRAS DNA. Pyrosequencing offered significantly greater sensitivity across a variety of mutant/wild-type mixtures for DNA derived from paraffin-embedded tissue.
WGA procedures have the potential to provide an unlimited source of DNA for large-scale genetic studies. We have optimized our Pyrosequencing methods so that we can use whole-genome-amplified DNA from paraffin-embedded tissue. By allele quantification, we demonstrated that fractions of mutant KRAS slightly decreased in whole-genome-amplified DNA samples, indicating that WGA reactions amplify the KRAS wild-type allele slightly better than the KRAS mutant allele (Figure 5). However, allele quantification on whole-genome-amplified DNA samples was still very precise, and mutation detection rates for whole-genome-amplified DNA samples were not significantly different from DNA samples that did not undergo WGA. Pyrosequencing showed superior sensitivities to dideoxy sequencing for both whole-genome-amplified DNA samples and those without WGA, including DNA samples from paraffin-embedded tissue. We have used PCR-based random oligonucleotide primed WGA (also known as primer extension preamplification) described by Zhang et al.12 Dietmaier et al8 showed that this method could effectively be used for paraffin-embedded tissue. Of note, Dean et al13 also described WGA by multiple displacement amplification (MDA) using phi 29 DNA polymerase that works optimally at room temperature and can amplify even high molecular weight DNA.13 WGA by MDA has a potential for use in common clinical molecular assays.14 However, the MDA WGA method failed to efficiently amplify DNA extracted from formalin-fixed paraffin-embedded tissue (unpublished data). Regardless of techniques, the use of WGA is potentially useful in both research and clinical settings, because it spares original samples and because many biomarkers can be tested even when the amounts of samples are limited.
A number of alternative KRAS mutation detection methods have been described in the literature. Similar to Pyrosequencing, single-nucleotide extension methods such as solid-phase minisequencing15,16,17 can accurately measure mutant allele relative to wild-type allele. However, the use of radioisotope greatly limits clinical usefulness of solid-phase minisequencing assay. Moreover, the technique is limited to single-nucleotide reading. Pyrosequencing represents a similar nucleotide extension technology with nonradioisotopic fluorescence detection platform that can read up to 40 to 50 nucleotides.
In addition, a variety of nonsequencing methods for KRAS mutation detection have been developed. Such methods include PCR-reverse dot blot,18 allele-specific oligonucleotide hybridization,19 activated RAS-GTP specific biosensor,20 real-time allele specific PCR,21 PCR followed by denaturing HPLC,22 oligonucleotide ligation assay,23 PCR-SSCP,24 PCR-DGGE,25 restriction endonuclease-mediated selective PCR,26 and PCR followed by HPLC-electrospray ionization tandem mass spectrometry.27 Innovative approaches for the detection of somatic mutations in general have been reviewed.28,29,30,31 Although extreme sensitivity can be achieved, a general problem with nonsequencing methods is that it is difficult or impossible to confirm independently the existence of any mutations that are identified.32 In this regard, sequencing methods to determine the exact identity of a specific mutation have a definitive advantage. Compared with nonsequencing methods, sequencing methods such as dideoxy sequencing and Pyrosequencing can serve as confirmatory tests to identify a specific mutation, even a rare variant. In fact, nonsequencing methods may have difficulty in detecting rare mutations, including two mutations in one allele, unless one uses an expanded panel of primers or probes to detect those rare variants. Furthermore, sequencing methods can demonstrate DNA sequence around specific nucleotide(s) of interest, providing a quality assurance measure that is especially important in clinical settings.
Digital PCR described by Vogelstein and Kinzler32 does offer the capability of quantifying mutant allele very accurately. However, its application to a large-scale study is currently limited because of the cost and labor-intensive nature of this technology. Alternatively, the array-based KRAS mutation detection system described by Prix et al33 uses peptide nucleic acid-mediated PCR clamping followed by biochip array hybridization. Nonetheless, this method may fail to detect some rare mutations. Moreover, the applicability of this technique to paraffin-embedded tissue and its cost effectiveness and limited throughput remain important issues.
One considerable advantage of Pyrosequencing is its ability to accurately quantify the amount of each allele. This is particularly useful in assay validation, quality control, and quality assurance, as we validated our KRAS Pyrosequencing by demonstrating good assay precision and reproducibility in quantifying the KRAS mutant allele in each one of the DNA mixtures. Allele quantification by Pyrosequencing allows objective quality control and quality assurance, and assay performance can be easily monitored by appropriate controls.
Another advantage of Pyrosequencing is that it can read a nucleotide sequence starting from the first nucleotide right next to a Pyrosequencing primer, so that one can design relatively small PCR products. Designing smaller PCR products is especially useful for degraded DNA samples, particularly DNA derived from paraffin-embedded tissue, in which DNA is typically fragmented into short fragments.
Compared with dideoxy sequencing, we believe that the Pyrosequencing assay offers simplicity and cost effectiveness, particularly in the setting of large-scale projects and clinical assays. Whereas dideoxy (BigDye Terminator) sequencing requires purification of PCR products, there is no need for a separate purification step with Pyrosequencing after PCR products are obtained. Thus, KRAS Pyrosequencing may be more cost effective than dideoxy (BigDye Terminator) sequencing in terms of both reagents and labor time.
As an alternative to Pyrosequencing, laser capture microdissection technique can collect pure population of tumor cells to increase sensitivity for DNA sequencing. However, performing laser capture microdissection is time consuming and labor intensive and yields less of DNA than manual tissue dissection, thereby limiting a menu of biomarkers that can be investigated.
Previous Pyrosequencing methods for NRAS mutation detection showed a detection limit of 15% of mutation-to-wild type ratio (ie, ∼13% of mutant allele among total alleles).34 This detection limit cannot be simply compared with that of our KRAS Pyrosequencing, because the detection limit is influenced by many different factors including specimens, the sequence to be analyzed (eg, KRAS vs. NRAS), primer design, the number of PCR cycles, and PCR bias. In fact, we showed evidence of a slight WGA PCR bias in favor of the wild-type KRAS allele. Consistent PCR bias may have some impact on gene copy number determinations in quantitative molecular genetic assays.35 Nonetheless, because of our carefully designed Pyrosequencing primer set, our KRAS Pyrosequencing assay achieves remarkable sensitivity of detecting approximately 5% of mutant KRAS alleles. Our data indicate that Pyrosequencing offers a superior analytical sensitivity when compared with dideoxy (BigDye Terminator) sequencing.
In conclusion, we developed a simple, robust, and sensitive sequencing assay for KRAS mutation detection using high-throughput Pyrosequencing technology. Our assay can be used for paraffin-embedded archival tissue in clinical settings and large population-based studies. The use of WGA can significantly limit the amounts of DNA required for analysis and allows many biomarkers to be tested on the same sample set.
Note Added in Proof
Nucleotide dispensation orders have been further optimized as follows (from 5′ to 3′): ATCG ATCG … for the KRAS-PF1 and KRAS-PF2 primers, and G ATGC ATGC … for the KRAS-PF3 primer.
Footnotes
Supported by National Institutes of Health grants P01-9483703, P01-9467802, and R01-9485602 and by Biotage AB and Biosystems.
References
- Fakhrai-Rad H, Pourmand N, Ronaghi M. Pyrosequencing: an accurate detection platform for single nucleotide polymorphisms. Hum Mutat. 2002;19:479–485. doi: 10.1002/humu.10078. [DOI] [PubMed] [Google Scholar]
- Chen DC, Saarela J, Nuotio I, Jokiaho A, Peltonen L, Palotie A. Comparison of GenFlex Tag array and Pyrosequencing in SNP genotyping. J Mol Diagn. 2003;5:243–249. doi: 10.1016/S1525-1578(10)60481-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan JA, Butchko AR, Beth Durso M. Use of Pyrosequencing of 16S rRNA fragments to differentiate between bacteria responsible for neonatal sepsis. J Mol Diagn. 2005;7:105–110. doi: 10.1016/s1525-1578(10)60015-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- Uhlmann K, Brinckmann A, Toliat MR, Ritter H, Nurnberg P. Evaluation of a potential epigenetic biomarker by quantitative methyl-single nucleotide polymorphism analysis. Electrophoresis. 2002;23:4072–4079. doi: 10.1002/elps.200290023. [DOI] [PubMed] [Google Scholar]
- Tost J, Dunker J, Gut IG. Analysis and quantification of multiple methylation variable positions in CpG islands by Pyrosequencing. Biotechniques. 2003;35:152–156. doi: 10.2144/03351md02. [DOI] [PubMed] [Google Scholar]
- Fuchs CS, Willett WC, Colditz GA, Hunter DJ, Stampfer MJ, Speizer FE, Giovannucci EL. The influence of folate and multivitamin use on the familial risk of colon cancer in women. Cancer Epidemiol Biomarkers Prev. 2002;11:227–234. [PubMed] [Google Scholar]
- Dietmaier W, Hartmann A, Wallinger S, Heinmoller E, Kerner T, Endl E, Jauch KW, Hofstadter F, Ruschoff J. Multiple mutation analyses in single tumor cells with improved whole genome amplification. Am J Pathol. 1999;154:83–95. doi: 10.1016/S0002-9440(10)65254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaghi M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 2001;11:3–11. doi: 10.1101/gr.11.1.3. [DOI] [PubMed] [Google Scholar]
- Garcia CA, Ahmadian A, Gharizadeh B, Lundeberg J, Ronaghi M, Nyren P. Mutation detection by Pyrosequencing: sequencing of exons 5–8 of the p53 tumor suppressor gene. Gene. 2000;253:249–257. doi: 10.1016/s0378-1119(00)00257-2. [DOI] [PubMed] [Google Scholar]
- Ahmadian A, Lundeberg J, Nyren P, Uhlen M, Ronaghi M. Analysis of the p53 tumor suppressor gene by Pyrosequencing. Biotechniques. 2000;28:140–144, 146–147. doi: 10.2144/00281rr02. [DOI] [PubMed] [Google Scholar]
- Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc Natl Acad Sci USA. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, Driscoll M, Song W, Kingsmore SF, Egholm M, Lasken RS. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra R, Medeiros LJ. Isothermal multiple displacement amplification: a highly reliable approach for generating unlimited high molecular weight genomic DNA from clinical specimens. J Mol Diagn. 2004;6:236–242. doi: 10.1016/S1525-1578(10)60516-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihalainen J, Siitari H, Laine S, Syvanen AC, Palotie A. Towards automatic detection of point mutations: use of scintillating microplates in solid-phase minisequencing. Biotechniques. 1994;16:938–943. [PubMed] [Google Scholar]
- Ihalainen J, Taavitsainen M, Salmivaara T, Palotie A. Diagnosis of pancreatic lesions using fine needle aspiration cytology: detection of K-ras point mutations using solid phase minisequencing. J Clin Pathol. 1994;47:1082–1084. doi: 10.1136/jcp.47.12.1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunio T, Reima I, Syvanen AC. Preimplantation diagnosis by whole-genome amplification, PCR amplification, and solid-phase minisequencing of blastomere DNA. Clin Chem. 1996;42:1382–1390. [PubMed] [Google Scholar]
- Albitar M, Wu WI, Feltz E, Jin G, Hirsch-Ginsberg C, Kantarjian H, Beran M. Simplified reverse dot blot analyses for detecting of ras oncogene mutations. Mol Diagn. 1997;2:169–176. doi: 10.1054/MODI00200169. [DOI] [PubMed] [Google Scholar]
- Marchetti A, Buttitta F, Pellegrini S, Chella A, Bertacca G, Filardo A, Tognoni V, Ferreli F, Signorini E, Angeletti CA, Bevilacqua G. Bronchioloalveolar lung carcinomas: k-ras mutations are constant events in the mucinous subtype. J Pathol. 1996;179:254–259. doi: 10.1002/(SICI)1096-9896(199607)179:3<254::AID-PATH589>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Becker CF, Hunter CL, Seidel RP, Kent SB, Goody RS, Engelhard M. A sensitive fluorescence monitor for the detection of activated Ras: total chemical synthesis of site-specifically labeled Ras binding domain of c-Raf1 immobilized on a surface. Chem Biol. 2001;8:243–252. doi: 10.1016/s1074-5521(01)00003-5. [DOI] [PubMed] [Google Scholar]
- Clayton SJ, Scott FM, Walker J, Callaghan K, Haque K, Liloglou T, Xinarianos G, Shawcross S, Ceuppens P, Field JK, Fox JC. K-ras point mutation detection in lung cancer: comparison of two approaches to somatic mutation detection using ARMS allele-specific amplification. Clin Chem. 2000;46:1929–1938. [PubMed] [Google Scholar]
- Lilleberg SL, Durocher J, Sanders C, Walters K, Culver K. High sensitivity scanning of colorectal tumors and matched plasma DNA for mutations in APC, TP53, K-RAS, and BRAF genes with a novel DHPLC fluorescence detection platform. Ann NY Acad Sci. 2004;1022:250–256. doi: 10.1196/annals.1318.039. [DOI] [PubMed] [Google Scholar]
- Rothschild CB, Brewer CS, Loggie B, Beard GA, Triscott MX. Detection of colorectal cancer K-ras mutations using a simplified oligonucleotide ligation assay. J Immunol Methods. 1997;206:11–19. doi: 10.1016/s0022-1759(97)00078-1. [DOI] [PubMed] [Google Scholar]
- Emanuel JR, Damico C, Ahn S, Bautista D, Costa J. Highly sensitive nonradioactive single-strand conformational polymorphism: detection of Ki-ras mutations. Diagn Mol Pathol. 1996;5:260–264. doi: 10.1097/00019606-199612000-00006. [DOI] [PubMed] [Google Scholar]
- Keohavong P, Zhu D, Whiteside TL, Swalsky P, Bakker A, Elder EM, Siegfried JM, Srivastava S, Finkelstein SD. Detection of infrequent and multiple K-ras mutations in human tumors and tumor-adjacent tissues. Anal Biochem. 1997;247:394–403. doi: 10.1006/abio.1997.2100. [DOI] [PubMed] [Google Scholar]
- Ward R, Hawkins N, O’Grady R, Sheehan C, O’Connor T, Impey H, Roberts N, Fuery C, Todd A. Restriction endonuclease-mediated selective polymerase chain reaction: a novel assay for the detection of K-ras mutations in clinical samples. Am J Pathol. 1998;153:373–379. doi: 10.1016/S0002-9440(10)65581-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lleonart ME, Ramon y Cajal S, Groopman JD, Friesen MD. Sensitive and specific detection of K-ras mutations in colon tumors by short oligonucleotide mass analysis. Nucleic Acids Res. 2004;32:e53. doi: 10.1093/nar/gnh051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcombe D, Newton CR, Little S. Advances in approaches to DNA-based diagnostics. Curr Opin Biotechnol. 1998;9:602–608. doi: 10.1016/s0958-1669(98)80137-7. [DOI] [PubMed] [Google Scholar]
- Makrigiorgos GM. PCR-based detection of minority point mutations. Hum Mutat. 2004;23:406–412. doi: 10.1002/humu.20024. [DOI] [PubMed] [Google Scholar]
- Jurinke C, Oeth P, van den Boom D. MALDI-TOF mass spectrometry: a versatile tool for high-performance DNA analysis. Mol Biotechnol. 2004;26:147–164. doi: 10.1385/MB:26:2:147. [DOI] [PubMed] [Google Scholar]
- Gut IG. DNA analysis by MALDI-TOF mass spectrometry. Hum Mutat. 2004;23:437–441. doi: 10.1002/humu.20023. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci USA. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prix L, Uciechowski P, Bockmann B, Giesing M, Schuetz AJ. Diagnostic biochip array for fast and sensitive detection of K-ras mutations in stool. Clin Chem. 2002;48:428–435. [PubMed] [Google Scholar]
- Sivertsson A, Platz A, Hansson J, Lundeberg J. Pyrosequencing as an alternative to single-strand conformation polymorphism analysis for detection of N-ras mutations in human melanoma metastases. Clin Chem. 2002;48:2164–2170. [PubMed] [Google Scholar]
- Ogino S, Wilson RB. Quantification of PCR bias caused by a single nucleotide polymorphism in SMN gene dosage analysis. J Mol Diagn. 2002;4:185–190. doi: 10.1016/S1525-1578(10)60702-7. [DOI] [PMC free article] [PubMed] [Google Scholar]