Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells (original) (raw)
Abstract
Abasic (AP)-endonuclease (APE) is responsible for repair of AP sites, and single-strand DNA breaks with 3′ blocking groups that are generated either spontaneously or during repair of damaged or abnormal bases via the DNA base excision repair (BER) pathway in both nucleus and mitochondria. Mammalian cells express only one nuclear APE, 36 kDa APE1, which is essential for survival. Mammalian mitochondrial (mt) BER enzymes other than mtAPE have been characterized. In order to identify and characterize mtAPE, we purified the APE activity from beef liver mitochondria to near homogeneity, and showed that the mtAPE which has 3-fold higher specific activity relative to APE1 is derived from the latter with deletion of 33 N-terminal residues which contain the nuclear localization signal. The mtAPE-sized product could be generated by incubating 35S-labeled APE1 with crude mitochondrial extract, but not with cytosolic or nuclear extract, suggesting that cleavage of APE1 by a specific mitochondria-associated N-terminal peptidase is a prerequisite for mitochondrial import. The low abundance of mtAPE, particularly in cultured cells might be the reason for its earlier lack of detection by western analysis.
INTRODUCTION
ROS-induced damage in DNA includes a plethora of oxidized bases, abasic (AP) sites and DNA strand breaks all of which are repaired via the base excision repair (BER) pathway. Repair of damaged bases is initiated with excision of a damaged or abnormal base by a DNA glycosylase thereby leaving a non-coding AP site. Oxidized base-specific mammalian DNA glycosylases, such as NTH1 and OGG1 further cleave the AP site via lyase reaction to generate 3′ α, β unsaturated deoxyribose (1). ROS also directly attacks deoxyribose and cleaves the DNA strand to produce 3′-glycolate termini (2,3). Both AP sites and 3′ blocking groups are processed by abasic (AP)-endonuclease (APE) to generate a 3′ OH terminus which serves as the primer for gap-filling DNA synthesis by a DNA polymerase. All APEs have both AP site-specific endonuclease and 3′ phosphodiesterase activities (4). Unlike Escherichia coli or yeast, mammalian cells express only one APE, 36 kDa APE1, whose sequence is highly conserved among various mammalian species. The human and bovine APE1s have 93% sequence identity. APE1 belongs to the E.coli Xth family and has significant homology with APN2 in yeast (4–6).
The mitochondrial genome is much more susceptible to endogenous, oxidative damage than the nuclear genome presumably because of both proximity to the site of ROS generation (in mitochondrial respiratory complexes), and the lack of associated histones (7,8). Oxidative damage to the mitochondrial genome has been implicated in various human degenerative diseases, and in aging (9,10). DNA repair in mitochondria should thus be extremely important, particularly for non-dividing cells (11). Although the mitochondria lack the nucleotide excision repair system (12), repair of oxidative damage via the BER pathway in the mitochondria has been demonstrated for a number of cell types (13–19). Uracil-DNA glycosylase (UDG) is the first DNA glycosylase to be identified in mitochondria (20,21). Nuclear and mitochondria-specific DNA glycosylases are encoded by the same nuclear genes. These mitochondrial enzymes lack the nuclear localization signal (NLS), and contain N-terminal mitochondrial targeting sequence (MTS) (22–24).
While most BER enzymes in mitochondria have been characterized, the nature of mtAPE remains unclear. APE activity, which was believed to be due to APE1, was demonstrated in Xenopus oocyte mitochondria (13). Tell et al. (25) showed APE1's localization in rat cell mitochondria. Because APE1 does not appear to possess N-terminal MTS, the targeting of APE1 to the mitochondria was difficult to explain, although APE1 was suggested to diffuse through the mitochondrial permeability transition pore in oxidatively stressed cells (26,27).
The presence of APE activity in mouse cell mitochondria was first demonstrated by Tomkinson et al. (28) who partially purified the enzyme, and observed APE1 antibody cross-reacting 66–68 kDa bands. However, these larger proteins were not further characterized. Subsequently a homolog of APE1, named APE2, was identified in the human genome database which encodes a 62 kDa protein (29). Although APE2 with putative MTS was shown to be present in both the nucleus and mitochondria, it has no detectable APE activity (29,30).
One problem in studying mitochondrial proteins, not appreciated earlier, is the difficulty of removing endoplasmic reticulum (ER) and cytosolic contaminants from purified mitochondria (31). Some proteins, originally shown to be present in the mitochondrial matrix, could in fact be adventitiously associated with the mitochondrial outer membrane and ER (32). However, such extraneous proteins are susceptible to trypsin which does not degrade the matrix proteins (33).
Because of the uncertainty about the nature of mtAPE, we decided to identify the mammalian mtAPE by purifying the enzyme, based on activity. We have shown here that mtAPE is derived from APE1 by proteolytic cleavage of 33 N-terminal residues.
MATERIALS AND METHODS
Purification of mitochondria
Mitochondria from beef liver (4 lbs) were purified by fractionating cell lysate via differential centrifugation with slight modification of a published procedure (20). The mitochondrial pellet was resuspended in 50 ml buffer A (10 mM HEPES–KOH, pH 7.4, 250 mM sucrose, 10 mM DTT, 0.5 mM EGTA and 2 m EDTA). The final purification step involved centrifugation in sucrose step gradient containing 1 and 1.5 M sucrose in 10 mM HEPES–KOH (pH 7.4) and 1 mM EDTA. The mitochondria were harvested from the sucrose band interphase, washed twice with buffer A, and then lysed with a buffer containing 20 mM HEPES–KOH (pH 7.4), 1 mM EDTA, 1 mM DTT, 300 mM KCl, 5% glycerol, protease inhibitor cocktail (Roche) and 0.5% Triton X-100. The lysate was centrifuged in a microfuge for 15 min, and the supernatant was stored at −80°C.
Trypsin treatment of mitochondria
Intact mitochondria from beef liver or mouse NIH3T3 cells (1 mg/ml) resuspended in buffer A were treated with trypsin (10 µg/ml) for 20 min at room temperature, followed by the addition of an equivalent amount of bovine trypsin inhibitor (Invitrogen) to inactivate the trypsin (33,34). Protease and mock-treated mitochondrial suspensions were washed twice in the same buffer before lysis.
Purification of mitochondrial APE from beef liver
The mitochondrial lysates were dialyzed against buffer B containing 25 mM Tris–HCl (pH 7.5), 50 mM KCl 0.1 mM EDTA, 1 mM DTT and 10% glycerol at 4°C for 3 h to remove Triton X-100, and then loaded on 50 ml HiTrap Q and SP–Sepharose columns (Amersham) connected in tandem. After washing with buffer B, the HiTrap-SP column was disconnected, from which the proteins were eluted with buffer B containing a linear gradient of KCl (0.05–1.0 M). Fractions with high-APE activity were pooled, dialyzed and then loaded onto a 5 ml HiTrap SP column. After washing, a 40 ml linear gradient of KCl (0.1–0.6 M) in buffer B was used, and the APE activity was eluted at 250–270 mM KCl. The pooled active fractions were similarly processed twice more using a 1 ml HiTrap-SP column. The peak activity present in two fractions from the last column was further purified by chromatography on a 1 ml HiTrap-Heparin column, using a 0.1–0.6 M KCl gradient. APE-containing fractions were pooled, snap-frozen and stored at −80°C.
Mass spectrometry and peptide sequence analysis
Protein bands in SDS–PAGE stained with Coomassie Blue were excised from the gel, digested with trypsin and then analyzed by MALDI-TOF (Applied Biosystems) for identification. The proteins were also independently identified by automated N-terminal sequencing after transferring to PVDF membrane.
Expression and purification of full-length and NΔ33 recombinant human APE1
The coding sequence for full-length human APE1 (hAPE1) was inserted in the pET15b vector (Novagen) at NdeI/XhoI sites for expression of APE1 in E.coli. The NΔ33 hAPE1 mutant, lacking 33 N-terminal residues, was similarly expressed using the same vector. Both plasmids encoded 20 additional amino acid residues including His6 sequence tag at the N-terminus. E.coli BL21(DE3) cells transformed with the APE1 plasmid were induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside at 0.6 _A_600 and then grown at 16°C overnight. After harvesting and then resuspending the bacteria in buffer C (20 mM Tris–HCl, pH 8.0 and 0.5 M NaCl), the cells were sonicated, filtered and then applied to a 3 ml Ni-NTA Superflow (Qiagen) column. After washing successively with 30 ml of buffer C and then 30 ml buffer C containing 40 mM imidazole, APE1 was eluted in 1 ml fractions with 10 ml buffer C containing 100 mM imidazole. After pooling active fractions and dialysis in a buffer containing 20 mM Tris–HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.1 mM DTT and 10% glycerol, the solution was treated with thrombin (10 U) before SP-Sepharose chromatography. APE1 was eluted with a linear gradient of NaCl (0.1–0.6 M) in the previous buffer. Thrombin removed all but two additional N-terminal residues in APE1 or NΔ33 APE1. The active fractions of both enzymes were stored at −20°C in a buffer containing 20 mM Tris–HCl (pH 8.0), 300 mM NaCl, 1 mM EDTA, 1 mM DTT and 50% glycerol.
AP-endonuclease assay
A 43mer oligo duplex containing tetrahydrofuran (THF; Midland) at position 31 in the sequence 5′-GATCTGATTCCCCATCTCCTCAGTTTCACTXCTGCACCGCATG-3′(X: THF) was 5′-terminally labeled using [γ-32P]ATP and T4 polynucleotide kinase (PNK). After annealing to the complementary strand with A opposite THF and subsequent purification by PAGE, the duplex oligo (500 nM) was incubated with mitochondrial fractions or recombinant APE1 at 37°C for 10 min in a 10 µl reaction mixture containing 50 mM KCl, 2 mM MgCl2, 0.5 mM DTT, 0.1 mM EDTA and 100 µg/ml BSA at pH 9.2 (50 mM AMPSO) or at pH 7.5 (50 mM Tris–HCl). After stopping the reaction with 80% formamide/10 mM NaOH containing 0.05% xylene cyanol, the oligos were separated by denaturing gel electrophoresis in 20% polyacrylamide containing 8 M urea. Radioactivity in the separated DNA bands was quantified in a PhosphorImager using Imagequant software (Molecular Dynamics). Preliminary enzyme activity assays were carried out to ensure linearity of product formation with respect to both time of incubation and the amount of extract.
The _K_m and _k_cat values of the NΔ33 APE1 and the full-length APE1 were determined after incubating 0.01 nM enzyme with various concentration of THF-containing oligo substrate at pH 9.2, or with 0.05 nM enzyme at pH 8.0, at 37°C for 4 min in the same reaction buffer as stated before.
DNA 3′ phosphodiesterase assay
To measure the DNA 3′ end-cleaning activity, a 26mer oligodeoxynucleotide with U at the 5′-terminus was labeled with [γ-32P]ATP by T4 PNK. This oligo was annealed with a 51mer oligo containing a non-complementary 25 nt extension at the 3′ end (30). Subsequent annealing of a complementary 25mer oligonucleotide and treatment with T4 DNA ligase generated a 51mer duplex with U at position 26 in the labeled strand. The 51mer duplex oligonucleotide was treated with E.coli, Udg (35) and Fpg (36) to generate 5′ 32P-labeled 3′ phosphate after excision of U and DNA strand cleavage owing to the βδ lyase activity of Fpg. In a parallel reaction, the duplex oligonucleotide was treated with Udg (35) and Nth (36) to generate 3′ 32P-phospho α,β-unsaturated aldehyde. The oligonucleotides were purified and treated with APE1 or PNK as described earlier (30) and the radioactivity in the free phosphate or phosphoaldehyde was quantified by PhosphoImager analysis.
Western analysis
The proteins after SDS–PAGE were transferred to a nitrocellulose membrane (BioRad) and blocked with TBST (20 mM Tris–HCl), pH 7.5, 500 mM NaCl with 0.1% Tween 20 containing 5% non-fat dry milk (37). The membranes were subsequently probed with rabbit anti-hAPE1 IgG (38) or with anti-lamin b antibody (Santa Cruz Biotechnology), also in TBST containing 5% non-fat dry milk. The bands were visualized using ECL (Amersham Biosciences) and analyzed with Imagequant software (Molecular Dynamics).
Synthesis of the 35S-labeled APE1
[35S]hAPE1 was synthesized in vitro via coupled transcription-translation using TnT-coupled Reticulocyte Lysate System (Promega). APE1 cDNA cloned in pRSET B vector (1 µg) was incubated in 50 µl reaction mixture containing [35S]methionine (20 µCi) according to the manufacturer's protocol. Aliquots of 1 µl were individually incubated with 25 µg extracts of nucleus, cytoplasm or mitochondria purified from beef liver in buffer A (20 µl) for 30 min at 37°C. After stopping the reaction with the loading buffer, the proteins were separated by SDS–PAGE and visualized with PhosphorImager.
Intracellular localization of full-length and truncated hAPE1
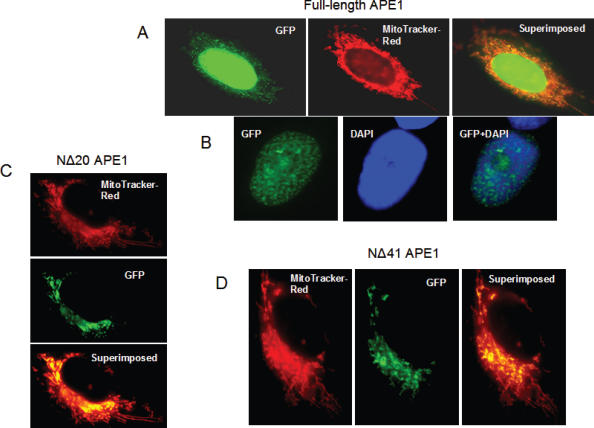
293 cells grown on cover slips in 35 mm dishes were transfected for 6 h with 0.5 µg plasmid DNA (full-length, NΔ20 and NΔ41 APE1-EGFP), using Lipofectamine 2000 and OptiMEM (Invitrogen), and 18 h later, the live cells were treated with MitoTracker Red (20 nM). In selected experiments, cells were fixed in methanol: acetone (1:1) and stained with DAPI. Fluorescent images were captured using a Photometrix Cool-SNAP Fx digital camera mounted on a NIKON Eclipse TE 200 UV microscope.
RESULTS
Identification of NΔ33 APE1 as the major mitochondrial AP-endonuclease
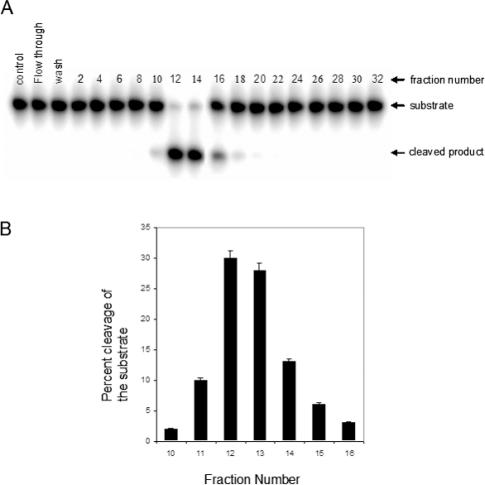
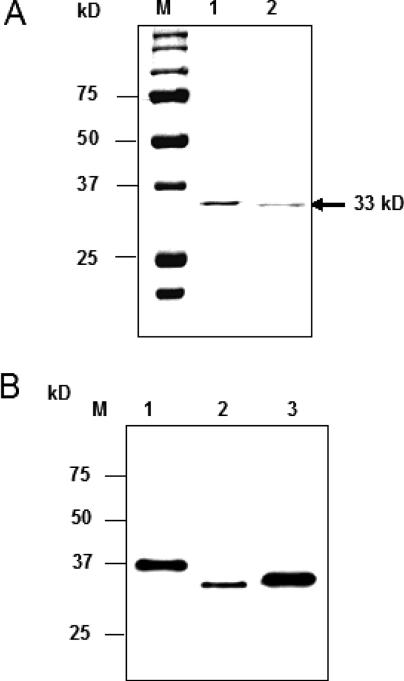
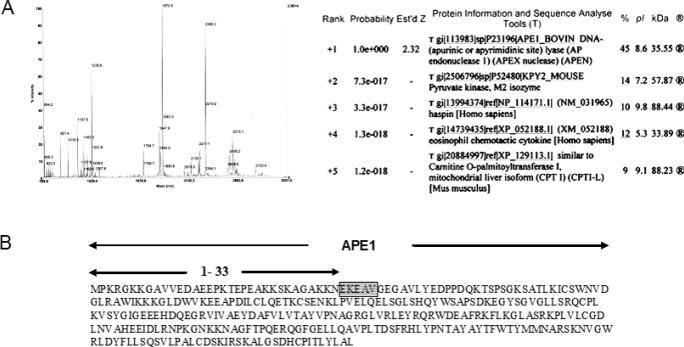
We purified APE activity from beef liver mitochondria as described in Materials and Methods. Final fractions 12–14 from Heparin–Sepharose chromatography contained the most APE activity (Figure 1). SDS–PAGE Analysis of fraction 12 indicated a 33 kDa major protein band (Figure 2A) which showed strong cross-reaction with hAPE1 antibody suggesting that the 33 kDa species is closely related to APE1 (Figure 2B). MALDI-TOF and electrospray mass spectrometry after trypsin digestion of the excised band (∼1 µg) identified the peptides as being derived from APE1 (Figure 3A). Subsequent automated Edman degradation showed the sequences to be EKEAV at the N-terminus of the band, corresponded to residues 34–38 in APE1. Thus mtAPE is derived from APE1 after deletion of 33 N-terminal residues (Figure 3B).
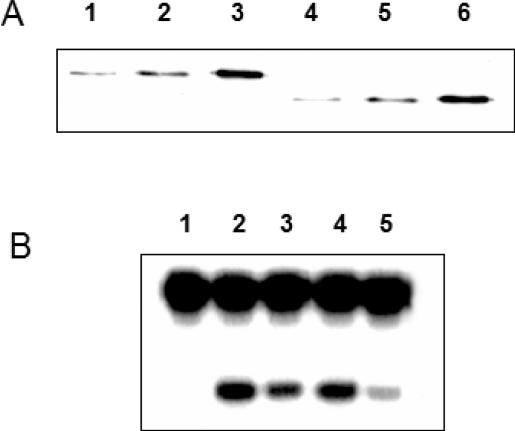
Figure 1.
APE activity of Heparin–Sepharose fractions. (A) APE activity of fractions was measured at 37°C for 10 min with 1:200 diluted fractions and 500 nM substrate. (B) Quantitative representation of APE activity in active fractions (10–16) after 1:500 dilution.
Figure 2.
Identification of purified bovine mtAPE. (A) Coomassie staining of fraction 12 (25 µl; Figure 1). Lane 1, Recombinant NΔ33 hAPE1. Lane 2, fraction 12; M, molecular weight markers. (B) Western blot of fraction 12 with APE1 antibody. Lane 1, 20 ng recombinant hAPE1; Lane 2, fraction 12 (2 µl); Lane 3, recombinant NΔ20 APE1 (25 ng).
Figure 3.
Confirmation of mtAPE as NΔ33 APE1. (A) MS analysis of trypsin-digested mtAPE band (fraction 12). (B) N-terminal sequence of mtAPE of EKEAV (shown in the box).
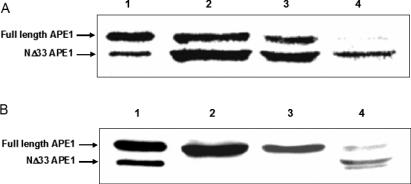
Comparative properties of recombinant human APE1 (hAPE1) and bovine mtAPE
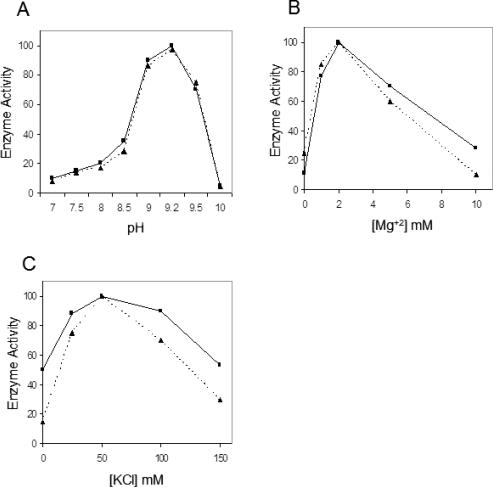
We compared the enzymatic parameters of purified, recombinant hAPE1 and bovine mtAPE. The pH dependence of APE activity showed the pH optimum to be 9.2, and the highest activity was observed at 50 mM KCl and 2 mM MgCl2, for both enzymes (Figure 4). We then compared the specific activity of full-length hAPE1 and bovine mtAPE by using equimolar amounts of hAPE1 and purified bovine mtAPE (Heparin–Sepharose fraction 12). The amount of the bovine enzyme was estimated by comparison with hAPE1 by quantitative western analysis (Figure 5A). Figure 5B shows that comparable cleavage of THF-oligo (500 nM) required 100 pM hAPE1 and about a third as much bovine enzyme. Thus the specific activity of bovine mtAPE appeared to be about 3-fold higher than of full-length hAPE1.
Figure 4.
Kinetic parameters for recombinant hAPE1 (closed square) and bovine mtAPE (closed triangle). Dependence on (A) pH, (B) Mg2+ and (C) KCl.
Figure 5.
Comparative endonuclease activity of recombinant hAPE1 and bovine mtAPE (fraction 12). (A) Quantification of hAPE1 and bovine mtAPE from western analysis. Lanes 1–3, 4, 8 and 16 ng of APE1, respectively; lanes 4–6, 0.5, 1 and 2 µl of fraction 12, respectively. (B) THF-oligo substrate was incubated with hAPE1 or fraction 12 as described in Materials and Methods. Lane 1, no enzyme; lane 2, 1 fmol hAPE1; lane 3, 0.5 fmol hAPE1; lanes 4–5, fraction 12, at 1000- and 2500-fold dilution, respectively.
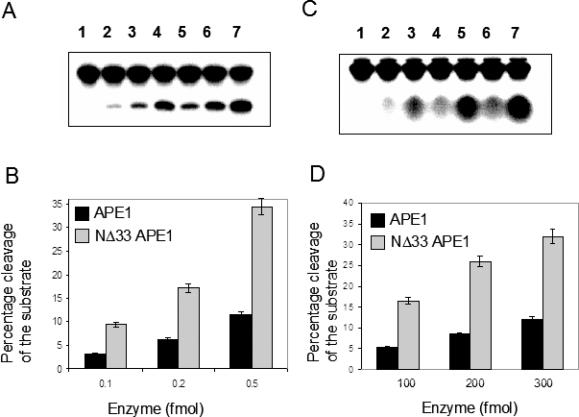
Enhanced endonuclease Activity of NΔ33 APE1 is due to increased enzyme turnover
In order to eliminate the possibility that the higher turnover of bovine mtAPE relative to hAPE1 is due to intrinsic difference of human and bovine APEs, we purified recombinant NΔ33 hAPE1 in the same way as the full-length hAPE1. We confirmed that the NΔ33 hAPE1 was indeed three times more active than the full-length hAPE1 (Figure 6A and B). We then compared the 3′ phosphoesterase activities of full-length and truncated enzymes with DNA containing 3′ phospho α,β unsaturated aldehyde. This 3′ phosphodiesterase activity was ∼100-fold lower compared with the endonuclease activity of APE1, in confirmation of earlier observations (4). Nevertheless, NΔ33 hAPE1 had 3-fold higher specific activity than the full-length APE1 (Figure 6C and D).
Figure 6.
Relative activity of NΔ33 and full-length hAPE1. (A) Endonuclease activity, lane 1, control; lanes 2–4, 0.1, 0.2 and 0.5 fmol full-length APE1; lanes 5–7, 0.1, 0.2 and 0.5 fmol NΔ33 APE1. (C) 3′ Phosphodiesterase activity, lane 1, control; lanes 2, 4 and 6; 100, 200 and 300 fmol APE1; lanes 3, 5and 7; 100, 200 and 300 fmol NΔ33 APE1. (B and D) Graphical representation of the results in (A) and (C), respectively.
On the other hand, full-length and NΔ33 hAPE1 showed comparable _K_m (∼18 nM) with the THF-oligo substrate similar to that observed earlier (39). This indicates N-33 deletion of APE1 does not affect its substrate affinity. However, the _k_cat of NΔ33 hAPE1 was 3-fold higher than that of the full length enzyme (Table 1). Although APE1's specific activity was 5-fold higher at pH 9.2 than at pH 7.4, a similar difference in specific activities of full-length and truncated APE1 was maintained at pH 7.4.
Table 1.
Kinetic parameters of full-length and truncated hAPE1
| APE1 | NΔ33 APE1 | |
|---|---|---|
| _K_m (nM) | 17 | 19 |
| _K_cat (min−1) | 68 | 200 |
| _K_cat/_K_m (min−1 nM−1) | 4 | 10.5 |
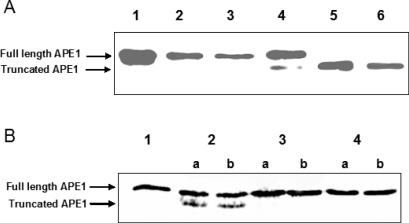
NΔ33 APE1 is the predominant APE in beef liver mitochondria
That the APE species with N-33 deletion is indeed present in the mitochondrial matrix, and was not generated artifactually from full-length APE1 during its purification, was ensured by immunoblotting extracts of crude, purified and trypsin-treated mitochondria. Controlled trypsin treatment cleaves most proteins bound to the mitochondrial outer membrane but not the matrix proteins. Western analysis of extracts of mitochondria purified through two cycles of sucrose gradient centrifugation and subsequent trypsin treatment, showed the presence of only the NΔ33 APE1 species, and not the full-length protein (Figure 7A). However, considerable loss of the total APE in trypsin-treated mitochondria was observed, probably because of damage to mitochondrial membrane during purification. Nuclear contamination of mitochondrial preparations as tested by the presence of lamin B was negligible (data not shown). Similarly, a very low level of cytoplasmic lactic dehydrogenase (<1.5% of the cytoplasmic extract) in the mitochondrial extract indicated only slight contamination with cytosolic proteins. In any case, the presence of NΔ33 APE1 as the major form of AP endonuclease in the mitochondrial matrix is evident, in spite of degradation of both truncated and full-length APE1 after prolonged trypsin treatment (Figure 7A, lane 4). To ensure exclusive mitochondrial localization of NΔ33 APE1, nuclear, cytoplasmic and mitochondrial extracts were used for western blotting with APE1 antibody using equal amount of protein. Figure 7B shows the presence of NΔ33 APE1 only in the mitochondria.
Figure 7.
Presence of NΔ33 APE1 in beef liver mitochondria. (A) Lane 1, full-length and NΔ33 APE1 markers; lane 2, mitochondrial extract (50 µg); lane 3, extract (50 µg) of sucrose density gradient-purified mitochondria; lane 4, 25 µg extract after trypsin treatment for 20 min. (B) Lane 1, APE1 and NΔ33 APE1 markers; lanes 2–4, extract (25 µg) of nucleus, cytoplasm and mitochondria, respectively.
In order to establish that mtAPE is similarly generated in other mammals by N-terminal cleavage of APE1, we examined mitochondrial APE in mouse NIH 3T3 cells. Figure 8A demonstrates the presence of only the truncated form of APE1 after trypsin treatment of the intact mitochondria (lane 5). The higher level of mtAPE-specific band in lane 5 compared with that in lane 4 was due to larger amount of mitochondrial extract in lane 5 and not due to conversion of the full-length enzyme to the truncated form.
Figure 8.
Presence of NΔ33 APE1 in NIH3T3 cells. (A) Western analysis. Lane 1, recombinant full length hAPE1; lane 2, 20 µg nuclear extract; lane 3, 20 µg cytoplasmic fraction; lane 4, 50 µg mitochondrial extract; lane 5, 50 µg mitochondrial extract after trypsin treatment for 20 min; lane 6, recombinant NΔ33 APE1. (B) Specific cleavage of full-length 35S-labeled hAPE1 by mitochondrial extract. Lane 1, APE1 control; lanes 2–4, treatment with mitochondrial, nuclear and cytoplasmic extracts, respectively as described in Materials and Methods. Full-length and cleaved products in duplicates (a and b) are indicated by arrows.
Generation of mtAPE from APE1 by a peptidase in mitochondrial extracts
To test whether a site-specific peptidase cleaves 33 N-terminal residues, we incubated 35S-labeled hAPE1 with extracts of nuclei, mitochondria and cytoplasm purified from the beef liver. Figure 8B shows that the peptidase activity is associated with the mitochondria, but is unlikely to be the mitochondrial matrix peptidase which is responsible for cleaving the MTS of most mitochondrial proteins.
Intracellular localization of full-length and truncated APE1
The experiments described so far suggest that mtAPE does not have an N-terminal MTS, and it appears to be generated outside the matrix. The NΔ33 APE1 lacking the NLS is imported into the mitochondrial matrix possibly via an internal MTS as with cytochrome C. We tested this possibility by examining subcellular distribution of ectopic full-length, NΔ20 and NΔ41 hAPE1 C-terminally fused to EGFP. The full-length APE1 was mostly localized to the nucleus in live cells as observed earlier (40) and confirmed by colocalization with DAPI in fixed cells (Figure 9A and B). The truncated proteins were predominantly distributed in the cytosol, as well as mitochondria as confirmed by colocalization with Mitotracker Red (Figure 9C and D). These results are consistent with our conclusion that the absence of NLS is a prerequisite for mitochondrial import of APE1.
Figure 9.
Intracellular localization of full-length and truncated hAPE1 with C-terminal EGFP tag. (A) Nuclear as well as cytoplasmic and mitochondrial localization of full-length APE1-EGFP in live cells. (B) Nuclear localization of full-length APE1 in fixed cells with nuclear DAPI staining. (C and D) Colocalization of NΔ20 APE1-EGFP and NΔ41 APE1-EGFP with MitoTracker Red showing their presence in the mitochondria.
DISCUSSION
Since the discovery of robust BER activity in mammalian mitochondria several years ago (13,19,41), most mitochondrial BER enzymes were identified and characterized. However, the identity of APE, a key BER enzyme was not established. Several recent studies indicated the presence of nuclear APE1 in the mitochondria, suggesting that APE1 is not altered after its mitochondrial import. However, this was unexpected because of the apparent absence of N-terminal MTS in APE1. We decided to adopt the ab initio approach for establishing the identity of mtAPE, based on activity. We have shown here that the mtAPE lacks the N-terminal 33 amino acid residues including the NLS required for nuclear import. We have further shown that the extract of mitochondria but not of nuclei or cytosol, cleaves recombinant hAPE1 to generate a mtAPE-sized product. Our recent studies suggest that a serine protease associated with mitochondria/ER is responsible for this activity (B. Szczesny, unpublished data). These results imply that the loss of NLS is a prerequisite for mitochondrial import of APE. Most mitochondrial proteins contain N-terminal MTS which is cleaved off by the mitochondrial processing peptidase localized within the matrix (42). In the absence of a candidate MTS at the N-terminus, it appears that mtAPE, like a few other mitochondrial proteins, has an alternative, internal MTS which is not cleaved for mitochondrial import. In any event, the level of mtAPE is generally low, and could not be detected in many cell lines by Western analysis of total mitochondrial extracts (B. Szczesny, unpublished data). The mtAPE could be distinguished from APE1 only by size difference in Western blots, while immunostaining with APE1 antibody will show the presence of the protein in both nucleus and mitochondria (25). This may explain failure to observe mtAPE as a distinct species in some earlier studies. We would like to point out that the truncated NΔ33 APE1 was present at a significant level only in beef liver (Figure 7, lane 2). In contrast, we could detect primarily the full-length APE1 with a trace of mtAPE-sized band in variety of human cell lines. We observed the mtAPE in the mitochondrial extract of mouse NIH 3T3 cells only after trypsin treatment (Figure 8A, lane 5). Our results are thus consistent with earlier observation that the full-length APE1 is the major species in cultured cells.
It is interesting that deleting 33 N-terminal amino acid residues increases the specific activity of mtAPE by 3-fold, primarily because of reduced affinity for the product relative to the substrate. Product inhibition of early BER enzymes, i.e. DNA glycosylases and APE has been observed before, leading to the concept of repair-coordination where the consecutive steps in the BER pathway could be coupled (43–45). The disordered N-terminal domain of APE1 is involved in interaction with other BER proteins including DNA polymerase β and XRCC1 (46,47). The absence of some N-terminal residues in mtAPE suggests that such coordination of BER may not be critical in mitochondrial DNA repair. At the same time, we have shown that up to 61 N-terminal amino acid residues in hAPE1 are dispensable for during in vitro repair activity (48). It is not known whether residues 34–61 present in mtAPE have a regulatory role during in vivo repair of mitochondrial genomes. We should also note that APE1 has two distinct roles in transcriptional regulation. It acts as a reductive activator of many transcription factors, e.g. C-Jun and p53, and was named Ref1 for this activity (49,50). Cys65 in APE1 was shown to be the active residue for Ref1 function which requires 127 N-terminal residues (51). APE1 was subsequently shown to act directly as a transcriptional repressor of renin, parathyroid and possibly other genes by participating in protein complexes which bind to the negative Ca2+ response elements (nCaRE) (52,53). We have shown the involvement of Lys6/Lys7 acetylation in this process (54). Thus the mtAPE lacks the acetylation-dependant regulatory activity, while retaining the Cys65-dependant redox function.
We have examined age-dependent changes in the intracellular distribution of APE1, whose cytosolic distribution in many cell types is surprising. Analysis of APE activity in nuclear, cytosolic and mitochondrial fractions of hepatocytes from various ages of BALB/C mice showed that the total APE activity in hepatocyte extracts does not vary significantly, but its intracellular redistribution occurs with age (31). The specific activity of APE is ∼4-fold higher in the nuclei of old hepatocytes, relatively to the cells obtained from young liver. The mtAPE level increases even more in old mouse livers. We hypothesized that chronic oxidative stress associated with aging is responsible for targeting of APE1 to the nucleus and mitochondria. Oxidative stress in cultured cells also increases the levels of APE in mitochondria and nuclei, in support of our hypothesis (38).
Lieberman and her collaborators have shown that APE1 is a component of the SET complex in cytotoxic T lymphocytes and natural killer cells, and is inactivated by the protease granzyme A which cleaves 31 N-terminal residues from APE1 (55). We could not explain these results because deletion of up to 61 N-terminal residues does not affect APE1's enzymatic activity (48). In fact, deletion of 33 N-terminal residues enhances APE1's repair activity as shown here. It is however possible that, N-terminal truncation leads to APE1's degradation in those lymphocytes.
Finally, we have shown recently that APE1 is essential for survival of APE1 conditional null mutant mouse embryo fibroblasts (56). Others have shown that APE1 downregulation by siRNA causes apoptosis of both untransformed and tumor cells (57,58). APE1 is generally believed to be essential for repairing nuclear DNA damage. However, in view of our earlier studies raising the possibility of APE1-independent BER in the nucleus (59), It is tempting to speculate that APE1's essentiality is due to its role as a precursor of mtAPE, and that the lack of mtDNA repair triggers apoptosis in APE1 deficient cells. It should now be possible to test this hypothesis using our APE1 conditional mutant cells.
Acknowledgments
We acknowledge Dr R. Roy's initial studies in purification of mitochondria and characterization of APE1 antibody cross reacting proteins. We also thank Dr David Konkel for critical editing of this manuscript, and Ms. Wanda Smith for expert secretarial assistance. We are grateful to A. Kurosky and S. S. Smith of the Biomolecular Resource Facility for protein characterization. This work was supported by USPHS grants R01 CA53791, ES08457, P01 AG021830 (S.M.), CA98664 (T.I.), and NIEHS Center Grant ES06676. Funding to pay the Open Access publication charges for this article was provided by R01 CA53791.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dodson M.L., Michaels M.L., Lloyd R.S. Unified catalytic mechanism for DNA glycosylases. J. Biol. Chem. 1994;269:32709–32712. [PubMed] [Google Scholar]
- 2.Giloni L., Takeshita M., Johnson F., Iden C., Grollman A.P. Bleomycin-induced strand-scission of DNA. Mechanism of deoxyribose cleavage. J. Biol. Chem. 1981;256:8608–8615. [PubMed] [Google Scholar]
- 3.Henner W.D., Grunberg S.M., Haseltine W.A. Enzyme action at 3′ termini of ionizing radiation-induced DNA strand breaks. J. Biol. Chem. 1983;258:15198–15205. [PubMed] [Google Scholar]
- 4.Demple B., Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 5.Johnson R.E., Torres-Ramos C.A., Izumi T., Mitra S., Prakash S., Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ribar B., Izumi T., Mitra S. The major role of human AP-endonuclease homolog Apn2 in repair of abasic sites in Schizosaccharomyces pombe. Nucleic Acids Res. 2004;32:115–126. doi: 10.1093/nar/gkh151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hudson E.K., Hogue B.A., Souza-Pinto N.C., Croteau D.L., Anson R.M., Bohr V.A., Hansford R.G. Age-associated change in mitochondrial DNA damage. Free Radic. Res. 1998;29:573–579. doi: 10.1080/10715769800300611. [DOI] [PubMed] [Google Scholar]
- 8.Yakes F.M., Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl Acad. Sci. USA. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace D.C. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 10.Shanske A.L., Shanske S., DiMauro S. The other human genome. Arch. Pediatr. Adolesc. Med. 2001;155:1210–1216. doi: 10.1001/archpedi.155.11.1210. [DOI] [PubMed] [Google Scholar]
- 11.Kang D., Hamasaki N. Maintenance of mitochondrial DNA integrity: repair and degradation. Curr. Genet. 2002;41:311–322. doi: 10.1007/s00294-002-0312-0. [DOI] [PubMed] [Google Scholar]
- 12.Clayton D.A., Doda J.N., Friedberg E.C. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc. Natl Acad. Sci. USA. 1974;71:2777–2781. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinz K.G., Bogenhagen D.F. Efficient repair of abasic sites in DNA by mitochondrial enzymes. Mol. Cell. Biol. 1998;18:1257–1265. doi: 10.1128/mcb.18.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bohr V.A., Stevnsner T., de Souza-Pinto N.C. Mitochondrial DNA repair of oxidative damage in mammalian cells. Gene. 2002;286:127–134. doi: 10.1016/s0378-1119(01)00813-7. [DOI] [PubMed] [Google Scholar]
- 15.Karahalil B., Hogue B.A., de Souza-Pinto N.C., Bohr V.A. Base excision repair capacity in mitochondria and nuclei: tissue-specific variations. FASEB J. 2002;16:1895–1902. doi: 10.1096/fj.02-0463com. [DOI] [PubMed] [Google Scholar]
- 16.LeDoux S.P., Wilson G.L., Beecham E.J., Stevnsner T., Wassermann K., Bohr V.A. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis. 1992;13:1967–1973. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- 17.Driggers W.J., LeDoux S.P., Wilson G.L. Repair of oxidative damage within the mitochondrial DNA of RINr 38 cells. J. Biol. Chem. 1993;268:22042–22045. [PubMed] [Google Scholar]
- 18.Shen C.C., Wertelecki W., Driggers W.J., LeDoux S.P., Wilson G.L. Repair of mitochondrial DNA damage induced by bleomycin in human cells. Mutat. Res. 1995;337:19–23. doi: 10.1016/0921-8777(95)00008-8. [DOI] [PubMed] [Google Scholar]
- 19.LeDoux S.P., Wilson G.L. Base excision repair of mitochondrial DNA damage in mammalian cells. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:273–284. doi: 10.1016/s0079-6603(01)68106-6. [DOI] [PubMed] [Google Scholar]
- 20.Domena J.D., Mosbaugh D.W. Purification of nuclear and mitochondrial uracil-DNA glycosylase from rat liver. Identification of two distinct subcellular forms. Biochemistry. 1985;24:7320–7328. doi: 10.1021/bi00346a045. [DOI] [PubMed] [Google Scholar]
- 21.Nilsen H., Otterlei M., Haug T., Solum K., Nagelhus T.A., Skorpen F., Krokan H.E. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 1997;25:750–755. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Croteau D.L., ap Rhys C.M., Hudson E.K., Dianov G.L., Hansford R.G., Bohr V.A. An oxidative damage-specific endonuclease from rat liver mitochondria. J. Biol. Chem. 1997;272:27338–27344. doi: 10.1074/jbc.272.43.27338. [DOI] [PubMed] [Google Scholar]
- 23.Tomkinson A.E., Bonk R.T., Kim J., Bartfeld N., Linn S. Mammalian mitochondrial endonuclease activities specific for ultraviolet-irradiated DNA. Nucleic Acids Res. 1990;18:929–935. doi: 10.1093/nar/18.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfanner N. Protein sorting: recognizing mitochondrial presequences. Curr. Biol. 2000;10:R412–R415. doi: 10.1016/s0960-9822(00)00507-8. [DOI] [PubMed] [Google Scholar]
- 25.Tell G., Crivellato E., Pines A., Paron I., Pucillo C., Manzini G., Bandiera A., Kelley M.R., Di Loreto C., Damante G. Mitochondrial localization of APE/Ref-1 in thyroid cells. Mutat. Res. 2001;485:143–152. doi: 10.1016/s0921-8777(00)00068-9. [DOI] [PubMed] [Google Scholar]
- 26.Takao M., Aburatani H., Kobayashi K., Yasui A. Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucleic Acids Res. 1998;26:2917–2922. doi: 10.1093/nar/26.12.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frossi B., Tell G., Spessotto P., Colombatti A., Vitale G., Pucillo C. H(2)O(2) induces translocation of APE/Ref-1 to mitochondria in the Raji B-cell line. J. Cell Physiol. 2002;193:180–186. doi: 10.1002/jcp.10159. [DOI] [PubMed] [Google Scholar]
- 28.Tomkinson A.E., Bonk R.T., Linn S. Mitochondrial endonuclease activities specific for apurinic/apyrimidinic sites in DNA from mouse cells. J. Biol. Chem. 1988;263:12532–12537. [PubMed] [Google Scholar]
- 29.Tsuchimoto D., Sakai Y., Sakumi K., Nishioka K., Sasaki M., Fujiwara T., Nakabeppu Y. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res. 2001;29:2349–2360. doi: 10.1093/nar/29.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiederhold L., Leppard J.B., Kedar P., Karimi-Busheri F., Rasouli-Nia A., Weinfeld M., Tomkinson A.E., Izumi T., Prasad R., Wilson S.H., et al. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Szczesny B., Mitra S. Effect of aging on intracellular distribution of abasic (AP) endonuclease 1 in the mouse liver. Mech. Ageing Dev. 2005;126:1071–1078. doi: 10.1016/j.mad.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Szczesny B., Hazra T.K., Papaconstantinou J., Mitra S., Boldogh I. Age-dependent deficiency in import of mitochondrial DNA glycosylases required for repair of oxidatively damaged bases. Proc. Natl Acad. Sci. USA. 2003;100:10670–10675. doi: 10.1073/pnas.1932854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gordon D.M., Wang J., Amutha B., Pain D. Self-association and precursor protein binding of Saccharomyces cerevisiae Tom40p, the core component of the protein translocation channel of the mitochondrial outer membrane. Biochem. J. 2001;356:207–215. doi: 10.1042/0264-6021:3560207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulke N., Sepuri N.B., Gordon D.M., Saxena S., Dancis A., Pain D. A multisubunit complex of outer and inner mitochondrial membrane protein translocases stabilized in vivo by translocation intermediates. J. Biol. Chem. 1999;274:22847–22854. doi: 10.1074/jbc.274.32.22847. [DOI] [PubMed] [Google Scholar]
- 35.Lindahl T., Ljungquist S., Siegert W., Nyberg B., Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J. Biol. Chem. 1977;252:3286–3294. [PubMed] [Google Scholar]
- 36.Boiteux S. Properties and biological functions of the NTH and FPG proteins of Escherichia coli: two DNA glycosylases that repair oxidative damage in DNA. J. Photochem. Photobiol. B. 1993;19:87–96. doi: 10.1016/1011-1344(93)87101-r. [DOI] [PubMed] [Google Scholar]
- 37.Boldogh I., Ramana C.V., Chen Z., Biswas T., Hazra T.K., Grosch S., Grombacher T., Mitra S., Kaina B. Regulation of expression of the DNA repair gene O6-methylguanine-DNA methyltransferase via protein kinase C-mediated signaling. Cancer Res. 1998;58:3950–3956. [PubMed] [Google Scholar]
- 38.Ramana C.V., Boldogh I., Izumi T., Mitra S. Activation of apurinic/apyrimidinic endonuclease in human cells by reactive oxygen species and its correlation with their adaptive response to genotoxicity of free radicals. Proc. Natl Acad. Sci. USA. 1998;95:5061–5066. doi: 10.1073/pnas.95.9.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson D.M., 3rd, Takeshita M., Grollman A.P., Demple B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J. Biol. Chem. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- 40.Jackson E.B., Theriot C.A., Chattopadhyay R., Mitra S., Izumi T. Analysis of nuclear transport signals in the human apurinic/apyrimidinic endonuclease (APE1/Ref1) Nucleic Acids Res. 2005;33:3303–3312. doi: 10.1093/nar/gki641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dianov G.L., Souza-Pinto N., Nyaga S.G., Thybo T., Stevnsner T., Bohr V.A. Base excision repair in nuclear and mitochondrial DNA. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:285–297. doi: 10.1016/s0079-6603(01)68107-8. [DOI] [PubMed] [Google Scholar]
- 42.Rehling P., Brandner K., Pfanner N. Mitochondrial import and the twin-pore translocase. Nature Rev. Mol. Cell Biol. 2004;5:519–530. doi: 10.1038/nrm1426. [DOI] [PubMed] [Google Scholar]
- 43.Hill J.W., Hazra T.K., Izumi T., Mitra S. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 2001;29:430–438. doi: 10.1093/nar/29.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mol C.D., Izumi T., Mitra S., Tainer J.A. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected] Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 45.Wilson S.H., Kunkel T.A. Passing the baton in base excision repair. Nature Struct. Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 46.Bennett R.A., Wilson D.M., III, Wong D., Demple B. Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway. Proc. Natl Acad. Sci. USA. 1997;94:7166–7169. doi: 10.1073/pnas.94.14.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vidal A.E., Boiteux S., Hickson I.D., Radicella J.P. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Izumi T., Mitra S. Deletion analysis of human AP-endonuclease: minimum sequence required for the endonuclease activity. Carcinogenesis. 1998;19:525–527. doi: 10.1093/carcin/19.3.525. [DOI] [PubMed] [Google Scholar]
- 49.Xanthoudakis S., Miao G., Wang F., Pan Y.C., Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xanthoudakis S., Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker L.J., Robson C.N., Black E., Gillespie D., Hickson I.D. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol. Cell. Biol. 1993;13:5370–5376. doi: 10.1128/mcb.13.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuchs S., Philippe J., Corvol P., Pinet F. Implication of Ref-1 in the repression of renin gene transcription by intracellular calcium. J. Hypertens. 2003;21:327–335. doi: 10.1097/00004872-200302000-00024. [DOI] [PubMed] [Google Scholar]
- 53.Okazaki T., Chung U., Nishishita T., Ebisu S., Usuda S., Mishiro S., Xanthoudakis S., Igarashi T., Ogata E. A redox factor protein, ref1, is involved in negative gene regulation by extracellular calcium. J. Biol. Chem. 1994;269:27855–27862. [PubMed] [Google Scholar]
- 54.Bhakat K.K., Izumi T., Yang S.H., Hazra T.K., Mitra S. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J. 2003;22:6299–6309. doi: 10.1093/emboj/cdg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan Z., Beresford P.J., Zhang D., Xu Z., Novina C.D., Yoshida A., Pommier Y., Lieberman J. Cleaving the oxidative repair protein Ape1 enhances cell death mediated by granzyme A. Nature Immunol. 2003;4:145–153. doi: 10.1038/ni885. [DOI] [PubMed] [Google Scholar]
- 56.Izumi T., Brown D.B., Naidu C.V., Bhakat K.K., Macinnes M.A., Saito H., Chen D.J., Mitra S. Two essential but distinct functions of the mammalian abasic endonuclease. Proc. Natl Acad. Sci. USA. 2005;102:5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vasko M.R., Guo C., Kelley M.R. The multifunctional DNA repair/redox enzyme Ape1/Ref-1 promotes survival of neurons after oxidative stress. DNA Repair (Amst.) 2005;4:367–379. doi: 10.1016/j.dnarep.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 58.Fung H., Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol. Cell. 2005;17:463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 59.Mokkapati S.K., Wiederhold L., Hazra T.K., Mitra S. Stimulation of DNA glycosylase activity of OGG1 by NEIL1: functional collaboration between two human DNA glycosylases. Biochemistry. 2004;43:11596–11604. doi: 10.1021/bi049097i. [DOI] [PubMed] [Google Scholar]