Molecular Analysis of Fungal Microbiota in Samples from Healthy Human Skin and Psoriatic Lesions (original) (raw)
Abstract
Psoriasis, a common cutaneous disease of unknown etiology, may be triggered by infections, including those due to fungi. Since the fungal community of human skin is poorly characterized, we aimed to analyze the mycological microbiota in healthy skin and psoriatic lesions. Twenty-five skin samples from five healthy subjects (flexor forearm) and three patients with psoriasis were analyzed using broad-range 18S ribosomal DNA (rDNA) and 5.8S rDNA/internal transcribed spacer 2 (ITS2) _Malassezia_-specific PCR primers. Broad-range PCR analysis indicated that most organisms resembled _Malassezia. Malassezia_-specific 5.8S/ITS2 analysis of 1,374 clones identified five species and four unknown phylotypes, potentially representing new species. The species distribution appears largely host specific and conserved in different sites of healthy skin. In three subjects, the Malassezia microbiota composition appeared relatively stable over time. Samples of Malassezia microbiota from healthy skin and psoriatic lesions were similar in one patient but substantially different in two others. These data indicate the predominance of Malassezia organisms in healthy human skin, host-specific variation, stability over time, and as yet, no consistent patterns differentiating psoriatic skin from healthy skin.
Human skin is colonized by diverse microbiota, including bacteria and fungi, that can be pathogenic under particular circumstances (14, 16). Traditionally, microorganisms have been identified by culture-dependent methods; however, many species are fastidious and underrepresented in cultures from mixed microbial communities (13), whereas others cannot be cultivated under known conditions (2). Therefore, culture-independent molecular techniques have been used for the identification of microbial species within ecosystems (2, 9, 27, 42).
Such methods, particularly the analysis of rRNA genes, have been employed to characterize bacterial and fungal communities associated with diverse human body sites, including intestine (11), gingiva (28, 33, 43), esophagus (45), vagina (65), and outer ear canal (13). As predicted, these studies revealed greater diversity, including previously undescribed organisms, than did previous analyses based on culture-dependent techniques.
The application of molecular techniques has been advocated to characterize the microbiota in both healthy and diseased skin (14). To date, rRNA data have been used to identify species associated with fungal dermatoses (21, 29, 38, 39) and PCR-based diagnostic tests have been developed (15, 26, 62). Psoriasis, a common dermatosis affecting about 3% of the population in industrialized countries (3), is characterized by erythrosquamous cutaneous lesions associated with abnormal patterns of keratinocyte growth and differentiation (35). Although of unknown etiology, trigger factors, including physical trauma and streptococcal infections, may provoke clinical manifestations (51). Fungal organisms, including Candida albicans (63) and Malassezia furfur (3), have also been associated with the development of psoriatic skin lesions, and differences have been observed in the Malassezia species distributions in healthy subjects and patients with psoriasis (23, 24, 46).
The aims of this study were to use molecular methods to identify the fungal species present in human skin, understand specificity by host and time, and compare the populations present in health and in psoriatic lesions.
MATERIALS AND METHODS
Subjects and sample collection.
Five healthy subjects (two males, three females; age 21 to 54; mean, 35.2 ± 11.3 years) and three subjects with psoriasis (three males; age 34 to 55; mean, 47.7 ± 11.8 years) were analyzed. All subjects provided written informed consent approved by the NYU Institutional Review Board. From each healthy subject, at least two samples were obtained from the left and right forearms and, for two subjects, another sample was obtained from each forearm 10 months after the first. From each patient with psoriasis, at least three skin samples, including unaffected skin and two or three samples from psoriatic lesions, were studied. From a patient with psoriasis (designated patient 1P), samples included two from the same digital lesion obtained six months apart and one from an elbow lesion. For the other patients, two or three separate lesions from different body sites were analyzed (for patient 2P, arm, leg, and forearm; for patient 3P, elbow and leg). Lesions differing in the extent of erythema, swelling, and scaling were chosen. No patient had ever received therapy for psoriasis. Samples were obtained in a DNA-free clean room by rubbing the skin using two sterile cotton swabs soaked in ST solution (0.15 M NaCl with 0.1% Tween 20). The head of each swab was aseptically cut from the handle, placed into a microcentrifuge tube containing 100 μl of ST solution, centrifuged for 5 min, and then removed. To detect possible contamination, negative controls were prepared using cotton swabs in ST solution without any contact with skin and then subjected to the above-mentioned procedures.
DNA isolation.
Total genomic DNA was extracted by adding an equal volume of 10 mM Tris-HCl (pH 8.0), 10 mM EDTA, 150 mM NaCl, 2% sodium dodecyl sulfate, and 0.5 mg/ml proteinase K to the centrifuged ST solution. After an overnight incubation at 55°C, proteinase K was inactivated by boiling for 5 min, glass beads (1.0 mm diameter) were added, samples were vortex mixed for 3 min, and phenol-chloroform DNA extraction was performed (49). The DNA was precipitated by incubation with 2.5 volumes of absolute ethanol at −20°C for >3 h and then centrifuging for 20 min. The DNA pellets were washed once with 70% ethanol, allowed to dry, and resuspended in 15 μl sterile distilled water.
PCR amplification.
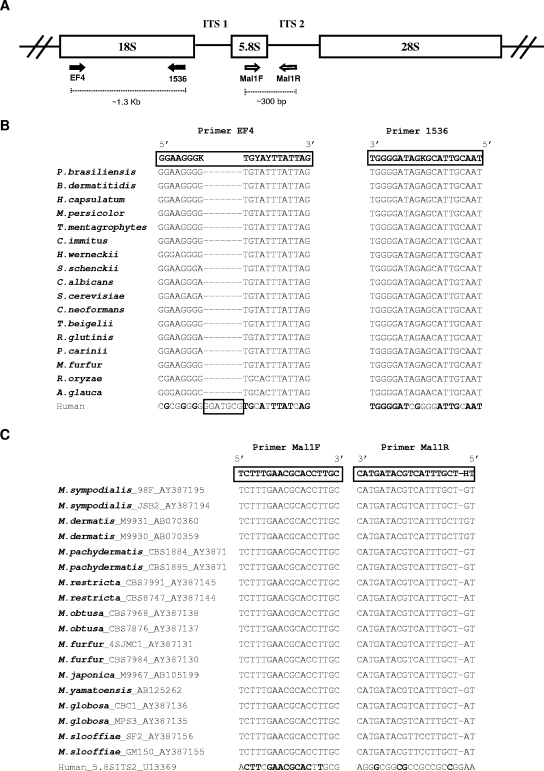
Universal PCR primers for fungi were used to amplify the 18S rRNA gene (Fig. 1A). Broad-range fungal primers were chosen based on comparison between 18S ribosomal DNA (rDNA) sequences from humans and from 17 fungal species representing clinically relevant groups from three different phyla (Fig. 1B) aligned using CLUSTAL W, version 1.81 (61). Primer EF4 anneals to a region containing a 7-nucleotide insertion in the human sequence, reducing the likelihood of amplifying human DNA. To specifically study the genus Malassezia, a region containing 5.8S rDNA and internal transcribed spacer 2 (ITS2) was amplified using _Malassezia_-specific PCR primers (Fig. 1A), based on pairwise alignment between the rRNA gene cluster (including ITS1, 5.8S, and ITS2) from humans and 18 Malassezia strains, representing 10 species (Fig. 1C). The PCRs were performed using 3.5 mM MgCl2, 0.4 μg/μl bovine serum albumin, 0.25 mM of each deoxynucleoside triphosphate, 20 pmol of each primer, 2.5 U Taq DNA polymerase (QIAGEN, Valencia, CA), and 5 μl of extracted DNA in a final 50-μl volume. The PCR conditions were 5 min at 94°C, 30 cycles of 30 s at 94°C, 30 s at 53°C (18S rDNA) or 55°C (5.8S rDNA/ITS2), and 30 s at 72°C, followed by 10 min at 72°C. The PCR products were analyzed by electrophoresis on 1% (wt/vol) Tris-acetate-EDTA-agarose gels containing ethidium bromide and visualized under UV light.
FIG. 1.
Fungal loci and sequences related to this study. (A) Schematic representation of the fungal ribosomal gene cluster, with PCR primers indicated (arrows). (B) Pairwise alignment between fragments of the 18S rRNA gene from human and representative fungal species, showing the sequences of primers EF4 and 1536 (long boxes). The human sequence contains a 7-bp insertion within the region represented by primer EF4 (small box). K, A/G; Y, T/C. (C) Pairwise alignment between fragments related to the 5.8S rRNA gene and ITS2 from humans and Malassezia species, showing the sequence of primers Mal1F and Mal1R. Bold letters in the human sequences refer to conserved nucleotides with the fungal sequences. H, T/C/A.
Construction of libraries, screening, and sequencing.
Each of the PCR products, whether obtained using universal primers or _Malassezia_-specific primers, were excised from Tris-acetate-EDTA-agarose gels and purified using the QIAquick gel extraction kit (QIAGEN), following the manufacturer's instructions. The purified fragments were cloned into pGEM-T easy vector (Promega, Madison, WI) and used to transform Escherichia coli DH5α or XL1-Blue cells. Inserts from transformed cells were PCR amplified using vector-specific primers (SP6 and T7) in a 50-μl mixture containing 1.5 mM MgCl2, 0.2 mM of each deoxynucleoside triphosphate, 5 pmol of each primer, and 1.25 U Taq DNA polymerase (QIAGEN). The PCR conditions were 2 min at 94°C, followed by 35 cycles of 45 s at 94°C, 30 s at 50°C, and 90 s at 72°C and then 20 min at 72°C, and the products were subject to purification (QIAGEN) according to the manufacturer's instructions. Five-microliter aliquots from the purified PCR products from the 18S rDNA libraries were digested with HaeIII (New England Biolabs, Ipswich, MA) for 1 h at 37°C, the restriction fragments were separated on 1% agarose gels, and restriction fragment length polymorphism (RFLP) types were identified. A second aliquot of representative PCR products from each one of the RFLP types was used to sequence ∼700 bp using primer 1536 (Fig. 1B). The analyses were performed on an ABI 3730 automatic sequencer (Applied Biosystems, Foster City, CA). To identify the species Malassezia restricta and Malassezia globosa from 5.8S rDNA/ITS2 libraries, PCR products were digested with DraI and XbaI, respectively. Nonrestricted fragments were sequenced using primer SP6 to identify other Malassezia species.
Phylogenetic analysis.
The 18S rDNA and 5.8S rDNA/ITS2 sequences were compared to those in the NCBI GenBank database by using BLASTN algorithm versions 2.2.8 to 2.2.11 (1) (http://www.ncbi.nlm.nih.gov/BLAST/), and sequences were aligned using CLUSTAL_X, version 1.8 (60). Chimeric and human sequences, corresponding to 5.9% of the clones, were not included in the alignments. Phylogenetic analyses were performed using Mega2 software, version 2.1 (34) (http://www.megasoftware.net). Matrices of distance were calculated using the Jukes-Cantor algorithm (31), and the neighbor-joining method (48) was used to generate phylogenetic trees. To identify the closest relatives of the cloned sequences, phylogenetic trees based on 18S rDNA data also included 16 known fungal species, representing clinically relevant groups. For phylogenetic analysis based on 5.8S rDNA and ITS2 to identify Malassezia, 10 sequences corresponding to known Malassezia species were included. The reliability of the tree topologies was tested by bootstrap resampling with 500 replicates. Novel phylotypes were defined as sequences with <97% similarity to known species in the public databases.
Estimation of species richness.
The species richness of normal and psoriatic skin samples was evaluated with a nonparametric richness estimator, Chao 1 (6), using EstimateS, version 7 (http://purl.oclc.org/estimates).
Double-principal coordinate analysis and cluster analysis.
Double-principal coordinate analysis (DPCoA) (44) was used to evaluate the sample diversity and analyze the relationship among samples. This method uses the phylotype dissimilarity matrix of samples to calculate the sample diversity. In this analysis, the dissimilarities between different phylotypes are defined as one, and the resulting sample Rao's diversity coefficient is therefore identical to the Gini-Simpson index. To facilitate the visualization of diversity, the first two orthogonal principal axes were obtained based on the sample dissimilarity and plotted to show the distribution of samples in a two-dimensional space. Unsupervised hierarchical clustering techniques were also used to evaluate similarity among samples based on the sample dissimilarity matrix. To assess the robustness of the clustering, bootstrap resampling was done with 1,000 simulations. The diversity information can be decomposed into within- and between-samples diversity values. This allowed the use of a “pseudo F” statistic (the ratio of within-cluster diversity and between-cluster diversity) to examine the possible clustering phenomena, and significance was evaluated by permutation tests (11).
Nucleotide sequence accession numbers.
The rDNA sequences of clones representing novel phylotypes were deposited in the GenBank database with accession numbers DQ119888 through DQ119891.
RESULTS
Broad-range PCR analysis.
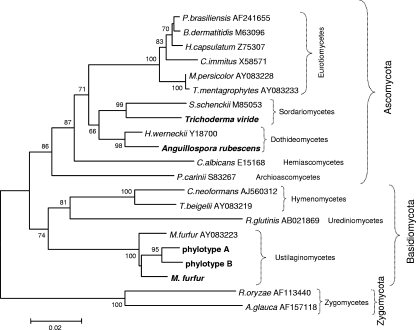
We first sought to analyze the complete fungal microbiota in the sampled skin by using broad-range primers to amplify fungal 18S rDNA. Taking advantage of a 7-nucleotide insertion present within the human 18S rDNA sequence but absent from the fungal sequences, the modification of primer EF4 (52), together with primer 1536 (5), was used to amplify expected 1.3-kb 18S rDNA PCR products (Fig. 1A and B) and create libraries of cloned fragments from specimens obtained from normal forearm skin and psoriatic lesions. Clones were screened from three separate libraries using RFLP analysis, and partial (∼700 bp) sequences were obtained from representative clones of each RFLP type (defined on the basis of the HaeIII restriction pattern). For each RFLP type, at least two clones were sequenced, which indicated high sequence similarity within each type (data not shown). After phylogenetic analyses were performed, sequences with ≤97% similarity with known GenBank sequences were designated phylotypes, defined based on RFLP type and nearest relative (as determined by BLASTN search) and by phylogenetic affiliation. Sequences included in the same phylotype had ≥97% identity to each other. More than 95% of the clones obtained from the libraries from each of the three skin samples were grouped into two unknown phylotypes (designated A and B) (Table 1), phylogenetically related to Malassezia furfur (Fig. 2), but with similarity < 97%. Although detected in both normal and psoriatic skin samples, their prevalence differed among the samples (Table 1). From each sample, only one sequence was similar (98%) to M. furfur (Table 1). Sequences with similarity to only Anguillospora rubescens and Trichoderma viride were found in single clones from a single psoriasis lesion (Table 1). Anguillospora rubescens is a freshwater hyphomycete (19) not previously reported as either belonging to the commensal human microbiota or responsible for infections. The genus Trichoderma comprises saprophytes, although some species, including T. viride (30), have been associated with opportunistic infections in immunocompromised patients (64). Both genera belong to the phylum Ascomycota, whereas the majority of the organisms detected in the samples are in the phylum Basidiomycota (Fig. 2).
TABLE 1.
Fungal species and phylotypes identified in 190 clones from normal skin and psoriatic lesions from patient 1P
| Organism identified (% similarity) | % Clonesa | ||
|---|---|---|---|
| Normal skin (n = 76)b | Psoriasis 1 (n = 61) | Psoriasis 2 (n = 53) | |
| Anguillospora rubescens (99) | 0 | 1.6 | 0 |
| Trichoderma viride (99) | 0 | 1.6 | 0 |
| Malassezia furfur (98) | 1.3 | 1.6 | 1.9 |
| Undeterminedc phylotype A | 44.7 | 8.2 | 58.5 |
| Undetermined phylotype B | 53.9 | 86.9 | 39.6 |
FIG. 2.
Neighbor-joining tree based on partial 18S rDNA sequences. The matrix of distances was calculated using the Jukes-Cantor algorithm. Bootstrap values are based on 500 replicates (values of at least 50% are shown). Organisms identified in this study are shown in bold. Codes correspond to GenBank accession numbers.
Malassezia microbiota.
Although the most prevalent sequences detected in these skin samples, belonging to phylotype A or B, were phylogenetically related to the genus Malassezia (Fig. 2), no species could be assigned because, except for that of M. furfur, GenBank does not contain sufficient 18S rDNA Malassezia sequences to distinguish among species. Therefore, Malassezia sequences in GenBank were evaluated to identify DNA regions that could be used for species-level identification. Sequences corresponding to the internal transcribed spacers (ITS1 and ITS2) and the 5.8S rRNA gene from Malassezia organisms are available and contain regions that differ between species but are conserved within a species. By designing primers to specifically amplify a fragment containing the 5.8S rRNA gene and ITS2 from Malassezia species, and not from humans (Fig. 1A and C), we studied the Malassezia microbiota in skin samples from five healthy subjects and three patients with psoriasis.
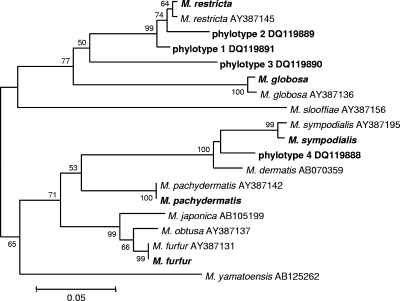
Among the healthy subjects, five different Malassezia species were identified, as well as four phylotypes distinct from the currently recognized species (Table 2). Malassezia restricta was the most commonly detected species in both samples from two subjects (designated 1N and 3N), whereas M. globosa predominated in one subject (2N). Phylotypes 1, 2, and 3, most closely related to M. restricta, and phylotype 4, related to Malassezia sympodialis (Fig. 3), may in fact represent new Malassezia species. Similar to results obtained with the healthy subjects, in the patients with psoriasis, three different species and one unknown phylotype were identified (Table 3); however, the relative clone abundances differed.
TABLE 2.
Malassezia species and phylotypes identified in 824 clones from normal skin samples from five subjectsa
| Organism identified (% similarity) | % Clones | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1N | 2N | 3N | 4N | 5N | Total (n = 824) | |||||||||||
| L-Jan (n = 72)b | L-Nov (n = 49) | R-Jan (n = 70) | R-Nov (n = 50) | L-Jan (n = 46) | L-Nov (n = 53) | R-Jan (n = 49) | R-Nov (n = 52) | L (n = 74) | R (n = 73) | L (n = 52) | R (n = 69) | L (n = 65) | R (n = 50) | |||
| Malassezia restricta (99-100) | 77.8 | 87.7 | 42.9 | 82.0 | 4.3 | 32.1 | 18.4 | 40.4 | 51.4 | 45.2 | 0 | 33.3 | 13.8 | 22.0> | 22.0 | 40.4 |
| Malassezia globosa (98-100) | 13.9 | 4.1 | 21.4 | 14.0 | 89.1 | 62.3 | 46.9 | 57.7 | 14.9 | 2.7 | 0 | 7.2 | 6.2 | 22.0 | 23.5 | |
| Malassezia sympodialis (99-100) | 1.4 | 4.1 | 28.6 | 2.0 | 0 | 3.8 | 16.3 | 1.9 | 4.1 | 8.2 | 96.2 | 7.2 | 15.4 | 30.0 | 15.0 | |
| Malassezia pachydermatis (97-99) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.7 | 0 | 1.4 | 40.0 | 0 | 3.5 | |
| Malassezia furfur (100) | 0 | 2.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.1 | |
| Undeterminedc phylotype 1 | 0 | 2.0 | 1.4 | 2.0 | 6.5 | 1.9 | 8.2 | 0 | 28.4 | 41.1 | 3.8 | 50.7 | 24.6 | 26.0 | 15.5 | |
| Undetermined phylotype 2 | 6.9 | 0 | 4.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.0 | |
| Undetermined phylotype 3 | 0 | 0 | 1.4 | 0 | 0 | 0 | 0 | 0 | 1.3 | 0 | 0 | 0 | 0 | 0 | 0.2 | |
| Undetermined phylotype 4 | 0 | 0 | 0 | 0 | 0 | 0 | 10.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.6 |
FIG. 3.
Neighbor-joining tree based on 5.8S rDNA and ITS2 sequences, showing the relationships among Malassezia organisms. The matrix of distances was calculated using the Jukes-Cantor algorithm. Bootstrap values are based on 500 replicates (values of at least 50% are shown). Organisms identified in this study are shown in bold. Codes correspond to GenBank accession numbers.
TABLE 3.
Malassezia species and phylotypes identified in 550 clones from healthy skin and psoriatic lesions from three patients with psoriasisa
| Organism identified (% similarity) | % Clones | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1P | 2P | 3P | |||||||||
| Normal skin (n = 72)b | Psoriasis 1A (n = 76) | Psoriasis 1B (n = 50) | Psoriasis 2 (n = 67) | Normal skin (n = 53) | Psoriasis 1 (n = 38) | Psoriasis 2 (n = 43) | Psoriasis 3 (n = 48) | Normal skin (n = 26) | Psoriasis 1 (n = 35) | Psoriasis 2 (n = 42) | |
| Malassezia restricta (98-100) | 30.5 | 50.0 | 56.0 | 38.8 | 0 | 15.8 | 13.9 | 0 | 0 | 25.7 | 9.5 |
| Malassezia globosa (98-100) | 38.9 | 15.8 | 6.0 | 31.3 | 39.6 | 23.7 | 9.3 | 0 | 100 | 54.3 | 14.3 |
| Malassezia sympodialis (99-100) | 12.5 | 2.6 | 0 | 26.9 | 0 | 36.8 | 20.9 | 2.1 | 0 | 2.9 | 47.6 |
| Undeterminedc phylotype 1 | 18.0 | 31.6 | 38.0 | 3.0 | 60.4 | 23.7 | 55.8 | 97.9 | 0 | 17.1 | 28.6 |
Negative controls, prepared using cotton swabs without any skin contact, did not generate detectable signals of amplification by electrophoresis; however, gel fragments were excised at locations where products should migrate, and libraries were constructed and screened. Several insert-containing colonies were isolated, ranging from approximately 3-fold to 100-fold fewer colonies, compared to those of the actual skin samples, with identified species corresponding to some of the common organisms found in skin samples (data not shown). Although probably due to cross-contamination between samples, such contamination has low impact on the results, considering the high ratio between numbers of colonies from skin samples and from negative controls. Contamination by human DNA was a function of the source of the specimen. Only 2 (0.2%) of 826 clones from normal skin samples of healthy hosts had human sequences, and similar results were observed in normal skin samples from patients with psoriasis (0.7% of 152 clones). In contrast, 49 (10.8%) of 454 clones from psoriatic lesion samples contained human DNA (P < 0.0001, compared to normal skin samples).
To determine the extent to which the data reflect the diversity of the samples, the species richness of normal and psoriatic skin samples was evaluated using the Chao 1 estimator (6). Based on these analyses, 100% of the estimated species number was detected in both normal and psoriatic skin samples, indicating the strength of the methodology.
Variability of the Malassezia microbiota.
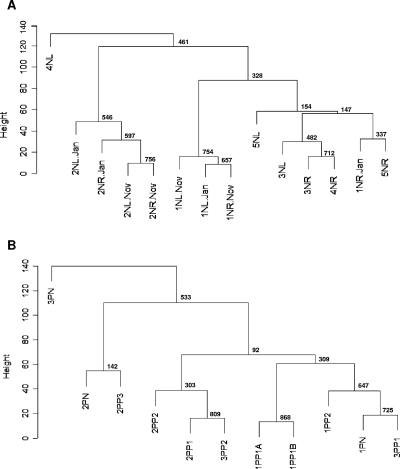
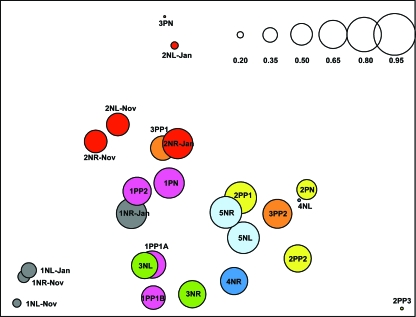
To evaluate similarities in the distributions of Malassezia organisms between skin samples, cluster analyses were performed. The samples were grouped based on the similarities in percentages of the Malassezia species, following a procedure used for analysis of gene expression data (12), and the reliability of clustering results was assessed by bootstrap resampling. The skin samples were also compared using DPCoA. Samples from the same body site in the same host obtained 6 or 10 months apart clustered together in four of five cases (Fig. 4A and B), indicating that the Malassezia microbiota are relatively stable over that time period. The proximity between samples from the same body site within an individual obtained 6 or 10 months apart was also verified by DPCoA (Fig. 5) and confirmed by statistical hypothesis testing (P = 0.05). In most of the healthy subjects, samples from the left and right forearms were closely related to each other (Fig. 4A and 5), suggesting that the distribution of Malassezia organisms is conserved in parallel skin sites and host specific. However, samples from subject 4N grouped in separate clusters, consistent with substantial differences in species distribution (Table 2). Samples from psoriasis patient 1P were closely related to each other, despite originating in healthy or psoriasis-involved skin (Fig. 4B and 5), but greater differences were observed among samples from patients 2P and 3P. Considering both the healthy subjects and patients, hypothesis testing confirmed that samples from an individual subject are closer together than samples from different subjects (P < 0.0001). In addition, DPCoA revealed that samples from healthy skin largely overlap with those from psoriatic lesions (Fig. 5) and a separation between these groups could not be confirmed by hypothesis testing (P = 0.78).
FIG. 4.
Consensus tree of hierarchical clustering of skin samples from healthy subjects (A) and patients with psoriasis (B). Height corresponds to Euclidian distance between samples. The number at each node represents the bootstrap value, based on 1,000 iterations. (A) 1N through 5N, healthy subjects; L, left arm; R, right arm. Samples L-Jan/R-Jan and L-Nov/R-Nov were obtained from the same site, 10 months apart. (B) 1P through 3P, patients with psoriasis; N, samples from normal skin; P1 through P3, samples from psoriatic lesions. Samples 1PP1A and 1PP1B were obtained from the same lesion, 6 months apart.
FIG. 5.
Scatterplot of the first two orthogonal principal axes based on the sample dissimilarity matrix. The samples from each subject are represented with the same color, and the sizes of the circles are proportional to the sample diversity, as determined by Rao's analysis. The diversity scale is shown in the upper right corner.
DISCUSSION
Here we report a broad-range analysis of the fungal microbiota in healthy forearm skin and psoriatic lesions by culture-independent methods. Initially, using universal primers to amplify fungal 18S rDNA from skin samples from a patient with psoriasis, most sequences appeared to represent the genus Malassezia. This genus has been described as part of the commensal skin microbiota, but it also has been associated with pityriasis versicolor, seborrheic dermatitis, atopic dermatitis, folliculitis, and psoriasis (3, 8, 16, 20). Patients with psoriasis, but not healthy subjects, have been reported to have serum antibodies to Malassezia proteins (36, 53) and _M. furfur-_induced, Th1-related cytokines in peripheral blood mononuclear cells (32). Malassezia furfur can invade cultured human keratinocytes, modulate proinflammatory and immunomodulatory cytokine synthesis, and affect the expression of cutaneous proteins (4), especially those related to cell migration and proliferation, potentially enhancing inflammation (3).
Because it was not possible to identify Malassezia at the species level in our broad-range analysis due to the limited 18S rDNA Malassezia sequences available, Malassezia species were analyzed using primers to amplify a fragment containing 5.8S rDNA and ITS2. Although the fragment analyzed in this study containing 5.8S rDNA and ITS2 is only ∼300 bp, the level of polymorphism between species is sufficient to allow their identification. Using restriction analysis to identify two common species decreases the number of clones to be sequenced, facilitating throughput and lowering costs associated with analysis of the biota. The skin swab technique is simple, allowing large areas to be sampled, and can be used repetitively in serial studies. The contamination by human DNA was more substantial in samples from psoriatic lesions, reflecting the enhanced cellular proliferation due to inflammation (35), but it represented a small fraction (10%) of the clones. Studies of species richness provide evidence that our sampling of Malassezia species in normal and psoriatic skin is at or very near completion. In total, the techniques used may have wider application in studies to determine the distribution of Malassezia species in human skin.
In the 25 skin samples from the eight subjects examined, five Malassezia species and four unidentified phylotypes were found. Based on the total number of clones, M. restricta and M. globosa were the most prevalent organisms identified. Both species have previously been detected by molecular (55, 56) and culture-dependent (23, 24, 46, 47) approaches in skin samples from healthy subjects and in patients with dermatoses. Malassezia sympodialis, also identified in the samples we studied, has been observed in both healthy subjects and patients with dermatoses (8, 23, 24, 41, 50, 56). Malassezia pachydermatis, associated with infections in dogs and other carnivores (10, 18), has been detected in human newborns (7) and in healthy dog owners (40). Although not often isolated from humans, our detection in samples from three (of five) healthy subjects suggests that M. pachydermatis may be substantially more common than was previously believed.
Using culture-dependent techniques, M. restricta, M. globosa, and M. sympodialis have been recovered in 38 to 55% of healthy persons and those with psoriasis (23, 24, 46), but their presence in all eight subjects we analyzed suggests a higher prevalence. Malassezia furfur, described as common in normal skin microbiota (37), has been associated with psoriasis (3). Using universal fungal primers, only 3 (1.5%) of 190 clones had sequences ≥97% similar to M. furfur (Table 1) and analysis using _Malassezia_-specific primers showed only 1 clone (0.07%) of the 1,374 analyzed. It was not detected in a sample from the same body site obtained 10 months later, confirming its low representation at the sites we sampled.
The results of hierarchical clustering analyses suggest that, in the majority of the samples, the Malassezia species distributions remained relatively stable over time, were similar among samples from the same healthy subject, and tended to be host specific. Among the three patients with psoriasis, the Malassezia microbiota were relatively conserved in samples from one patient, whether from healthy or psoriatic skin, whereas substantial differences were detected among samples from the other two patients, possibly reflecting variation in the microbiota at different body sites. Samples obtained from leg (designated 2PP2 and 3PP2) as well as samples from elbow (1PP2 and 3PP1) grouped together (Fig. 4B), which is consistent with this hypothesis and with a prior culture-based study (23). Although the same species were detected in both healthy and diseased skin from patients with psoriasis, the compositions of the communities may still differ. Genotypes of M. globosa (54) and M. restricta (56) isolated from patients with atopic dermatitis have been found to differ from those from healthy subjects, suggesting the possibility of genotype-specific roles in pathogenesis that could not be detected by our species-level analyses.
In conclusion, we provide evidence that most fungal organisms in the normal healthy human forearm skin and in psoriatic lesions belong to the genus Malassezia. The use of culture-independent methods to identify Malassezia organisms is important, since they are fastidious (15, 21, 22). Eleven Malassezia species are currently accepted (17, 25, 57-59), with four described since 2002. The four novel Malassezia phylotypes detected in this study possibly correspond to new species and should be further characterized, ideally with growth as pure cultures. Analysis of the clones and longitudinal studies indicate host specificity of the distribution of Malassezia species. Thus far, the data do not support the hypothesis that Malassezia populations differ in psoriasis and healthy skin.
Acknowledgments
We thank Jackie Eisenberg and David Alevi for their technical assistance.
This work was supported in part by the Senior Scholar Award from the Ellison Medical Foundation, the Filomena D'Agostino Foundation, the Diane Belfer Program in Human Microbial Ecology in Health and Disease, and the Michael Saperstein Medical Scholars Research Fund.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25**:**3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59**:**143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baroni, A., I. Paoletti, E. Ruocco, M. Agozzino, M. A. Tufano, and G. Donnarumma. 2004. Possible role of Malassezia furfur in psoriasis: modulation of TGF-β1, integrin, and HSP70 expression in human keratinocytes and in the skin of psoriasis-affected patients. J. Cutan. Pathol. 31**:**35-42. [DOI] [PubMed] [Google Scholar]
- 4.Baroni, A., B. Perfetto, I. Paoletti, E. Ruocco, N. Canozo, M. Orlando, and E. Buommino. 2001. Malassezia furfur invasiveness in a keratinocyte cell line (HaCat): effects on cytoskeleton and on adhesion molecule and cytokine expression. Arch. Dermatol. Res. 293**:**414-419. [DOI] [PubMed] [Google Scholar]
- 5.Borneman, J., and R. J. Hartin. 2000. PCR primers that amplify fungal rRNA genes from environmental samples. Appl. Environ. Microbiol. 66**:**4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao, A. 1987. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 43**:**783-791. [PubMed] [Google Scholar]
- 7.Chryssanthou, E., U. Broberger, and B. Petrini. 2001. Malassezia pachydermatis fungaemia in a neonatal intensive care unit. Acta Paediatr. 90**:**323-327. [PubMed] [Google Scholar]
- 8.Crespo Erchiga, V., and V. Delgado Florencio. 2002. Malassezia species in skin diseases. Curr. Opin. Infect. Dis. 15**:**133-142. [DOI] [PubMed] [Google Scholar]
- 9.Dawson, S. C., and N. R. Pace. 2002. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. USA 99**:**8324-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorogi, J. 2002. Pathological and clinical aspects of the diseases caused by Malassezia species. Acta Microbiol. Immunol. Hung. 49**:**363-369. [DOI] [PubMed] [Google Scholar]
- 11.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308**:**1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95**:**14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank, D. N., G. B. Spiegelman, W. Davis, E. Wagner, E. Lyons, and N. R. Pace. 2003. Culture-independent molecular analysis of microbial constituents of the healthy human outer ear. J. Clin. Microbiol. 41**:**295-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fredricks, D. N. 2001. Microbial ecology of human skin in health and disease. J. Investig. Dermatol. Symp. Proc. 6**:**167-169. [DOI] [PubMed] [Google Scholar]
- 15.Gemmer, C. M., Y. M. DeAngelis, B. Theelen, T. Boekhout, and T. L. Dawson, Jr. 2002. Fast, noninvasive method for molecular detection and differentiation of Malassezia yeast species on human skin and application of the method to dandruff microbiology. J. Clin. Microbiol. 40**:**3350-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gueho, E., T. Boekhout, H. R. Ashbee, J. Guillot, A. Van Belkum, and J. Faergemann. 1998. The role of Malassezia species in the ecology of human skin and as pathogens. Med. Mycol. 36(Suppl. 1)**:**220-229. [PubMed] [Google Scholar]
- 17.Gueho, E., G. Midgley, and J. Guillot. 1996. The genus Malassezia with description of four new species. Antonie Leeuwenhoek 69**:**337-355. [DOI] [PubMed] [Google Scholar]
- 18.Guillot, J., and R. Bond. 1999. Malassezia pachydermatis: a review. Med. Mycol. 37**:**295-306. [DOI] [PubMed] [Google Scholar]
- 19.Gulis, V., and L. Marvanova. 1999. Three new scolecosporous hyphomycetes from waters in Belarus. Mycotaxon 72**:**237-250. [Google Scholar]
- 20.Gupta, A. K., R. Batra, R. Bluhm, T. Boekhout, and T. L. Dawson, Jr. 2004. Skin diseases associated with Malassezia species. J. Am. Acad. Dermatol. 51**:**785-798. [DOI] [PubMed] [Google Scholar]
- 21.Gupta, A. K., T. Boekhout, B. Theelen, R. Summerbell, and R. Batra. 2004. Identification and typing of Malassezia species by amplified fragment length polymorphism and sequence analyses of the internal transcribed spacer and large-subunit regions of ribosomal DNA. J. Clin. Microbiol. 42**:**4253-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta, A. K., Y. Kohli, and R. C. Summerbell. 2000. Molecular differentiation of seven Malassezia species. J. Clin. Microbiol. 38**:**1869-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta, A. K., Y. Kohli, R. C. Summerbell, and J. Faergemann. 2001. Quantitative culture of Malassezia species from different body sites of individuals with or without dermatoses. Med. Mycol. 39**:**243-251. [DOI] [PubMed] [Google Scholar]
- 24.Hernandez Hernandez, F., L. J. Mendez Tovar, E. Bazan Mora, A. Arevalo Lopez, A. Valera Bermejo, and R. Lopez Martinez. 2003. Species of Malassezia associated with various dermatoses and healthy skin in the Mexican population. Rev. Iberoam. Micol. 20**:**141-144. (In Spanish.) [PubMed] [Google Scholar]
- 25.Hirai, A., R. Kano, K. Makimura, E. R. Duarte, J. S. Hamdan, M. A. Lachance, H. Yamaguchi, and A. Hasegawa. 2004. Malassezia nana sp. nov., a novel lipid-dependent yeast species isolated from animals. Int. J. Syst. Evol. Microbiol. 54**:**623-627. [DOI] [PubMed] [Google Scholar]
- 26.Hu, S., W. H. Chung, S. I. Hung, H. C. Ho, Z. W. Wang, C. H. Chen, S. C. Lu, T. T. Kuo, and H. S. Hong. 2003. Detection of Sporothrix schenckii in clinical samples by a nested PCR assay. J. Clin. Microbiol. 41**:**1414-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180**:**4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hutter, G., U. Schlagenhauf, G. Valenza, M. Horn, S. Burgemeister, H. Claus, and U. Vogel. 2003. Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiology 149**:**67-75. [DOI] [PubMed] [Google Scholar]
- 29.Jackson, C. J. 2001. Molecular identification and strain typing of dermatophyte fungi. Nippon Ishinkin Gakkai Zasshi 42**:**7-10. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs, F., B. Byl, N. Bourgeois, J. Coremans-Pelseneer, S. Florquin, G. Depre, J. Van de Stadt, M. Adler, M. Gelin, and J. P. Thys. 1992. Trichoderma viride infection in a liver transplant recipient. Mycoses 35**:**301-303. [DOI] [PubMed] [Google Scholar]
- 31.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 32.Kanda, N., K. Tani, U. Enomoto, K. Nakai, and S. Watanabe. 2002. The skin fungus-induced Th1- and Th2-related cytokine, chemokine and prostaglandin E2 production in peripheral blood mononuclear cells from patients with atopic dermatitis and psoriasis vulgaris. Clin. Exp. Allergy 32**:**1243-1250. [DOI] [PubMed] [Google Scholar]
- 33.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96**:**14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17**:**1244-1245. [DOI] [PubMed] [Google Scholar]
- 35.Lebwohl, M. 2003. Psoriasis. Lancet 361**:**1197-1204. [DOI] [PubMed] [Google Scholar]
- 36.Liang, Y. S., H. Q. Wen, and R. Xiao. 2003. Serum levels of antibodies for IgG, IgA, and IgM against the fungi antigen in psoriasis vulgaris. Hunan Yi Ke Da Xue Xue Bao 28**:**638-640. (In Chinese.) [PubMed] [Google Scholar]
- 37.Ljubojevic, S., M. Skerlev, J. Lipozencic, and A. Basta-Juzbasic. 2002. The role of Malassezia furfur in dermatology. Clin. Dermatol. 20**:**179-182. [DOI] [PubMed] [Google Scholar]
- 38.Machouart-Dubach, M., C. Lacroix, M. F. de Chauvin, I. Le Gall, C. Giudicelli, F. Lorenzo, and F. Derouin. 2001. Rapid discrimination among dermatophytes, Scytalidium spp., and other fungi with a PCR-restriction fragment length polymorphism ribotyping method. J. Clin. Microbiol. 39**:**685-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makimura, K. 2001. Species identification system for dermatophytes based on the DNA sequences of nuclear ribosomal internal transcribed spacer 1. Nippon Ishinkin Gakkai Zasshi 42**:**61-67. [DOI] [PubMed] [Google Scholar]
- 40.Morris, D. O. 2005. Malassezia pachydermatis carriage in dog owners. Emerg. Infect. Dis. 11**:**83-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakabayashi, A., Y. Sei, and J. Guillot. 2000. Identification of Malassezia species isolated from patients with seborrhoeic dermatitis, atopic dermatitis, pityriasis versicolor and normal subjects. Med. Mycol. 38**:**337-341. [DOI] [PubMed] [Google Scholar]
- 42.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276**:**734-740. [DOI] [PubMed] [Google Scholar]
- 43.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183**:**3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pavoine, S., A. B. Dufour, and D. Chessel. 2004. From dissimilarities among species to dissimilarities among communities: a double principal coordinate analysis. J. Theor. Biol. 228**:**523-537. [DOI] [PubMed] [Google Scholar]
- 45.Pei, Z., E. J. Bini, L. Yang, M. Zhou, F. Francois, and M. J. Blaser. 2004. Bacterial biota in the human distal esophagus. Proc. Natl. Acad. Sci. USA 101**:**4250-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prohic, A. 2003. Identification of Malassezia species isolated from scalp skin of patients with psoriasis and healthy subjects. Acta Dermatovenerol. Croat. 11**:**10-16. [PubMed] [Google Scholar]
- 47.Rendic, E., C. Diaz, and F. Fich. 2003. Characterization of species of the gender Malassezia in patients with seborrheic dermatitis and subjects without skin lesions. Rev. Med. Chile 131**:**1295-1300. (In Spanish.) [PubMed] [Google Scholar]
- 48.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4**:**406-425. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. E. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Sandstrom Falk, M. H., M. Tengvall Linder, C. Johansson, J. Bartosik, O. Back, T. Sarnhult, C. F. Wahlgren, A. Scheynius, and J. Faergemann. 2005. The prevalence of Malassezia yeasts in patients with atopic dermatitis, seborrhoeic dermatitis and healthy controls. Acta Derm. Venereol. 85**:**17-23. [DOI] [PubMed] [Google Scholar]
- 51.Schon, M. P., and W. H. Boehncke. 2005. Psoriasis. N. Engl. J. Med. 352**:**1899-1912. [DOI] [PubMed] [Google Scholar]
- 52.Smit, E., P. Leeflang, B. Glandorf, J. D. van Elsas, and K. Wernars. 1999. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 65**:**2614-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Squiquera, L., R. Galimberti, L. Morelli, L. Plotkin, R. Milicich, A. Kowalckzuk, and J. Leoni. 1994. Antibodies to proteins from Pityrosporum ovale in the sera from patients with psoriasis. Clin. Exp. Dermatol. 19**:**289-293. [DOI] [PubMed] [Google Scholar]
- 54.Sugita, T., M. Kodama, M. Saito, T. Ito, Y. Kato, R. Tsuboi, and A. Nishikawa. 2003. Sequence diversity of the intergenic spacer region of the rRNA gene of Malassezia globosa colonizing the skin of patients with atopic dermatitis and healthy individuals. J. Clin. Microbiol. 41**:**3022-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sugita, T., H. Suto, T. Unno, R. Tsuboi, H. Ogawa, T. Shinoda, and A. Nishikawa. 2001. Molecular analysis of Malassezia microflora on the skin of atopic dermatitis patients and healthy subjects. Nippon Ishinkin Gakkai Zasshi 42**:**217-218. (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 56.Sugita, T., M. Tajima, M. Amaya, R. Tsuboi, and A. Nishikawa. 2004. Genotype analysis of Malassezia restricta as the major cutaneous flora in patients with atopic dermatitis and healthy subjects. Microbiol. Immunol. 48**:**755-759. [DOI] [PubMed] [Google Scholar]
- 57.Sugita, T., M. Tajima, M. Takashima, M. Amaya, M. Saito, R. Tsuboi, and A. Nishikawa. 2004. A new yeast, Malassezia yamatoensis, isolated from a patient with seborrheic dermatitis, and its distribution in patients and healthy subjects. Microbiol. Immunol. 48**:**579-583. [DOI] [PubMed] [Google Scholar]
- 58.Sugita, T., M. Takashima, M. Kodama, R. Tsuboi, and A. Nishikawa. 2003. Description of a new yeast species, Malassezia japonica, and its detection in patients with atopic dermatitis and healthy subjects. J. Clin. Microbiol. 41**:**4695-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sugita, T., M. Takashima, T. Shinoda, H. Suto, T. Unno, R. Tsuboi, H. Ogawa, and A. Nishikawa. 2002. New yeast species, Malassezia dermatis, isolated from patients with atopic dermatitis. J. Clin. Microbiol. 40**:**1363-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25**:**4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22**:**4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turin, L., F. Riva, G. Galbiati, and T. Cainelli. 2000. Fast, simple and highly sensitive double-rounded polymerase chain reaction assay to detect medically relevant fungi in dermatological specimens. Eur. J. Clin. Investig. 30**:**511-518. [DOI] [PubMed] [Google Scholar]
- 63.Waldman, A., A. Gilhar, L. Duek, and I. Berdicevsky. 2001. Incidence of Candida in psoriasis—a study on the fungal flora of psoriatic patients. Mycoses 44**:**77-81. [DOI] [PubMed] [Google Scholar]
- 64.Walsh, T. J., and A. H. Groll. 1999. Emerging fungal pathogens: evolving challenges to immunocompromised patients for the twenty-first century. Transpl. Infect. Dis. 1**:**247-261. [DOI] [PubMed] [Google Scholar]
- 65.Zhou, X., S. J. Bent, M. G. Schneider, C. C. Davis, M. R. Islam, and L. J. Forney. 2004. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology 150**:**2565-2573. [DOI] [PubMed] [Google Scholar]