Use of Electrochemical DNA Biosensors for Rapid Molecular Identification of Uropathogens in Clinical Urine Specimens (original) (raw)
Abstract
We describe the first species-specific detection of bacterial pathogens in human clinical fluid samples using a microfabricated electrochemical sensor array. Each of the 16 sensors in the array consisted of three single-layer gold electrodes—working, reference, and auxiliary. Each of the working electrodes contained one representative from a library of capture probes, each specific for a clinically relevant bacterial urinary pathogen. The library included probes for Escherichia coli, Proteus mirabilis, Pseudomonas aeruginosa, Enterocococcus spp., and the Klebsiella-Enterobacter group. A bacterial 16S rRNA target derived from single-step bacterial lysis was hybridized both to the biotin-modified capture probe on the sensor surface and to a second, fluorescein-modified detector probe. Detection of the target-probe hybrids was achieved through binding of a horseradish peroxidase (HRP)-conjugated anti-fluorescein antibody to the detector probe. Amperometric measurement of the catalyzed HRP reaction was obtained at a fixed potential of −200 mV between the working and reference electrodes. Species-specific detection of as few as 2,600 uropathogenic bacteria in culture, inoculated urine, and clinical urine samples was achieved within 45 min from the beginning of sample processing. In a feasibility study of this amperometric detection system using blinded clinical urine specimens, the sensor array had 100% sensitivity for direct detection of gram-negative bacteria without nucleic acid purification or amplification. Identification was demonstrated for 98% of gram-negative bacteria for which species-specific probes were available. When combined with a microfluidics-based sample preparation module, the integrated system could serve as a point-of-care device for rapid diagnosis of urinary tract infections.
Urinary tract infection (UTI) is the most common urological disease in the United States and the second most common bacterial infection of any organ system (12, 32). UTI is a major cause of patient morbidity and health care expenditure for all age groups, accounting for over 7 million office visits and more than 1 million hospital admissions per year (39). Catheter-associated UTI accounts for 40% of all nosocomial infections and more than 1 million cases per year (22, 41, 49). The total cost of UTI to the United States health care system in the year 2000 was approximately $3.5 billion (13, 19, 20). An important component of these health care costs involves the processing of urine specimens by clinical microbiology laboratories. Urine is the type of body fluid most frequently submitted to clinical microbiology laboratories for culture. A major drawback of microbiological culture systems is the time lag of approximately 2 days between specimen collection and pathogen identification. The primary cause of the delay between specimen collection and pathogen identification is the time to colony formation after the specimen is plated on solid culture media. In the absence of an expeditious microbiological diagnosis, clinicians must decide whether to initiate empirical antimicrobial treatment without supportive clinical laboratory data. Injudicious use of antimicrobial agents contributes to the incidence of adverse drug reactions and the emergence of resistant pathogens. A rapid molecular approach to accurate and reliable detection and identification of clinically significant concentrations of bacterial pathogens in clinical urine specimens would be of considerable benefit.
Recent engineering advances have enabled the development of electrochemical DNA biosensors with molecular diagnostic capabilities (2, 8, 18, 33, 47). Electrochemical DNA biosensors offer several advantages compared to alternative molecular detection approaches, including the ability to analyze complex body fluids, high sensitivity, compatibility with microfabrication technology, a low power requirement, and compact instrumentation compatible with portable devices (18, 48). Electrochemical DNA sensors consist of a recognition layer containing oligonucleotide probes and an electrochemical signal transducer. A well-established electrochemical DNA sensor strategy involves “sandwich” hybridization of target nucleic acids by capture and detector probes (5, 7, 46, 50). In this strategy, the target is anchored to the sensor surface by the capture probe and detected by hybridization with a detector probe linked to a reporter function. Detector probes coupled to oxidoreductase reporter enzymes allow amperometric detection of redox signals by the sensor electrodes (28, 34). When a fixed potential is applied between the working and reference electrodes, enzyme-catalyzed redox activity is detected as a measurable electrical current (11, 16, 27). The current amplitude is a direct reflection of the number of target-probe-reporter enzyme complexes anchored to the sensor surface. Because the initial step in the electrochemical detection strategy is nucleic acid hybridization rather than enzyme-based target amplification, electrochemical sensors are able to directly detect target nucleic acids in clinical specimens, an advantage over nucleic acid amplification techniques, such as PCR.
Ultrasensitive target detection by amperometric sensors requires electrodes with surface properties that provide an extremely high signal-to-noise ratio. Advances in microfabrication technology have allowed production of a novel electrochemical sensor array consisting of 16 sensors with optical-grade surface characteristics. The electrodes in each sensor are deposited onto a plastic support in the form of a 50-nM-thick gold film with a minimum of surface irregularities (17). The smoothness of the gold allows the sensor surface to be coated with a densely packed chemical self-assembled monolayer (SAM) that insulates the electrode from background noise (6). In this study, a panel of oligonucleotide capture and detector probe pairs was developed for the sensor array to function as a “UTI chip” for detection and identification of uropathogens. This culture-independent electrochemical sensor strategy is rapid, requiring approximately 45 min from sample acquisition to data readout, and does not require labeling or amplification of the 16S rRNA target. A blinded clinical study involving urine specimens from patients at risk of UTI demonstrated that the performance of the UTI chip was comparable to that of routine clinical microbiology studies. This report confirms the feasibility of direct detection and species-specific identification of bacterial pathogens in clinical specimens using an electrochemical DNA biosensor.
MATERIALS AND METHODS
Uropathogen isolates and clinical urine specimens.
Uropathogen isolates and clinical urine specimens were obtained from the University of California—Los Angeles (UCLA) Clinical Microbiology Laboratory with approval from the UCLA and Veterans’ Affairs institutional review boards and appropriate Health Insurance Portability and Accountability Act exemptions. The isolates were received in vials containing brucella broth with 15% glycerol (BBL, Maryland) and were stored at −70°C. Overnight bacterial cultures were freshly inoculated into Luria broth (LB) and grown to logarithmic phase as measured by the optical density at 600 nm. Concentrations in the logarithmic-phase specimens were determined by serial plating, typically yielding 107 to 108 bacteria/ml. The uropathogens grown in LB were stored as frozen pellets at −70°C until the time of experimentation. Appropriate dilutions were made when the target specificities of the probes were determined so that the different uropathogen numbers were within 1 order of magnitude of each other.
Probe design.
Species- and group-specific capture and detector probe pairs were designed using a bioinformatics-based approach (M. A. Suchard, J. C. Liao, M. Mastali, M. M. Kelley, and D. A. Haake, unpublished data) that compared 16S rRNA gene sequences obtained from the NCBI database (Bethesda, MD) and from uropathogen isolates with the estimated hybridization accessibilities of 16S rRNA target sequences (14). In addition to species- and group-specific probe pairs, a universal probe pair was designed to hybridize with all bacterial 16S rRNA gene sequences. Both capture and detector probes were 27 to 35 bp in length, with their hybridization sites typically separated by a gap of 6 bp. Capture and detector probes were synthesized with 5′ biotin and 5′ fluorescein modifications, respectively (MWG, High Point, NC).
Electrochemical sensor array.
Electrochemical sensor arrays were provided by GeneFluidics (Monterey Park, CA). As shown in Fig. 1A, each sensor in the 16-sensor array consisted of a central working electrode surrounded by a reference electrode and an auxiliary electrode. The single-layer electrode design populated with alkanethiolate SAM surface modifications was described previously (16), with modifications in the electrode configuration and fabrication process. Sensor arrays used in the current study were batch fabricated by deposition of a 50-nm gold layer onto a plastic substrate. Forty microliters of 0.1 mM K3Fe(CN)6 (potassium hexacyanoferrate; Sigma, St. Louis, MO) was applied to each sensor, and cyclic voltammetry (3) was performed using a chip mounter (Fig. 1B) and a 16-channel potentiostat (GeneFluidics) as a quality control measure to characterize the alkanethiolate SAM on the sensor surface. Sensors found to have peak cyclic voltammetry currents of >100 nA were rejected to avoid sensors with incomplete SAM insulation, which would result in excessive background noise during amperometric measurement. This and each of the subsequent steps was followed by washing with a stream of deionized H2O applied to the sensor surface for approximately 2 to 3 s and drying for 5 s under a stream of nitrogen. The carboxyl termini of the SAM alkanethiols were activated and functionalized as previously described (17). In brief, each working electrode was incubated with 2.5 μl of 100 mM _N_-hydroxysuccinimide-400 mM _N_-3-dimethylaminopropyl-_N_-ethylcarbodiimide for 10 min. The activated sensors were incubated in biotin (5 mg/ml in 50 mM sodium acetate; Pierce, Rockford, IL) for 10 min. The biotinylated sensors were incubated in 4 μl of 0.5 U/ml of streptavidin in RNase-free H2O (catalog no. 821739; MP Biomedicals, Aurora, OH) for 10 min. The streptavidin-coated sensors were incubated with biotinylated capture probes (4 μl; 1 μM in 1 M phosphate buffer, pH 7.4) for 30 min.
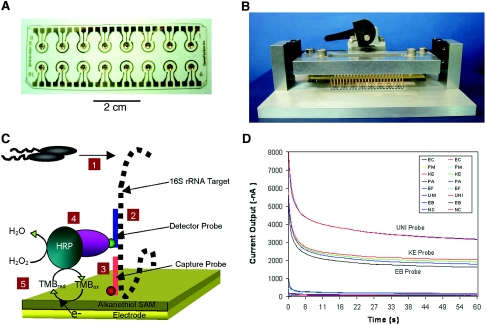
FIG. 1.
Components and performance of the electrochemical sensor. (A) The 16-sensor array (2.5 by 7.5 cm) was microfabricated with a thin, optical-grade layer of gold electrodes deposited on plastic. Each sensor in the array contained three electrodes: a central working electrode, a circumferential reference electrode, and a short auxiliary electrode. (B) The chip mounter with contact pins for simultaneous reading of the current output from each of the sensors in the array. (C) Detection strategy. (1) Bacterial lysis to release 16S rRNA target (black dashed line). (2) Hybridization of the target with the fluorescein (green circle)-labeled detector probe (blue line). (3) Hybridization of the target with the biotin (red circle)-labeled capture probe (orange line). (4) Binding of anti-fluorescein antibody conjugated with HRP to the target-probe sandwich. (5) Generation of current by transfer of electrons to the electron transfer mediator, TMB. (D) Current output in an experiment involving a clinical urine specimen containing K. pneumoniae showing signal stabilization from all 16 sensors in the array within 60 seconds. Probe results were obtained by averaging the log10 current outputs from duplicate sensor readings at 60 seconds.
Amperometric detection of bacterial 16S rRNA.
An overview of the electrochemical sensor detection strategy is presented in Fig. 1C. One milliliter of bacteria in Luria broth, inoculated clean urine, or a clinical urine sample was centrifuged at 10,000 × g for 5 min. The supernatant was discarded, and the bacteria were lysed by resuspension of the pellet in 10 μl of 1 M NaOH and incubation at room temperature for 5 min. In some experiments, the addition of 1 M NaOH was preceded by resuspension of the bacterial pellet in 10 μl of 0.1% Triton X-100, 2 mM EDTA, and 1 mg/ml lysozyme (Sigma) in 20 mM Tris-HCl, pH 8.0, and incubation at room temperature for 5 min. Fifty microliters of the detector probe (0.25 μM) in 2.5% bovine serum albumin (Sigma, St. Louis, MO)-1 M phosphate buffer, pH 7.4, was added to the bacterial lysate and incubated for 10 min at 65°C to allow target-probe hybridization. Four microliters of the bacterial-lysate-detector probe mixture was deposited on each of the working electrodes in the sensor array and incubated for 15 min at 65°C in a humidified chamber. After the array was washed and dried, 4 μl of 0.5 U/ml anti-fluorescein horseradish peroxidase (HRP) Fab fragments (Roche; diluted in 0.5% casein in 1 M phosphate-buffered saline, pH 7.4) was deposited on each of the working electrodes for 15 min. After the array was washed and dried, a prefabricated plastic well manifold (GeneFluidics, Monterey Park, CA) was bonded to the sensor array. The sensor array was put into the chip reader (Fig. 1B), and 50 μl of 3,3′5,5′-tetramethylbenzidine (TMB)-H2O2 solution (K-Blue Low Activity TMB Substrate; Neogen, Lexington, KY) was placed on each of the sensors in the array so as to cover all three electrodes of each sensor. Electrochemical measurements were immediately and simultaneously taken for all 16 sensors. For each array, negative control (NC) sensors were tested using the universal (UNI) capture and detector probes, and 2.5% bovine serum albumin (Sigma, St. Louis, MO) in 1 M phosphate buffer, pH 7.4, instead of bacterial lysate. The entire assay protocol was completed within 45 min from the time when bacterial lysis commenced. Amperometric current versus time was measured using a multichannel potentiostat (GeneFluidics). The voltage was fixed at −200 mV (versus the reference), and the electroreduction current was measured at 60 s after the HRP redox reaction reached steady state.
Clinical-validity study design.
Clinical urine specimens from routine urine cultures collected from inpatients and outpatients were received and submitted to the UCLA Clinical Microbiology Laboratory. Routine plating on trypticase soy agar with 5% sheep blood was performed on each specimen for phenotypic identification and colony counting, while an aliquot of each specimen was held at 4°C overnight. On the day after being plated, specimens were selected for inclusion in the study on the basis of a rapid indole test for the purpose of including uropathogens other than Escherichia coli in approximately one-half of the specimens. The other half of the specimens were divided between _E. coli_-containing specimens and specimens determined to have “no significant growth” or “no growth” (see definitions below). Because most UTIs involve a single uropathogen, specimens determined by the clinical microbiology laboratory to have more than one organism present were excluded. Blinded specimens selected for inclusion in the study were stripped of patient identifiers and any microbiological data before delivery to the research laboratory for testing with the electrochemical sensor array.
Experiments were performed on all specimens using the 16-sensor array “UTI chip,” in which the UNI, EB, EC, PM, KE, PA, and EF capture probes (defined in Tables 1 and 2) were tested in duplicate. The two remaining sensors served as negative controls (including capture and detector probes without bacterial lysate). The degree of variance in the electrochemical sensor measurements was determined by comparing duplicate measurements for all experiments. The background signal level was determined by averaging the log10 results of the two negative control sensors and the log10 results of the four lowest species-specific probe pairs (from among the EC, PM, KE, PA, and EF sensors). A receiver operating characteristic (ROC) curve analysis (29, 52) was performed to determine the optimal threshold for a positive result to maximize the weighted accuracy, where weighted accuracy was defined as (5 × “sensitivity” + “specificity”)/6 to account for the greater diagnostic importance of minimizing false-negative results than of minimizing false-positive results. Sensor results were determined in a three-step algorithm. First, the average of the log10 UNI results was compared with background to determine whether the specimen contained bacteria. Second, for specimens predicted to contain bacteria, the identity of the bacteria in the specimen was determined by comparing the average log10 result of the highest species-specific signal (from among the EC, PM, KE, PA, and EF sensors) with the background. Third, if no species-specific signal was positive, then the average log10 result of the EB probe pair was compared with the background to determine if the bacteria present in the specimen were members of the family Enterobacteriaceae.
TABLE 1.
Sequences of capture and detector probe pairs used with electrochemical sensor arraya
| Probe | Positionb (length) | Sequence (5′-3′) |
|---|---|---|
| Escherichia coli (EC) | ||
| Capture | 449 (35-mer) | GTCAATGAGCAAAGGTATTAACTTTACTCCCTTCC |
| Detector | 408 (35-mer) | CTGAAAGTACTTTACAACCCGAAGGCCTTCTTCAT |
| Proteus mirabilis (PM) | ||
| Capture | 202 (35-mer) | GGGTTCATCCGATAGTGCAAGGTCCGAAGAGCCCC |
| Detector | 162 (35-mer) | GGTCCGTAGACATTATGCGGTATTAGCCACCGTTT |
| Klebsiella & Enterobacter spp. (KE) | ||
| Capture | 449 (35-mer) | GTCAATCGMCRAGGTTATTAACCTYAHCGCCTTCC |
| Detector | 408 (35-mer) | CTGAAAGTGCTTTACAACCCGAAGGCCTTCTTCAT |
| Pseudomonas aeruginosa (PA) | ||
| Capture | 111 (35-mer) | CCCACTTTCTCCCTCAGGACGTATGCGGTATTAGC |
| Detector | 70 (35-mer) | TTCCGGACGTTATCCCCCACTACCAGGCAGATTCC |
| Enterococcus spp. (EF) | ||
| Capture | 207 (35-mer) | TTGGTGAGCCGTTACCTCACCAACTAGCTAATGCA |
| Detector | 165 (35-mer) | GTCCATCCATCAGCGACACCCGAAAGCGCCTTTCA |
| Enterobacteriaceae (EB) | ||
| Capture | 1241 (35-mer) | CGGACTACGACATACTTTATGAGGTCCGCTTGCTC |
| Detector | 1137 (35-mer) | GAGGTCGCTTCTCTTTGTATATGCCATTGTAGCAC |
| Universal bacterial (UNI) | ||
| Capture | 797 (27-mer) | CATCGTTTACGGCGTGGACTACCAGGG |
| Detector | 766 (31-mer) | TATCTAATCCTGTTTGCTCCCCACGCTTTCG |
TABLE 2.
Species specificities of the uropathogen probe pairs used with the electrochemical sensor array
| Probe pair | Species detecteda | Species not detecteda |
|---|---|---|
| EC | Ec | Cf, Ea, El, Eo, Ko, Kp, Pa, Pm |
| PM | Pm | Cf, Ea, Ec, El, Eo, Ko, Kp, Pa |
| KE | Ea, El, Ko, Kp | Cf, Ec, Pa, Pm |
| PA | Pa | Ec, Ef, El, Eo, Kp, Pm |
| EF | Ef, Eo | Ec, Kp, Pa, Pm, Sa |
| EB | Cf, Ea, Ec, El, Ko, Kp, Mm, Pm, | Ef, Eo, Pa, Sa, Ss |
| UNI | Cf, Ea, Ec, Ef, El, Eo, Ko, Kp, Mm, Pa, Pm, Ss, Sa | None |
“UTI chip” results were compared with clinical microbiology data after the electrochemical-sensor experiment was completed. Standard clinical microbiology laboratory procedures were followed for all specimens selected for testing. Gram-negative bacilli present in specimens at a concentration of ≥10,000 bacteria/ml and any species present at a concentration of ≥100,000 bacteria/ml were identified. Specimens with <10,000 bacteria/ml of 1 or 2 species or <50,000 bacteria/ml of >2 species were reported as “no significant growth.” Specimens with <1,000 bacteria/ml were reported as “no growth.”
RESULTS
Amperometric detection of 16S rRNA using the electrochemical sensor array.
When TMB and H2O2 were added to electrochemical sensors with surface-bound HRP-probe-target complexes bound to the surface, an amperometric signal was detected that rapidly increased to a steady-state level within 60 seconds. In the example shown in Fig. 1D, where Klebsiella pneumoniae 16S rRNA was bound in greater amounts to the UNI, EB, and KE sensors, the steady-state current level correlated with the amount of HRP enzyme present on the sensor surface. HRP is required for the production of the amperometric signal because it catalyzes the reactions in the redox relay system that ultimately result in the transfer of electrons from the electrode to H2O2, resulting in the formation of H2O. TMB was essential in functioning as an electron transfer mediator between the electrode and HRP (11, 16, 27). The interactions of HRP, TMB, and H2O2 on the electrode surface are illustrated in schematic form in Fig. 1C.
Development of probes for the “UTI chip.”
The electrochemical sensor was used to determine which probe pairs had the greatest sensitivity and specificity for binding to 16S rRNA in lysates of uropathogens. Capture and detector probes were designed to hybridize to species- and group-specific regions of the 16S rRNA molecule that are accessible to hybridization with oligonucleotide probes, as determined by prior flow cytometric analysis (14). Candidate probe pairs for E. coli, Proteus mirabilis, Pseudomonas aeruginosa, Enterococcus spp., Klebsiella spp., Enterobacter spp., and the Enterobacteriaceae group were tested for uropathogen detection sensitivity and specificity using the electrochemical sensor to arrive at the optimal probe set shown in Table 1.
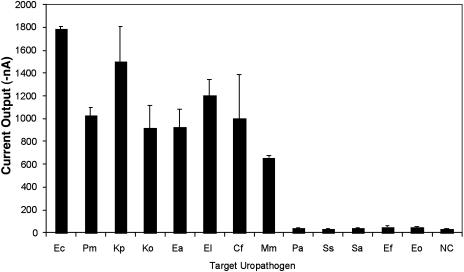
Table 2 summarizes the observed specificities of the probe pairs using the electrochemical sensor array. Significant sequence similarities between Klebsiella and Enterobacter sp. 16S sequences precluded the design of species-specific probes for these organisms. The Enterococcus probe pair (EF) was specific for both Enterococcus faecalis and Enterococcus faecium. The universal probe pair (UNI) detects all of the uropathogens tested. Figure 2 shows detection by the EB probe pair of all members of the family Enterobacteriaceae tested, but not equal numbers of P. aeruginosa, Staphylococcus spp., or Enterococcus spp. As shown in Table 2 and Fig. 2, both the UNI and EB probes detect less common uropathogens, such as Citrobacter spp. and Morganella spp., for which species-specific probes are not yet available.
FIG. 2.
Specificities of _Enterobacteriaceae_-specific probe pairs. Positive signals were seen for all Enterobacteriaceae species tested but not for gram-positive uropathogens (Eo, Ef, Ss, and Sa) or P. aeruginosa (Pa). (See the footnote to Table 2 for bacterial species abbreviations.) Means and standard deviations of experiments performed in duplicate are shown. NC refers to the negative control experiments performed with capture and detector probes but without bacterial lysate.
Species-specific detection of uropathogens in clinical urine specimens.
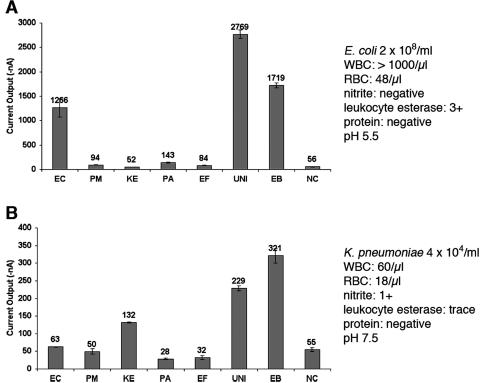
To determine analytic validity, bacteria in culture medium, inoculated urine, and clinical urine specimens were tested using the UTI chip and seven probe pairs with specificities relevant to the detection of the most prevalent uropathogens. Clinical urine specimens represented the most significant challenge to the electrochemical sensor detection strategy because they frequently also contained high concentrations of host proteins, white blood cells, red blood cells, and cellular debris, in addition to wide variations in pH. In pilot experiments, the UTI chip produced accurate results with clinical urine specimens representing a broad range of uropathogen species and concentrations, as well as a broad range of urine specimen parameters related to the host inflammatory response to UTI. Figure 3A illustrates that a high signal-to-noise ratio was maintained in the case of a clinical urine specimen from a patient with an E. coli UTI in which high numbers of white blood cells were present and the pH of the urine was 5.5. The high amperometric signals using the UNI, EB, and EC sensors indicated that the streptavidin coating on the sensor surface remained intact and the target-probe hybridization step of the electrochemical sensor protocol was not inhibited by host cells or changes in urinary pH. The low background signals with the remaining sensors indicated that the SAM layer was also unaffected by exposure to urine. Figure 3B illustrates the results obtained for a urine specimen from a patient with a K. pneumoniae UTI containing a relatively low bacterial concentration of 4 × 104 organisms/ml, demonstrating that the detection sensitivity of the sensor was maintained despite urine specimen conditions. Table 3 summarizes the UTI chip results for detection of uropathogens in clinical urine specimens containing these and several other bacterial species. Accurate results were obtained using the electrochemical sensor array despite high levels of protein, red blood cells, and white blood cells and variations in pH from 5.0 to 7.0.
FIG. 3.
Direct, species-specific detection of uropathogens in representative clinical urine specimens using the electrochemical sensor array. Current output for each of the probe pairs in the array are shown in nanoamperes. The mean current output of duplicate sensors is shown above each bar; the error bars represent the standard deviations. The probe pair designation is shown below each bar, and the species specificity is given in Table 2. The urinalysis and microbiological characteristics of each specimen are shown to the right of the bar graph. The background signal level was determined by averaging the log10 results of the NC sensors and the sensors with the four lowest species-specific probe pairs (from among EC, PM, KE, PA, and EF). As described in the text, significant signals were 0.30 log unit (5 standard deviations) above background. (A) E. coli in this clinical urine specimen produced significant signals in the UNI, EB, and EC sensors despite high numbers of white blood cells (WBC). RBC, red blood cells. (B) 16S rRNA from as few as 4 × 104 K. pneumoniae cells/ml in urine produced significant signal levels in the UNI, EB, and KE sensors.
TABLE 3.
Urinalysis and microbiological characteristics of representative clinical urine specimens tested with the electrochemical sensor array containing seven probe pairs (“UTI chip”)
| Uropathogen | Concn (CFU/ml) | Urinalysis resulta | Positive probes | |||
|---|---|---|---|---|---|---|
| pH | Protein | RBC/μl | WBC/μl | |||
| E. coli | 2.6 × 108 | 5.5 | Neg | 48 | >1,000 | UNI, EB, EC |
| P. mirabilis | >1.0 × 105 | 7.0 | 100 mg/dl | >1,000 | >1,000 | UNI, EB, PM |
| K. pneumoniae | 4.0 × 104 | 7.5 | Neg | 18 | 60 | UNI, EB, KE |
| E. aerogenes | 1.6 × 107 | 5.0 | Trace | 2 | 556 | UNI, EB, KE |
| Pseudomonas sp. | >1.0 × 105 | 7.0 | 30 mg/dl | 177 | 103 | UNI, PA |
| Enterococcus sp. | 1.3 × 106 | 7.0 | Neg | 2 | 26 | UNI, EF |
Clinical-validity study involving blinded clinical urine specimens.
A total of 89 blinded clinical urine specimens were received from the clinical microbiology laboratory. Eleven urine specimens were found to contain more than one organism and were excluded from further analysis. The remaining 78 specimens that were analyzed included 58 with bacteria identified to species level by the clinical microbiology laboratory, 8 specimens classified by the clinical microbiology laboratory as “no significant growth,” and 12 specimens that were “no growth.” The 58 positive specimens contained a broad diversity of uropathogens: 26 contained E. coli, 3 contained P. mirabilis, 8 contained K. pneumoniae, 1 contained Enterobacter cloacae, 5 contained Enterobacter aerogenes, 1 contained P. aeruginosa, 2 contained Citrobacter freundii, 1 contained Citrobacter koseri, 1 contained Serratia marcescens, 8 contained Enterococcus spp., and 2 contained Staphylococcus saprophyticus.
The 16-sensor array allowed each of eight electrochemical sensor measurements (seven probe pairs plus one negative control) to be performed in duplicate. Sensor-to-sensor variance in the clinical study was determined by comparing the results from testing of all 78 samples, which yielded 1,248 (2 × 8 × 78) paired results. The duplicate residual errors were found to have a log normal distribution. The standard deviations of the duplicates were roughly constant at a value of 0.06 log units for all sensors in the UTI chip array. A receiver operating characteristic curve analysis found that the optimal UTI chip weighted accuracy was maximal at 91% for a mean log positive over mean log background threshold of 0.25 to 0.33 log units. A 0.30-log-unit threshold roughly equal to 5 standard deviations above background was applied for all sensor pairs in the array. As shown in Table 4, this approach yielded an overall sensitivity for detection of uropathogens in clinical urine specimens by the UNI probe pair of 54/58 (93%; standard error, ±3.3%). UNI probe specificity was estimated at 10/12 (83%; standard error, ±10.8%), although this number could not be determined with great accuracy because there were only 12 “no-growth” specimens in the sample. Because the signal interpretation algorithm was designed for specimens containing no more than one organism, it is difficult to make further estimates of specificity. However, we do note that there was only one incorrect species identification: one specimen containing S. marcescens was falsely positive by the PA probe pair.
TABLE 4.
Results of a blinded study of clinical urine specimens showing a high level of sensitivity for detection of gram-negative bacteria by the “UTI Chip”a
| Clinical microbiology (species or result) | UNI sensors (positive/total = %) [mean (range)]b | EB sensors (positive/total = %) [mean (range)]b | Species sensors (positive/total = %) [mean (range)]b |
|---|---|---|---|
| Gram-negative bacteria | 48/48 = 100 [1.32 (0.40-1.84)] | 46/47 = 98c [1.17 (0.46-1.60)] | 43/44 = 98d [1.00 (0.34-1.59)] |
| Escherichia coli | 26/26 = 100 [1.30 (0.54-1.77)] | 25/26 = 96 [1.17 (0.46-1.60)] | Ec 26/26 = 100 [1.06 (0.48-1.59)] |
| Klebsiella pneumoniae | 8/8 = 100 [1.22 (0.40-1.84)] | 8/8 = 100 [1.13 (0.46-1.13)] | KE 7/8 = 88 [1.16 (0.76-1.50)] |
| Enterobacter aerogenes | 5/5 = 100 [1.35 (0.93-1.65)] | 5/5 = 100 [1.20 (0.82-1.52)] | KE 5/5 = 100 [0.86 (0.37-1.26)] |
| Enterobacter cloacae | 1/1 = 100 [1.43] | 1/1 = 100 [1.08] | KE 1/1 = 100 [0.34] |
| Proteus mirabilis | 3/3 = 100 [1.41 (1.16-1.66)] | 3/3 = 100 [1.14 (0.92-1.45)] | Pm 3/3 = 100 [0.74 (0.48-0.96)] |
| Citrobacter freundii | 2/2 = 100 [1.49 (1.43-1.55)] | 2/2 = 100 [1.32 (1.25-1.38)] | 0/2 = 0 [NA] |
| Citrobacter koseri | 1/1 = 100 [1.23] | 1/1 = 100 [1.29] | 0/1 = 0 [NA] |
| Serratia marcescens | 1/1 = 100 [1.67] | 1/1 = 100 [1.36] | False positive Pa 0.36 |
| Enterobacteriaceae | 47/47 = 100 [1.32 (0.40-1.84)] | 46/47 = 98 [1.17 (0.46-1.60)] | 46/47 = 98 [1.02 (0.34-1.59)] |
| Pseudomonas aeruginosa | 1/1 = 100 [1.33] | 0/1 = 0 [NA]e | Pa 1/1 = 100 [0.55] |
| Gram-positive bacteria | 6/10 = 60 [0.95 (0.33-1.78)] | 0/10 = 0 [NA] | 2/8 = 25d [0.99 (0.63-1.36)] |
| Enterococcus species | 5/8 = 63 [0.92 (0.33-1.78)] | 0/8 = 0 [NA] | Ef 2/8 = 25 [0.99 (0.63-1.36)] |
| S. saprophyticus | 1/2 = 50 [1.10] | 0/2 = 0 [NA] | 0/2 = 0 [NA] |
| All specimens with bacteria | 54/58 = 93 [1.28 (0.33-1.84)] | 46/47 = 98c [1/17 (0.46-1.60)] | 45/52 = 87d [1.00 (0.34-1.59)] |
| No significant growth | 5/8 = 63 [0.42 (0.31-0.57)] | 0/8 = 0 [NA] | Ef 1/8 = 13 [0.31] |
| No growth | 2/12 = 17 [0.38 (0.34-0.42)] | 0/12 = 0 [NA] | 0/12 = 0 [NA] |
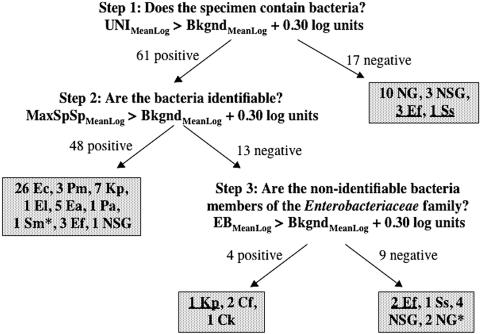
The sensitivity of the UNI probe for detection of gram-negative bacteria in clinical urine specimens was 48/48 (100%), because all four false-negative specimens contained gram-positive bacteria. Both the Enterobacteriaceae and species-specific probe pairs demonstrated similar 98% sensitivities for detection of gram-negative bacteria in clinical urine specimens. Figure 4 illustrates the three-step UTI chip signal interpretation algorithm, which showed the high level of accuracy for detection and identification of gram-negative bacteria in clinical urine specimens. Of 44 specimens containing gram-negative bacteria for which there were species-specific probes, only one specimen containing K. pneumoniae was falsely negative using the KE probe pair. The sensitivity of the “UTI chip” was lower for gram-positive organisms; three specimens containing enterococci and one specimen containing S. saprophyticus were falsely negative using the UNI probe pair.
FIG. 4.
“UTI Chip” signal interpretation algorithm. The three-step algorithm used to interpret the results of electrochemical-sensor experiments on 78 specimens that met inclusion criteria is shown. Positive signals were those with a mean log of greater than 0.30 log unit (5 standard deviations) over background. “UNI” and “EB” are eubacterial- and _Enterobacteriaceae_-specific probe signals, respectively. “Bkgnd” is the background signal, as defined in the text. “MaxSpSp” refers to the maximum species-specific signal. Clinical microbiology results are given in shaded boxes. Two-letter species abbreviations are given in the footnote of Table 2. NSG indicates “no significant growth.” NG indicates “no growth.” An asterisk indicates false-positive results, and false-negative results are underlined.
DISCUSSION
In this study, we describe a novel electrochemical sensor array platform that uses oligonucleotide capture and detector probes for detection and identification of bacterial uropathogens in clinical urine specimens. Amperometric biosensors for detection of in vitro cultivated bacteria using antibody capture or nucleic acid hybridization approaches have been described, but not for detection of bacteria in clinical specimens (1, 4, 9, 10, 15, 21, 26, 30, 35, 40, 43). A convergence of technological innovations from several disciplines, including microfabrication, materials science, electrochemistry, and molecular microbiology, contributed to the design of the electrochemical sensor array and the detection strategy utilized in the current study (16, 17, 42). The resulting biosensor demonstrated a high signal-to-noise ratio and low variance between duplicate sensors that was maintained despite contact with bacteria obtained directly from clinical urine specimens. This is the first report we are aware of describing the detection of bacterial pathogens in human body fluids using an electrochemical sensor, as well as the clinical validity of this approach.
The sensor technique utilized here is an electrochemical sandwich assay in which target 16S rRNA is bound by both a capture and a detector probe (16). The capture probe anchors the target to the sensor, and the detector probe provides a means for recognizing target bound on the sensor surface. This sandwich strategy has been successfully employed in several types of electrochemical sensors (5, 7, 46, 50). In our system, as in most electrochemical sandwich assays, the detector probe is linked directly or indirectly to HRP for amperometric detection of redox current (5, 7, 50). An alternative electrochemical sandwich assay approach involving a ferrocene-modified detector probe has been described (46). When the detector probe is hybridized to the target on the sensor surface, the ferrocene moieties mediate electron transfer to the gold electrode via a phenylacetylene molecular wire embedded in the electrode's SAM. In any microfabricated electrochemical sensor, the SAM is essential for reducing background current by insulating the working electrode when a potential difference is applied between the working and reference electrodes (3). When our electrochemical sensor results are to be read, the sensor is placed in a potentiostat and a voltage of −200 mV is applied between the working and auxiliary electrodes, resulting in polarization of the working electrode with negative charges. HRP substrates, such as TMB, then serve to transfer electrons from the electrode surface to the HRP across the SAM (11, 16, 27).
Electrochemical sensors directly detect nucleic acid targets by hybridization, so that sensitivity and specificity problems associated with nucleic acid amplification in the presence of biological inhibitors are avoided. The accuracy of the “UTI chip” was demonstrated for samples with significant amounts of somatic cells and urinary protein and ranges of pH. In contrast, PCR detection assays for urine specimens are subject to false-negative results due to DNA polymerase inhibitors, which may not be removed even after a nucleic acid purification step (24, 25). Application of PCR assays to complex mixtures of nucleic acids can produce biased target amplification, resulting in problems with specificity (36, 44). We and others have encountered sensitivity and specificity problems related to sample contamination and/or urinary inhibitors in our attempts to use PCR for detection of uropathogens in clinical urine specimens (38, 45). Electrical and fluidics systems can be miniaturized, so electrochemical sensors are potentially less expensive and more portable than sophisticated optical detection systems currently being used in PCR detection assays. These intrinsic advantages may be critical when sensor technology is eventually applied in an automated point-of-care device.
We developed a library of species-specific probes that recognize over 90% of uropathogens submitted to the clinical microbiology laboratory. 16S rRNA was chosen as the sensor target because it exists in high copy numbers in bacterial cells and is an essential component of ribosomes. 16S rRNA gene sequences of the relevant species of bacteria are well characterized and contain regions of diversity and conservation that are useful for molecular diagnostic purposes (37). Similar to probes used for 16S rRNA-based fluorescence in situ hybridization assays, the oligonucleotide probes that were developed for use with the electrochemical sensor array hybridize with species-specific and surface-accessible regions of the 16S rRNA target molecule. The panel of probes described in the clinical-feasibility study was able to detect and identify a broad range of gram-negative uropathogens. The absence of a positive signal from the UNI probe effectively rules out a gram-negative bacterial UTI. Our detection system had reduced sensitivity for gram-positive uropathogens, such as Enterococcus species and S. saprophyticus. The most likely explanation for this problem is resistance of the gram-positive cell wall to the alkaline lysis method used in our study. Development of alternative lysis methods that would be applicable to all potential uropathogens is an area of active investigation in our laboratory.
A short time from specimen collection to readout is essential to an approach intended for a point-of-care application. Our current detection strategy requires approximately 45 min: bacterial lysis for 5 min, probe hybridization for 25 min, and enzyme amplification for 15 min. The amperometric reading is currently being measured at 60 s, by which point the current flow has reached steady state (Fig. 1D). The reaction kinetics in each step of the protocol is limited by passive diffusion (concentration of molecules versus time). We anticipate that the sample preparation time can be further reduced by optimization of bacterial-lysis efficiency and hybridization kinetics.
The optical-grade surface characteristics of the gold electrodes in our electrochemical sensor array allowed the formation of pinhole-free SAMs. Highly insulating SAMs improve sensitivity by reducing sensor background and increasing the signal-to-noise ratio. Sensitivity was also improved by integrating liquid phase detector probe-target hybridization for maximum signal detection efficiency and solid-phase probe-sensor immobilization for maximum target capture efficiency (16). The most commonly used microbiologic criterion for UTI is greater than 105 CFU/ml from a clean-catch voided urine sample (23, 51), although the actual concentrations of uropathogens in clinical urine specimens are frequently higher. A robust uropathogen diagnostic system should be able to detect and quantify bacteria over a wide spectrum of bacterial concentrations and urine parameters. The studies presented here indicate that the UTI chip is able to detect uropathogens over wide range of clinical urine characteristics (Table 3) and at bacterial concentrations as low as 4 × 104 CFU/ml (Fig. 3B). The results of the clinical-feasibility study proved this to be an appropriate level of sensitivity for detection of clinically relevant concentrations of bacteria in urine. Given that only 4 μl of the 60-μl lysate-probe mixture, or 1/15 of the total, is applied to the sensor surface, the ability to detect as few as 4.0 × 104 CFU/ml (Fig. 3B) translates to a total of 2,600 bacteria. E. coli contain between 5 × 103 and 2 × 104 copies of 16S rRNA per cell (31). Therefore, we estimate that the rRNA detection limit of the sensor is within the femtomolar (3 × 10−16) range, which compares favorably with other electrochemical DNA sensors (8). This level of sensitivity is achieved using raw bacterial lysates from actual body fluids and represents an important advance compared to previous studies.
The studies presented here demonstrate the analytical and clinical validity of an electrochemical DNA sensor for quantitative, species-specific detection of uropathogens. The culture- and PCR-independent molecular identification was achieved in 45 min. The ability of the sensor to provide genotypic identification of uropathogens and to rapidly differentiate between bacterial pathogens is clearly superior to current clinical microbiology approaches, which are limited by the growth rates of bacteria and typically require at least 48 h from sample collection to reporting. The sensor array and the detection assay, in their present forms, are not yet ready for widespread application. Strategies for improving the sensitivity of the electrochemical assay for detection of uropathogens, particularly gram-positive bacteria, are being examined. Improvements in sensitivity should allow amperometric detection of clinically significant uropathogen concentrations below 10,000 CFU/ml, which would otherwise be considered “no significant growth” by current clinical microbiology standards. It is anticipated that detection of multiple uropathogens in a single clinical specimen will also be feasible. Eventually, testing of larger numbers of clinical specimens will be required to accurately determine the sensitivity and specificity of the electrochemical-sensor approach. The electrochemical sensor and the simplicity of its sample preparation requirements are compatible with eventual integration with an automated microfluidics-based sample preparation module. Concentration of bacteria in the urine sample, coupled with active mixing of reagents instead of passive diffusion, would significantly reduce overall sample preparation time and enhance sensitivity. Our studies lay the foundation for analyses of the clinical utility of our UTI chip. Rapid detection and identification of uropathogens at the point of care will have a profound impact on clinical decision making when managing a patient with suspected UTI.
Acknowledgments
We thank the other members of the UCLA Urosensor Bioengineering Research Partnership for their insights and helpful suggestions: Chih-Ming Ho, Linda McCabe, Yao Hua Zhang, Chien-Pin Sun, Yanbao Ma, and Linda Gibson. We also thank James Matsunaga of UCLA/VAGLAHS; Susan K. Haake of the UCLA School of Dentistry; and John Kibler, Shu-ching Ma, and Ashish Pradhan of GeneFluidics for expert assistance.
This study was supported by Bioengineering Research Partnership grant EB00127 (to B.M.C.) from the National Institute of Biomedical Imaging and Bioengineering and in part by the Wendy and Ken Ruby Fund for Excellence in Pediatric Urology Research. J.C.L. is the recipient of an American Foundation for Urologic Disease (AFUD) fellowship. We thank Frank W. Clark, Jr., for his early generous support.
REFERENCES
- 1.Abdel-Hamid, I., D. Ivnitski, P. Atanasov, and E. Wilkins. 1999. Flow-through immunofiltration assay system for rapid detection of E. coli O157:H7. Biosens. Bioelectron. 14**:**309-316. [DOI] [PubMed] [Google Scholar]
- 2.Albers, J., T. Grunwald, E. Nebling, G. Piechotta, and R. Hintsche. 2003. Electrical biochip technology—a tool for microarrays and continuous monitoring. Anal. Bioanal. Chem. 377**:**521-527. [DOI] [PubMed] [Google Scholar]
- 3.Bard, A. J., and L. R. Faulkner. 2001. Electrochemical methods fundamentals and applications, 2nd ed. John Wiley & Sons, Inc., Hoboken, N.J.
- 4.Basu, M., S. Seggerson, J. Henshaw, J. Jiang, R. del A Cordona, C. Lefave, P. J. Boyle, A. Miller, M. Pugia, and S. Basu. 2004. Nano-biosensor development for bacterial detection during human kidney infection: use of glycoconjugate-specific antibody-bound gold NanoWire arrays (GNWA). Glycoconj. J. 21**:**487-496. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, C. N., D. Gal, N. Cristler, C. Banditrat, and A. Heller. 2002. Enzyme-amplified amperometric sandwich test for RNA and DNA. Anal. Chem. 74**:**158-162. [DOI] [PubMed] [Google Scholar]
- 6.Chaki, N. K., and K. Vijayamohanan. 2002. Self-assembled monolayers as a tunable platform for biosensor applications. Biosens. Bioelectron. 17**:**1-12. [DOI] [PubMed] [Google Scholar]
- 7.Dequaire, M., and A. Heller. 2002. Screen printing of nucleic acid detecting carbon electrodes. Anal. Chem. 74**:**4370-4377. [DOI] [PubMed] [Google Scholar]
- 8.Drummond, T. G., M. G. Hill, and J. K. Barton. 2003. Electrochemical DNA sensors. Nat. Biotechnol. 21**:**1192-1199. [DOI] [PubMed] [Google Scholar]
- 9.Ercole, C., M. Del Gallo, M. Pantalone, S. Santucci, L. Mosiello, C. Laconi, and A. Lepidi. 2002. A biosensor for Escherichia coli based on a potentiometric alternating biosensing (PAB) transducer. Sens. Actuators B 83**:**48-52. [Google Scholar]
- 10.Ertl, P., M. Wagner, E. Corton, and S. R. Mikkelsen. 2003. Rapid identification of viable Escherichia coli subspecies with an electrochemical screen-printed biosensor array. Biosens. Bioelectron. 18**:**907-916. [DOI] [PubMed] [Google Scholar]
- 11.Fanjul-Bolado, P., M. B. Gonzalez-Garcia, and A. Costa-Garcia. 2005. Amperometric detection in TMB/HRP-based assays. Anal. Bioanal. Chem. 382**:**297-302. [DOI] [PubMed] [Google Scholar]
- 12.Foxman, B., R. Barlow, H. D'Arcy, B. Gillespie, and J. D. Sobel. 2000. Urinary tract infection: self-reported incidence and associated costs. Ann. Epidemiol. 10**:**509-515. [DOI] [PubMed] [Google Scholar]
- 13.Freedman, A. L. 2005. Urologic Diseases in North America Project: trends in resource utilization for urinary tract infections in children. J. Urol. 173**:**949-954. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs, B. M., G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64**:**4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabig-Ciminska, M., H. Andresen, J. Albers, R. Hintsche, and S. O. Enfors. 2004. Identification of pathogenic microbial cells and spores by electrochemical detection on a biochip. Microb. Cell Fact. 3**:**2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gau, J. J., E. H. Lan, B. Dunn, C. M. Ho, and J. C. Woo. 2001. A MEMS based amperometric detector for E. coli bacteria using self-assembled monolayers. Biosens. Bioelectron. 16**:**745-755. [DOI] [PubMed] [Google Scholar]
- 17.Gau, V., S. C. Ma, H. Wang, J. Tsukuda, J. Kibler, and D. A. Haake. 2005. Electrochemical molecular analysis without nucleic acid amplification. Methods _3_7**:**73-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gooding, J. J. 2002. Electrochemical DNA hybridization biosensor. Electroanalysis 14**:**1149-1156. [Google Scholar]
- 19.Griebling, T. L. 2005. Urologic Diseases in America Project: trends in resource use for urinary tract infections in men. J. Urol. 173**:**1288-1294. [DOI] [PubMed] [Google Scholar]
- 20.Griebling, T. L. 2005. Urologic Diseases in America Project: trends in resource use for urinary tract infections in women. J. Urol. 173**:**1281-1287. [DOI] [PubMed] [Google Scholar]
- 21.Ivnitski, D., E. Wilkins, H. T. Tien, and A. Ottova. 2000. Electrochemical biosensor based on supported planar lipid bilayers for fast detection of pathogenic bacteria. Electrochem. Commun. 2**:**457-460. [Google Scholar]
- 22.Kunin, C. M. 1997. Care of the urinary catheter, p. 227-299. In C. M. Kunin (ed.), Urinary tract infections: detection, prevention, and management, 5th ed. Williams & Wilkins, Baltimore, Md.
- 23.Kunin, C. M., L. V. White, and T. H. Hua. 1993. A reassessment of the importance of “low-count” bacteriuria in young women with acute urinary symptoms. Ann. Intern. Med. 119**:**454-460. [DOI] [PubMed] [Google Scholar]
- 24.Land, S., S. Tabrizi, A. Gust, E. Johnson, S. Garland, and E. M. Dax. 2002. External quality assessment program for Chlamydia trachomatis diagnostic testing by nucleic acid amplification assays. J. Clin. Microbiol. 40**:**2893-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahony, J., S. Chong, D. Jang, K. Luinstra, M. Faught, D. Dalby, J. Sellors, and M. Chernesky. 1998. Urine specimens from pregnant and nonpregnant women inhibitory to amplification of Chlamydia trachomatis nucleic acid by PCR, ligase chain reaction, and transcription-mediated amplification: identification of urinary substances associated with inhibition and removal of inhibitory activity. J. Clin. Microbiol. 36**:**3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazenko, R. S., F. Rieders, and J. D. Brewster. 1999. Filtration capture immunoassay for bacteria: optimization and potential for urinalysis. J. Microbiol. Methods 36**:**157-165. [DOI] [PubMed] [Google Scholar]
- 27.Mecheri, B., L. Piras, L. Ciotti, and G. Caminati. 2004. Electrode coating with ultrathin films containing electroactive molecules for biosensor applications. IEEE Sens. J. 4**:**171-179. [Google Scholar]
- 28.Mehrvar, M., and M. Abdi. 2004. Recent developments, characteristics, and potential applications of electrochemical biosensors. Anal. Sci. 20**:**1113-1126. [DOI] [PubMed] [Google Scholar]
- 29.Metz, C. E. 1986. ROC methodology in radiologic imaging. Investig. Radiol. 21**:**720-733. [DOI] [PubMed] [Google Scholar]
- 30.Mittelmann, A. S., E. Z. Ron, and J. Rishpon. 2002. Amperometric quantification of total coliforms and specific detection of Escherichia coli. Anal. Chem. 74**:**903-907. [DOI] [PubMed] [Google Scholar]
- 31.Neidhardt, F., and H. Umbarger. 1996. Chemical composition of Escherichia coli, p. 13-16. In F. Neidhardt et al. (ed.), Escherichia coli and Salmonella typhimurium, 2nd ed., vol. I. ASM Press, Washington, D.C. [Google Scholar]
- 32.Nicolle, L. 2001. Epidemiology of urinary tract infection. Infect. Med. 18**:**153-162. [Google Scholar]
- 33.Palecek, E., and F. Jelen. 2002. Electrochemistry of nucleic acids and development of DNA sensors. Crit. Rev. Anal. Chem. 3**:**261-270. [Google Scholar]
- 34.Pearson, J. E., A. Gill, and P. Vadgama. 2000. Analytical aspects of biosensors. Ann. Clin. Biochem. 37**:**119-145. [DOI] [PubMed] [Google Scholar]
- 35.Perez, F. G., M. Mascini, I. E. Tothill, and A. P. Turner. 1998. Immunomagnetic separation with mediated flow injection analysis amperometric detection of viable Escherichia coli O157. Anal. Chem. 70**:**2380-2386. [DOI] [PubMed] [Google Scholar]
- 36.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64**:**3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Relman, D. A. 1999. The search for unrecognized pathogens. Science 284**:**1308-1310. [DOI] [PubMed] [Google Scholar]
- 38.Rosenstraus, M., Z. Wang, S. Y. Chang, D. DeBonville, and J. P. Spadoro. 1998. An internal control for routine diagnostic PCR: design, properties, and effect on clinical performance. J. Clin. Microbiol. 36**:**191-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schappert, S. M. 1999. Ambulatory care visits to physician offices, hospital outpatient departments, and emergency departments: United States, 1997. Vital Health Stat. 13**:**i-iv, 1-39. [PubMed] [Google Scholar]
- 40.Sippy, N., R. Luxton, R. J. Lewis, and D. C. Cowell. 2003. Rapid electrochemical detection and identification of catalase positive micro-organisms. Biosens. Bioelectron. 18**:**741-749. [DOI] [PubMed] [Google Scholar]
- 41.Stamm, W. E. 1991. Catheter-associated urinary tract infections: epidemiology, pathogenesis, and prevention. Am. J. Med. 91**:**65S-71S. [DOI] [PubMed] [Google Scholar]
- 42.Sun, C. P., J. C. Liao, Y. H. Zhang, V. Gau, M. Mastali, J. T. Babbitt, W. S. Grundfest, B. M. Churchill, E. R. McCabe, and D. A. Haake. 2005. Rapid, species-specific detection of uropathogen 16S rDNA and rRNA at ambient temperature by dot-blot hybridization and an electrochemical sensor array. Mol. Genet. Metab. 84**:**90-99. [DOI] [PubMed] [Google Scholar]
- 43.Susmel, S., G. G. Guilbault, and C. K. O'Sullivan. 2003. Demonstration of labeless detection of food pathogens using electrochemical redox probe and screen printed gold electrodes. Biosens. Bioelectron. 18**:**881-889. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62**:**625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toye, B., W. Woods, M. Bobrowska, and K. Ramotar. 1998. Inhibition of PCR in genital and urine specimens submitted for Chlamydia trachomatis testing. J. Clin. Microbiol. 36**:**2356-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Umek, R. M., S. W. Lin, J. Vielmetter, R. H. Terbrueggen, B. Irvine, C. J. Yu, J. F. Kayyem, H. Yowanto, G. F. Blackburn, D. H. Farkas, and Y. P. Chen. 2001. Electronic detection of nucleic acids: a versatile platform for molecular diagnostics. J. Mol. Diagn. 3**:**74-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, J. 2002. Electrochemical nucleic acid biosensors. Anal. Chim. Acta 469**:**63-71. [Google Scholar]
- 48.Wang, J. 2002. Portable electrochemical systems. Trends Anal. Chem. 21**:**226-232. [Google Scholar]
- 49.Warren, J. W. 1991. The catheter and urinary tract infection. Med. Clin. N. Am. 75**:**481-493. [DOI] [PubMed] [Google Scholar]
- 50.Williams, E., M. I. Pividori, A. Merkoci, R. J. Forster, and S. Alegret. 2003. Rapid electrochemical genosensor assay using a streptavidin carbon-polymer biocomposite electrode. Biosens/ Bioelectron. 19**:**165-175. [DOI] [PubMed] [Google Scholar]
- 51.Wilson, M. L., and L. Gaido. 2004. Laboratory diagnosis of urinary tract infections in adult patients. Clin. Infect. Dis. 38**:**1150-1158. [DOI] [PubMed] [Google Scholar]
- 52.Zhou, X., N. Obuchowski, and D. McClish. 2002. Statistical methods in diagnositic medicine. Wiley & Sons Interscience, New York, N.Y.