Role of lgtC in Resistance of Nontypeable Haemophilus influenzae Strain R2866 to Human Serum (original) (raw)
Abstract
We are investigating a nontypeable Haemophilus influenzae (NTHI) strain, R2866, isolated from a child with meningitis. R2866 is unusually resistant to killing by normal human serum. The serum 50% inhibitory concentration (IC50) for this strain is 18%, approaching that of encapsulated H. influenzae. R3392 is a derivative of R2866 that was found to have increased sensitivity to human serum (IC50, 1.5%). Analysis of tetrameric repeat regions within lipooligosaccharide (LOS) biosynthetic genes in both strains indicated that the glycosyltransferase gene lgtC was out of frame (“off”) in most colonies of R3392 but in frame with its start codon (“on”) in most colonies of the parent. We sought antigenic and biochemical evidence for modification of the LOS structure. In a whole-cell enzyme-linked immunosorbent assay, strain R3392 displayed reduced binding of the Galα1,4Gal-specific monoclonal antibody 4C4. Mass spectrometry analysis of LOS from strain R2866 indicated that the primary oligosaccharide glycoform contained four heptose and four hexose residues, while that of R3392 contained four heptose and three hexose residues. We conclude that the R2866 lgtC gene encodes a galactosyltransferase involved in synthesis of the 4C4 epitope, as in other strains, and that expression of lgtC is associated with the high-level serum resistance that has been observed for this strain. This is the first description of the genetic basis of high-level serum resistance in NTHI, as well as the first description of LOS composition in an NTHI strain for which the complete genome sequence has been determined.
The gram-negative bacterium Haemophilus influenzae is a common commensal and occasional pathogen of the upper respiratory tract. Prevalence studies have shown that H. influenzae strains isolated from the upper airway are predominately unencapsulated (nontypeable H. influenzae [NTHI]). While most disease associated with NTHI is limited to mucosal sites, a small proportion of NTHI infections are systemic. Many of these are in elderly adults, occurring as a complication of pneumonia. In the past decade, there have been several reports of NTHI strains or strains with capsules of types other than b isolated from the blood or cerebrospinal fluid (CSF) of apparently normal children who had been immunized with an H. influenzae type b (Hib) vaccine (7, 8, 10, 38). To facilitate development of vaccines able to prevent all H. influenzae infections, an understanding of the virulence of invasive NTHI is critical.
A 1996 case report was one of the first to describe NTHI meningitis in a child with normal immunological and anatomical status (36). We found that R2866, the strain isolated from this child, is nearly as resistant as a virulent type b strain, E1a, to the bactericidal effects of normal adult human serum. We demonstrated that R2866 lacks the genes for capsule synthesis and secretion. Using flow cytometry, we showed that when R2866 is incubated in 40% normal human serum, the classical pathway of complement activation is initiated normally, but the deposition of component C3 is delayed. In contrast, the avirulent, unencapsulated laboratory strain Rd KW20 rapidly bound C3 in our assays and was killed shortly after exposure to human serum (53). As described here, fortuitous identification of a serum-sensitive variant of R2866 allowed us to study the molecular basis of the high-level serum resistance of this strain. During our early evaluation of the virulence of R2866 in experimental infections, we retained a single colony, designated R3392, isolated from the blood of an infant rat. We subsequently found that R3392 had modestly increased virulence for infant rats (not shown). The phenotype of R3392 that is most important for the studies described here is that it was much more sensitive to human serum than R2866. We do not know whether infant rat passage generally selects for variants with increased sensitivity to human serum or whether R3392 is an anomaly.
It has long been known that for pneumococci and certain other pathogens, the virulence of laboratory-passaged strains can be increased by passage through an appropriate laboratory animal (41). The increased virulence in some cases persists through subsequent passages on artificial media. This old observation makes sense in light of our present knowledge that several mucosal pathogens, including pneumococci, Neisseria spp., and H. influenzae, are subject to phase-variable expression of surface molecules (4, 37, 48). Following infection of an animal with a mixed population of phase variants, selective pressure may allow outgrowth of variants that were minor components in the inoculum.
In H. influenzae, translational on-off switching of several genes occurs stochastically, through slipped-strand mispairing of a variable number of nucleotide repeats, which are usually tetrameric and located after the start codon of the gene (49). These loci have been termed “contingency genes,” whose expression permits selective expansion of clones with the surface molecule(s) most favorable for the immediate environment. These phase-variable surface molecules include antigenically important structures such as lipooligosaccharide (LOS) and adhesins (4). Because of the high frequency of alteration in repeat length, any population of H. influenzae is a mixture of variants that differ in expression of the contingency genes. As organisms move from one environment, such as the nasopharynx, to another, such as the middle ear or the bloodstream, a different mixture of variants may be selected.
We describe here our investigation of the molecular basis of the serum sensitivity of R3392, leading to the conclusion that expression of the phase-variable glycosyltransferase gene lgtC is a critical factor in the high-level serum resistance of R2866. There have been several reported examples of specific LOS modifications that modulate serum resistance and other virulence-related phenotypes of otitis media isolates of NTHI (24, 51). However, this is the first report of the role of LOS composition in the high-level serum resistance of an NTHI bloodstream isolate. We used a combination of genetic, immunochemical, and biochemical techniques to characterize the phase-variable genes, LOS, and the serum resistance of R2866 and R3392.
MATERIALS AND METHODS
Bacterial strains and culture media.
All bacterial strains used in this study are listed in Table 1. _H. influenzae s_trains were stored at −80°C in 50% defibrinated horse blood (PML Microbiologics) and 50% autoclaved skim milk. Strains were grown at 37°C in Difco brain heart infusion broth (Becton Dickinson, Sparks, MD) supplemented with 10 μg each of hemin, l-histidine, and β-NAD+ (Sigma, St. Louis, MO) per ml (sBHI). Difco Bacto agar (Becton Dickinson) was added at 15 g/liter to sBHI for solid medium. Strains were also cultured on enriched chocolate agar (GC medium base containing 1% hemoglobin and 1% Isovitalex). Ribostamycin (Sigma) was added to sBHI at a concentration of 25 μg/ml when selection for the TSTE cassette was performed. The Escherichia coli cells used for subcloning of lic3A knockout vectors were Max Efficiency DH5-α competent cells (Gibco BRL). All E. coli strains were grown in Luria-Bertani (LB) broth or on LB agar plates at 37°C.
TABLE 1.
Characteristics of the bacterial strains used in this study
| Strain | Genotype or relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli DH5α | F− φ80 lacZ_Δ_M15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rk− mk+) phoA supE44 thi-1 gyrA96 relA1 tonA | Gibco BRL |
| H. influenzae | ||
| R2866 | Invasive nontypeable clinical isolate | 36 |
| R3392 | Isolated after rat infection with R2866 | This study |
| R3521 | R2866 Δ_lic3A_, ribostamycin resistant | This study |
| R3522 | Isolated after rat infection with R3521 | This study |
| 2019 | Nontypeable sputum isolate | 6 |
| Neisseria gonorrhoeae | ||
| 1291 | LOS contains lacto-_N_-neotetraose | 27 |
| 1291 pgm | Truncated LOS | 56 |
| PID-2 | Reference strain for LOS SDS-PAGE analysis | 3 |
Animal model of infection.
The infant rat model of H. influenzae infection (45) was used as previously described (36). The variant strain designated R3392 was propagated from a single colony recovered after infection of infant rats with strain R2866, and strain R3522 was similarly derived from a single colony recovered after infection with strain R3521. These experiments were carried out at the University of Missouri—Columbia, and the infection protocol was reviewed and approved by that institution's Institutional Animal Care and Use Committee.
MLST.
To verify that H. influenzae recovered from infant rat blood were derived from the strains used for inoculation, they were typed using a published multilocus sequence typing (MLST) system (33). Approximately 450-bp internal fragments of seven housekeeping genes (adk, atpG, frdB, fucK, mdh, pgi, and recA) were amplified and sequenced for strains R2866, R3392, R3521, and R3522. The sequences were identical for all four strains, and sequence type 99 was assigned by the curator of the H. influenzae MLST database (http://haemophilus.mlst.net).
Determination of on/off state of phase-variable genes by fragment size analysis of tetrameric repeat regions.
Genomic DNA was prepared using the DNeasy kit (QIAGEN). DNA from individual colonies was obtained by using a toothpick to sample a colony and suspend bacteria in 100 μl of a 10% suspension (wt/vol) of Chelex resin (Bio-Rad, Hercules, CA). After vortexing the suspension for 15 s, the samples were heated to 100°C for 10 min, placed on ice for 2 min, and centrifuged for 2 min at 8,000 × g. Fifty microliters of supernatant was saved and used as a PCR template. Primers were designed using the Rd KW20 or R2866 genome sequences, available through the microbial genome database of NCBI (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi); primer sequences are listed in Table 2. All primers were synthesized by Integrated DNA Technologies (Coralville, IA).
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′→3′) |
|---|---|
| Loci for amplification of tetrameric repeat regiona | |
| lic1A | |
| Forward | TATTACATAATCTTTCAGCTAACCG |
| Reverse | TACTTCCTCAGGCTTGTAGCAAG |
| lic2A | |
| Forward | GGAAAATGCAACTGAACGTCGC |
| Reverse | CTTATTCCATAATAAGAAATGGC |
| lic3A | |
| Forward | GCTAAGATACAACGGAGATCG |
| Reverse | CGGAAAACATCATAATCTTTAGG |
| lgtC | |
| Forward | ATCGAGCAAAGGCATTGAGT |
| Reverse | TCATTGTCTGACTGACAGTC |
| lex2A | |
| Forward | GGCGGAATTATGTTAATCAC |
| Reverse | GCTTGCATATAAGCTTTTCG |
| oafA | |
| Forward | TTCCAGAATTACTTGTAGGATCTTTG |
| Reverse | CATTAAAAACAAGCAGGAAAATAATAG |
| Generation of lic3A deletion mutantb | |
| lic3del-R | TAGGGCTATGgatcCAGTTGTACGATCTCCG |
| lic3del-F | caactggatcCATAGCCCTAAATATGAGAG |
| XhoI-lic3a-4kb-F | ttgcctcgaggcCTTCAGATGAACC |
| SacI-lic3a-4kb-R | ttcggagctcCATTTGCGACAATAC |
For each phase-variable gene of R2866 studied, a region including the tetranucleotide repeat region was amplified from genomic DNA, sequenced, and translated to determine what numbers of repeat units resulted in the gene being in frame with its start codon (“on”). The PCR conditions were 95°C for 12 min; 10 cycles of 94°C for 15 s, 50°C for 15 s, and 72°C for 15 s; and 20 cycles of 89°C for 15 s, 50°C for 15 s, 72°C for 15 s, and 72°C for 10 min. The same region was amplified using a primer set in which the forward primer was labeled with 6-carboxyfluorescein (6-FAM) to allow detection of products within gels by fluorescence. Analysis of these PCR products using the GeneScan or GeneMapper systems (Applied Biosystems, Inc., Foster City, CA) resulted in accurate size determination, with a precision of 1 bp.
In order to evaluate the heterogeneity of a population of bacteria, PCR was performed on DNA prepared from multiple individual colonies, using the fluorescently labeled forward primers as described above. Amplicons that differed from genomic DNA by 4 bp or 8 bp could readily be distinguished, and the on or off state of the corresponding gene could be determined by comparison of these product sizes with the size of the genomic DNA product that had been sequenced.
Construction of lic3A deletion mutant of R2866.
To generate a lic3A deletion, a mega-primer mutagenesis strategy was used. Primer sequences are listed in Table 2. Primers lic3del-R and XhoI-4kb-lic3a-F were used to amplify a 1.57-kb region of the R2866 chromosome just upstream of the lic3A gene, and primers lic3del-F and SacI-4kb-lic3a-R were used to amplify a 1.41-kb region immediately downstream of the lic3A gene. Primers lic3del-F and lic3del-R include a 20-base complementary region containing a BamHI restriction site. The two amplicons were mixed together, denatured, and allowed to anneal and then subjected to an extension step. The resulting 3-kb fragment of double-stranded DNA was amplified using primers XhoI-4kb-lic3a-F and SacI-4kb-lic3a-R and cloned into pBS SK+. A TSTE cassette (44) was inserted into this construct at the BamHI site, and the resulting plasmid was named plic3aΔTSTE. This plasmid was linearized with SacI and transformed into R2866 using the M-IV transformation protocol (18) with ribostamycin selection. Transformants growing on ribostamycin were screened by PCR using combinations of XhoI-lic3a-F, SacI-lic3a-R, and primers internal to the TSTE cassette. Colonies with amplicons confirming deletion of lic3A were retained, and one colony was expanded and retained as strain R3521.
The details of the procedure are as follows. PCR was carried out using the Expand Long Template PCR kit (Roche). The DNA amplification steps used the following protocol: 94°C for 1 min (Taq polymerase was added after thermal cycler reached the specific temperature); 10 cycles of 94°C for 10 s, 50°C for 30 s, and 68°C for 6 min; followed by 20 cycles of 94°C for 10 s, 50°C for 30 s, and 6 min plus 20 s longer for each cycle at 68°C; and a final extension step of 12 min at 68°C. The amplification products were purified using the QIAGEN PCR cleanup kit with an elution volume of 30 μl. Equimolar amounts of the two products were mixed and subjected to the following elongation protocol: 95°C for 1 min and 3 cycles of 95°C for 30 s, 50°C for 30 s, and 68°C for 1 min. At the end of the last extension step, a hold at 68°C was used to add 1 μl of a mixture of 9 μM each of XhoI-lic3a-4kb-F and SacI-lic3a-4kb-R to the PCR batch. The second round of amplification used the same PCR protocol as the first.
Serum bactericidal assay.
Blood was obtained from 7 to 10 healthy adult volunteers under protocols approved by the University of Missouri—Columbia Institutional Review Board and the Institutional Review Board of the Seattle Biomedical Research Institute. Blood was allowed to clot at room temperature, and human serum was isolated aseptically, pooled, and stored at −80°C in 1-ml aliquots.
The microtiter plate serum bactericidal assay has been described previously (12). Log-phase bacteria (2,000 CFU/ml) were incubated for 30 min at 37°C with pooled normal human serum serially diluted in 10 mM phosphate-buffered saline containing 4 mM KCl and 0.1% gelatin and then plated to determine bacterial survival. The concentration of serum that killed 50% of bacteria was calculated after fitting the data to a Boltzmann sigmoidal curve, using the program XLfit 4.1 (ID Business Solutions, Guildford, United Kingdom), and is referred to as the 50% inhibitory concentration (IC50) of the serum for that strain. Treatment of bacteria with 40% serum to select for variants with increased resistance was carried out as described previously (11).
Whole-cell ELISA.
A whole-cell enzyme-linked immunosorbent assay (ELISA) was performed using the monoclonal antibodies (MAbs) 3F11, 6E4, and 4C4 by a method modified from Abdillahi and Poolman (1). Antibody 4C4 (kindly provided by Eric Hansen, University of Texas Southwestern Medical Center) was raised against H. influenzae membranes (29) and was later shown to react with Galα1,4Gal (20). Antibody 3F11 was raised against Neisseria gonorrhoeae cells (3) and was subsequently shown to react with lacto-_N_-neotetraose (30). Antibody 6E4 was raised against H. influenzae cells and reacts with 2-keto-3-deoxyoctulosonic acid (KDO) (46). Prior to incubation with the monoclonal antibodies, half of the wells were treated with 0.05 U neuraminidase/ml from Vibrio cholerae (Roche Molecular Biochemicals) in neuraminidase buffer (50 mM sodium acetate, 100 mM NaCl, 4 mM CaCl2, pH 5.5), and the other half were incubated in neuraminidase buffer alone for 6 h at 37°C in a humidified chamber. The plates were washed again, and 100 μl of each monoclonal antibody in antibody buffer (150 mM NaCl, 10 mM Tris, pH 7.4, 0.3% Tween 20) was serially diluted twofold from a starting dilution of 1:40. The plates were incubated at 25°C overnight. The plates were washed, and 100 μl of a 1:2,000 dilution of goat anti-mouse immunoglobulin M phosphatase-conjugated secondary antibody (Kirkegaard and Perry Laboratories) was added to each well and allowed to incubate at 25°C for 1 h. The plates were washed, and 100 μl of developer (1 mg/ml _p_-nitrophenyl phosphate [Sigma], 0.96% diethanolamine, 2 mM MgCl2, pH 10.0) was added to each well. The samples were allowed to develop for 30 min, and 50 μl of 4 N NaOH was added to each well to quench the reaction. The plates were read at 405 nm with a Bio-Tec Instruments EL 311SX microplate reader. Assays were repeated six times, and data are presented as the means of the absorbance obtained at an antibody dilution of 1:160.
LOS preparation and neuraminidase treatment.
Organisms were grown on sBHI agar in the presence or absence of supplemental Neu5Ac (100 μg/ml), and LOS was prepared by hot phenol-water extraction as described previously (2). For neuraminidase treatment, LOS was resuspended at 1 mg/ml in water and 10 μg was digested with 5 mU of neuraminidase in neuraminidase buffer and incubated at 37°C for 2 h.
SDS-PAGE analysis of LOS.
LOS was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a 4% (wt/vol) polyacrylamide stacking gel and a 15% (wt/vol) polyacrylamide Tris-glycine resolving gel as previously described (25, 26). Electrophoresis was run in an SE 600 vertical gel apparatus (Hoefer Scientific Instruments, San Francisco, CA) at 4°C by using precooled running buffer consisting of 0.25 M Tris and 1.92 M glycine, pH 8.0, and placing the entire unit in a refrigerator at 4°C. Electrophoresis was carried out at 9 mA/gel constant current until samples passed through both stacking gels. The current was then raised to 12 mA/gel until the tracking dye reached 1 cm from the bottom of the resolving gel. LOS was visualized using the protocol accompanying the Rapid-Ag-Stain kit (ICN Pharmaceuticals, Costa Mesa, CA).
MALDI MS.
LOS samples prepared from H. influenzae R2866 and R3392 were O deacylated for 35 min at 37°C with 50 μl anhydrous hydrazine. The reaction was then cooled and quenched with 200 μl ice-cold acetone (15), drop dialyzed for 30 min, and dried. In order to dephosphorylate the deacylated LOS (_O_-LOS), the samples were reconstituted in aqueous hydrofluoric acid (HF [48%]) and left at 4°C for 16 h. Samples were dried as previously described (39). Dried samples were dissolved in 30 μl MilliQ water (Millipore). Dowex 50 beads (100 to 200 mesh, NH4+ form; Bio-Rad, Hercules, Calif.) were added to a mixture containing equal volumes of dephosphorylated _O_-LOS and a saturated solution of 2,5-dihydroxybenzoic acid (Aldrich) in 4:1 acetone-water (Aldrich). Samples were spotted onto a stainless steel matrix-assisted laser desorption ionization (MALDI) target and analyzed on a Voyager DESTR-Plus time-of-flight instrument (Applied Biosystems) with an N2 laser (337 nm) in negative-ion mode with linear optics (15). The delay time was 165 ns, and the grid voltage was 94% of the full acceleration voltage (20 kV). Spectra were acquired, averaged, and mass calibrated with an external calibrant consisting of an equimolar mixture of angiotensin I, ACTH 18-39 and ACTH 7-38 (Bachem, Torrance, CA). Tandem mass spectrometry (MS/MS) data were acquired on a Finnigan LTQ linear ion trap coupled to a vMALDI ion source (Thermo Electron, San Jose, Calif.). The vMALDI ion source operates at 170 mTorr and uses an SI nitrogen laser (337.7 nm). Samples were mixed 1:1 with a 50-mg/ml solution of 2,5-dihydroxybenzoic acid in 50% acetonitrile and then spotted onto a 96-well sample plate. Tandem mass spectrometry (MS/MS) data were collected in positive-ion mode using a precursor ion isolation width of 3 m/z, normalized collision energy of 40% (radio frequency amplitude in percentage used to fragment ions), activation Q of 0.25, and an activation time of 30 ms. Spectra were recorded using automated gain control to control the number of laser shots and the automatic spectrum filter tool.
Statistics.
Groups were compared using the Mann-Whitney test (Fig. 1) or the Fisher exact probability test (Table 3), calculated online using the VassarStats Website for Statistical Computation (http://faculty.vassar.edu/lowry/VassarStats.html).
FIG. 1.
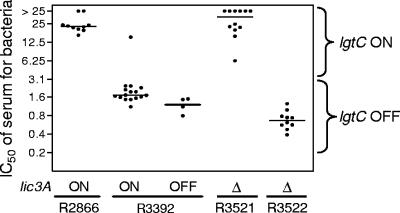
IC50 of normal human serum for cultures derived from individual colonies of R2866, R3392, R3521, and R3522. DNA preparations from the same colonies were used for the data shown in Table 3. For strain R3392, data for _lic3A-_on and _lic3A-_off colonies are plotted separately. All R3392 colonies are lgtC off, with the exception of the single colony with an IC50 of 15%, which is lgtC on. For R3521, all colonies are lgtC on. Serum IC50 was significantly higher for colonies of R2866 than for colonies of R3392 (P < 0.0001); serum IC50 was significantly higher for colonies of R3521 than for colonies of R3522 (P < 0.0001).
TABLE 3.
Analysis of tetrameric repeat regions within LOS biosynthetic genes
| Phase-state status | Result (no. of colonies) for bacterial straina: | ||||
|---|---|---|---|---|---|
| lic3A+ | Δ_lic3A_ | ||||
| R2866 | R3392 | Serum-treated R3392 | R3521 | R3522 | |
| lic1A | |||||
| On | 11 | 19 | 10b | 11 | 11 |
| Off | 0 | 0 | 0 | 0 | 0 |
| lic2A | |||||
| On | 11 | 20 | 11 | 7 | 7 |
| Off | 0 | 0 | 0 | 0 | 0 |
| lic3A | |||||
| On | 11 | 15 | 8 | NA | NA |
| Off | 0 | 5 | 3 | NA | NA |
| lex2A | |||||
| On | 10 | 8 | 11 | 11 | 1 |
| Off | 1 | 1 | 0 | 0 | 10 |
| lgtC | |||||
| On | 9 | 1 | 7b | 10 | 0 |
| Off | 2 | 19 | 3 | 1 | 11 |
| oafA | |||||
| On | 0 | 0 | 0 | 0 | 0 |
| Off | 11 | 20 | 11 | 7 | 7 |
RESULTS
R3392 and R3522 have increased sensitivity to human serum.
We found previously that strain R2866 was unusually resistant to normal human serum: the IC50 of human serum for R2866 was approximately 12.5%, within twofold that for the type b strain E1a and 15 times that for Rd KW20 (12). In contrast, the variant strain R3392, derived from R2866, is substantially more sensitive to human serum than its parent, with a serum IC50 of 1.6%. The results of multilocus sequence typing were identical for R2866 and R3392, making it very unlikely that R3392 is a contaminant. We prepared cultures from individual colonies of R2866 and R3392 and determined their resistance to human serum (Fig. 1). Ten colonies of R2866 displayed high-level resistance (IC50 of >12.5%), while all except 1 of 20 R3392 colonies were much more sensitive (IC50 of 1.1% to 2.3%). This suggested that serum resistance of R2866 is phase variable. Consistent with this, when we treated strain R3392 with human serum and cultured the surviving bacteria, the serum-treated population was substantially increased in resistance to serum (IC50 of 8.4%; geometric mean of two replicate assays).
Previous studies have identified several aspects of LOS structure that vary within a strain and can affect serum resistance. These include sialylation, acetylation, decoration with phosphorylcholine, and expression of various glycosyl structures, including a digalactosyl moiety that is recognized by MAb 4C4 (14, 17, 24, 49, 51). As each laboratory uses different methods to quantify serum resistance, we were unable to determine from these reports whether any of these structural features is thought to make as large a difference in serum resistance as the difference between R2866 and R3392. Of three known H. influenzae sialyltransferases, lic3A is the only one reported to be phase variable. A lic3A deletion mutant, R3521, was found to be as resistant to human serum as the parent strain, R2866. We isolated a serum-sensitive variant of R3521, designated R3522, and found it to be at least as sensitive to serum as R3392 (Fig. 1). While this does not rule out a role for sialic acid in the serum resistance of R2866, it is unlikely that the difference between R2866 and R3392 in serum resistance can be explained simply by phase-variable sialylation. The potential role of other phase-variable genes was evaluated by determining whether they were on or off in R2866 and its derivatives.
lgtC is on in R2866 and R3521 but off in R3392 and R3522.
We used fragment analysis to evaluate the on/off state of phase-variable genes in individual colonies (Table 3). The genes chosen included those identified in the Rd KW20 genome as phase-variable genes involved in LOS biosynthesis (21, 23, 49): the phosphorylcholine transferase gene lic1A, the glycosyltransferase genes lic2A and lgtC, and the sialyltransferase gene lic3A. Also included were the recently identified acetyltransferase gene oafA (14) and lex2A, a phase-variable LOS biosynthetic gene (9) that is not present in Rd KW20 (13).
For each gene to be studied, the tetranucleotide repeat region was amplified by PCR, and its exact size (within 1 bp) was determined by gel electrophoresis using the Genescan system. The size of each product was compared to the sizes of PCR products that had been analyzed in the same system and that had also been sequenced by standard methods. This method allowed accurate calculation of the number of tetranucleotide repeat units in the DNA from each colony, which in turn allowed determination of whether the gene was in frame (on), allowing gene expression, or out of frame (off) relative to its start codon. (In these studies, the term “gene expression” refers solely to the on/off state of the gene; it does not imply any knowledge of the level of gene transcription or of protein production.)
We saw a correlation between serum resistance and the on/off state of the lgtC gene: lgtC was on in most colonies of the serum-resistant strains R2866 and R3521 and was off in most colonies of the serum-sensitive strains R3392 and R3522 (Table 3). Notably, the IC50 for the single colony of R3392 with lgtC on was 15% (Fig. 1). No such correlation was seen for any other gene studied (Table 3). oafA was off in all colonies studied. lic1A and lic2A were on in most or all colonies of R2866, R3392, R3521, and R3522. lex2A was on in most colonies of R2866, R3392, and R3521 and off in most colonies of R3522. For R3392, we identified both _lic3A_-off and _lic3A_-on colonies and found little if any difference between these groups in serum resistance (Fig. 1).
The correlation between serum resistance and lgtC expression was further supported by finding that lgtC was on in most colonies recovered from R3392 after incubation with serum (Table 3). Of 11 colonies analyzed, lgtC was on in 7 colonies and off in 3; 1 colony was a mixture of on and off.
These data suggest that expression of lgtC plays a critical role in the high-level resistance of R2866 and related strains to the bactericidal activity of normal human serum. Consistent with this, a mutant of R2866 in which lgtC was inactivated was as sensitive to human serum as R3392 (Ho et al., submitted for publication). The most likely function of lgtC in R2866 is in LOS biosynthesis, and we confirmed this by comparing the LOS structures of strains R2866 and R3392.
LOS analysis by SDS-PAGE.
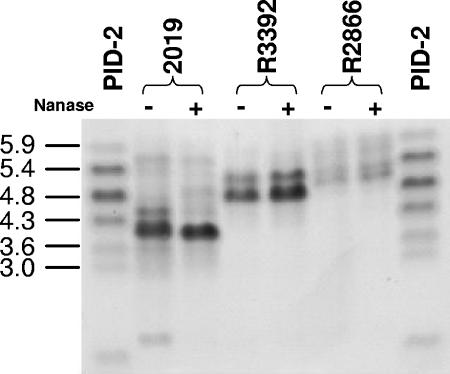
Previous work has shown that LOS structure affects virulence-associated phenotypes such as serum resistance, mucosal persistence, and bloodstream survival (16, 17, 50, 51). We analyzed LOS from R2866 and R3392 by SDS-PAGE. LOS from strain R3392 displayed increased electrophoretic mobility compared to R2886, suggesting a smaller mass. Sialylation of R2866 LOS does not appear to be responsible for this difference, as migration on the gel was not affected by treatment with neuraminidase (Fig. 2).
FIG. 2.
SDS-PAGE analysis of nontypeable H. influenzae treated (+) or not treated (−) with neuraminidase (Nanase). PID-2, reference LOS from N. gonorrhoeae; 2019, reference strain, included as a neuraminidase control. Although the R2866 lanes are underloaded relative to the other samples, it is apparent that R3392 LOS has greater mobility than that of R2866 and that neuraminidase treatment does not affect the mobility of the LOS prepared from either of these two strains.
Whole-cell ELISA.
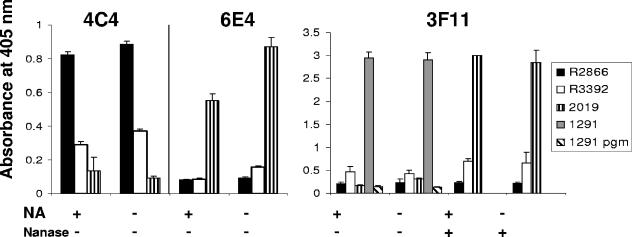
To further characterize differences between LOS of R2866 and R3392, we used an ELISA to measure binding to whole bacteria of three monoclonal antibodies whose specificity for LOS epitopes has been well characterized (Fig. 3). H. influenzae strain 2019 and gonococcal strain 1291 were used as controls for antibody binding. The greatest difference between R2866 and R3392 was in their binding of antibody 4C4: R2866 optical density (OD) readings for 4C4 were two to three times greater than that of 4C4 with R3392. In separate experiments (data not shown), we found that signal strength of strains R2866 and R3392 with 4C4 and 6E4 was unaffected by neuraminidase treatment in the ELISA. Antibody 4C4 has been shown to recognize Galα1,4Gal (19). This disaccharide structure is referred to below as the 4C4 epitope.
FIG. 3.
Whole-cell ELISA of nontypeable H. influenzae strains 2019, R3392, and R2866 with monoclonal antibodies 6E4, 3F11, and 4C4. Bacteria were grown in the presence or absence of neuraminic acid (NA), and selected wells were treated with neuraminidase (Nanase) before exposure to antibody. Strains 1291 and 1291 pgm were used as positive and negative controls, respectively, for 3F11 reactivity. The absorbance at 405 nm indicates the relative amount of antigen exposed for antibody detection. Representative data are shown and expressed as means of six replicates ± standard deviation.
Antibody 3F11 recognizes a terminal _N_-acetyllactosamine structure (54, 56) that can be a substrate for sialylation. Strain 1291, which contains this structure, was strongly reactive with 3F11, while a pgm mutant of 1291 that produces a severely truncated LOS did not react. Strain 2019 was strongly reactive with 3F11 only after treatment with neuraminidase. These observations were true even for bacteria grown on medium without added Neu5Ac. While 3F11 gave a stronger signal with strain R3392 than with R2866, the OD reading was much less than those of strains 2019 and 1291. OD readings for strain R3392 with 3F11 were approximately doubled by neuraminidase treatment. These data suggest that a minority of R3392 LOS molecules may contain _N_-acetyllactosamine and that some of those molecules may be sialylated.
Antibody 6E4, which recognizes KDO (46), did not bind to either R2866 or R3392, although reactivity with strain 2019 was readily demonstrated. It is unlikely that R2866 and related strains lack KDO; binding of 6E4 by these strains may be sterically hindered by glycosyl chains that differ in position or structure from those of strain 2019.
LOS analysis by MALDI-MS.
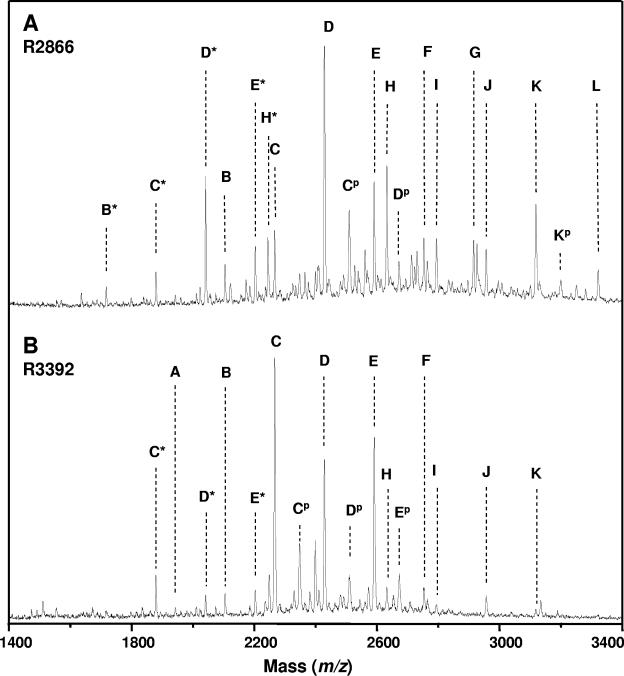
We utilized MALDI-MS to identify the specific differences in sugar composition between the LOS of strains R2866 and R3392 (Table 4 and Fig. 4). LOS samples were treated with anhydrous hydrazine to remove O-linked acyl chains in the lipid A region, and the O-LOS were then dephosphorylated with 48% aqueous HF prior to analysis by MALDI-MS. The major dephosphorylated O-LOS glycoform (peak C in Fig. 4) in strain R3392 had an abundant deprotonated molecular ion, [M-H]− at m/z 2,267.3, consistent with the structure lipid A′-KDO-Hep4-Hex3, where lipid A′ refers to the di-N-acyl-dephosphorylated form. Other abundant LOS-glycoforms in R3392 include those containing 1-2 and 4-6 hexoses. Trace amounts of LOS glycoforms containing a single HexNAc and up to seven hexoses were also observed. In the case of strain R2866, the major dephosphorylated O-LOS glycoform (peak D in Fig. 4) has the proposed composition of lipidA-KDO-Hep4-Hex4 ([M-H]− at m/z 2,429.4), differing from that seen in R3392 by a single hexose. Furthermore, in strain R2866, the diversity of glycoforms is greater than in strain R3392, with glycoforms containing up to seven hexoses and two _N_-acetylhexosamines. Peaks corresponding to glycoforms containing a single phosphate were observed in both strains R2866 and R3392, suggesting that dephosphorylation was incomplete. Also of note are peaks resulting from the loss of half of the lipid A. These glycosidic fragments are likely a by-product of the HF treatment of the O-LOS and were not observed in the mass spectra of the O-LOS prior to HF treatment (data not shown). Comparison of mass spectra of the desphosphorylated O-LOS with spectra of the O-LOS prior to dephosphorylation indicate that the major glycoforms from both strains were modified with five phosphate moieties and two phosphoethanolamine moieties (data not shown). Additionally, mass spectra taken prior to aqueous HF dephosphorylation failed to show the presence of silaic acid-containing glycoforms in either R2866 or R3392. Thus, the predominant glycoform of R3392 contains one hexose less than that of R2866, and the distribution of the minor glycoforms of the LOS of strain R3392 is less diverse. The presence of four heptoses in both strains was confirmed by tandem mass spectrometry. The tandem mass spectrometry showed for both R2866 and R3392 that the predominant isoform of the major glycoforms contained both terminal hexose (loss of 162 Da from molecular ion) and terminal heptose (loss of 192 Da from molecular ion).
TABLE 4.
LOS glycoforms observed after HF treatment of O-deacylated LOS
| Glycoform | Composition (no. of Hex/HexNAc molecules)a | [M-H]− (m/z)b | |||
|---|---|---|---|---|---|
| Calculated | Observed | ||||
| Hex | HexNAc | R2866 | R3392 | ||
| Normal | |||||
| A | 1 | 0 | 1,942.9 | 1,943.3 | |
| B | 2 | 0 | 2,105.1 | 2,105.0 | 2,105.0 |
| C | 3 | 0 | 2,267.3 | 2,267.2 | 2,267.3 |
| D | 4 | 0 | 2,429.4 | 2,429.4 | 2,429.4 |
| E | 5 | 0 | 2,591.6 | 2,591.6 | 2,591.7 |
| F | 6 | 0 | 2,753.7 | 2,753.5 | 2,753.7 |
| G | 7 | 0 | 2,915.8 | 2,915.8 | |
| H | 4 | 1 | 2,632.6 | 2,632.7 | 2,632.9 |
| I | 5 | 1 | 2,794.8 | 2,794.7 | 2,794.4 |
| J | 6 | 1 | 2,956.9 | 2,956.9 | 2,957.3 |
| K | 7 | 1 | 3,119.0 | 3,119.1 | 3,118.7 |
| L | 7 | 2 | 3,322.2 | 3,322.4 | |
| Loss of 1/2 lipid Ac | |||||
| B* | 2 | 0 | 1,717.7 | 1,717.5 | |
| C* | 3 | 0 | 1,879.8 | 1,879.5 | 1,879.5 |
| D* | 4 | 0 | 2,041.9 | 2,041.7 | 2,041.8 |
| E* | 5 | 0 | 2,204.1 | 2,203.8 | 2,203.8 |
| H* | 4 | 1 | 2,245.1 | 2,245.0 | |
| Phosphorylatedd | |||||
| Cp | 3 | 0 | 2,348.3 | 2,348.8 | |
| Dp | 4 | 0 | 2,510.4 | 2,510.4 | 2,511.2 |
| Ep | 5 | 0 | 2,673.5 | 2,672.8 | 2,673.3 |
| Kp | 7 | 1 | 3,200.0 | 3,200.3 |
FIG. 4.
Negative-ion MALDI time-of-flight mass spectra of dephosphorylated _O_-LOS from H. influenzae stains R2866 (A) and R3392 (B). Note the differences between the two panels in peaks C (representing a glycoform with three hexoses and four heptoses) and D (four hexoses and four heptoses). See Table 4 for molecular weights and proposed compositions. An asterisk indicates the loss of half the lipid A moiety from the O-deacylated LOS, and p indicates the presence of a phosphate moiety.
DISCUSSION
This study adds to our understanding of the complex relationship between LOS structure and serum resistance in H. influenzae. Previous studies in this area have used encapsulated strains, the laboratory isolate Rd KW20, or clinical NTHI isolates that are not well characterized. This is the first study of LOS composition in an NTHI strain for which the genome sequence is available, as well as the first study of the LOS of an NTHI strain isolated from systemic disease. Our studies identified a critical role for the lgtC gene in serum resistance for strain R2866. This gene encodes a glycosyltransferase that adds the distal sugar in the digalactose structure recognized by monoclonal antibody 4C4. Use of this antibody allows us to compare our data with those of several other laboratories.
A role for the 4C4 epitope in resistance to human serum is clearly established. Weiser and Pan reported that a 4C4-positive variant of nontypeable strain H233 was relatively resistant to human serum, although sensitive to rat serum (50). They proposed that this is a result of the frequent presence of the 4C4 epitope, Galα1,4Gal, on human cells as part of the Pk blood group antigen; thus antibodies to this structure rarely occur in humans. Griffin et al. introduced the lex2A gene into strain Rd KW20, allowing expression of the 4C4 and 5G8 epitopes, and found a modest increase in resistance to human serum (IC60 of approximately 1.5%, compared with a serum IC60 of approximately 0.8% for the parent strain) (17). Although it is difficult to compare such data from one laboratory to another, it appears that even the most resistant derivative of Rd KW20 was much more sensitive to serum than our strain R2866 (serum IC50 of >12.5%, as reported in this study and previously) (12). Thus far, mutation of Rd KW20 to produce altered LOS structures that increase serum resistance has not generated mutants with high-level serum resistance such as that seen in R2866.
Both of the studies noted above are consistent with our data in that expression of the 4C4 epitope appears to increase resistance to normal human serum. However, the magnitude of the effect differs substantially from strain to strain. The effect of this epitope on the serum IC50 of strain Rd KW20 is approximately 2-fold, as noted above, but the IC50 of normal human serum for the invasive strain R2866 is decreased 10-fold when the single gene lgtC is switched off or inactivated. We reported recently that a different blood isolate, R3063, is also capable of a wide range of serum resistance, mediated by the on/off state of a phase-variable glycosyltransferase gene, losA (11). It is not known whether commensal isolates and those cultured from ear aspirates are similarly subject to wide variation in serum resistance.
Other studies have identified two additional LOS substituents that affect resistance to human serum, phosphorylcholine, and sialic acid. Decoration of LOS by phosphorylcholine increases the sensitivity of bacteria to human serum by binding C-reactive protein, leading to the activation of complement (51, 52). We found lic1A to be on for both R2866 and R3392, suggesting that they are equally capable of adding phosphorylcholine. We found previously that R2866 bacteria are heterogeneous with regard to decoration by phosphorylcholine and that depletion of C-reactive protein from pooled normal adult serum had little effect on killing of R2866 (53).
Sialylation of LOS can increase both resistance to human serum and virulence in the chinchilla model of otitis media (5, 43). We found little direct evidence for Neu5Ac in the LOS of R2866. Neu5Ac was not identified by mass spectrometry, and neuraminidase treatment had little effect on antibody binding. It is possible that the predominant glycoforms in R2866 LOS are not substrates for sialylation. Our observations that both R2866 and R3392 bound small amounts of antibody 3F11 and that binding of 3F11 to R3392 was increased by neuraminidase treatment suggest that a minor glycoform contains a lacto-_N_-neotetraose structure that may be sialylated. Deletion of the sialyltransferase gene lic3A, which sialylates glycosyl chains substituting heptose III, did not have a major effect on resistance to human serum. The other two sialyltransferase genes, siaA and lsgB (28), have not yet been studied in strain R2866. Strain R2866 may be similar to Moraxella catarrhalis in being highly resistant to human serum without sialylation of LOS; in that organism, the Galα1,4Gal structure is thought to be critical for maximum expression of serum resistance (55).
We cannot be certain that the only difference between R2866 and R3392 is the on/off state of lgtC. Other, undetected mutations might contribute to the difference in serum resistance. However, the finding that two independently isolated _lgtC_-off variants (R3392 and R3522) were both substantially increased in susceptibility to serum, as well as the demonstration that a single _lgtC_-on colony derived from R3392 is serum resistant (Fig. 1), suggests strongly that lgtC expression state affects serum resistance.
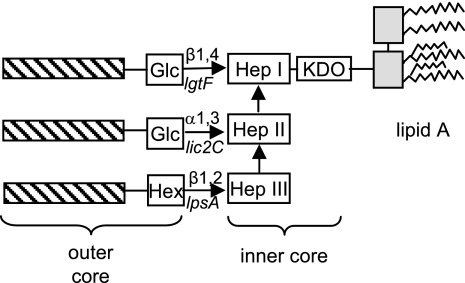
It is not possible to predict the structure of the principal LOS glycoform of R2866 from the glycosyl composition reported here and from our current knowledge of the genetic basis of the LOS structures of other strains. Based on analysis of LOS structures from numerous strains of H. influenzae, Hood and colleagues (22) have reported that all strains investigated have a common inner core consisting of a chain of three heptose residues attached to KDO. As shown in the schematic drawing in Fig. 5, the outer core of LOS consists of glycosyl chains containing one or several sugars, usually hexose or _N_-acetylhexosamine, attached to one or more of the inner-core heptose residues. These glycosyl chains are sometimes modified by the addition of acetate, phosphoethanolamine, phosphorylcholine, or other residues.
FIG. 5.
Schematized structure for H. influenzae LOS, based on data summarized in reference 22. The three hatched bars represent glycosyl chains (one or more residues) that may be present. Nonsugar substituents such as phosphorylcholine, acetate, glycine, and phosphoethanolamine are not shown. Hep, heptose; Glc, glucose; Hex, hexose.
Our data indicate that the major glycoform in R2866 LOS contains four heptose and four hexose residues attached to a single KDO that is linked to lipid A. We suspect that three of the four heptoses form the standard inner core structure and that the fourth heptose is in the outer core, as seen previously in NTHI strains 981, 1207, 1209, 1233, and 9274 (31, 32, 40) and in Haemophilus ducreyi (34, 35, 42). In H. ducreyi, addition of the outer core heptose is catalyzed by LosB (LbgB), and modification of the heptose by the addition of a galactose is catalyzed by LosA (LbgA), an enzyme encoded by the gene adjacent to losB (15, 47). The genome of strain R2866 contains genes that are homologous to H. ducreyi losA and losB (predicted amino acid sequences are 77% and 69% similar, respectively). This losB homolog is the best candidate for a gene encoding an outer-core heptosyltransferase.
In addition to the losA homolog, R2866 contains five additional potential hexosyltransferase genes: lgtF, lpsA, lic2A, lex2A, and lgtC. Of these, lgtC, lic2A, and lex2A are phase variable, and all three were found to be on in R2866 (Table 3). Data described above are consistent with a role for LgtC in synthesis of the principal glycoform of R2866 LOS; it is almost certain that LgtC catalyzes the addition of the distal galactose of a Galα1,4Gal disaccharide (the 4C4 epitope). We cannot say with certainty which of the other five putative hexosyltransferases is involved in addition of the remaining three hexoses detected in the principal glycoform of R2866. The proximal galactose of the 4C4 epitope might be added either by the LosA homolog noted above or by Lic2A, as reported for Rd KW20 (20) and for other NTHI strains (50). The presence of lgtF and lpsA suggests that heptose I and heptose III are each substituted for by at least one hexose. Alternatively, the principal glycoform of R2866 LOS may be similar to the 4-heptose, 5-hexose structure reported for strain 981, which lacks a glycosyl substituent on heptose III (31). This is consistent with our finding that the predominant isoform of the major glycoforms of R2866 and R3392 contained both terminal hexose and terminal heptose. As R2866 contains no lic2B homolog (12), it is likely that heptose II is unsubstituted.
The major glycoform of R3392 LOS was found to contain four heptose residues, like R2866, but only three hexose residues, one fewer than R2866. Since we found that lgtC is off in R3392, it is likely that the “missing” hexose in R3392 is the distal galactose of the 4C4 epitope. This interpretation is consistent with the reduced binding of antibody 4C4 by R3392. Although we discuss the predominant glycoform here, it must be emphasized that the LOS isolated from strains R2866 and R3392 are a diverse mixture of glycoforms, with structures containing between one and seven hexose moieties and up to two _N_-acetylhexosamine moieties. It is possible that Lic2A and Lex2A are involved in synthesis of these minor structures. We have no data on R3522 that would allow us to interpret the observation that lex2A is off in this strain.
While strain R2866 was the first NTHI strain found to have human serum resistance approaching that of encapsulated strains, it is not unique in this regard. Our recent survey of 53 clinical isolates identified five, in addition to R2866, for which serum IC50 is greater than 10% (12). Nothing is known of the LOS structures of these additional strains. The current data indicate that for R2866, the _lgtC_-dependent 4C4 epitope is essential for resistance to human serum, but it is not likely to be sufficient. It will be of interest to determine whether a heptose-containing outer core is found in other strains with high-level resistance to human serum.
In summary, we have identified the lgtC gene, required for synthesis of the 4C4 epitope, as a critical mediator of the high-level serum resistance of strain R2866. Studies are currently under way to characterize the LOS of R2866 and its derivatives more fully and to identify the genes involved in the synthesis of alternative structures. An important aspect of this work is characterization of the specific interactions between complement and the bacterial surface, which may be mediated by the LOS differences described above. These studies will be described in a forthcoming paper (Ho et al., submitted for publication).
Acknowledgments
This work was supported by NIH grants AI 44002, AI 46512, and DC 005833 (A.L.S.); AI 24616 (M.A.A. and B.G.); and the M. J. Murdock Charitable Trust (A.L.S.).
We would like to thank Thermo Electron for the generous use of the use of their vMALDI-LTQ linear ion trap mass spectrometer. We thank Jennifer Geelhood for technical assistance and for critical reading of the manuscript.
REFERENCES
- 1.Abdillahi, H., and J. T. Poolman. 1988. Typing of group-B Neisseria meningitidis with monoclonal antibodies in the whole-cell ELISA. J. Med. Microbiol. 26**:**177-180. [PubMed] [Google Scholar]
- 2.Allen, S., A. Zaleski, J. W. Johnston, B. W. Gibson, and M. A. Apicella. 2005. Novel sialic acid transporter of Haemophilus influenzae. Infect. Immun. 73**:**5291-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apicella, M. A., K. M. Bennett, C. A. Hermerath, and D. E. Roberts. 1981. Monoclonal antibody analysis of lipopolysaccharide from Neisseria gonorrhoeae and Neisseria meningitidis. Infect. Immun. 34**:**751-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayliss, C. D., D. Field, and E. R. Moxon. 2001. The simple sequence contingency loci of Haemophilus influenzae and Neisseria meningitidis. J. Clin. Investig. 107**:**657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouchet, V., D. W. Hood, J. Li, J. R. Brisson, G. A. Randle, A. Martin, Z. Li, R. Goldstein, E. K. Schweda, S. I. Pelton, J. C. Richards, and E. R. Moxon. 2003. Host-derived sialic acid is incorporated into Haemophilus influenzae lipopolysaccharide and is a major virulence factor in experimental otitis media. Proc. Natl. Acad. Sci. USA 100**:**8898-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campagnari, A. A., M. R. Gupta, K. C. Dudas, T. F. Murphy, and M. A. Apicella. 1987. Antigenic diversity of lipooligosaccharides of nontypable Haemophilus influenzae. Infect. Immun. 55**:**882-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campos, J., M. Hernando, F. Roman, M. Pérez-Vázquez, B. Aracil, J. Oteo, E. Lazaro, F. de Abajo, and Group of Invasive Haemophilus Infections of the Autonomous Community of Madrid, Spain. 2004. Analysis of invasive Haemophilus influenzae infections after extensive vaccination against H. influenzae type b. J. Clin. Microbiol. 42**:**524-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerquetti, M., M. L. Ciofi degli Atti, G. Renna, A. E. Tozzi, M. L. Garlaschi, P. Mastrantonio, and HI Study Group. 2000. Characterization of non-type B Haemophilus influenzae strains isolated from patients with invasive disease. J. Clin. Microbiol. 38**:**4649-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cope, L. D., R. Yogev, J. Mertsola, J. L. Latimer, M. S. Hanson, G. H. McCracken, Jr., and E. J. Hansen. 1991. Molecular cloning of a gene involved in lipooligosaccharide biosynthesis and virulence expression by Haemophilus influenzae type B. Mol. Microbiol. 5**:**1113-1124. [DOI] [PubMed] [Google Scholar]
- 10.Cuthill, S. L., M. M. Farley, and L. G. Donowitz. 1999. Nontypable Haemophilus influenzae meningitis. Pediatr. Infect. Dis. J. 18**:**660-662. [DOI] [PubMed] [Google Scholar]
- 11.Erwin, A. L., P. J. Bonthuis, J. L. Geelhood, K. L. Nelson, K. W. McCrea, J. R. Gilsdorf, and A. L. Smith. 2006. Heterogeneity in tandem octanucleotides within Haemophilus influenzae lipopolysaccharide biosynthetic gene losA affects serum resistance. Infect. Immun. 74**:**3408-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erwin, A. L., K. L. Nelson, T. Mhlanga-Mutangadura, P. J. Bonthuis, J. L. Geelhood, G. Morlin, W. C. Unrath, J. Campos, D. W. Crook, M. M. Farley, F. W. Henderson, R. F. Jacobs, K. Mühlemann, S. W. Satola, L. van Alphen, M. Golomb, and A. L. Smith. 2005. Characterization of genetic and phenotypic diversity of invasive nontypeable Haemophilus influenzae. Infect. Immun. 73**:**5853-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269**:**496-512. [DOI] [PubMed] [Google Scholar]
- 14.Fox, K. L., H. H. Yildirim, M. E. Deadman, E. K. Schweda, E. R. Moxon, and D. W. Hood. 2005. Novel lipopolysaccharide biosynthetic genes containing tetranucleotide repeats in Haemophilus influenzae, identification of a gene for adding O-acetyl groups. Mol. Microbiol. 58**:**207-216. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, B. W., J. J. Engstrom, C. M. Johm, W. Hines, and A. M. Falick. 1997. Characterization of bacterial lipooligosaccharides by delayed extraction, matrix-assisted laser desorption time-of-flight mass spectrometry. J. Am. Soc. Mass Spectrom. 8**:**645-658. [Google Scholar]
- 16.Griffin, R., C. D. Bayliss, M. A. Herbert, A. D. Cox, K. Makepeace, J. C. Richards, D. W. Hood, and E. R. Moxon. 2005. Digalactoside expression in the lipopolysaccharide of Haemophilus influenzae and its role in intravascular survival. Infect. Immun. 73**:**7022-7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffin, R., A. D. Cox, K. Makepeace, J. C. Richards, E. R. Moxon, and D. W. Hood. 2005. Elucidation of the monoclonal antibody 5G8-reactive, virulence-associated lipopolysaccharide epitope of Haemophilus influenzae and its role in bacterial resistance to complement-mediated killing. Infect. Immun. 73**:**2213-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herriott, R. M., E. M. Meyer, and M. Vogt. 1970. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J. Bacteriol. 101**:**517-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.High, N. J., M. E. Deadman, D. W. Hood, and E. R. Moxon. 1996. The identification of a novel gene required for lipopolysaccharide biosynthesis by Haemophilus influenzae RM7004, using transposon Tn_916_ mutagenesis. FEMS Microbiol. Lett. 145**:**325-331. [DOI] [PubMed] [Google Scholar]
- 20.High, N. J., M. E. Deadman, and E. R. Moxon. 1993. The role of a repetitive DNA motif (5′-CAAT-3′) in the variable expression of the Haemophilus influenzae lipopolysaccharide epitope α Gal(1-4)β Gal. Mol. Microbiol. 9**:**1275-1282. [DOI] [PubMed] [Google Scholar]
- 21.Hood, D. W., M. E. Deadman, T. Allen, H. Masoud, A. Martin, J. R. Brisson, R. Fleischmann, J. C. Venter, J. C. Richards, and E. R. Moxon. 1996. Use of the complete genome sequence information of Haemophilus influenzae strain Rd to investigate lipopolysaccharide biosynthesis. Mol. Microbiol. 22**:**951-965. [DOI] [PubMed] [Google Scholar]
- 22.Hood, D. W., M. E. Deadman, A. D. Cox, K. Makepeace, A. Martin, J. C. Richards, and E. R. Moxon. 2004. Three genes, lgtF, lic2C and lpsA, have a primary role in determining the pattern of oligosaccharide extension from the inner core of Haemophilus influenzae LPS. Microbiology 150**:**2089-2097. [DOI] [PubMed] [Google Scholar]
- 23.Hood, D. W., M. E. Deadman, M. P. Jennings, M. Bisercic, R. D. Fleischmann, J. C. Venter, and E. R. Moxon. 1996. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 93**:**11121-11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hood, D. W., K. Makepeace, M. E. Deadman, R. F. Rest, P. Thibault, A. Martin, J. C. Richards, and E. R. Moxon. 1999. Sialic acid in the lipopolysaccharide of Haemophilus influenzae: strain distribution, influence on serum resistance and structural characterization. Mol. Microbiol. 33**:**679-692. [DOI] [PubMed] [Google Scholar]
- 25.Inzana, T. J. 1983. Electrophoretic heterogeneity and interstrain variation of the lipopolysaccharide of Haemophilus influenzae. J. Infect. Dis. 148**:**492-499. [DOI] [PubMed] [Google Scholar]
- 26.Inzana, T. J., and M. A. Apicella. 1999. Use of a bilayer stacking gel to improve resolution of lipopolysaccharides and lipooligosaccharides in polyacrylamide gels. Electrophoresis 20**:**462-465. [DOI] [PubMed] [Google Scholar]
- 27.John, C. M., J. M. Griffiss, M. A. Apicella, R. E. Mandrell, and B. W. Gibson. 1991. The structural basis for pyocin resistance in Neisseria gonorrhoeae lipooligosaccharides. J. Biol. Chem. 266**:**19303-19311. [PubMed] [Google Scholar]
- 28.Jones, P. A., N. M. Samuels, N. J. Phillips, R. S. Munson, Jr., J. A. Bozue, J. A. Arseneau, W. A. Nichols, A. Zaleski, B. W. Gibson, and M. A. Apicella. 2002. Haemophilus influenzae type b strain A2 has multiple sialyltransferases involved in lipooligosaccharide sialylation. J. Biol. Chem. 277**:**14598-14611. [DOI] [PubMed] [Google Scholar]
- 29.Kimura, A., and E. J. Hansen. 1986. Antigenic and phenotypic variations of Haemophilus influenzae type b lipopolysaccharide and their relationship to virulence. Infect. Immun. 51**:**69-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandrell, R. E., J. M. Griffiss, and B. A. Macher. 1988. Lipooligosaccharides (LOS) of Neisseria gonorrhoeae and Neisseria meningitidis have components that are immunochemically similar to precursors of human blood group antigens. Carbohydrate sequence specificity of the mouse monoclonal antibodies that recognize crossreacting antigens on LOS and human erythrocytes. J. Exp. Med. 168**:**107-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Månsson, M., D. W. Hood, E. R. Moxon, and E. K. Schweda. 2003. Structural characterization of a novel branching pattern in the lipopolysaccharide from nontypeable Haemophilus influenzae. Eur. J. Biochem. 270**:**2979-2991. [DOI] [PubMed] [Google Scholar]
- 32.Månsson, M., D. W. Hood, E. R. Moxon, and E. K. Schweda. 2003. Structural diversity in lipopolysaccharide expression in nontypeable Haemophilus influenzae. Identification of L-glycerol-D-manno-heptose in the outer-core region in three clinical isolates. Eur. J. Biochem. 270**:**610-624. [DOI] [PubMed] [Google Scholar]
- 33.Meats, E., E. J. Feil, S. Stringer, A. J. Cody, R. Goldstein, J. S. Kroll, T. Popovic, and B. G. Spratt. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 41**:**1623-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melaugh, W., A. A. Campagnari, and B. W. Gibson. 1996. The lipooligosaccharides of Haemophilus ducreyi are highly sialylated. J. Bacteriol. 178**:**564-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melaugh, W., N. J. Phillips, A. A. Campagnari, M. V. Tullius, and B. W. Gibson. 1994. Structure of the major oligosaccharide from the lipooligosaccharide of Haemophilus ducreyi strain 35000 and evidence for additional glycoforms. Biochemistry 33**:**13070-13078. [DOI] [PubMed] [Google Scholar]
- 36.Nizet, V., K. F. Colina, J. R. Almquist, C. E. Rubens, and A. L. Smith. 1996. A virulent nonencapsulated Haemophilus influenzae. J. Infect. Dis. 173**:**180-186. [DOI] [PubMed] [Google Scholar]
- 37.Novak, R., and E. Tuomanen. 1999. Pathogenesis of pneumococcal pneumonia. Semin. Respir. Infect. 14**:**209-217. [PubMed] [Google Scholar]
- 38.O'Neill, J. M., J. W. St. Geme III, D. Cutter, E. E. Adderson, J. Anyanwu, R. F. Jacobs, and G. E. Schutze. 2003. Invasive disease due to nontypeable Haemophilus influenzae among children in Arkansas. J. Clin. Microbiol. 41**:**3064-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phillips, N. J., M. A. Apicella, J. M. Griffiss, and B. W. Gibson. 1992. Structural characterization of the cell surface lipooligosaccharides from a nontypable strain of Haemophilus influenzae. Biochemistry 31**:**4515-4526. [DOI] [PubMed] [Google Scholar]
- 40.Rahman, M. M., X. X. Gu, C. M. Tsai, V. S. Kolli, and R. W. Carlson. 1999. The structural heterogeneity of the lipooligosaccharide (LOS) expressed by pathogenic non-typeable Haemophilus influenzae strain NTHi 9274. Glycobiology 9**:**1371-1380. [DOI] [PubMed] [Google Scholar]
- 41.Saladino, R. A., A. M. Stack, G. R. Fleisher, C. M. Thompson, D. E. Briles, L. Kobzik, and G. R. Siber. 1997. Development of a model of low-inoculum Streptococcus pneumoniae intrapulmonary infection in infant rats. Infect. Immun. 65**:**4701-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schweda, E. K. H., J. A. Jonasson, and P.-E. Jansson. 1995. Structural studies of lipooligosaccharides from Haemophilus ducreyi ITM 5535, ITM 3147, and a fresh clinical isolate, ACY1: evidence for intrastrain heterogeneity with the production of mutually exclusive sialylated or elongated glycoforms. J. Bacteriol. 177**:**5316-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Severi, E., G. Randle, P. Kivlin, K. Whitfield, R. Young, R. Moxon, D. Kelly, D. Hood, and G. H. Thomas. 2005. Sialic acid transport in Haemophilus influenzae is essential for lipopolysaccharide sialylation and serum resistance and is dependent on a novel tripartite ATP-independent periplasmic transporter. Mol. Microbiol. 58**:**1173-1185. [DOI] [PubMed] [Google Scholar]
- 44.Sharetzsky, C., T. D. Edlind, J. J. LiPuma, and T. L. Stull. 1991. A novel approach to insertional mutagenesis of Haemophilus influenzae. J. Bacteriol. 173**:**1561-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, A. L., D. H. Smith, D. R. Averill, Jr., J. Marino, and E. R. Moxon. 1973. Production of Haemophilus influenzae b meningitis in infant rats by intraperitoneal inoculation. Infect. Immun. 8**:**278-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spinola, S. M., Y. A. Kwaik, A. J. Lesse, A. A. Campagnari, and M. A. Apicella. 1990. Cloning and expression in Escherichia coli of a Haemophilus influenzae type b lipooligosaccharide synthesis gene(s) that encodes a 2-keto-3-deoxyoctulosonic acid epitope. Infect. Immun. 58**:**1558-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tullius, M. V., N. J. Phillips, N. K. Scheffler, N. M. Samuels, R. S. Munson, Jr., E. J. Hansen, M. Stevens-Riley, A. A. Campagnari, and B. W. Gibson. 2002. The lbgAB gene cluster of Haemophilus ducreyi encodes a β-1,4-galactosyltransferase and an α-1,6-dd-heptosyltransferase involved in lipooligosaccharide biosynthesis. Infect. Immun. 70**:**2853-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Woude, M. W., and A. J. Baumler. 2004. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17**:**581-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiser, J. N. 2000. The generation of diversity by Haemophilus influenzae. Trends Microbiol. 8**:**433-435. [DOI] [PubMed] [Google Scholar]
- 50.Weiser, J. N., and N. Pan. 1998. Adaptation of Haemophilus influenzae to acquired and innate humoral immunity based on phase variation of lipopolysaccharide. Mol. Microbiol. 30**:**767-775. [DOI] [PubMed] [Google Scholar]
- 51.Weiser, J. N., N. Pan, K. L. McGowan, D. Musher, A. Martin, and J. Richards. 1998. Phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae contributes to persistence in the respiratory tract and sensitivity to serum killing mediated by C-reactive protein. J. Exp. Med. 187**:**631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiser, J. N., M. Shchepetov, and S. T. H. Chong. 1997. Decoration of lipopolysaccharide with phosphorylcholine: a phase-variable characteristic of Haemophilus influenzae. Infect. Immun. 65**:**943-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams, B. J., G. Morlin, N. Valentine, and A. L. Smith. 2001. Serum resistance in an invasive, nontypeable Haemophilus influenzae strain. Infect. Immun. 69**:**695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamasaki, R., W. Nasholds, H. Schneider, and M. A. Apicella. 1991. Epitope expression and partial structural characterization of F62 lipooligosaccharide (LOS) of Neisseria gonorrhoeae: IgM monoclonal antibodies (3F11 and 1-1-M) recognize non-reducing termini of the LOS components. Mol. Immunol. 28**:**1233-1242. [DOI] [PubMed] [Google Scholar]
- 55.Zaleski, A., N. K. Scheffler, P. Densen, F. K. N. Lee, A. A. Campagnari, B. W. Gibson, and M. A. Apicella. 2000. Lipooligosaccharide Pk (Galα1-4Galβ1-4Glc) epitope of Moraxella catarrhalis is a factor in resistance to bactericidal activity mediated by normal human serum. Infect. Immun. 68**:**5261-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, D., D. S. Stephens, B. W. Gibson, J. J. Engstrom, C. F. McAllister, F. K. Lee, and M. A. Apicella. 1994. Lipooligosaccharide biosynthesis in pathogenic Neisseria. Cloning, identification, and characterization of the phosphoglucomutase gene. J. Biol. Chem. 269**:**11162-11169. [PubMed] [Google Scholar]