Flanking sequences profoundly alter polyglutamine toxicity in yeast (original) (raw)
Abstract
Protein misfolding is the molecular basis for several human diseases. How the primary amino acid sequence triggers misfolding and determines the benign or toxic character of the misfolded protein remains largely obscure. Among proteins that misfold, polyglutamine (polyQ) expansion proteins provide an interesting case: Each causes a distinct neurodegenerative disease that selectively affects different neurons. However, all are broadly expressed and most become toxic when the glutamine expansion exceeds ≈39 glutamine residues. The disease-causing polyQ expansion proteins differ profoundly in the amino acids flanking the polyQ region. We therefore hypothesized that these flanking sequences influence the specific toxic character of each polyQ expansion protein. Using a yeast model, we find that sequences flanking the polyQ region of human huntingtin exon I can convert a benign protein to a toxic species and vice versa. Further, we observe that flanking sequences can direct polyQ misfolding to at least two morphologically distinct types of polyQ aggregates. Very tight aggregates always are benign, whereas amorphous aggregates can be toxic. We thereby establish a previously undescribed systematic characterization of the influence of flanking amino acid sequences on polyQ toxicity.
Keywords: huntingtin, Huntington's disease, neurodegenerative disease, protein misfolding
Proteins that misfold into toxic species are a common cause of diverse neurodegenerative disorders (1, 2). Protein misfolding and its toxic outcomes are presumed to be determined by the amino acid sequence of the particular protein. In several misfolding disorders, genetic mutations result in aberrant amino acid sequences of a protein and are responsible for its misfolding and toxicity. Despite intense study, little is known regarding how these intramolecular modulators of protein misfolding influence toxicity. This question is inherently difficult to attack because of the unrelated nature of the proteins whose misfolding causes disease and the plethora of environments and cell types in which they misfold.
Polyglutamine (polyQ) expansions provide an attractive opportunity for dissecting the relationships between protein misfolding and toxicity, because the same mutation in different contexts produces distinct pathologies. To date, eight distinct disorders are known to be caused by polyQ expansions: Huntington's disease (HD), spinal and bulbar muscular atrophy (SBMA), dentatorubral and pallidoluysian atrophy, and several spinocerebellar ataxias 1, 2, 3, 7, and 17 (3). In all of these diseases, the longer the polyQ expansion, the more severe the disease and the earlier its onset. Moreover, most of the pathologies are manifested when the expansion of the glutamine repeats reaches a similar critical threshold of ≈39 glutamine residues (3). Further, placing a polyQ expansion in a protein that normally does not contain one is sufficient to cause neurodegeneration in mice (4). Clearly, there must be a common underlying mechanism of pathology shared by all different polyQ disorders.
Notably, the different proteins containing polyQ expansions are otherwise unrelated, i.e., the polyQ regions are embedded in different amino acid sequences in each disease-causing polyQ protein (3). Moreover, the toxicity they produced first manifests in, and most severely affects, different neurons. For example, in HD, expansion of the polyQ region in the protein huntingtin primarily causes the demise of medium spiny neurons in the striatum (5, 6), whereas the polyQ expansion in the androgen receptor mostly affects sensory and motor neurons in patients with spinal and bulbar muscular atrophy (7, 8). The different pathologies these misfolded proteins produce, however, are not simply due to differences in their patterns and levels of expression. Many polyQ proteins are highly expressed in cell types in which they do not produce overt toxicity (9–12).
In vitro studies of proteins containing polyQ repeats establish that they form detergent-insoluble, amyloid-like aggregates only when the polyQ expansion is of disease-causing length (13, 14). These aggregates resemble those observed in neurons of patients suffering from polyQ diseases (15). However, although the production of microscopically salient aggregates is associated with the polyQ expansions, this alone does not determine toxicity. Aggregates composed of polyQ proteins are found in numerous cell types that have no apparent pathology, and some cells with pathology have no obvious aggregates (16–18). Indeed, even in the same original cell line, PC-12 rat pheochromocytoma cells, independent clones expressing similar polyQ-containing exon I fragments of huntingtin, all exhibited polyQ length-dependent aggregation, but some lines exhibited overt toxicity, whereas others did not (19–21). Thus, in addition to cell-type specific differences in proteome composition and stochastic variations in polyQ misfolding, the different amino acid sequences flanking the polyQ regions must constitute the molecular basis of polyQ toxicity.
We and others have developed yeast strains that express huntingtin exon I fragments with polyQ regions of different lengths (22–25). A similar huntingtin exon I fragment is found in huntingtin aggregates within neurons of HD patients and is itself sufficient to produce neurodegeneration in mouse models (26, 27). Several independent studies demonstrate that at a basic level, the yeast model faithfully recapitulates the molecular basis of polyQ length-dependent toxicity: (i) The molecular chaperone Hsp104 solubilzes huntingtin exon I fragments with expanded polyQ regions and reduces their toxicity in both a yeast and a mouse model for HD (28, 29). (ii) An intrabody directed against the amino-terminal part of huntingtin exon I decreased the amount of aggregated polyQ huntingtin and simultaneously antagonized polyQ length-dependent toxicity, both in yeast and neuronal tissue culture (30). (iii) A screen in yeast identified small compounds that inhibited toxicity and aggregation of polyQ huntingtin (31). The efficacy of these compounds was validated in neuronal tissue culture, cultured brain slices from HD transgenic mice, and a fly model (31).
An advantage of yeast cells is that they are not confounded by the perplexing variations in cellular proteomes that characterize distinct cell types in whole organisms or in cultured mammalian cells. This feature, along with their genetic tractability, provides an ideal system for dissecting the effects of sequences flanking the polyQ regions on the toxicity of polyQ expansion proteins. The few huntingtin exon I fragments studied in yeast to date faithfully recapitulate polyQ length-dependent aggregation (22–25, 32), but, surprisingly, in only one case is the fragment with a polyQ expansion toxic (25). The huntingtin constructs tested in these studies are related but differ in the nature of sequences flanking the polyQ region.
Here we establish that specific amino acid sequences flanking the polyQ region of huntingtin exon I greatly influence polyQ length-dependent toxicity: The commonly used FLAG-epitope (DYKDDDK) at the amino terminus of huntingtin exon I can unmask the toxicity of an otherwise benign polyQ protein, whereas the endogenous carboxyl-terminal polyproline region of huntingtin exon I can convert toxic proteins into nontoxic ones. Flanking sequences also strongly modulate the morphology of aggregates formed by polyQ proteins. Constructs that form one or two tight aggregates with a small surface per cell always were benign. Amorphous aggregates, however, have the capacity of being cytotoxic. This work establishes that within the same cellular environment, diverse intramolecular factors have the capacity to determine whether misfolded proteins are benign or lethal. In an accompanying study, we describe how intermolecular interactions together with the intramolecular factors described here modulate polyQ toxicity (33).
Results
Amino Acids Flanking the polyQ Region of Huntingtin Exon I Mask and Unmask polyQ Length-Dependent Toxicity.
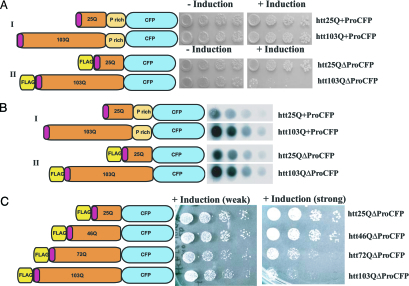
As described in ref. 22, when the exon I region of huntingtin is fused to GFP [or cyan fluorescent protein (CFP)] and expressed in yeast, it is not toxic, whether it contains a polyQ region of normal length or a disease-associated expansion (Fig. 1, construct I). However, idiosyncratically, another huntingtin exon I GFP (or CFP) fusion (25), with two modifications flanking the polyQ sequence, was toxic only when it contained the polyQ expansion (Fig. 1, construct II). In contrast to the nontoxic protein, the protein with Q length-dependent toxicity carried a synthetic peptide commonly used for antibody recognition (FLAG-tag, DYKDDDDK) at its amino terminus and also contained a deletion of the endogenous proline-rich region that immediately follows the polyQ sequence in the human huntingtin protein (Fig. 1, construct II).
Fig. 1.
A nontoxic huntingtin exon I construct and construct with graded polyQ length-dependent toxicity. (A) Yeast cells containing huntingtin constructs I (Upper) or II (Lower) with the indicated number polyQ repeats were spotted on plates that either induce (+Induction) or repress (−Induction) huntingtin exon I expression. Four serial 5-fold dilutions of cells are shown. Toxicity is reflected by the reduced growth of cells under inducing conditions. (B) Dot blots prepared with protein extracts from yeast cells expressing the indicated huntingtin constructs 8 h after induction. Four serial dilutions of the protein lysates are shown. Huntingtin constructs were detected with an anti-GFP antibody. (C) Yeast cells containing construct II with an increasing number of polyQ repeats were spotted on plates for either weak expression for constructs under control of the MET promoter (Center) or strong induction for constructs under the control of the GAL1 promoter (Right). (Left) A schematic representation of the huntingtin exon I proteins. The red insert in the polyQ region indicates the 16-aa-long amino-terminal sequence of human huntingtin.
To assess the toxicity of both constructs, we transformed identical yeast strains with both huntingtin constructs. Cells were pregrown in a medium that repressed expression of the huntingtin variant. We then plated them simultaneously onto two different media: one without inducer, to control for cell density, and another one with inducer, to test toxicity. Our results demonstrate that construct I with 103 glutamines did not produce toxicity, whereas construct II with 103 glutamines produced toxicity under exactly the same experimental conditions in yeast cells with identical genetic backgrounds (Fig. 1A).
Next, we examined the levels of accumulated protein after induction. Dot-blot analysis showed that constructs I and II accumulated at similar levels (Fig. 1B). We confirmed these results in three different yeast strains with different genetic backgrounds with at least three independent transformants for each strain (W303 as shown in Figs. 1–3; BY4741 and 74D-694, data not shown). Therefore, our findings rule out the possibility that the previously reported difference in toxicity for constructs I and II was caused by variations in genetic backgrounds of the yeast strains used by different laboratories, different culture conditions, or differences in the levels of accumulation of the proteins. We conclude that the sequences flanking the polyQ region can determine whether the same polyQ huntingtin-derived sequences will be toxic or benign. All experiments in this study were carried out in yeast cells that contain the protein Rnq1 in its prion conformation, which is a crucial prerequisite for polyQ aggregation and toxicity (25, 33).
Fig. 2.
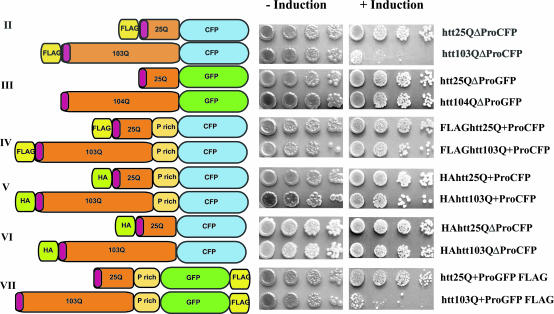
PolyQ toxicity depends on amino acids flanking the polyQ region. Yeast cells containing huntingtin constructs II–VII with either 25 or 103 polyQ repeats (except construct III with 104 repeats) were spotted on plates that either induce (+Induction) or repress (−Induction) huntingtin exon I expression (see Materials and Methods for details). Four serial 5-fold dilutions of cells are shown.
Fig. 3.
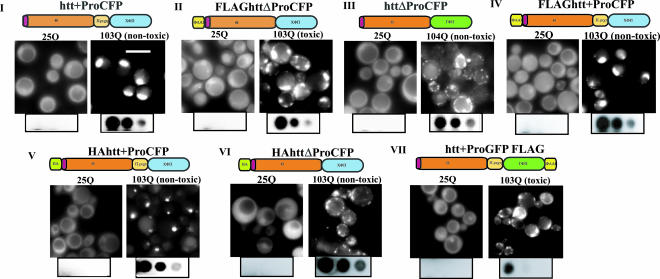
Flanking amino acids modulate polyQ aggregate morphology. Cells expressing the indicated huntingtin exon I CFP or GFP fusions proteins (constructs I–VII) were analyzed by fluorescence microscopy. (Scale bar: 5 μm.) Below the micrographs, filter retardation assays of protein lysates of cells expressing the indicated huntingtin exon I are shown. Three 5-fold dilutions of the protein extracts are shown.
We next asked whether construct II could exhibit the graded Q length-dependent toxicity that is the hallmark of polyQ expansion diseases. We constructed polyQ variants of 46 and 72 residues and compared them to the 25Q and 103Q variants. We detected no toxicity in cells expressing the 25Q plasmid, whereas the toxicity of the other constructs (46Q, 72Q, and 103Q) increased with the length of their polyQ regions (Fig. 1C). Also, the expression levels of these huntingtin fragments strongly influenced their toxicity: When the huntingtin constructs were controlled by the strong GAL1 promoter, toxicity of the constructs bearing 72 and 103 glutamine residues was extreme (Fig. 1C Right), whereas constructs under the weaker MET1 promoter resulted in mild toxicity (Fig. 1C Left). Thus, the expression of these proteins in yeast cells provides a model for the general length- and dosage-dependent toxicity of proteins with polyQ expansions observed in several organisms.
To determine whether it was the addition of the FLAG tag or the loss of the proline-rich region that is responsible for the Q length-dependent toxicity of construct II, we examined 25Q and 103Q proteins that either lacked the FLAG tag or had the endogenous proline-rich region restored (Fig. 2, constructs III and IV). Surprisingly, either change completely eliminated polyQ length-dependent toxicity: Cells expressing huntingtin exon I fusions without the FLAG tag and lacking the proline-rich region were fully viable (Fig. 2, construct III), as were cells expressing fusions containing both the FLAG tag and the proline-rich region (Fig. 2, construct IV).
These initial experiments were performed with single-copy expression constructs stably integrated into the genome and under the control of the galactose-inducible promoter GAL1. We also tested these constructs with a high copy 2μ plasmid to achieve very high levels of expression (data not shown). Even at these extreme levels of expression, no toxicity was detected if the proteins lacked the FLAG tag or contained the polyproline region. Thus, the strong differences in toxicity were solely due to the capacity of flanking sequences to completely alter the toxicity of the polyQ expansion.
Specificity of the FLAG Tag in polyQ Length-Dependent Toxicity.
To determine whether other amino-terminal extensions would confer polyQ length-dependent toxicity in the absence of the proline-rich region, we replaced the negatively charged FLAG tag (amino acid sequence DYKDDDDK) with a different, commonly used amino-terminal epitope tag of similar length: HA (the hemagglutinin epitope, YPYDVPDYA; Fig. 2, constructs V and VI). In contrast to the FLAG tag, proteins with the HA tag were not at all toxic (Fig. 2). The effects of altering even short sequences flanking the polyQ sequence in huntingtin exon I were surprising but reproducible. Again, each result was reproduced by using at least three independent transformants in three yeast strains with different genetic backgrounds in several independent experiments (data not shown).
FLAG-Induced Toxicity Is Independent of Its Position in the polyQ Protein.
We next asked whether the ability of the FLAG tag to unmask the toxicity of the polyQ sequence depended on its immediate proximity to the polyQ region. To do so, we moved the FLAG tag from the amino terminus of huntingtin exon I to the carboxyl terminus of GFP, separating the FLAG sequence from the polyQ region by the robustly folded and functional (i.e., fully fluorescent) GFP protein (Fig. 2, construct VII). In this position, the FLAG tag caused polyQ length-dependent toxicity even in the presence of the otherwise protective proline-rich region (Fig. 2). Again, this toxicity was sequence-specific because an HA tag placed carboxyl terminal of the polyQ tract did not induce toxicity (24). Thus, the protective effect of the proline-rich region depended on its proximity to the toxic FLAG tag.
Amino Acids Flanking the polyQ Region of Huntingtin Exon I Modulate Aggregate Morphology.
We now examined the relationship between solubility, aggregation, and toxicity for each of the huntingtin exon I variants. All of the 25Q huntingtin variants exhibited diffuse fluorescence (Fig. 3). All of the 103Q variants formed fluorescent foci and were insoluble in 2% SDS as demonstrated by filter retardation assays (Fig. 3). Thus, the formation of neither SDS insoluble aggregates nor fluorescent foci distinguishes toxic proteins from nontoxic ones.
Different 103Q huntingtin aggregates however, did have strikingly different morphologies. A subset of the proteins always formed one (occasionally two) tight fluorescent focus per cell (Fig. 3, constructs I, IV, and V). This morphology was observed only with nontoxic proteins that contained the proline-rich region. In other cases, the proteins formed several amorphous aggregates dispersed throughout the cytosol in every cell (Fig. 3, constructs II, III, VI, and VII). Both of the toxic proteins (II and VII) formed aggregates of this type. However, two nontoxic proteins (III and VI) formed aggregates that were morphologically indistinguishable from the aggregates formed by toxic huntingtin exon I constructs. Thus, the toxic proteins had a distinct amorphous pattern of aggregation, but morphology alone was not a toxicity-defining characteristic.
Discussion
We have taken advantage of the model organism Saccharomyces cerevisiae to explore one of the most puzzling phenomena in the field of protein misfolding: Diverse proteins containing polyQ expansions share, at some very basic level, a common mechanism of toxicity. In their endogenous setting they cause neurodegeneration once the polyQ expansion reaches a critical length of ≈39 residues. And yet, each protein exhibits a baffling cell-specific pattern of toxicity that does not correlate with its expression level and only poorly correlates with its spatial pattern of aggregation. A fragment of one of these polyQ expansion proteins, the exon I sequence of huntingtin on its own can cause neurodegenerative disease in transgenic mice (26). This fragment alone also can produce toxicity in yeast (25). Notably, we find that it did so in the graded, Q length-dependent manner that is the hallmark of polyQ diseases. Toxicity was profoundly influenced by sequences flanking the polyQ tract. Sequences flanking the polyQ region were sufficient to transform a polyQ expansion protein from benign to lethal and a lethal one to benign.
We created a series of 14 huntingtin exon I constructs, in each case making pairs of proteins with a normal polyQ tract (25Q) and a disease-related variant (103Q). The constructs carried different endogenous and heterologous amino acid sequences immediately flanking the polyQ region or distal to it. Fluorescent protein fusions allowed us to monitor aggregation and toxicity simultaneously in the same cells. In contrast to the often variable effects of polyQ sequences in animal models or mammalian cell lines (19, 21, 26, 34, 35), polyQ length-dependent toxicity in yeast was highly reproducible even when tested in yeast strains with different genetic backgrounds. This reproducibility allowed us to unambiguously establish the importance of amino acid sequences flanking the polyQ region to determine toxicity. In these experiments, we were greatly aided by the use of the most tightly regulated yeast promoter known (GAL1), which completely prevents expression and the associated complications of selective pressures, until the moment when glucose is removed and galactose added. Similarly, we took advantage of expression constructs stably integrated in the same yeast chromosomal site to maintain uniform expression in all cells in the culture.
It has been suggested that the toxicity of Q expanded huntingtin is due to its ability to sequester wild-type huntingtin protein, obliterating its essential function (36). It also has been proposed that huntingtin aggregates kill neurons by restricting the free passage of cargo in long neuronal processes (37). In addition, recent work suggests that the polyQ expansion in ataxin1 stabilizes the protein against degradation and may cause disease by a toxic increase of its normal function (38). Although all of these features may play an important part, none of them can explain the toxicity of huntingtin in our experiments. Considering that the fragment of huntingtin we expressed is but a small portion of the wild-type protein, it is very unlikely that it provides normal huntingtin function in yeast. Further, given that there is no huntingtin homolog in yeast, it cannot be sequestering wild-type protein. Thus, we establish that the misfolding of polyQ-expanded huntingtin can directly produce a toxic gain-of-function phenotype.
We found that amino acid sequences flanking the polyQ region have the capacity either to unmask the latent toxicity of an otherwise benign polyQ expansion protein or to abolish the toxicity of a lethal one. A FLAG tag (DYKDDDDK) unmasked the latent polyQ length-dependent toxicity in polyQ-expanded huntingtin exon I. The effect was highly specific because a tag of similar size (HA, YPYDVPDYA) had no effect. Strikingly, the FLAG tag was equally effective when located at the amino terminus of huntingtin exon I or separated from the polyQ region by a stably folded, functional protein (GFP). Thus, unlike other amino acid sequences flanking the polyQ region (39), the FLAG sequence probably does not transform the conformation of the polyQ region but rather induces toxic protein–protein interactions. Several studies document the amino-terminal modification of different polyQ proteins with the ubiquitin-like molecule SUMO (40, 41). Moreover, sumoylation of polyQ proteins can enhance their toxicity in different models (40). It is noteworthy that the carboxyl terminus of human SUMO is enriched in negatively charged amino acids (EEEDVIE), which is the most prominent feature of the FLAG epitope. Thus, it is tempting to speculate that the amino-terminal FLAG epitope in our toxic constructs mimics this sumoylation and modulates its toxicity in the context of polyQ expansion proteins.
We also found that the 50-aa proline-rich sequence carboxyl terminal of the polyQ region in huntingtin exon I could completely abrogate the toxicity of an otherwise lethal polyQ expansion protein. Many polyQ proteins, including several associated with human disease (e.g., atrophin, ataxin-1, and ataxin-7) have proline-rich regions adjacent to their polyQ tracts (42). The codon most commonly found in glutamine expansions (CAG) can change to a codon for proline (CCX) by the alteration of a single nucleotide, but polyQ cannot change to polyproline by a simple frameshift. We speculate that the proline-rich regions adjacent to Q rich regions have not arisen by a single chance event but by selection for their protective effects on polyQ toxicity. In a recent biochemical study, Wetzel et al. (42) reported that a oligoproline region attached to the carboxyl terminus of a polyQ protein can decrease the rate of formation of polyQ amyloid-like aggregates and also decrease the stability of aggregates when compared with polyQ proteins of the same glutamine repeat lengths without the oligoproline sequence. These results offer a possible kinetic explanation for the protective effect of the proline-rich region in our experiments. The huntingtin exon I constructs that contain the proline-rich region aggregate relatively slowly (unpublished data) and, thereby, may allow the cell to transform them into a nontoxic form. In contrast, huntingtin exon I fragments without the proline-rich region aggregate more efficiently (unpublished data) and produce toxic species by bypassing the cellular defense mechanism. Alternatively, the proline-rich region could enable specific proteins, such as prolyl isomerases or proteins with SH3 domains, to interact with the polyQ protein and, thereby, transform it into a toxicity-ameliorating species. In an accompanying paper (33), we establish that both the toxicity-inducing FLAG tag and the toxicity-ameliorating proline-rich region also can operate in trans. These results imply that the toxic and protective effects of amino acids flanking the polyQ region are mediated by the recruitment of other proteins to the polyQ expansion proteins.
Many studies report a correlation between the toxicity of polyQ expansion proteins and the formation of intracellular protein inclusions (6, 43–46), yet a number of recent reports question their relevance (47–51). Moreover, a recent study reported that the occurrence of diffuse polyQ protein strongly correlates with toxicity (52). We compared toxic and nontoxic huntingtin exon I variants in the same cell type, grown under the same conditions. Both toxic and nontoxic variants formed inclusions (as defined by fluorescence microscopy) and aggregates (as defined by SDS resistance in filter-retardation assays). Thus, aggregation per se cannot be the determinant of toxicity. However, in the experiments presented here and in an accompanying paper (33), toxicity always correlated with polyQ aggregation, and toxicity always was reduced when the polyQ-expansion proteins were soluble. Notably, toxic and nontoxic proteins produced aggregates with different morphologies: When cells contained one or two discrete inclusions with crisp boundaries per cell, no toxicity was observed. These inclusions were reminiscent of aggresomes, particular types of inclusions observed in animal cells, which are believed to be benign and might even perform a protective function (53, 54). Also, these aggregates have a small surface and, therefore, might abolish or at least minimize otherwise toxic protein interactions with the polyQ proteins. In contrast, all toxic huntingtin exon I constructs formed several amorphous aggregates per cell. This kind of aggregation displays a much larger surface of the polyQ proteins and, hence, might facilitate efficient toxic protein interactions. Two of our constructs (constructs III and VI), however, showed the amorphous aggregate morphology without toxicity, which demonstrates that aggregate morphology alone does not determine polyQ toxicity. Notably, in neuronal cell cultures, huntingtin fragments that do not contain the proline-rich region are found in aggregates with different morphologies and different coaggregating proteins when compared with huntingtin exon I fragments that contain the proline-rich region (8).
The yeast systems offers a unique platform to study protein misfolding because the strong and reproducible patterns of aggregation and toxicity in yeast allow the disentanglement of what might appear as minor, unreliable, or idiosyncratic differences in other systems. Our study provides a previously undescribed systematical characterization of the intramolecular requirements for polyQ toxicity. We demonstrate that short amino acid sequences flanking the polyQ region can determine the benign or toxic character of a polyQ expansion protein. In the accompanying article (33), we demonstrate that these flanking sequences also can execute their protective or toxic functions in trans underlining their significance to polyQ toxicity.
Materials and Methods
Materials.
Chemicals were purchased from Sigma–Aldrich and Fluka. All yeast strains used in this study are in the W303 (_MAT_α _can1_-_100 ade2_-_1 his3_-11, _15 trp1_-_1 ura3_-1 leu23,112) genetic background. Yeast shuttle vectors for genomic integration and ectopic expression were used throughout this work as described in ref. 55. Plasmids expressing huntingtin exon I fusions are summarized in Figs. 1 and 2. Plasmids were generated by using standard molecular biology techniques. The expression of huntingtin fusion proteins was controlled by the inducible GAL1 promoter unless noted otherwise. The anti-GFP antibody was purchased from Sigma-Aldrich. This antibody recognizes both GFP and CFP. Notably, GFP and CFP fusions produced identical results in regard to huntingtin exon I toxicity, aggregation, and aggregate morphology.
Yeast Methods.
Yeast media were prepared essentially as described in ref. 56 by using complete supplemental mixtures from BIO101. Transformation of yeast was performed by the lithium acetate method (57). Yeast integrating plasmids (pRS303 backbone) were linearized by restriction with BstXI before transformation. Induction of GAL1 promoter-controlled protein expression for fluorescence microcopy was achieved by growing yeast cultures in selective media containing raffinose (2%) as the sole carbon source to mid-log phase followed by growth in galactose (2%) containing selective media for 8 h. Alternatively, cells were grown in media containing glucose to mid-log phase washed three times in sterile water and then incubated in galactose containing media for 8–10 h.
Growth Assays.
Yeast transformants were grown overnight in selective media with glucose (2%) as a sole carbon source. After the OD600 was determined, cultures were diluted to equal concentrations. Cells were spotted in four 5-fold dilutions with the most concentrated spot containing ≈100,000 cells by using a spotter (Frogger; V&P Scientific, San Diego). Equal spotting was controlled by spotting the cells on yeast extract/peptone/dextrose (data not shown) and selective complete plates with glucose as carbon source in parallel.
Filter Retardation Assay and Dot Blots.
Filter retardation assays of aggregated material were performed essentially as described in ref. 23. Cells were harvested after 8 h of induction of the indicated huntingtin exon I fusions. Total protein concentrations of different samples were determined by Bradford assays and equalized to ensure equal loading. The huntingtin fusion proteins were detected by an anti-GFP antibody. Dot blot was performed as the filter retardation assays by using a poly(vinylidene difluoride) membrane instead of the cellulose acetate membrane.
Fluorescence Microscopy.
Fluorescence microscopy was carried out by using a Zeiss Axioplan II microscope and openlab (Improvision, Lexington, MA). Cells were imaged by using a ×100 objective after 8 h of induction. For visualization of GFP- and CFP-fusion proteins, GFP and cyan filters were used. metamorph (Universal Imaging, Downingtown, PA) was used to take pictures.
Acknowledgments
M.L.D. was supported by a postdoctoral fellowship from the Huntington's Disease Society of America. P.J.M. was supported by National Institutes of Health Grant NS0472371.
Abbreviations
CFP
cyan fluorescent protein
HD
Huntington's disease
polyQ
polyglutamine
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Ross C. A., Poirier M. A. Nat. Med. 2004;10(Suppl.):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 2.Thomas P. J., Qu B. H., Pedersen P. L. Trends Biochem. Sci. 1995;20:456–459. doi: 10.1016/s0968-0004(00)89100-8. [DOI] [PubMed] [Google Scholar]
- 3.Zoghbi H. Y., Orr H. T. Annu. Rev. Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 4.Ordway J. M., Tallaksen-Greene S., Gutekunst C. A., Bernstein E. M., Cearley J. A., Wiener H. W., Dure L. S., IV, Lindsey R., Hersch S. M., Jope R. S., et al. Cell. 1997;91:753–763. doi: 10.1016/s0092-8674(00)80464-x. [DOI] [PubMed] [Google Scholar]
- 5.Graveland G. A., Williams R. S., DiFiglia M. Science. 1985;227:770–773. doi: 10.1126/science.3155875. [DOI] [PubMed] [Google Scholar]
- 6.DiFiglia M., Sapp E., Chase K. O., Davies S. W., Bates G. P., Vonsattel J. P., Aronin N. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 7.La Spada A. R., Wilson E. M., Lubahn D. B., Harding A. E., Fischbeck K. H. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 8.Qin Z. H., Wang Y., Sapp E., Cuiffo B., Wanker E., Hayden M. R., Kegel K. B., Aronin N., DiFiglia M. J. Neurosci. 2004;24:269–281. doi: 10.1523/JNEUROSCI.1409-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharp A. H., Loev S. J., Schilling G., Li S. H., Li X. J., Bao J., Wagster M. V., Kotzuk J. A., Steiner J. P., Lo A., et al. Neuron. 1995;14:1065–1074. doi: 10.1016/0896-6273(95)90345-3. [DOI] [PubMed] [Google Scholar]
- 10.Schilling G., Sharp A. H., Loev S. J., Wagster M. V., Li S. H., Stine O. C., Ross C. A. Hum. Mol. Genet. 1995;4:1365–1371. doi: 10.1093/hmg/4.8.1365. [DOI] [PubMed] [Google Scholar]
- 11.Nishiyama K., Nakamura K., Murayama S., Yamada M., Kanazawa I. Ann. Neurol. 1997;41:599–605. doi: 10.1002/ana.410410508. [DOI] [PubMed] [Google Scholar]
- 12.Nishiyama K., Murayama S., Goto J., Watanabe M., Hashida H., Katayama S., Nomura Y., Nakamura S., Kanazawa I. Ann. Neurol. 1996;40:776–781. doi: 10.1002/ana.410400514. [DOI] [PubMed] [Google Scholar]
- 13.Scherzinger E., Sittler A., Schweiger K., Heiser V., Lurz R., Hasenbank R., Bates G. P., Lehrach H., Wanker E. E. Proc. Natl. Acad. Sci. USA. 1999;96:4604–4609. doi: 10.1073/pnas.96.8.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G. P., Davies S. W., Lehrach H., Wanker E. E. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 15.Diaz-Hernandez M., Moreno-Herrero F., Gomez-Ramos P., Moran M. A., Ferrer I., Baro A. M., Avila J., Hernandez F., Lucas J. J. J. Neurosci. 2004;24:9361–9371. doi: 10.1523/JNEUROSCI.2365-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuemmerle S., Gutekunst C. A., Klein A. M., Li X. J., Li S. H., Beal M. F., Hersch S. M., Ferrante R. J. Ann. Neurol. 1999;46:842–849. [PubMed] [Google Scholar]
- 17.Simeoni S., Mancini M. A., Stenoien D. L., Marcelli M., Weigel N. L., Zanisi M., Martini L., Poletti A. Hum. Mol. Genet. 2000;9:133–144. doi: 10.1093/hmg/9.1.133. [DOI] [PubMed] [Google Scholar]
- 18.Gutekunst C. A., Li S. H., Yi H., Mulroy J. S., Kuemmerle S., Jones R., Rye D., Ferrante R. J., Hersch S. M., Li X. J. J. Neurosci. 1999;19:2522–2534. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aiken C. T., Tobin A. J., Schweitzer E. S. Neurobiol. Dis. 2004;16:546–555. doi: 10.1016/j.nbd.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 20.Apostol B. L., Kazantsev A., Raffioni S., Illes K., Pallos J., Bodai L., Slepko N., Bear J. E., Gertler F. B., Hersch S., et al. Proc. Natl. Acad. Sci. USA. 2003;100:5950–5955. doi: 10.1073/pnas.2628045100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S. H., Cheng A. L., Li H., Li X. J. J. Neurosci. 1999;19:5159–5172. doi: 10.1523/JNEUROSCI.19-13-05159.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krobitsch S., Lindquist S. Proc. Natl. Acad. Sci. USA. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muchowski P. J., Schaffar G., Sittler A., Wanker E. E., Hayer-Hartl M. K., Hartl F. U. Proc. Natl. Acad. Sci. USA. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes R. E., Lo R. S., Davis C., Strand A. D., Neal C. L., Olson J. M., Fields S. Proc. Natl. Acad. Sci. USA. 2001;98:13201–13206. doi: 10.1073/pnas.191498198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meriin A. B., Zhang X., He X., Newnam G. P., Chernoff Y. O., Sherman M. Y. J. Cell Biol. 2002;157:997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S. W., Bates G. P. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 27.Mende-Mueller L. M., Toneff T., Hwang S. R., Chesselet M. F., Hook V. Y. J. Neurosci. 2001;21:1830–1837. doi: 10.1523/JNEUROSCI.21-06-01830.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cashikar A. G., Duennwald M., Lindquist S. L. J. Biol. Chem. 2005;280:23869–23875. doi: 10.1074/jbc.M502854200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vacher C., Garcia-Oroz L., Rubinsztein D. C. Hum. Mol. Genet. 2005;14:3425–3433. doi: 10.1093/hmg/ddi372. [DOI] [PubMed] [Google Scholar]
- 30.Colby D. W., Chu Y., Cassady J. P., Duennwald M., Zazulak H., Webster J. M., Messer A., Lindquist S., Ingram V. M., Wittrup K. D. Proc. Natl. Acad. Sci. USA. 2004;101:17616–17621. doi: 10.1073/pnas.0408134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X., Smith D. L., Meriin A. B., Engemann S., Russel D. E., Roark M., Washington S. L., Maxwell M. M., Marsh J. L., Thompson L. M., et al. Proc. Natl. Acad. Sci. USA. 2005;102:892–897. doi: 10.1073/pnas.0408936102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaffar G., Breuer P., Boteva R., Behrends C., Tzvetkov N., Strippel N., Sakahira H., Siegers K., Hayer-Hartl M., Hartl F. U. Mol. Cell. 2004;15:95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 33.Duennwald M. L., Jagadish S., Giorgini F., Muchowski P. J., Lindquist S. Proc. Natl. Acad. Sci. USA. 2006;103:11051–11056. doi: 10.1073/pnas.0604548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kazantsev A., Preisinger E., Dranovsky A., Goldgaber D., Housman D. Proc. Natl. Acad. Sci. USA. 1999;96:11404–11409. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slow E. J., Graham R. K., Osmand A. P., Devon R. S., Lu G., Deng Y., Pearson J., Vaid K., Bissada N., Wetzel R., et al. Proc. Natl. Acad. Sci. USA. 2005;102:11402–11407. doi: 10.1073/pnas.0503634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuccato C., Ciammola A., Rigamonti D., Leavitt B. R., Goffredo D., Conti L., MacDonald M. E., Friedlander R. M., Silani V., Hayden M. R., et al. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 37.Harjes P., Wanker E. E. Trends Biochem. Sci. 2003;28:425–433. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- 38.Tsuda H., Jafar-Nejad H., Patel A. J., Sun Y., Chen H. K., Rose M. F., Venken K. J., Botas J., Orr H. T., Bellen H. J., Zoghbi H. Y. Cell. 2005;122:633–644. doi: 10.1016/j.cell.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 39.Ignatova Z., Gierasch L. M. J. Biol. Chem. 2006;281:12959–12967. doi: 10.1074/jbc.M511523200. [DOI] [PubMed] [Google Scholar]
- 40.Steffan J. S., Agrawal N., Pallos J., Rockabrand E., Trotman L. C., Slepko N., Illes K., Lukacsovich T., Zhu Y. Z., Cattaneo E., et al. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- 41.Riley B. E., Zoghbi H. Y., Orr H. T. J. Biol. Chem. 2005;280:21942–21948. doi: 10.1074/jbc.M501677200. [DOI] [PubMed] [Google Scholar]
- 42.Bhattacharyya A., Thakur A. K., Chellgren V. M., Thiagarajan G., Williams A. D., Chellgren B. W., Creamer T. P., Wetzel R. J. Mol. Biol. 2006;355:524–535. doi: 10.1016/j.jmb.2005.10.053. [DOI] [PubMed] [Google Scholar]
- 43.Davies S. W., Turmaine M., Cozens B. A., DiFiglia M., Sharp A. H., Ross C. A., Scherzinger E., Wanker E. E., Mangiarini L., Bates G. P. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 44.Becher M. W., Kotzuk J. A., Sharp A. H., Davies S. W., Bates G. P., Price D. L., Ross C. A. Neurobiol. Dis. 1998;4:387–397. doi: 10.1006/nbdi.1998.0168. [DOI] [PubMed] [Google Scholar]
- 45.Ordway J. M., Cearley J. A., Detloff P. J. Philos. Trans. R. Soc. London B. 1999;354:1083–1088. doi: 10.1098/rstb.1999.0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang W., Dunlap J. R., Andrews R. B., Wetzel R. Hum. Mol. Genet. 2002;11:2905–2917. doi: 10.1093/hmg/11.23.2905. [DOI] [PubMed] [Google Scholar]
- 47.Saudou F., Finkbeiner S., Devys D., Greenberg M. E. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 48.Klement I. A., Skinner P. J., Kaytor M. D., Yi H., Hersch S. M., Clark H. B., Zoghbi H. Y., Orr H. T. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- 49.Cummings C. J., Reinstein E., Sun Y., Antalffy B., Jiang Y., Ciechanover A., Orr H. T., Beaudet A. L., Zoghbi H. Y. Neuron. 1999;24:879–892. doi: 10.1016/s0896-6273(00)81035-1. [DOI] [PubMed] [Google Scholar]
- 50.Shimohata T., Sato A., Burke J. R., Strittmatter W. J., Tsuji S., Onodera O. Neurosci. Lett. 2002;323:215–218. doi: 10.1016/s0304-3940(02)00162-3. [DOI] [PubMed] [Google Scholar]
- 51.Taylor J. P., Tanaka F., Robitschek J., Sandoval C. M., Taye A., Markovic-Plese S., Fischbeck K. H. Hum. Mol. Genet. 2003;12:749–757. doi: 10.1093/hmg/ddg074. [DOI] [PubMed] [Google Scholar]
- 52.Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 53.Johnston J. A., Ward C. L., Kopito R. R. J. Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kopito R. R. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 55.Sikorski R. S., Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaiser C., Michaelis S., Mitchell A. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 57.Guthrie C., Fink G., editors. Methods in Enzymology: Guide to Yeast Genetics and Molecular Biology. Vol. 169. San Diego, CA: Academic; 1991. [PubMed] [Google Scholar]