Intracellular trafficking of nucleic acids (original) (raw)
. Author manuscript; available in PMC: 2015 Mar 2.
Published in final edited form as: Expert Opin Drug Deliv. 2004 Nov;1(1):127–140. doi: 10.1517/17425247.1.1.127
Abstract
Until recently, the attention of most researchers has focused on the first and last steps of gene transfer, namely delivery to the cell and transcription, in order to optimise transfection and gene therapy. However, over the past few years, researchers have realised that the intracellular trafficking of plasmids is more than just a ‘black box’ and is actually one of the major barriers to effective gene delivery. After entering the cytoplasm, following direct delivery or endocytosis, plasmids or other vectors must travel relatively long distances through the mesh of cytoskeletal networks before reaching the nuclear envelope. Once at the nuclear envelope, the DNA must either wait until cell division, or be specifically transported through the nuclear pore complex, in order to reach the nucleoplasm where it can be transcribed. This review focuses on recent developments in the understanding of these intracellular trafficking events as they relate to gene delivery. Hopefully, by continuing to unravel the mechanisms by which plasmids and other gene delivery vectors move throughout the cell, and by understanding the cell biology of gene transfer, superior methods of transfection and gene therapy can be developed.
Keywords: cytoskeleton, gene delivery, non-viral gene therapy, nuclear transport, plasmid, trafficking, transfection
1. Introduction
In order to transfect a cell, either through viral or non-viral methods, a variety of both extracellular and intracellular barriers must be overcome before exogenous gene expression can occur. Included among these barriers are the plasma membrane, the cytoskeletal meshwork in the cytoplasm, and the nuclear envelope. Vectors must cross the plasma membrane, either by endocytosis and subsequent endosomal escape, or by direct penetration, move through the cytoplasm, and be transported into the nucleus before any transcription of their encoded genes can take place. Cellular entry of exogenous DNA is a topic that has been repeatedly examined and there are several reviews in this area covering transfection methods, such as cationic lipid-mediated delivery and electroporation [1–6]. This review will focus on two other areas that hinder successful gene therapy, namely cytoplasmic and nuclear trafficking of non-viral vectors.
2. Cytoplasmic trafficking
2.1 Does DNA diffuse through the cytoplasm?
Once DNA has entered the cell, it must somehow traverse the cytoplasm to reach and gain entry into the nucleus before it can be transcribed and the resulting mRNA eventually translated. Depending on cell shape and point of entry, the distance from the cell membrane to the cell nucleus can be quite large, ranging from µm in smaller cells to > 100s µm in larger eukaryotic cells. Indeed, specialised cells, such as neurons, even display cytoplasmic trafficking of solutes on the order of ≥ 1 mm [7].
The cytoplasm, described by R Chambers [8] in the 1940s as resembling a reversible gel-sol system, is now understood to be a complex system composed of multiple cytoskeletal elements, including microfilaments, microtubules and intermediate filaments (Figure 1). These elements are organised into a complex, crowded latticework that is constantly remodelling itself in response to a variety of internal and external stimuli. Cartoon representations of the cytoplasm of yeast as drawn to scale by Goodsell depict an extremely crowded compartment, with little room for solutes to travel through unhindered [9]. Translational diffusion measurements in cells suggest that the crowdedness of the cytoplasm in fibroblasts resembles a 12 – 13% dextran or Ficoll® solution [10], which would correlate to half of the total cytoplasmic protein being structural and not in solution [11]. Due to the interactions and interdependencies of cytoskeletal filaments with one another, the cytoplasmic volume may be better described as an aqueous gel rather than a homogeneous solution [11].
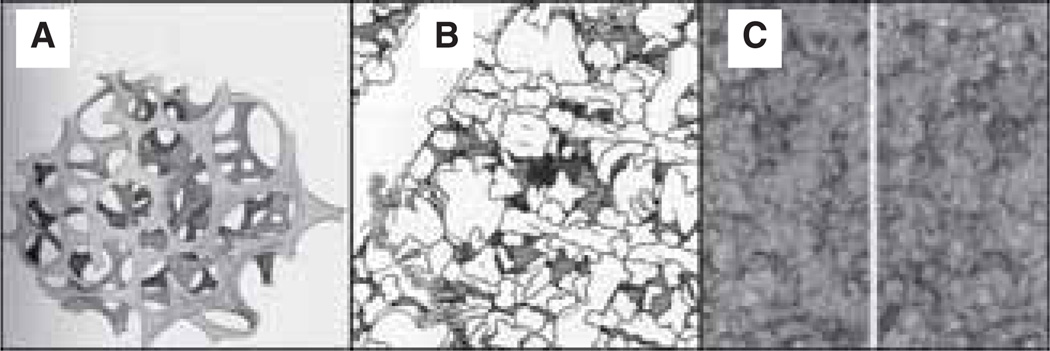
Figure 1. Drawings and micrographs demonstrating the cytoplasmic crowding caused by cytoskeletal elements.
(A) Drawing from the early 1900’s illustrating the reticular theory of protoplasmic arrangement. Reprinted from [120]. (B) Illustration demonstrating the crowdedness of baker’s yeast cytoplasm (1,000,000 × magnification), with components drawn to scale and at the correct concentrations. Microtubules (large rod in upper left corner), actin (smaller rods running horizontally throughout the panel) and intermediate filaments (medium-sized rod running diagonally along the right hand side) are all represented. Reprinted with permission from The machinery of life. GOODSELL DS, figure 5.2, page 68, (1993), Copyright © Springer-Verlag [9]. (C) High-voltage stereo electron micrographs (80,000 × magnification) depicting the structure of the cytoplasmic matrix in a thin margin of a cultured NRK cell. Reproduced from The Journal of Cell Biology, 1984, vol. 99(1,2), 3s–12s, figure 9, by copyright permission of The Rockefeller University Press [22].
NRK: Normal rat kidney.
The fluid-phase viscosity of the cytoplasm is 1.1 – 1.4 cP, as measured by time-resolved fluorescence anisotropy [12] or ratio imaging of a viscosity-sensitive fluorophore [13]. This is only slightly more dense than water. However, passive diffusion of either double-stranded DNA fragments [14] or macromolecule-sized solutes [15] in HeLa cells and fibroblasts is essentially non-existent. Microinjected DNA fragments > 2000 bp in length, or any macromolecule > 2000 kDa, show no translational diffusion through the cytoplasm; the ratio of the effective cytoplasmic diffusion coefficient to that in water (D/Do) for these molecules is < 0.01. The diffusion coefficient for a 2000 bp DNA fragment in water (Do) is in the order of 3 × 10−8 cm/s [15]. Work from Verkman’s [15] group determined that this decrease in macromolecular mobility was not due to solute sieving from scaffolding structures such as actin and microtubules. This is a surprising result given the previous data suggesting that molecular sieving of molecules increases as they get larger [16,17]. In contrast to the studies by Luby-Phelps [16,18], Verkman’s studies found that the degree of slowing in the cytoplasm was not decreased for solutes with a radius of gyration < 300 Å, which would correspond to a fluorescein-5-isothiocyanate–dextran molecule < 580 kDa. This result led them to conclude that there were no scaffolding structures with characteristic dimensions < ~ 250 Å. This agrees with measurements made on the smallest principle component of the cytoskeleton, actin, which has been shown to have a characteristic length of 1 µm and a persistence length of 7 – 20 µm, as measured by other groups [19–21]. This is also supported by whole-mount electron microscopy studies, which suggest that the cytoskeletal network has a mesh radius in the range of 350 – 500 Å to fill the cytoplasmic space [22]. For larger dextran molecules, Popov and Poo [17] found that they encountered disproportionately higher viscous resistances, and that these larger resistances were diminished when actin filaments were disrupted. However, actin filament disruption did not affect the diffusion rates of smaller dextrans. Taken together, these results certainly suggest that while molecular crowding may cause significant changes in the diffusion rates of small solutes in the cytoplasm, the diffusion of larger solutes, such as DNA, is significantly impeded by the cytoskeleton.
If plasmids are not moving through the cytoplasm by simple diffusion, they must travel toward the nucleus via some sort of active, or facilitative, process. Two potential pathways that have been studied are the routing of the DNA through lysosomes and endosomes to the nuclear periphery, and the directed movement via some interactions with the molecular motors associated with the cytoskeleton. In either case, exogenous DNA must be transported efficiently to the nucleus to avoid cytoplasmic degradation and, more importantly, to be expressed.
2.2 Intracellular transport of plasmid DNA taken up via endocytosis
Unprotected DNA is degraded within minutes by nucleases present in normal plasma [23]. Thus, plasmid DNA must be protected by encapsulation in viral or non-viral particles, or by condensation into tightly packed particulates formed by polycations, such as proteins or cationic lipid vesicles [24]. In this regard, liposomes both protect the DNA and mediate internalisation through endocytosis. As depicted in Figure 2, after capture, the plasmid must escape the endolysosomes by destabilising the membrane of the endolysosomal compartment [25]. The DNA must then dissociate from the lipid/DNA complex before it can enter the nucleus through the nuclear pore complex (NPC) [23]. Coonrod and colleagues [26] found that within 1 h of transfection using calcium phosphate precipitation, the DNA accumulated in the perinuclear region of both primary fibroblasts and HeLa cells. Similar results were obtained using lipofectin or electroporation, although nuclear accumulation occurred over a longer time scale. This data agrees with the work from Zabner’s [27] group, who found that lipid–DNA complexes aggregated into large perinuclear complexes that often showed a highly ordered tubular structure. Although both groups determined that the DNA aggregates into the perinuclear region of the cell, they have different opinions on how the DNA travels through the endolysosomal pathway. Coonrod and colleagues [26], using linear double-stranded DNA, found that regardless of the method of transfection (i.e., calcium phosphate precipitation, lipofection or electroporation), all of the exogenous DNA was routed through the endosomes and lysosomes, as determined by colocalisation of the labelled DNA with lysosomal markers [26]. They also found that inhibition of either endosomal fusion or lysosomal translocation was sufficient to prevent gene expression. This counteracts the theory proposed by Huang and others [27–29], which states that DNA escapes from the endosomes prior to fusion with the lysosome and then travels toward the nucleus. In this model, lipofectin acts as a destabilising agent aiding in the escape of DNA from the endosome prior to fusion with the lysosome. Without this intervention, the DNA would enter the lysosome resulting in massive DNA damage. Although there is some debate regarding the details of the endolysosomal pathway, it is clear that the liberation of the DNA–liposome complex is a rather inefficient operation, leaving a majority of the complex in the lysosome where it is degraded [30]. Regardless of the pathway for endocytic trafficking, the end result is that the DNA must escape from the vesicles and be liberated into the cytoplasm before nuclear import can occur. The only distinction between the models is the distance this ‘free’ DNA must travel in the cytoplasm to reach the nucleus.
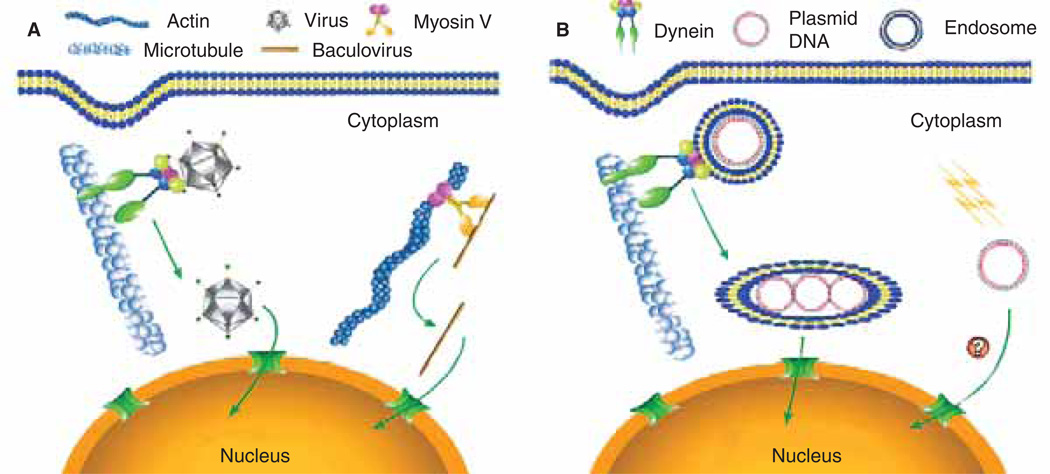
Figure 2. Models of cytoplasmic movement of exogenous DNA.
(A) Viruses such as adenovirus, HIV, parvovirus and HSV-1 use molecular motors such as the microtubule-based motor dynein to move their genomes toward the host cell’s nucleus, whereas others, such as baculovirus, use the actin cytoskeleton, perhaps through interactions with myosin family members. (B) Plasmid DNA complexed with liposomes is endocytosed and the endosomes are trafficked through interactions between the endosome and dynein, resulting in accumulation at the perinuclear region [26,27]. ‘Naked’ plasmid DNA entering the cytoplasm directly by either electroporation or microinjection is transported to the nucleus using an as yet unidentified pathway.
2.3 Cytoplasmic trafficking of viral DNA
Throughout evolution, viruses have developed many strategies to target their genomes to the nucleus of host cells (Figure 2) [31,32]. As with plasmids, once certain viruses enter their hosts, they too must traverse the cytoplasm to enter the nucleus for replication. Studies in recent years have pointed to the virus’ use of the cytoskeleton to facilitate transport toward the nucleus following endocytosis-mediated cell entry [33]. By fluorescently labelling viral particles or capsid proteins, several laboratories have been able to track viral movement through the cytoplasm toward the nucleus. For example, Hope and colleagues [32,34] tracked HIV though MAGI and HeLa cells by fusing viral protein R with green fluorescent protein and following the movement of the virion through the cytoplasm. They observed that the virus travelled by a curvilinear path and accumulated in the perinuclear region of the cell. Counter-staining of cytoskeletal filaments, or injecting the cells with fluorescently-labelled filament monomers, revealed that there was a high level of co-localisation between the virus particles and microtubules, but not actin filaments [34]. This movement was reduced by a monoclonal antibody that inhibits the dynein motor complex, suggesting that HIV uses the retrograde microtubule motor dynein to facilitate its transport toward the nucleus. Similar studies have been conducted by other groups, showing that a variety of viruses, such as adenovirus [35,36], canine parvovirus [37], and herpes simplex virus type 1 [38], all use microtubule networks and the dynein motor complex to transport their genomes toward the nucleus. In each case, disruption of the microtubule network through the use of depolymerising drugs, or inhibition of the retrograde motor dynein through the addition of an antibody against either an intermediate chain of the motor complex or dynactin, prevented accumulation of the virus particles at the nucleus of the host cell. Even though linear movement along the microtubule networks via molecular motors has been detected at speeds of ≤ 3 µm/s [36], it would appear that in the case of adenovirus, the particles do not travel via a direct route toward the nucleus, but rather are constantly switching between plus- and minus-end-directed motilities that ultimately leads to a net negative (toward the nucleus) movement along the microtubule at 0.06 µm/s.
Although microtubules appear to be the dominant highways for viruses, they are by no means the exclusive route. Van Loo and colleagues [39] showed that baculovirus uses the actin cytoskeletal network exclusively to traffic its genome toward the nucleus. Furthermore, although Hope [34] showed that HIV moved along microtubules via dynein, disruption of the microtubule network was not sufficient to stop the movement of the virus toward the nucleus. It was only when they disrupted both the microtubule and the actin network that they were able to abolish viral movement. Glotzer and colleagues [40] made similar findings with adenovirus.
2.4 Cytoplasmic trafficking of ‘free’ DNA
Although it is widely accepted that DNA complexed with a lipid vehicle enters the cell through endocytosis, and that it is possible that some, or all, of the trafficking through the cytoplasm is done via endosomes and/or lysosomes, it is unclear how plasmids move through the cytoplasm as non-membrane-bound molecules. Based on the above discussions, it is unlikely that the ‘free’ DNA diffuses through to the cytoplasm to any meaningful extent. However, it has not been determined whether this ‘free’ DNA can utilise the various cytoskeletal networks for directed movement towards the nucleus. What is known, however, is that once DNA is released into the cytoplasm, either following escape from the endosomes and the polymers with which it is complexed, or after direct entry by electroporation or a similar technique, the DNA does not remain ‘free’ for very long. The cytoplasm is filled with DNA- and RNA-binding proteins, as well as polyamines and other polycations, that will rapidly complex with the released DNA. This could serve several functions. First, the charge neutralisation could aid in condensation of the plasmid to reduce its size and perhaps allow some amount of limited diffusion. Second, the interactions may also shield the DNA from rapid degradation in the cytoplasm which would otherwise occur [23]. Finally, it is also possible that some of the proteins that bind to the DNA may in fact act as adapters to microtubule- and/or actin-based motors. As cytoplasmic cytoskeletal motor proteins do not bind to DNA, the only way that the plasmid could utilise the motors would be through intermediary proteins that can bind to the plasmid. This remains to be determined, but similar models for DNA-binding protein-mediated transport of plasmids into the nucleus support this possibility [41–46].
Taken together, these data suggest that cytoplasmic trafficking of viruses, and possibly other large macromolecules, results from a combination of motor-driven directed transport and molecular sieving. Thus, the fewer crosslinks and actin fibres that are in the way, the faster a particle can move along microtubules or vice versa. Whether plasmids that are free of viral capsids and proteins use similar pathways to reach the nucleus is unclear, but based on the extremely low rates of DNA diffusion in the cytoplasm, it is likely that some form of directed movement is used.
3. Nuclear trafficking
3.1 Nuclear envelope as a barrier
Once plasmids traverse the cytoplasm, they are confronted by the nuclear envelope, which regulates all traffic of macromolecules, including proteins, DNAs, RNAs and oligonucleotides, between the cytosol and the nucleus. Although some researchers have used cytoplasmic expression systems that rely on bacteriophage transcriptional machinery to express exogenous genes without the need for nuclear translocation of the plasmids [47–49], non-viral gene therapy is greatly limited by the low efficiency of plasmid DNA nuclear entry. Hence, it is important to understand, and exploit, the mechanisms by which plasmids enter the nucleus.
In non-dividing cells, the nuclear envelope remains intact and hence is a major barrier to gene transfer. It is well known that non-dividing cells (i.e., growth-arrested, confluent and primary cells) are difficult to transfect. In 1980, Capecchi and others [50] found that plasmids microinjected into the cytoplasm were incapable of gene expression, whereas those injected into the nucleus had high gene expression. When Graessman [51] injected 1000 – 2000 copies of a plasmid into the cytoplasm of cells, gene expression was < 3% of that seen when the nuclei of cells were injected with the same number of plasmids. Similar results have been seen in a number of different cell types by various groups [52,53]. The same is true for many viral genomes. Direct nuclear injection of DNA genomes from SV40, or reverse-transcribed retroviral genomes, results in the production of 10 – 100-fold more infectious virus particles than if the genomes are injected into the cytoplasm [54–56]. As gene expression or DNA replication must follow nuclear localisation of the DNA, these results suggested that nuclear translocation of DNA was a rate-limiting step in the gene transfer process.
In contrast to what is seen in non-dividing cells, it has been shown that in actively-dividing cells, the nuclear envelope is not a major hindrance to gene transfer. For example, in actively dividing primary human airway epithelial cells, gene expression following liposome-mediated transfection was 10 times greater, compared with non-dividing cells (as determined by BrdU Inc.) [57]. It has also been reported that cells transfected in the G2 or G2–M phase expressed 50 – 300-fold more gene product than those transfected in G1[58], suggesting that although efficient gene transfer and expression are regulated by the cell cycle, dividing cells in any phase of the cell cycle can be relatively efficiently transfected.
During transfection, only a fraction of the plasmids that enter the cytoplasm successfully enter the nucleus where gene expression is initiated. When HeLa and CV1 cells were transfected with fluorescently labelled plasmids using liposomes (~ 106 plasmids/cell), 2000 copies of plasmid were found to be intracellular in either cell type within 2 h [59]. Of the intracellular plasmids, 60% localised to the nuclei of HeLa cells, whereas only 30% of the plasmids did so in CV1 cells at the same time point. It should be pointed out that the plasmids used in these experiments were capable of targeting to the nucleus due to the inclusion of the SV40 enhancer (see below).
All these data show that the nuclear envelope is a major barrier to DNA nuclear import especially in non-dividing cells, but nature, or luck, has developed ways to break through this barrier.
3.2 DNA nuclear entry in the absence of cell division
Despite the barrier provided by the nuclear envelope, many viruses that utilise the nucleus during their life cycles are able to infect non-dividing cells, and plasmids have been delivered to non-dividing cells, or tissue made up of cells, that do not divide, or divide slowly. Thus, there must be mechanisms in place for this nuclear targeting.
Most retroviruses enter the nucleus during mitosis when the nuclear envelope temporarily breaks down, and, consequently, can only productively infect dividing cells, but many more viruses infect non-dividing cells. These include HIV, SV40, adenovirus, herpes simplex virus and hepatitis B virus, among others [31,60–63]. Furthermore, even certain bacterial pathogens have the ability to transfer DNA elements into the nuclei of non-dividing tissues, such as Agrobacterium tumefaciens in tobacco and other plants [64]. Each pathogen uses a slightly different approach to target their genomes to the nuclei of target cells, yet they all have the ability to get their DNA into the nucleus and all utilise the nuclear pore complex in a signal-mediated manner.
Plasmids are also routinely delivered to non-dividing cells in culture and to various tissues that are made up of cells that do not divide or divide very slowly [41,65–69]. Nuclear entry of plasmids in the absence of cell division was first directly observed after plasmids were microinjected into the cytoplasm of primary myotubes [65]. Wolff and colleagues [65] showed that the biotin-labelled plasmids were able to enter nuclei of myotubes and to direct gene expression. The nuclear entry of plasmids was a facilitated process that used nuclear pores. Nuclear localisation of the DNA, as followed by gene expression, was inhibited by co-injection of wheat germ agglutinin, a lectin that blocks nuclear transport of karyophilic proteins through the nuclear pore complex. This suggested either that karyophilic proteins containing nuclear localisation signals (NLSs) enabled the injected plasmids to enter nucleus, or that the DNA itself may have an intrinsic signal for nuclear pore recognition and entry.
3.3 Sequence-specific nuclear import of plasmid DNA
Using microinjection, Dean [41] found that nuclear import of plasmids in non-dividing cells is sequence-specific. When protein-free, purified SV40 DNA (5243 bp) was microinjected into the cytoplasm of TC7 African Green monkey kidney cells, the majority of plasmids were detected in cell nuclei by 6 – 8 h after injection by in situ hybridisation. In these non-dividing cells, whereas SV40 DNA localised in the nuclei with 8 h, other plasmids, including pBR322, pUC19 and pGL3-basic (Promega Corp.), did not enter the nuclei until the cells were allowed to divide [41,44]. This result suggestes that SV40 DNA containes some element that allows preferential nuclear import of the DNA. Indeed, when as little as 50 bp of the SV40 enhancer was cloned into these other plasmids, they were able to enter nuclei with the same kinetics as the entire SV40 genome [41,42]. This finding supports earlier studies by Graessman [51] in which the SV40 enhancer was postulated to have a ‘helper’ activity for nuclear localisation apart from the classical transcriptional activity attributed to it. Such sequence-specific nuclear targeting activity of the SV40 enhancer has been seen in all cells tested so far, including cultured cell lines derived from monkey, rat, mouse, hamster and human origin, and non-transformed primary cells from rat, chicken and human tissues [41]. This sequence from the SV40 genome has been termed the SV40 DNA nuclear targeting sequence, or SV40 DTS.
In vivo studies have also demonstrated that the SV40 DTS greatly increases gene transfer and reporter gene expression in muscle and the vasculature [67–70]. Incorporation of the SV40 enhancer into a plasmid delivered to mouse muscle by electroporation increased reporter gene expression < 20-fold [67,68]. Furthermore, gene expression was more persistent in mouse muscle with the plasmid containing the SV40 enhancer [68]. In the vasculature, Young et al. [71] showed that the presence of the SV40 DTS increased gene expression by 40 – 200-fold at different times following delivery by electroporation. Moreover, this increased gene expression was the result of increased DNA nuclear import. In situ hybridisation of the transferred plasmid confirmed that the SV40 sequence, containing the plasmid localised to the nuclei of cells in the vasculature by as early as 8 h following gene delivery, whereas plasmids without the SV40 DTS failed to localise to the nucleus and were cleared from the tissue within 48 h [71].
Despite these results, which suggests that the SV40 DTS is required in cell culture for nuclear import in the absence of cell division, and greatly increases nuclear localisation and gene expression in vivo in non-dividing tissues, plasmids lacking the SV40 enhancer are able to direct gene expression in skeletal muscle after either injection alone, or injection and electroporation [72–74]. Similarly, in studies in the vasculature, low levels of gene transfer and expression are detected with plasmids lacking the SV40 DTS [71]. The reason for this is likely to be that when large numbers of plasmids lacking a DTS are delivered to tissues, some of the intracellular plasmids can enter the nucleus with low efficiency and, thus, lead to gene expression. Indeed, it has been shown that when 100,000 DTS-lacking plasmids are delivered to the cytoplasm of a myotube in vivo, no gene expression is observed, but when 1,000,000 plasmids are injected, the cells express the gene product [75].
3.4 DNA nuclear targeting sequences
The mechanism by which the SV40 enhancer sequence causes nuclear import of DNA is not fully understood. The nuclear-targeting ability of SV40 enhancer is probably due to its binding sites for a number of general transcription factors, such as AP1, AP2, AP3, NF-κB, Oct-1, TEF-I and TEF-II [76–78]. These transcription factors are synthesised in the cytoplasm and all contain NLSs that facilitate their translocation from the cytoplasm to the nucleus. When exogenous SV40 DTS-containing DNA is present in the cytoplasm, newly synthesised transcription factors may bind to their binding sites on the plasmid to form a DNA–protein complex. Because transcription factors contain NLSs, they can function as ‘adaptors’ between the plasmid DNA and the protein nuclear import machinery to promote DNA nuclear import (Figure 3).
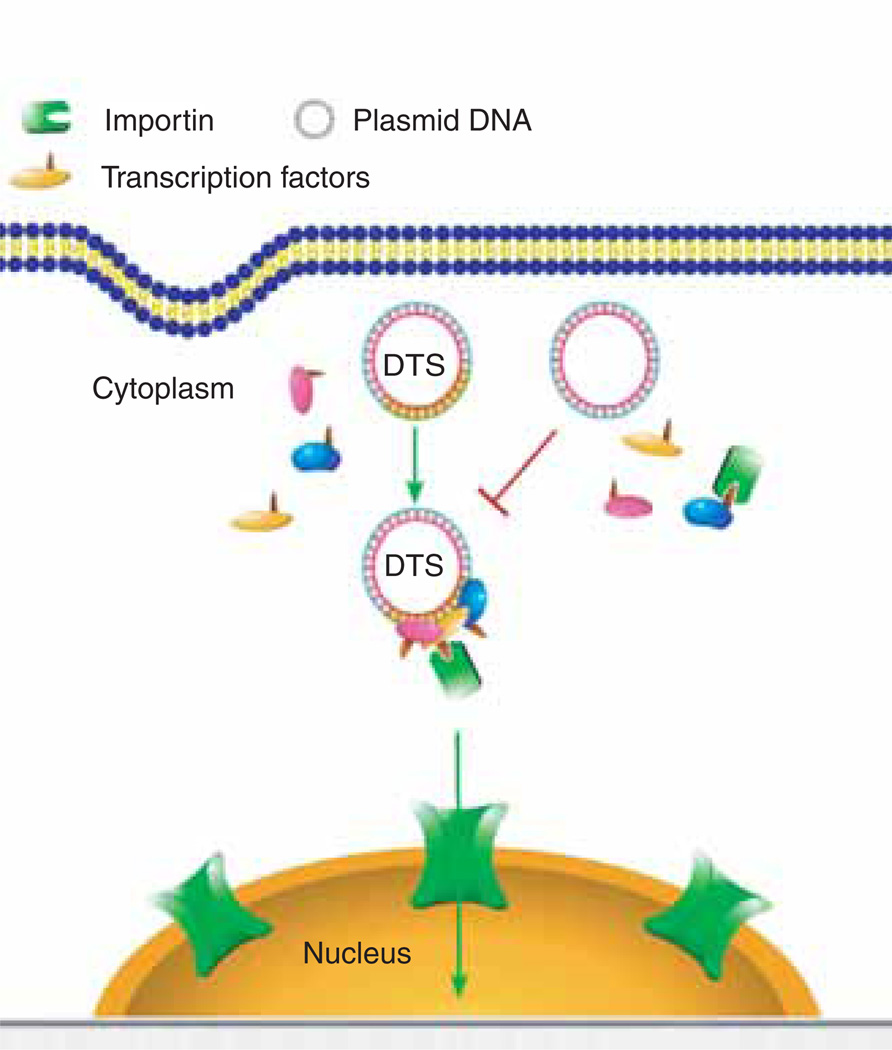
Figure 3. Model of sequence-specific plasmid nuclear import.
Plasmids containing a DTS (highlighted in yellow) bind to newly synthesised transcription factors and form a three-dimensional complex in cytoplasm. The nuclear localisation signals on these transcription factors can be recognised by members of the importin family to mediate plasmid nuclear entry. By contrast, plasmids lacking a DTS fail to form import-competent complexes.
DTS: DNA nuclear targeting sequence.
For transcription factors to be nuclear import ‘adaptors’, it is important that their NLS are still exposed after forming a DNA–protein complex. Otherwise, the complex would be unable to interact with importins, which are the NLS receptors. For example, zinc-finger transcription factors such as SP-1 would not be able to mediate DNA nuclear import because the NLS is embedded within the DNA-binding zinc-finger domain. Similarly, the NLS of the c-Jun component of the AP1 transcription factor would also be unable to bind to the importins because its NLS is immediately adjacent to the DNA binding region of the protein.
Based on this model, one possible prediction is that any eukaryotic promoter or enhancer will act as a DTS. However, this is not the case. Several strong viral promoters have been tested for their nuclear targeting ability, but none of them showed such activity. The immediate-early promoter and enhancer from cytomegalovirus, the Rous sarcoma virus long terminal repeats (LTR), the Moloney murine leukaemia virus LTR, and the herpes simplex virus thymidylate kinase promoter, are unable to mediate plasmid nuclear import in non-dividing cells [42,44,51]. By comparing the transcription factor binding sites on the SV40 enhancer and these other promoters, it was found that the only identified transcription factor that binds to the SV40 enhancer, but not to the other sequences, is AP2, suggesting that AP2 may play a major role in mediating plasmid nuclear import. However, plasmids containing a synthetic AP2 binding site, or an endogenous promoter containing an AP2 site, failed to localise to the nuclei of all cell types [44]. Hence, it appears that DNA nuclear import is not facilitated by a single transcription factor. Import is more likely to require multiple transcription factors bound to the plasmid to form a proper three-dimensional structure with the DNA sequence, so as to be recognised by the NLS-dependent, protein nuclear import machinery.
Binding sites for NF-κB may be another possible nuclear targeting sequence. When five repetitive NF-κB binding sites were cloned into a luciferase-expressing plasmid, gene expression increased 12-fold in transfected cells [45]. This increase was further enhanced when the NF-κB activator TNF-α was present. Nuclear import of the plasmid was also assessed in these experiments using a fluorescently-labelled peptide nucleic acid (PNA) bound to the plasmid. Similar to the enhancement in gene expression, nuclear localisation of the plasmids containing repetitive NF-κB binding sites was increased compared with that of the parent plasmids. In the presence of TNF-α, the plasmids containing repetitive NF-κB binding sites were mostly localised in nuclei, whereas TNF-α did not change the localisation pattern of the parent plasmids significantly. Because TNF-α activates NF-κB nuclear translocation, these data suggested a role of NF-κB binding sites in nuclear import of the modified plasmids. However, the plasmids used in this study (pGL3-enhancer, Promega) also contained the SV40 enhancer region. Hence, it is not clear if NF-κB bindings sites alone are sufficient to promote nuclear import.
The _ori_P sequence from Epstein-Barr virus (EBV) has also been proposed to act as a DTS, although only in combination with the EBV-encoded protein, EBV nuclear antigen (EBNA)-1 [46]. EBNA-1 is a DNA-binding protein, encoded by the virus that binds to a specific sequence of DNA present in the origin region of the virus. As with other transcription factors, EBNA-1 contains an NLS and binds to a unique DNA sequence. When a plasmid containing the _ori_P sequence was microinjected into the cytoplasm of EBNA-1-expressing cells, increased gene expression was detected compared with similar injections in EBNA-1 non-expressing cells. This led researchers to propose that the EBNA-1 protein was facilitating nuclear import of the plasmid. However, because expression was also increased, albeit to a lesser extent, in EBNA-1-positive cells injected in the nuclei, it is unclear exactly what contribution the potential increased nuclear import played.
3.5 Cell-specific DNA nuclear impor
Based on the DNA transcription factor nuclear import model proposed by Dean [42], it was postulated that cell-specific nuclear targeting sequences may exist. It is possible that binding of cell-specific transcription factors to a plasmid could lead to cell-specific nuclear import in cells, in which these particular transcription factors are present. At present, several such sequences have been identified, including one that acts in smooth muscle cells and another in endothelial cells [44,79].
One of the identified cell-specific nuclear targeting sequences is the smooth muscle γ-actin (SMGA) promoter. The SMGA gene is expressed in smooth muscle cells and is regulated at the transcriptional level by at least two transcription factors, serum response factor (SRF) and Nkx3 [80–82]. When injected into smooth muscle cells, plasmids carrying the SMGA promoter are imported into the nuclei [44]. By contrast, in non-smooth muscle cell types, such as endothelial or epithelial cells, the SMGA promoter plasmids are restricted to the cytoplasm. Thus, the SMGA promoter can function as a smooth muscle cell-specific DTS [44].
The minimal sequence of the SMGA promoter necessary for nuclear transport encompasses the first ~ 400 bp of the promoter from −404 to +25. This region contains binding sites for other general (C/EBP and AP2) and smooth muscle specific transcription factors (SRF and Nkx3)[80–82]. When a plasmid containing the 400 bp SMGA promoter was injected into the cytoplasm of non-smooth muscle cells (CV1 cells) that expressed SRF, some of the plasmids were able to localise to the nuclei of the cells, whereas they remained completely cytoplasmic in the parent, SRF-lacking CV1 cells [44]. However, the level of nuclear import was less than that seen in smooth muscle cells, suggesting that additional smooth muscle transcription factors, other than SRF, are needed for maximal nuclear import of the plasmid DNA. Indeed, detailed analysis has demonstrated that mutation of either of the two SRF or Nkx3 binding sites within the SMGA DTS abolishes nuclear import of the plasmids. Thus, it appears that both transcription factors, and possibly accessory binding proteins, are needed for optimal nuclear import of the DNA.
3.6 DNA nuclear import in permeabilised cells
In order to characterise the molecular mechanism(s) of DNA nuclear import and to identify the proteins involved, digitonin-permeabilised cells have been used by several groups [43,83,84]. Digitonin preferentially solubilises cholesterol, which is in greatest abundance in the plasma membrane and to a much lesser extent in the nuclear envelope. By washing detergent-treated cells, the endogenous cytoplasm can be removed, and by adding fractionated extracts and purified recombinant transport proteins into the system, the requirements for nuclear import can be determined [85].
Using this approach, Wolff and colleagues [83] first demonstrated that linear fragments of double-stranded DNA are transported into the nucleus in an energy- and temperature-dependent manner. DNA nuclear translocation was inhibited by wheat germ agglutinin, suggesting that the transport of DNA is mediated through the NPC. DNA nuclear import was also saturable, but was not competitively inhibited by excess NLS-containing proteins, suggesting that the linear DNA fragments entered the nucleus through a pathway distinct from that of classic NLS-containing proteins. Moreover, unlike all other macromolecules studied thus far, the nuclear import of these fragments was found to be independent of the addition of cytoplasmic extracts (and hence any NLS-machinery). The fluorescently-labelled linear DNA fragments that localised to the nuclei of permeabilised cells failed to localise to the nuclei of microinjected cells. Furthermore, in contrast to the observations that 5 – 15 kb plasmids can be imported into nuclei of microinjected cells [41,42,65], Hagstrom and colleagues [65] found that nuclear uptake of the linear DNA fragments in permeabilised cells was size dependent; fragments < 1000 bp localised to the nuclei, but larger fragments were excluded. There are two likely reasons for these findings. First, the method used to label the DNA fragments resulted in large numbers of fluorophores being attached randomly throughout the length of the DNA fragments. Thus, such saturation-labelling techniques may alter the ability of proteins to bind to the DNA that may normally mediate nuclear import in vivo. Perhaps by using different labelling techniques, a better correlation between microinjected and permeabilised cells would be achieved. Alternatively, it is possible that linear fragments of DNA behave differently to plasmids.
In separate experiments using plasmids, either fluorescently tagged with a triplex-forming PNA, or labelled at a distinct site by nick-translation, it was demonstrated that plasmid nuclear uptake was time-, energy-, and temperature-dependent [43,86]. The nuclear import was mediated by the NPC, as the addition of wheat germ agglutinin, or an antibody that inhibits NLS-dependent transport through the NPC (mAb414), completely abolished plasmid nuclear import. Nuclear entry of plasmids, as well as NLS-containing proteins was dependent on the addition of cytoplasmic extracts. When using purified importin-α, importin-β and Ran (proteins that mediate and control NLS-containing protein nuclear import), plasmids were excluded from the nuclei unless nuclear extract was also provided [43]. Moreover, plasmid nuclear import was also dependent on the presence of a DTS within the plasmid, as seen in microinjected cells [43]. The probable reason for this nuclear extract requirement is that the extract provides a source of transcription factors. Because purified importin-α, importin-β and Ran do not bind to DNA directly, DNA-binding, NLS-containing proteins must also be added to act as adapters between the DNA and the importins. Thus, using a method to label DNA at one distinct locus on the plasmid, DNA-binding proteins are able to bind to all other regions of the plasmid, including the SV40 DTS, to form a DNA–protein complex that is then transported into the nucleus. Taken together, these results support the DNA–protein complex model for plasmid nuclear import.
3.7 DNA/nuclear localisation signal–peptide conjugate and peptide nucleic acid technology
Even though the SV40 DTS uses the NLSs of endogenous transcription factors for its nuclear entry, exogenous proteins and peptides containing various NLSs have also been used to facilitate plasmid nuclear entry through the NPC-mediated pathway (Figure 4) [87,88]. High mobility group (HMG)-1, HMG-17 and histone H1 have been reported to increase gene expression when used as DNA packaging agents [89]. Similarly, a recombinant histone H1, containing the SV40 large T-antigen NLS, increased gene expression using cationic [90] or anionic [91] liposomes to transfect cells. However, when the NLS–H1–DNA complexes were microinjected into the cytoplasm, they showed no greater expression than DNA alone [90], suggesting that the effect of incorporation of NLS–H1 is to increase internalisation of the DNA–protein complex, rather than to increase nuclear import.
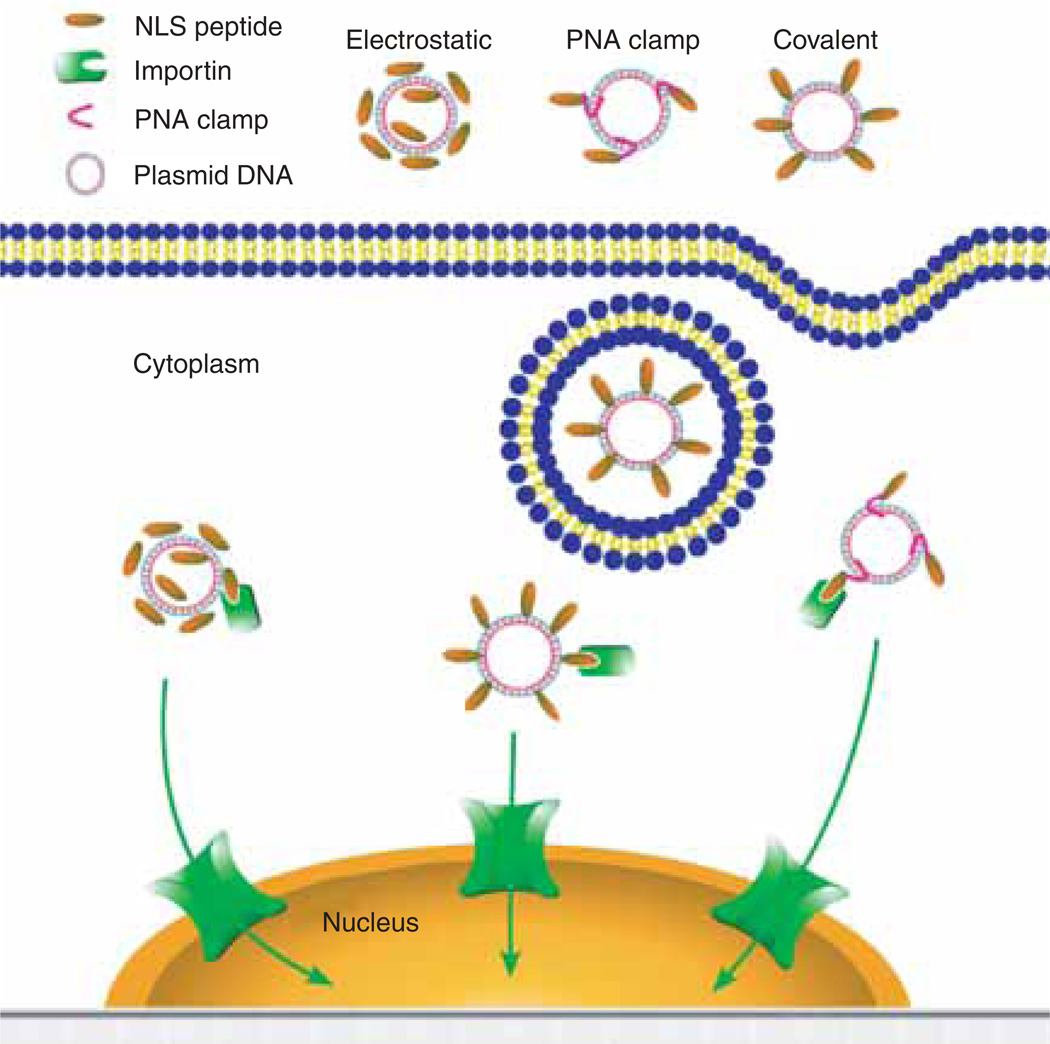
Figure 4. NLS peptide-mediated plasmid nuclear import.
Plasmids can be complexed with NLS peptides using different methods, including electrostatic interaction, PNA clamps or covalent conjugation. The various plasmid–NLS peptide complexes can be delivered to and internalised into cells by a variety of methods. Once in the cytoplasm, the NLS peptides complexed or bound to the plasmids can be recognised by the importin family members to facilitate plasmid nuclear entry.
NLS: Nuclear localisation signal; PNA: Peptide nucleic acid.
NLS peptides have also been used to increase nuclear import of plasmids, with varying success. Synthetic NLS peptides are easier to prepare and use compared with proteins, and thus, would be superior for drug delivery tools. The most commonly used nuclear targeting peptide is the SV40 T-antigen NLS. When synthetic SV40 T-antigen NLS peptides were complexed with plasmid DNA and microinjected into the cytoplasm of zebrafish embryos, increased nuclear localisation of the plasmids was observed, as was increased integration and expression of the transgene [92–95]. In a cell-free system, Collas and Alestrom [95] went on to demonstrate that the nuclear import of DNA–NLS peptide complexes is a two-step process involving binding to, and translocation across, the nuclear envelope. This process is energy-dependent, inhibited by agents that block the NPC, and requires cytoplasmic extracts. In more recent studies, NLS peptides have also been shown to increase gene expression of plasmids delivered to mammalian cells [96–100].
Because NLS peptides complexed to plasmids by electrostatic interactions (i.e., non-covalently attached) could become disassociated from the DNA in the cytoplasm, several groups have developed novel methods to covalently link NLS peptides to plasmids. Sebestyen et al. [84] observed an increase in nuclear localisation of plasmids covalently linked to an SV40 large T-antigen NLS peptide in digitonin-permeabilised cells. However, this increased nuclear localisation could not be recapitulated in cultured HeLa cells that were cytoplasmically microinjected. This could be explained by the heavy conjugation of peptides to the DNA that may interfere with the interaction with cellular proteins. Almost 200 NLS peptides were conjugated to a 4.8 kb plasmid, which could hinder plasmid nuclear import, rather than promoting it. Furthermore, covalently linked NLS peptides could also interfere with transcription. Using an indirect conjugating method or fewer peptides/plasmid, failed to increase gene expression significantly [101,102]. Another study, using linear-capped DNA, showed that a single NLS peptide is sufficient to increase nuclear translocation [98]. Their DNA construct was linked to an NLS via a base within the hairpin cap at one end of the DNA, and led to a 10- to 1000-fold increase in gene expression in different cell types. The fold difference in rapidly dividing cells was higher than that in non-dividing cells, suggesting that the NLS–DNA conjugate may increase aspects of the gene transfer and expression process other than by increased nuclear import alone.
PNAs are nucleic acid analogs in which the phosphodiester backbone has been replaced with a polyamide backbone made up of repeating _N_-(2-amnioethyl)glycine units [103,104]. The major advantage of this backbone substitution is that the resulting nucleic acid is resistant to both nucleases and proteases. One advantage is that PNAs can also form a triplex with specific sequences in DNA with high affinity (10−6 – 10−9 M)[103]. Another advantage is that the PNA-binding site can be placed anywhere in a plasmid (i.e., away from the promoter or DTS), so that it is less likely to interfere with transcription, or nuclear import, of the plasmid. As binding of PNAs to a plasmid does not change its supercoiled conformation, they are a highly attractive tool with which to attach peptides and other molecules to DNA [105,106].
Using PNA technology to link NLS peptides to plasmids has shown great potential to improve DNA delivery. When PNA–NLS peptides were complexed with plasmids and transfected into cells by polyethylenimine (PEI), they increased gene expression over uncomplexed DNA by up to eightfold [107]. The investigators concluded that the increase was due to the increase in DNA nuclear import. More recently, Branden et al. [108] hybridised the same PNA–NLS complex with fluorescently-labelled oligonucleotides and successfully delivered them to different tissues in mice. Other investigators have also shown the potential of PNAs to enhance gene delivery in different cell lines [109,110].
Although it is proposed that these NLS-containing proteins and peptides increase gene expression by increasing nuclear import of the plasmid–protein complex, no definitive experiments have been performed to test this. Consequently, as with the NLS–H1 and hairpin-attached NLS peptide examples [90,98], it is possible that they may act at other stages of the gene delivery process, such as DNA condensation, membrane attachment, or endosome release, or may reduce plasmid complex particle size to facilitate cytoplasmic movement, rather than directly increasing nuclear import.
3.8 Alternative routes of plasmid nuclear entry
Although most data suggest that plasmids enter the nucleus via a signal-mediated process through the NPC, several alternative pathways have been suggested. Most of the previous discussion has been concerned with the fate of free DNA released into the cytoplasm following either plasma membrane translocation (i.e., electroporation or microinjection) or endosomal release (i.e., following liposome- or polymermediated transfection). Central to this discussion is the assumption that the DNA is largely free from the complexing agent, especially in the case of chemical transfection methods. Indeed, some evidence suggests that interactions of cationic lipid–DNA complexes with free heparin may lead to release upon cell binding or internalisation [111], whereas other data point to interactions of the lipid complexes with the endosomal membrane for complex liberation [25]. Zabner demonstrated that when cells were transfected with DNA complexed with cationic lipids, most of the DNA–lipid complexes localised to large membrane-bound structures at the nuclear periphery [27]. Although not detected directly, Zabner concluded that at least some of the DNA must be released into the cytoplasm, based on the fact that gene expression was detected in these cells. They also injected liposome–DNA complexes directly into the cytoplasm or nucleus and found that at the ratio of lipid to DNA that gave optimal transfection, no gene expression could be detected when the complexes were injected into either cellular compartment. This further supports the assumption that at least some of the DNA is freed from the complexes once in the cytoplasm, and that this fraction of free DNA is likely to enter the nucleus and direct gene expression.
The polymer PEI has been used extensively to transfect a variety of cells. Several reports have looked at the intracellular trafficking of the PEI–DNA complexes to determine how the complexes localise to the nucleus [112–115]. In one report, using directly-labelled DNA and labelled PEI, researchers found that in contrast to what is typically seen with liposome complexes, PEI and DNA remain complexed in the cell and appear to localise to the nucleus coordinately [113]. Coupled with recent electron microscopy studies that suggest that PEI–DNA complexes can enter the nuclei as crystalline arrays, it is possible that PEI enters the nucleus by novel, unknown mechanisms [112–115].
Traub and colleagues [116] found that the cytoplasmic intermediate filament protein vimentin, can migrate to the nucleus when bound to single- or double-stranded oligonucleotides, facilitating their nuclear delivery. Similarly, when supercoiled plasmids, regardless of sequence, were bound to vimentin through the cryptic DNA-binding activity of this protein, they caused the rapid movement of the protein into the nucleus, suggesting that the supercoiled DNAs may have the ability to rapidly migrate into the nucleus [117]. However, others have not detected this activity [41–44,66].
Finally, theories regarding fusion of endosomes and/or lipid–DNA complexes with the nuclear envelope have been proposed over the years as a mechanism for nuclear entry of transfected DNA [118]. Similar mechanisms have also been suggested for the nuclear targeting of various viruses [119]. However, apart from the fact that transfected DNA appears to localise to the perinuclear region following endocytosis [26,27], there is no compelling evidence for entry of the DNA into the nucleus via direct nuclear envelope fusion. Given the double-membrane structure of the nuclear envelope, even if fusion between an endosomal membrane and the outer nuclear membrane bilayer occurred, the DNA would, at best, be delivered to the space residing between the two nuclear envelope bilayers, and would still be trapped ‘outside’ the nucleus.
4. Expert opinion and conclusion
Although the intracellular trafficking of exogenous DNA may not be a normal event in the cell, mechanisms do exist for its transport. Some of these have evolved over billions of years, as viruses and other pathogens have perfected ways to invade the host, even though others appear to be fortuitous piracy, as in the case of the SV40 enhancer, which binds to proteins on its way to the nucleus. Although it is unclear how plasmids move through the cytoplasm to the nuclear envelope, based on the viscosity and mesh-like structure of the cytoplasm, it is highly unlikely that this is a random event. More likely than not, it may turn out that plasmids, as with the genomes of viruses, bind to proteins that enable them to use the cytoskeletal highways and their associated motor proteins for transport, as depicted in Figure 2B. Once delivered to the nuclear envelope, NLS-containing proteins, provided by the host, that are bound to the DNA allow it to be translocated across the nuclear envelope into the nucleus. Whether these proteins bind before, during, or after cytoplasmic trafficking remains to be seen. Regardless, the end result is that the DNA is targeted to the nucleus where gene expression can occur. The goal of all gene therapy approaches is to target enough DNA to the nuclei of cells to obtain sufficient expression for a therapeutic effect. By characterising and understanding the mechanisms of the cytoplasmic trafficking and nuclear import of plasmids, we can overcome our relative inability to target substantial amounts of DNA to the nucleus, thus increasing transfection efficiency and, ultimately, gene therapy.
Acknowledgements
The authors’ research was supported in part by NIH grants HL59956, EY12962, HL71643 and HL76139 (RCG), and a grant from the Sandler Programme for Asthma Research.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Guy J, Drabek D, Antoniou M. Delivery of DNA into mammalian cells by receptor-mediated endocytosis and gene therapy. Mol. Biotechnol. 1995;3(3):237–248. doi: 10.1007/BF02789334. [DOI] [PubMed] [Google Scholar]
- 2.Bally MB, Harvie P, Wong FM, et al. Biological barriers to cellular delivery of lipid-based DNA carriers. Adv. Drug Deliv. Rev. 1999;38(3):291–315. doi: 10.1016/s0169-409x(99)00034-4. [DOI] [PubMed] [Google Scholar]
- 3.Weaver JC, Chizmadzhev YA. Theory of electroporation: a review. Bioelectrochemistry and Bioenergetics. 1996;41(2):135–160. [Google Scholar]
- 4.Niidome T, Huang L. Gene therapy progress and prospects: nonviral vectors. Gene Ther. 2002;9(24):1647–1652. doi: 10.1038/sj.gt.3301923. [DOI] [PubMed] [Google Scholar]
- 5.Lechardeur D, Lukacs GL. Intracellular barriers to non-viral gene transfer. Curr. Gene Ther. 2002;2(2):183–194. doi: 10.2174/1566523024605609. [DOI] [PubMed] [Google Scholar]
- 6.Wiethoff CM, Middaugh CR. Barriers to nonviral gene delivery. J. Pharm. Sci. 2003;92(2):203–217. doi: 10.1002/jps.10286. [DOI] [PubMed] [Google Scholar]
- 7.Vanden Berghe P, Hennig GW, Smith TK. Characteristics of intermittent mitochondrial transport in guinea pig enteric nerve fibers. Am. J. Physiol. Gastrointest Liver Physiol. 2004;286(4):671–682. doi: 10.1152/ajpgi.00283.2003. [DOI] [PubMed] [Google Scholar]
- 8.Chambers R. The micromanipulation of living cells. In: Moulton FR, editor. The cell and protoplasm. Washington, DC, USA: Science Press; 1940. pp. 20–30. [Google Scholar]
- 9.Goodsell DS. The machinery of life. New York: Springer-Verlag; 1993. p. 140. •• This book effectively illustrates the 3-dimensional complexity of cellular proteins and their interactions with one another.
- 10.Hou L, Lanni F, Luby-Phelps K. Tracer diffusion in F-actin and Ficoll mixtures. Toward a model for cytoplasm. Biophys. J. 1990;58(1):31–43. doi: 10.1016/S0006-3495(90)82351-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luby-Phelps K. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int. Rev. Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. •• This is a comprehensive review of the rheological properties of the cytoplasm that discusses the classical assumptions made about the cytoplasm and experimental results from several authors in this field.
- 12.Fushimi K, Verkman AS. Low viscosity in the aqueous domain of cell cytoplasm measured by picosecond polarization microfluorimetry. J. Cell Biol. 1991;112(4):719–725. doi: 10.1083/jcb.112.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luby-Phelps K, Mujumdar S, Mujumdar RB, et al. A novel fluorescence ratiometric method confirms the low solvent viscosity of the cytoplasm. Biophys. J. 1993;65(1):236–242. doi: 10.1016/S0006-3495(93)81075-0. • A novel technique using two indocyanine dyes to determine solute viscosity that is insensitive to other parameters such as temperature, hydrogen bonds, and most importantly large macromolecules. These experiments showed that the solvent viscosity of the cytoplasm is not radically different from water.
- 14.Lukacs GL, Haggie P, Seksek O, et al. Size-dependent DNA mobility in cytoplasm and nucleus. J. Biol. Chem. 2000;275(3):1625–1609. doi: 10.1074/jbc.275.3.1625. • A rigorous examination of DNA mobility through the cytoplasm, indicating that larger DNA fragments are unable to diffuse through the cytoplasm passively.
- 15.Seksek O, Biwersi J, Verkman AS. Translational diffusion of macromolecule-sized solutes in cytoplasm and nucleus. J. Cell Biol. 1997;138(1):131–412. doi: 10.1083/jcb.138.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luby-Phelps K, Castle PE, Taylor DL, Lanni F. Hindered diffusion of inert tracer particles in the cytoplasm of mouse 3T3 cells. Proc. Natl. Acad. Sci. USA. 1987;84(14):4910–4913. doi: 10.1073/pnas.84.14.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popov S, Poo MM. Diffusional transport of macromolecules in developing nerve processes. J. Neurosci. 1992;12(1):77–85. doi: 10.1523/JNEUROSCI.12-01-00077.1992. • This article indicates that, at least in the case of neurons, the cytoskeleton, specifically actin, can affect diffusion of large macromolecules.
- 18.Provance DW, Jr, Mcdowall A, Marko M, Luby-Phelps K. Cytoarchitecture of size-excluding compartments in living cells. J. Cell Sci. 1993;106(2):565–577. doi: 10.1242/jcs.106.2.565. [DOI] [PubMed] [Google Scholar]
- 19.Riveline D, Wiggins CH, Goldstein RE, Ott A. Elastohydrodynamic study of actin filaments using fluorescence microscopy. Physical Review E. 1997;56(2):1330–1333. [Google Scholar]
- 20.Isambert H, Maggs AC. Dynamics and rheology of actin solutions. Macromolecules. 1996;29(3):1036–1040. [Google Scholar]
- 21.Boal DH. Mechanics of the cell. Cambridge, UK: Cambridge University Press; 2002. p. 406. [Google Scholar]
- 22.Porter KR. The cytomatrix: a short history of its study. J. Cell Biol. 1984;99(1,2):3s–12s. doi: 10.1083/jcb.99.1.3s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lechardeur D, Sohn KJ, Haardt M, et al. Metabolic instability of plasmid DNA in the cytosol: a potential barrier to gene transfer. Gene Ther. 1999;6:482–497. doi: 10.1038/sj.gt.3300867. [DOI] [PubMed] [Google Scholar]
- 24.Hope MJ, Mui B, Ansell S, Ahkong QF. Cationic lipids, phosphatidylethanolamine and the intracellular delivery of polymeric, nucleic acid-based drugs (review) Mol. Membr. Biol. 1998;15(1):1–14. doi: 10.3109/09687689809027512. [DOI] [PubMed] [Google Scholar]
- 25.Xu Y, Szoka FCJ. Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry. 1996;35:5616–5623. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- 26.Coonrod A, Li FQ, Horwitz M. On the mechanism of DNA transfection: efficient gene transfer without viruses. Gene Ther. 1997;4(12):1313–1321. doi: 10.1038/sj.gt.3300536. •• A novel finding with evidence that gene delivery through the endosomal–lysosomal pathway may actually travel through the lysosome, offering an alterative pathway for plasmid trafficking through the cytoplasm.
- 27.Zabner J, Fasbender AJ, Moninger T, Poellinger KA, Welsh MJ. Cellular and molecular barriers to gene transfer by a cationic lipid. J. Biol. Chem. 1995;270:18997–19007. doi: 10.1074/jbc.270.32.18997. •• This is an outstanding article that was one of the first to dissect the gene transfer process of lipid-mediated gene transfer in cultured cells. A variety of techniques were employed to identify and partially characterise the rate-limiting steps in the gene transfer process.
- 28.Zhou X, Huang L. DNA transfection mediated by cationic liposomes containing lipopolylysine: characterization and mechanism of action. Biochim. Biophys. Acta. 1994;1189(2):195–203. doi: 10.1016/0005-2736(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 29.El Ouahabi A, Thiry M, Pector V, et al. The role of endosome destabilizing activity in the gene transfer process mediated by cationic lipids. FEBS Lett. 1997;414(2):187–192. doi: 10.1016/s0014-5793(97)00973-3. [DOI] [PubMed] [Google Scholar]
- 30.Zauner W, Kichler A, Schmidt W, Mechtler K, Wagner E. Glycerol and polylysine synergize in their ability to rupture vesicular membranes: a mechanism for increased transferrin-polylysine-mediated gene transfer. Exp. Cell Res. 1997;232(1):137–145. doi: 10.1006/excr.1997.3486. [DOI] [PubMed] [Google Scholar]
- 31.Whittaker GR. Virus nuclear import. Adv. Drug Deliv. Rev. 2003;55(6):733–747. doi: 10.1016/s0169-409x(03)00051-6. [DOI] [PubMed] [Google Scholar]
- 32.Campbell EM, Hope TJ. Role of the cytoskeleton in nuclear import. Adv. Drug Deliv. Rev. 2003;55(6):761–771. doi: 10.1016/s0169-409x(03)00049-8. • An excellent review on the cytoskeleton and its role in nuclear import of viral genomes.
- 33.Stidwill RP, Greber UF. Intracellular virus trafficking reveals physiological characteristics of the cytoskeleton. News Physiol. Sci. 2000;15:67–71. doi: 10.1152/physiologyonline.2000.15.2.67. [DOI] [PubMed] [Google Scholar]
- 34.Mcdonald D, Vodicka MA, Lucero G, et al. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 2002;159(3):441–452. doi: 10.1083/jcb.200203150. • An excellent article demonstrating the intracellular trafficking of HIV through cells. These experiments showed the dependence of HIV on the retrograde motor dynein for translocation through the cytoplasm to the nucleus.
- 35.Suomalainen M, Nakano MY, Keller S, et al. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 1999;144(4):657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leopold PL, Kreitzer G, Miyazawa N, et al. Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum. Gene Ther. 2000;11(1):151–165. doi: 10.1089/10430340050016238. • An excellent examination of microtubule motor dynamics and how various cytoskeletal inhibitors and stabilising agents affect adenoviral movement through the cytoplasm of A549 cells.
- 37.Suikkanen S, Aaltonen T, Nevalainen M, et al. Exploitation of microtubule cytoskeleton and dynein during parvoviral traffic toward the nucleus. J. Virol. 2003;77(19):10270–10279. doi: 10.1128/JVI.77.19.10270-10279.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sodeik B, Ebersold MW, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 1997;136(5):1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Loo ND, Fortunati E, Ehlert E, et al. Baculovirus infection of nondividing mammalian cells: mechanisms of entry and nuclear transport of capsids. J. Virol. 2001;75(2):961–970. doi: 10.1128/JVI.75.2.961-970.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glotzer JB, Michou AI, Baker A, Saltik M, Cotten M. Microtubule-independent motility and nuclear targeting of adenoviruses with fluorescently labeled genomes. J. Virol. 2001;75:2421–2434. doi: 10.1128/JVI.75.5.2421-2434.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dean DA. Import of plasmid DNA into the nucleus is sequence specific. Exp. Cell Res. 1997;230:293–302. doi: 10.1006/excr.1996.3427. • This was the first paper to demonstrate that the nuclear import of plasmids is a sequence-specific process.
- 42.Dean DA, Dean BS, Muller S, Smith LC. Sequence requirements for plasmid nuclear entry. Exp. Cell Res. 1999;253:713–722. doi: 10.1006/excr.1999.4716. •• This work characterised the DNA sequence elements that are required for plasmid nuclear import in non-dividing cells, and showed that whereas transcription factor binding sites within the SV40 enhancer are needed for nuclear import, not all promoters or enhancers containing binding sites for transcription factors can function to target plasmids to the nucleus.
- 43.Wilson GL, Dean BS, Wang G, Dean DA. Nuclear import of plasmid DNA in digitonin-permeabilized cells requires both cytoplasmic factors and specific DNA sequences. J. Biol. Chem. 1999;274:22025–22032. doi: 10.1074/jbc.274.31.22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vacik J, Dean BS, Zimmer WE, Dean DA. Cell-specific nuclear import of plasmid DNA. Gene Ther. 1999;6:1006–1014. doi: 10.1038/sj.gt.3300924. • This was the first paper to identify and characterise a cell-specific DNA nuclear import sequence that functions uniquely in one cell type.
- 45.Mesika A, Grigoreva I, Zohar M, Reich Z. A regulated, NFkappaB-assisted import of plasmid DNA into mammalian cell nuclei. Mol. Ther. 2001;3(5):653–657. doi: 10.1006/mthe.2001.0312. [DOI] [PubMed] [Google Scholar]
- 46.Langle-Rouault F, Patzel V, Benavente A, et al. Up to 100-fold increase of apparent gene expression in the presence of Epstein-Barr virus oriP sequences and EBNA1: implications of the nuclear import of plasmids. J. Virol. 1998;72(7):6181–6185. doi: 10.1128/jvi.72.7.6181-6185.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brisson M, Tseng WC, Almonte C, Watkins S, Huang L. Subcellular trafficking of the cytoplasmic expression system. Hum. Gene Ther. 1999;10(16):2601–2613. doi: 10.1089/10430349950016645. [DOI] [PubMed] [Google Scholar]
- 48.Brisson M, He Y, Li S, Yang JP, Huang L. A novel T7 RNA polymerase autogene for efficient cytoplasmic expression of target genes. Gene Ther. 1999;6(2):263–270. doi: 10.1038/sj.gt.3300827. [DOI] [PubMed] [Google Scholar]
- 49.Chen X, Li Y, Xiong K, Wagner TE. A self-initiating eukaryotic transient gene expression system based on contransfection of bacteriophage T7 RNA polymerase and DNA vectors containing a T7 autogene. Nucleic Acids Res. 1994;22(11):2114–2120. doi: 10.1093/nar/22.11.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Capecchi MR. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell. 1980;22(2 Pt 2):479–488. doi: 10.1016/0092-8674(80)90358-x. • This seminal work demonstrated that plasmids injected into the nuclei of cells express robustly, whereas the same plasmids injected into the cytoplasm of non-dividing cells fail to express their gene product. This was the first paper to demonstrate truly that transport of plasmids into the nucleus is perhaps the rate limiting step in gene delivery.
- 51.Graessman M, Menne J, Liebler M, Graeber I, Graessman A. Helper activity for gene expression, a novel function of the SV40 enhancer. Nucleic Acids Res. 1989;17:6603–6612. doi: 10.1093/nar/17.16.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mirzayans R, Remy AA, Malcom PC. Differential expression and stability of foreign genes introduced into human fibroblasts by nuclear versus cytoplasmic microinjection. Mutation Res. 1992;281:115–122. doi: 10.1016/0165-7992(92)90045-j. [DOI] [PubMed] [Google Scholar]
- 53.Thornburn AM, Alberts AS. Efficient expression of miniprep plasmid DNA after needle micro-injection into somatic cells. Biotechniques. 1993;1(4):356–358. [PubMed] [Google Scholar]
- 54.Graessman M, Graessman A. Regulation of SV40 gene expression. Adv. Cancer Res. 1981;35:111–149. doi: 10.1016/s0065-230x(08)60910-0. [DOI] [PubMed] [Google Scholar]
- 55.Kopchick JJ, Ju G, Skalka AM, Stacey DW. Biological activity of cloned retroviral DNA in microinjected cells. Proc. Natl. Acad. Sci. USA. 1981;78(7):4383–4387. doi: 10.1073/pnas.78.7.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakanishi A, Clever J, Yamada M, Li PL, Kasamatsu H. Association with capsid proteins promotes nuclear targeting of simian virus 40. Proc. Natl. Acad. Sci. USA. 1996;93:96–100. doi: 10.1073/pnas.93.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fasbender A, Zabner J, Zeiher BG, Welsh MJ. A low rate of cell proliferation and reduced DNA uptake limit cationic lipid-mediated gene transfer to primary cultures of ciliated human airway epithelia. Gene Ther. 1997;4:1173–1180. doi: 10.1038/sj.gt.3300524. [DOI] [PubMed] [Google Scholar]
- 58.Brunner S, Sauer T, Carotta S, et al. Cell cycle dependence of gene transfer by lipoplex, polyplex and recombinant adenovirus. Gene Ther. 2000;7:401–407. doi: 10.1038/sj.gt.3301102. • This paper demonstrates the cell cycle dependence of gene transfer.
- 59.James MB, Giorgio TD. Nuclear-associated plasmid, but not cell-associated plasmid, is correlated with transgene expression in cultured mammalian cells. Mol. Ther. 2000;1(4):339–346. doi: 10.1006/mthe.2000.0054. • This paper takes a quantitative approach to define the relative cellular and nuclear uptake of plasmids to begin to develop quantitative models for gene delivery.
- 60.Clever JL, Dean DA, Kasamatsu H. Identification of a DNA-binding domain within the simian virus 40 capsid proteins Vp2 and Vp3. J. Biol. Chem. 1993;268:20877–20883. [PubMed] [Google Scholar]
- 61.Kasamatsu H, Nakanishi A. How do animal DNA viruses get to the nucleus? Ann. Rev. Microbiol. 1998;52:627–686. doi: 10.1146/annurev.micro.52.1.627. [DOI] [PubMed] [Google Scholar]
- 62.Whittaker GR, Helenius A. Nuclear import and export of viruses and virus genomes. Virology. 1998;246(1):1–23. doi: 10.1006/viro.1998.9165. [DOI] [PubMed] [Google Scholar]
- 63.Cullen BR. Journey to the center of the cell. Cell. 2001;105:697–700. doi: 10.1016/s0092-8674(01)00392-0. [DOI] [PubMed] [Google Scholar]
- 64.Zupan J, Muth TR, Draper O, Zambryski P. The transfer of DNA from agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J. 2000;23(1):11–28. doi: 10.1046/j.1365-313x.2000.00808.x. [DOI] [PubMed] [Google Scholar]
- 65.Dowty ME, Williams P, Zhang G, Hagstrom JE, Wolff JA. Plasmid DNA entry into postmitotic nuclei of primary rat myotubes. Proc. Natl. Acad. Sci. USA. 1995;92:4572–4576. doi: 10.1073/pnas.92.10.4572. •• This was the first paper to demonstrate that plasmids can enter the nuclei of non-dividing cells by way of the nuclear pore complex.
- 66.Dean BS, Byrd JN, Jr, Dean DA. Nuclear targeting of plasmid DNA in human corneal cells. Cur. Eye Res. 1999;19:66–75. doi: 10.1076/ceyr.19.1.66.5344. [DOI] [PubMed] [Google Scholar]
- 67.Li S, Maclaughlin FC, Fewell JG, et al. Muscle-specific enhancement of gene expression by incorporation of the SV40 enhancer in the expression plasmid. Gene Ther. 2001;8:494–497. doi: 10.1038/sj.gt.3301419. [DOI] [PubMed] [Google Scholar]
- 68.Blomberg P, Eskandarpour M, Xia S, Sylven C, Islam KB. Electroporation in combination with a plasmid vector containing SV40 enhancer elements results in increased and persistent gene expression in mouse muscle. Biochem. Biophys. Res. Commun. 2002;298(4):505–510. doi: 10.1016/s0006-291x(02)02486-5. [DOI] [PubMed] [Google Scholar]
- 69.Young JL, Dean DA. Non-viral gene transfer strategies for the vasculature. Microcirculation Res. 2002;9:35–50. doi: 10.1038/sj/mn/7800120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martin JB, Young JL, Benoit JN, Dean DA. Gene transfer to intact mesenteric arteries by electroporation. J. Vasc. Res. 2000;37(5):372–380. doi: 10.1159/000025753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Young JL, Benoit JN, Dean DA. Effect of a DNA nuclear targeting sequence on gene transfer and expression of plasmids in the intact vasculature. Gene Ther. 2003;10(17):1465–1470. doi: 10.1038/sj.gt.3302021. • This paper showed that sequence-specific DNA nuclear import occurs in living animals, not just in cultured cells.
- 72.Wolff JA, Malone RW, Williams P, et al. Direct gene transfer into mouse muscle in vivo . Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. •• This work was the first to show that naked, uncomplexed plasmids could be injected into mouse skeletal muscle to drive robust gene expression. This finding and paper provided the basis for all subsequent studies on the use of naked DNA injection into muscle for DNA vaccines and other uses.
- 73.Wolff JA, Ludtke JJ, Acsadi G, Williams P, Jani A. Long-term persistence of plasmid DNA and foreign gene expression in mouse muscle. Hum. Mol. Genetics. 1992;1:363–369. doi: 10.1093/hmg/1.6.363. • This paper demonstrated that once in the nuclei of skeletal muscle cells, plasmids are retained in an episomal form and continue to express for up to 19 months in mice following direct DNA injection into muscle.
- 74.Wolff JA, Dowty ME, Jiao S, et al. Expression of naked plasmids by cultured myotubes and entry into T tubules and caveolae of mammalian skeletal muscle. J. Cell Sci. 1992;103:1249–1253. doi: 10.1242/jcs.103.4.1249. [DOI] [PubMed] [Google Scholar]
- 75.Utvik JK, Nja A, Gundersen K. DNA injection into single cells of intact mice. Hum. Gene Ther. 1999;10(2):291–300. doi: 10.1089/10430349950019075. [DOI] [PubMed] [Google Scholar]
- 76.Dynan WS, Chervitz SA. Characterization of a minimal simian virus 40 late promoter: enhancer elements in the 72-base-pair repeat not required. J. Virol. 1989;63:1420–1427. doi: 10.1128/jvi.63.3.1420-1427.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dynan WS, Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- 78.Wildeman AG. Regulation of SV40 early gene expression. Biochem. Cell Biol. 1988;66(6):567–577. doi: 10.1139/o88-067. [DOI] [PubMed] [Google Scholar]
- 79.Dean DA. Nucleocytoplasmic trafficking. In: Mahato RI, editor. Pharmaceutical perspectives of nucleic acid-based therapeutics. London, UK: Harwood Academic Publishers; 2002. pp. 229–260. [Google Scholar]
- 80.Browning CL, Culberson DE, Aragon IV, et al. The developmentally regulated expression of serum response factor plays a key role in the control of smooth muscle-specific genes. Dev. Biol. 1998;194(1):18–37. doi: 10.1006/dbio.1997.8808. [DOI] [PubMed] [Google Scholar]
- 81.Carson JA, Fillmore RA, Schwartz RJ, Zimmer WE. The smooth muscle gamma-actin gene promoter is a molecular target for the mouse bagpipe homologue, mNkx3-1, and serum response factor. J. Biol. Chem. 2000;275(50):39061–39072. doi: 10.1074/jbc.M006532200. [DOI] [PubMed] [Google Scholar]
- 82.Kovacs AM, Zimmer WE. Cell specific transcription of the smooth muscle γ-actin gene requires both positive and negative acting cis-elements. Gene Exp. 1998;7:115–129. [PMC free article] [PubMed] [Google Scholar]
- 83.Hagstrom JE, Ludtke JJ, Bassik MC, et al. Nuclear import of DNA in digitonin-permeabilized cells. J. Cell Sci. 1997;110:2323–2331. doi: 10.1242/jcs.110.18.2323. [DOI] [PubMed] [Google Scholar]
- 84.Sebestyén MG, Ludtke JL, Bassik MC, et al. DNA vector chemistry: the covalent attachment of signal peptides to plasmid DNA. Nature Biotech. 1998;16:80–85. doi: 10.1038/nbt0198-80. [DOI] [PubMed] [Google Scholar]
- 85.Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. • This paper established the permeabilised cell system for studying signal-mediated protein, RNA and DNA nuclear import and export.
- 86.Colin M, Moritz S, Fontanges P, et al. The nuclear pore complex is involved in nuclear transfer of plasmid DNA condensed with an oligolysine-RGD peptide containing nuclear localisation properties. Gene Ther. 2001;8(21):1643–1653. doi: 10.1038/sj.gt.3301572. [DOI] [PubMed] [Google Scholar]
- 87.Hebert E. Improvement of exogenous DNA nuclear importation by nuclear localization signal-bearing vectors: a promising way for non-viral gene therapy? Biol. Cell. 2003;95(2):59–68. doi: 10.1016/s0248-4900(03)00007-8. [DOI] [PubMed] [Google Scholar]
- 88.Escriou V, Carriere M, Scherman D, Wils P. NLS bioconjugates for targeting therapeutic genes to the nucleus. Adv. Drug Deliv. Rev. 2003;55(2):295–306. doi: 10.1016/s0169-409x(02)00184-9. [DOI] [PubMed] [Google Scholar]
- 89.Bottger M, Zaitsev SV, Otto A, Haberland A, Vorob’ev VI. Acid nuclear extracts as mediators of gene transfer and expression. Biochim. Biophys. Acta. 1998;1395(1):78–87. doi: 10.1016/s0167-4781(97)00128-0. [DOI] [PubMed] [Google Scholar]
- 90.Fritz JD, Herweijer H, Zhang G, Wolff JA. Gene transfer into mammalian cells using histone-condensed plasmid DNA. Hum. Gene Ther. 1996;7(12):1395–1404. doi: 10.1089/hum.1996.7.12-1395. [DOI] [PubMed] [Google Scholar]
- 91.Hagstrom JE, Sebestyen MG, Budker V, et al. Complexes of non-cationic liposomes and histone H1 mediate efficient transfection of DNA without encapsulation. Biochim. Biophys. Acta. 1996;1284(1):47–55. doi: 10.1016/0005-2736(96)00106-x. [DOI] [PubMed] [Google Scholar]
- 92.Collas P, Alestrom P. Nuclear localization signal of SV40 T antigen directs import of plasmid DNA into sea urchin male pronuclei in vitro . Mol. Reprod. Dev. 1996;45(4):431–438. doi: 10.1002/(SICI)1098-2795(199612)45:4<431::AID-MRD4>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 93.Collas P, Husebye H, Alestrom P. The nuclear localization sequence of the SV40 T antigen promotes transgene uptake and expression in zebrafish embryo nuclei. Transgenic Res. 1996;5(6):451–458. doi: 10.1007/BF01980210. [DOI] [PubMed] [Google Scholar]
- 94.Collas P, Alestrom P. Nuclear localization signals: a driving force for nuclear transport of plasmid DNA in zebrafish. Biochem. Cell Biol. 1997;75(5):633–640. [PubMed] [Google Scholar]
- 95.Collas P, Alestrom P. Rapid targeting of plasmid DNA to zebrafish embryo nuclei by the nuclear localization signal of SV40 T antigen. Mol. Mar. Biol. Biotechnol. 1997;6(1):48–58. [PubMed] [Google Scholar]
- 96.Aronsohn AI, Hughes JA. Nuclear localization signal peptides enhance cationic liposome-mediated gene therapy. J. Drug Target. 1998;5(3):163–169. doi: 10.3109/10611869808995871. [DOI] [PubMed] [Google Scholar]
- 97.Ludtke JJ, Zhang G, Sebestyen MG, Wolff JA. A nuclear localization signal can enhance both the nuclear transport and expression of 1 kb DNA. J. Cell Sci. 1999;112(12):2033–2041. doi: 10.1242/jcs.112.12.2033. [DOI] [PubMed] [Google Scholar]
- 98.Zanta MA, Belguise-Valladier P, Behr JP. Gene delivery: a single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleus. Proc. Natl. Acad. Sci. USA. 1999;96(1):91–96. doi: 10.1073/pnas.96.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Subramanian A, Ranganathan P, Diamond SL. Nuclear targeting peptide scaffolds for lipofection of nondividing mammalian cells. Nat. Biotechnol. 1999;17(9):873–877. doi: 10.1038/12860. [DOI] [PubMed] [Google Scholar]
- 100.Chan CK, Jans DA. Enhancement of polylysine-mediated transferrinfection by nuclear localization sequences: polylysine does not function as a nuclear localization sequence. Hum. Gene Ther. 1999;10(10):1695–1702. doi: 10.1089/10430349950017699. [DOI] [PubMed] [Google Scholar]
- 101.Ciolina C, Byk G, Blanche F, et al. Coupling of nuclear localization signals to plasmid DNA and specific interaction of the conjugates with importin alpha. Bioconjug. Chem. 1999;10(1):49–55. doi: 10.1021/bc980061a. [DOI] [PubMed] [Google Scholar]
- 102.Neves C, Byk G, Scherman D, Wils P. Coupling of a targeting peptide to plasmid DNA by covalent triple helix formation. FEBS Lett. 1999;453(1–2):41–45. doi: 10.1016/s0014-5793(99)00674-2. [DOI] [PubMed] [Google Scholar]
- 103.Dean DA. Peptide nucleic acids: versatile tools for gene therapy strategies. Adv. Drug Deliv. Rev. 2000;44(2–3):81–95. doi: 10.1016/s0169-409x(00)00087-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koppelhus U, Nielsen PE. Cellular delivery of peptide nucleic acid (PNA) Adv. Drug Deliv. Rev. 2003;55(2):267–280. doi: 10.1016/s0169-409x(02)00182-5. [DOI] [PubMed] [Google Scholar]
- 105.Zelphati O, Liang X, Nguyen C, et al. PNA-dependent gene chemistry: stable coupling of peptides and oligonucleotides to plasmid DNA. Biotechniques. 2000;28:304–316. doi: 10.2144/00282rr01. • This paper demonstrated that peptides and other molecules could be efficiently attached to plasmids using PNA clamps to increase gene transfer or aid in the study of DNA trafficking.
- 106.Zelphati O, Liang X, Hobart P, Felgner PL. Gene chemistry: functionally and conformationally intact fluorescent plasmid DNA. Hum. Gene Ther. 1999;10(1):15–24. doi: 10.1089/10430349950019156. • This was the first paper to demonstrate that PNA clamps could be used to fluorescently tag plasmids without altering their structure, function or transcriptional activity. This was a major accomplishment and provided an excellent set of reagents for the study of DNA trafficking in cells.
- 107.Branden LJ, Mohamed AJ, Smith CI. A peptide nucleic acid-nuclear localization signal fusion that mediates nuclear transport of DNA. Nat. Biotechnol. 1999;17(8):784–787. doi: 10.1038/11726. [DOI] [PubMed] [Google Scholar]
- 108.Branden LJ, Christensson B, Smith CI. In vivo nuclear delivery of oligonucleotides via hybridizing bifunctional peptides. Gene Ther. 2001;8(1):84–87. doi: 10.1038/sj.gt.3301345. [DOI] [PubMed] [Google Scholar]
- 109.Liang KW, Hoffman EP, Huang L. Targeted delivery of plasmid DNA to myogenic cells via transferrin-conjugated peptide nucleic acid. Mol. Ther. 2000;1(3):236–243. doi: 10.1006/mthe.2000.0043. [DOI] [PubMed] [Google Scholar]
- 110.Morris MC, Chaloin L, Choob M, et al. Combination of a new generation of PNAs with a peptide-based carrier enables efficient targeting of cell cycle progression. Gene Ther. 2004;11(9):757–764. doi: 10.1038/sj.gt.3302235. [DOI] [PubMed] [Google Scholar]
- 111.Wiethoff CM, Smith JG, Koe GS, Middaugh CR. The potential role of proteoglycans in cationic lipid-mediated gene delivery. Studies of the interaction of cationic lipid-DNA complexes with model glycosaminoglycans. J. Biol. Chem. 2001;276(35):32806–32813. doi: 10.1074/jbc.M007940200. [DOI] [PubMed] [Google Scholar]
- 112.Pollard H, Remy JS, Loussouarn G, et al. Polyethylenimine but not cationic lipids promotes transgene delivery to the nucleus in mammalian cells. J. Biol. Chem. 1998;273(13):7507–7511. doi: 10.1074/jbc.273.13.7507. [DOI] [PubMed] [Google Scholar]
- 113.Godbey WT, Wu KK, Mikos AG. Tracking the intracellular path of poly(ethylenimine)/DNA complexes for gene delivery. Proc. Natl Acad. Sci. USA. 1999;96(9):5177–5181. doi: 10.1073/pnas.96.9.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Thomas M, Klibanov AM. Conjugation to gold nanoparticles enhances polyethylenimine’s transfer of plasmid DNA into mammalian cells. Proc. Natl Acad. Sci. USA. 2003;100(16):9138–9143. doi: 10.1073/pnas.1233634100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Oh YK, Suh D, Kim JM, et al. Polyethylenimine-mediated cellular uptake, nucleus trafficking and expression of cytokine plasmid DNA. Gene Ther. 2002;9(23):1627–1632. doi: 10.1038/sj.gt.3301735. [DOI] [PubMed] [Google Scholar]
- 116.Hartig R, Shoeman RL, Janetzko A, Grub S, Traub P. Active nuclear import of single-stranded oligonucleotides and their complexes with non-karyophilic macromolecules [In Process Citation] Biol. Cell. 1998;90(5):407–426. [PubMed] [Google Scholar]
- 117.Hartig R, Shoeman RL, Janetzko A, Tolstonog G, Traub P. DNA-mediated transport of the intermediate filament protein vimentin into the nucleus of cultured cells. J. Cell Sci. 1998;111(24):3573–3584. doi: 10.1242/jcs.111.24.3573. [DOI] [PubMed] [Google Scholar]
- 118.Friend DS, Papahadjopoulos D, Debs RJ. Endocytosis and intracellular processing accompanying transfection mediated by cationic liposomes. Biochim. Biophys. Acta. 1996;1278(1):41–50. doi: 10.1016/0005-2736(95)00219-7. [DOI] [PubMed] [Google Scholar]
- 119.Kartenbeck J, Stukenbrok H, Helenius A. Endocytosis of simian virus 40 into the endoplasmic reticulum. J. Cell Biol. 1989;109:2721–2729. doi: 10.1083/jcb.109.6.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dahlgren U, Kepner WA. Principles of animal histology. New York: Macmillan Company; 1908. p. 515. [Google Scholar]