Functional consequences of alteration of N-linked glycosylation sites on the neurokinin 1 receptor (original) (raw)
Abstract
The neurokinin 1 receptor (NK1R), a G protein-coupled receptor involved in diverse functions including pain and inflammation, has two putative N-linked glycosylation sites, Asn-14 and Asn-18. We studied the role of N-linked glycosylation in the functioning of the NK1R by constructing three receptor mutants: two single mutants (Asn → Gln-14 and Asn → Gln-18) and a double mutant, lacking both glycosylation sites. Using a lentiviral transfection system, the mutants were stably transfected into NCM 460 cells, a nontransformed human colonic epithelial cell line. We observed that the magnitude of glycosylation as estimated by changes in gel migration depends on the number of glycosylation sites available, with the wild-type receptor containing the greatest amount of glycosylation. All mutant receptors were able to bind to substance P and neurokinin A ligand with similar affinities; however, the double mutant, nonglycosylated NK1R showed only half the _B_max of the wild-type NK1R. In terms of receptor function, the ablation of both N-linked glycosylation sites did not have a profound effect on the receptors' abilities to activate the MAP kinase families (p42/p44, JNK, and p38), but did affect SP-induced IL-8 secretion. All mutants were able to internalize, but the kinetics of internalization of the double mutant receptor was more rapid, when compared with wild-type NK1R. Therefore, glycosylation of NK1R may stabilize the receptor in the plasma membrane. These results contribute to the ongoing elucidation of the role of glycosylation in G protein-coupled receptors and the study of the neurokinin receptors in particular.
Keywords: G protein-coupled receptor, substance P
The neurokinin 1 receptor (NK1R) mediates a range of proinflammatory and pain processes. For example, NK1R levels are up-regulated in sites of joint inflammation, asthma, inflammatory bowel disease, acute pancreatitis, and abdominal cell adhesion formation (1–6). It is also speculated to be involved in modulation of HIV infection (7). There is considerable interest in NK1R as a therapeutic target, and antagonists are currently being tested for treatment of depression, emesis, asthma, and breast cancer.
The NK1R was identified, cloned, and characterized as a member of the rhodopsin family of the G protein-coupled receptor (GPCR) superfamily (8–10). The NK1R has different binding affinities for its tachykinin ligands, which include substance P (SP), neurokinin A (NKA), NKA-like peptides, and neurokinin B. SP, first identified by von Euler and Gaddum (11) in 1931, was isolated by Chang and Leeman (12) in 1970, and later synthesized by Tregear et al. (13). SP was identified as a sialogogic peptide, but has also been shown to function as a neurotransmitter, a neuromodulator, and a key mediator of inflammatory processes (14). In nontransformed mucosal epithelial cells, SP can induce secretion of proinflammatory cytokines, such as IL-6, IL-8, and TNF-α, and this induction requires activation of the transcription factor NF-κB (15, 16). SP binding to the NK1R elicits inositol production (17), mobilizes Ca2+ (18), and activates the ERK1 and ERK2 members of the MAPK family (19). In fact, transactivation of the EGF receptor has been demonstrated upon SP stimulation (20), leading to cell proliferation that involves matrix metalloproteinases and TGF-α secretion (21).
Although SP is the preferred ligand for NK1R, several groups have shown that NKA binds NK1R with high enough affinity to elicit a biologic response (22–24). More recently, it has been found that benzoylphenylalanine3-SP, a photoactivatable ligand for the NK1R, covalently labeled the N-terminal region of the NK1R between residues 11 and 21 (25). Therefore, the binding region for SP encompasses the glycosylation region of the NK1R, and perturbations of the glycosylation sites not only provide insight on the receptor itself, but also on the ligand–receptor complex.
The neurokinin receptors contain putative N-linked glycosylation sites on their extracellular amino termini: residues 14 and 18 on NK1R, residues 11 and 19 on neurokinin-2 receptor, and residues 23, 50, and 73 on neurokinin-3 receptor. The role of these receptor glycosylation sites is unclear. Based on the particular receptor, carbohydrate moieties have been shown to be involved in various receptor functions, including receptor stability and activation state (EGF receptor) (26); receptor folding (luteinizing hormone receptor) (27); trafficking and expression (human δ opioid receptor, neurotensin receptor) (28, 29); and ligand binding and signal transduction (parathyroid hormone receptor, somatostatin subtype 3 receptor) (30, 31). It is apparent from these studies that glycosylation effects on GPCRs are empiric and specific to the receptor, requiring investigation in each receptor system.
Based on the evidence pointing to receptor glycosylation as critical in ligand binding, internalization dynamics, and ligand-directed downstream signaling, as well as our previous work suggesting that rat NK1R was highly glycosylated when expressed in CHO cells (32), we sought to understand the role of glycosylation in the NK1R. To do this, we created mutants of NK1R that lack one or both of its N-linked glycosylation sites.
Results
Extent of Glycosylation in Transfected NCM 460 Cells.
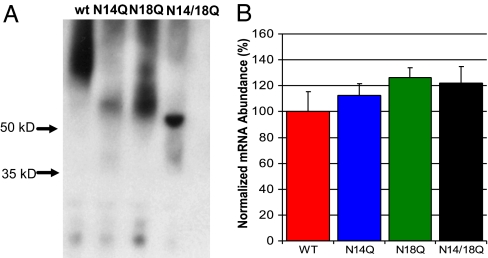
An NK1R immunoblot was performed to analyze the extent of receptor glycosylation in the transfected colonic epithelial NCM 460 cells. The immunoblot shows a greater relative mass of the wild-type NK1R as compared with the two single glycosylation mutants and the double glycosylation mutant (Fig. 1A). The single glycosylation mutants appear to migrate to the same extent as each other and also indicate a relative mass that is greater than the double mutant. The wild-type and single glycosylation mutants tend to have a more diffuse banding pattern as compared with the double glycosylation mutant, which runs in a sharp band very close to the NK1R predicted peptide mass of ≈50 kDa. These results confirm earlier evidence that the native NK1R is glycosylated, and that the mutations at N14Q and N18Q, as expected, lead to a decrease in the level of receptor glycosylation. Furthermore, the various cell lines were shown to express equivalent NK1R transcript, as shown by quantitative RT-PCR (Fig. 1B; α = 0.05, P > 0.05, null hypothesis accepted).
Fig. 1.
Extent of glycosylation and expression of wild-type and mutant receptors. (A) NK1R immunoblot of transfected NCM 460 cell lysates shows the nonglycosylated mutant receptors migrate at a lower relative mass than the wild-type NK1R, reflecting a loss of carbohydrate moieties. (B) Real-time PCR comparison of lentiviral mutants in NCM 460 cells, demonstrating equal NK1R message for the NCM 460 cells transfected with the various glycosylation mutants (α = 0.05, P > 0.05). NCM 460 NK1R mRNA levels were normalized to TATA-box binding protein mRNA. The values are expressed as a percentage of the wild-type NK1R mRNA abundance.
Binding Assays of NCM 460 Cells Transfected with NK1R.
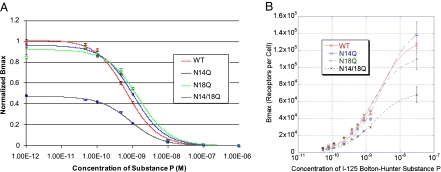
To observe what effect receptor glycosylation has on ligand binding, we performed competitive binding analysis of wild-type and glycosylation mutant NK1R expressed in NCM 460 cells. Unlabeled SP was used to compete with 125I Bolton Hunter-SP (125I-BH-SP) reagent in receptor binding assays, as detailed in Materials and Methods. These assays (Fig. 2A) demonstrated no effect of glycosylation on the _K_i of SP in the presence of 125I-BH-SP (data were normalized to wild-type values). The _K_i of SP in all cell lines was ≈1.0 nM, in agreement with published results of wild-type NK1R (33–35). These results indicate that the various glycosylation mutants have affinities for SP similar to that of the wild-type receptor. In contrast, the apparent _B_max depended on whether there was an N-linked glycosylation site available. The apparent _B_max of the single glycosylation mutants were similar to that of the wild-type receptor, but the apparent _B_max of the double mutant was only 48% of wild type. This finding suggested that the number of receptors expressed on the surface of the cells transfected with mutant, nonglycosylated receptor may be altered.
Fig. 2.
Competition binding and 125I-BH-SP saturation analyses of wild-type and glycosylation receptor mutants in NCM 460 cells. These analyses demonstrate that there are no effects of glycosylation on the _K_i of SP in the presence of 125I-BH-SP (A; competition binding analysis), but that there is a 48% decrease in the _B_max of the nonglycosylated mutant when compared with the other mutants and wild-type (B; saturation analysis).
As shown in Fig. 2B, confirming results from the competition binding assays above, 125I-BH-SP saturation analysis of wild-type and glycosylation mutant receptors demonstrates no effect of glycosylation on the _K_D of 125I-BH-SP. For saturation analysis experiments, cells were treated with increasing concentrations of 125I-BH-SP with and without 1 μM SP, representing nonspecific and total binding, respectively. The _K_D was ≈3 nM, which is within the range found in the literature, indicating that ligand affinity is not affected by receptor glycosylation. However, this saturation analysis demonstrates a dramatic reduction in the _B_max of the double glycosylation mutant, when compared with wild-type receptor (Fig. 2B). Whereas, the single glycosylation mutants did not vary significantly from the wild-type receptor, the _B_max of the nonglycosylated receptor was reduced significantly (≈2-fold). This result indicates that the amount of functional, fully nonglycosylated NK1R expressed in the plasma membrane is almost half that of wild-type NK1R and suggests that glycosylation may play a role in NK1R stabilization and dynamics in the membrane.
Receptor Internalization in NCM 460 Cells.
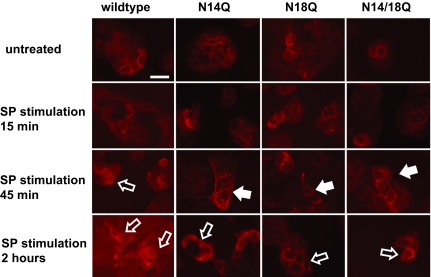
We next examined receptor dynamics by immunocytochemistry of NCM 460 cells stably transfected with wild-type and nonglycosylated NK1R. Cells were treated with 10 nM SP for 0 min to 2 h, probed with a polyclonal antibody raised against the H83 epitope of the NK1R, and visualized by confocal microscopy. As shown in Fig. 3, unstimulated cells have a ribbon-like NK1R immunoreactivity, reflecting a uniform NK1R distribution on the membrane surface of the cells. After SP stimulation, NK1R immunoreactivity becomes punctate, reflecting formation of vesicles and internalization. Much of this punctate immunoreactivity resolves by 2 h of continuous SP stimulation, indicating that receptor internalization has reached a steady state.
Fig. 3.
NK1R immunostaining of NCM 460 cells transfected with wild-type and mutant NK1 receptors, after stimulation by SP for the indicated durations. Untreated cells containing wild-type or mutant receptors show NK1R immunoreactivity on the cell surface, whereas SP stimulation triggers NK1R internalization, visualized as punctate staining (closed arrows). After longer SP stimulation, the mutant receptors are still retained within the cell, in contrast to the wild-type receptors that have returned to the cell surface (open arrows) by 45 min. Representative images of each cell line are shown. (Scale bar: 20 μm.)
Immunoreactivity of cells containing nonglycosylated NK1R indicates punctate staining within 15 min of SP stimulation similar to the wild-type receptors, suggesting that NK1Rs lacking glycosylation are still able to internalize and recycle back to the surface. Interestingly, the singly and doubly glycosylation mutant receptors show more punctate staining at 45 min of continuous SP stimulation, suggesting that the absence of carbohydrate moieties may affect receptor recycling back to the surface, after internalization.
Kinetics of Receptor Internalization in NCM 460 Cells.
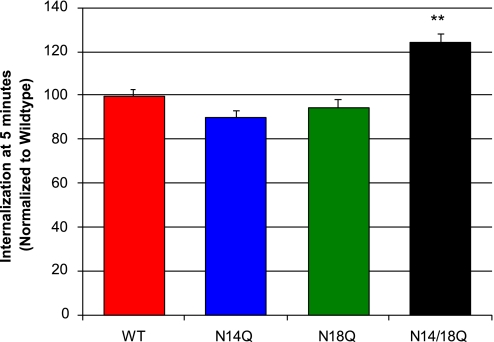
Our confocal data of receptor internalization and recycling showed that a fully deglycosylated receptor may internalize faster, but return to the cell surface more slowly, than wild-type receptors. We sought to further elucidate these processes by using acid wash and radioactive quantification as detailed in Materials and Methods (36). We found that 24% more of the double glycosylation receptor mutants (N14Q/N18Q) are internalized within 5 min, relative to the amount of internalized wild-type receptors (Fig. 4), indicating that receptors lacking glycosylation may internalize faster than wild-type receptors. These results are consistent with our confocal microscopy observations. One explanation for these results may be that receptors continuously oscillate and are in equilibria among different states that may favor internalization; glycosylation may shift these equilibria to internalization-favored states. The lack of steric hindrance conferred by the carbohydrate moieties may lower the threshold for conformational changes of the receptor because of the energy of the system.
Fig. 4.
Internalization kinetics of NK-1R mutants. Cells preincubated with 125I-BH-SP for 1 h at 4°C were warmed to 37°C for the indicated times. Cells were acid-washed to remove surface-bound ligand and treated with alkaline wash to determine the amount of internalized ligand by a gamma counter. The double glycosylation receptor mutant displays a significant 24% increase in the relative amount of internalized receptor when compared with the wild-type receptor (α = 0.01, P < 0.01).
MAPK Family Signaling in NCM 460 Cells Transfected with NK1Rs.
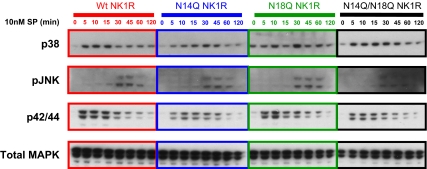
We next sought to characterize NK1R-induced signaling pathways in our various NK1R-expressing NCM 460 cell lines, because previous work has shown that NK1R can activate the MAPK families after 100 nM SP stimulation (20). Our cell lines containing the mutant NK1Rs were therefore compared after SP-induced phosphorylation of the MAPK pathways p38, p42/p44, and JNK. As shown by immunoblot in Fig. 5, p38 and p42/p44 are phosphorylated within 5 min of SP incubation, whereas JNK phosphorylation occurs within 30 min in cells with wild-type and mutant NK1Rs. To ensure equivalent loading of samples, total p42/p44, total p38, and total JNK were also assayed; cells did not show an increase in total (phosphorylated plus unphosphorylated) MAPK levels (Fig. 5 and data not shown).
Fig. 5.
Comparison of SP-induced phosphorylation of MAPK pathways p38, p42/p44, and JNK in cell lines transfected with NK1R mutants. Immunoblot shows the duration of SP-mediated activation of p38, p42/p44, and JNK in all cell lines. p38 and p42/p44 are phosphorylated within 5 min of SP incubation, whereas JNK phosphorylation occurs within 30 min. Wild-type and mutant receptors exhibit similar phosphorylation patterns. Total p42/p44 (shown), total p38, and total JNK were also assayed to assure loading equivalency. Representative immunoblots from independent experiments are shown.
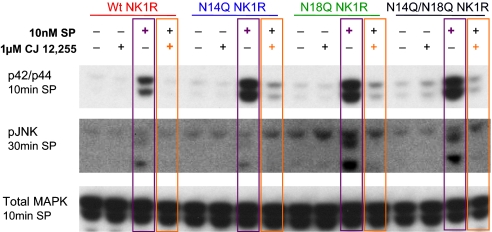
Furthermore, the observed MAPK phosphorylation is a downstream signaling event of the NK1R, because pretreatment with the NK1R antagonist CJ12,255 ablates p42/p44 and JNK phosphorylation induced by SP (Fig. 6, purple, SP alone; orange, antagonist plus SP) at 10 and 30 min of SP treatment for p42/44 and pJNK activation, respectively. Therefore, the results show that SP can activate the MAPK families p38, p42/p44, and JNK, and that this activation is specifically mediated by the NK1R.
Fig. 6.
Immunoblots demonstrating SP-specific effect of MAPK phosphorylation, using NK1R antagonist CJ12,255. Pretreatment of cells with the NK1R antagonist ablates p42/p44 and JNK phosphorylation induced by SP (purple: SP alone; orange: antagonist plus SP) at the indicated times, suggesting that SP acts solely through NK1R to activate the MAPK family.
IL-8 Production in NCM 460 Cells Transfected with NK1R.
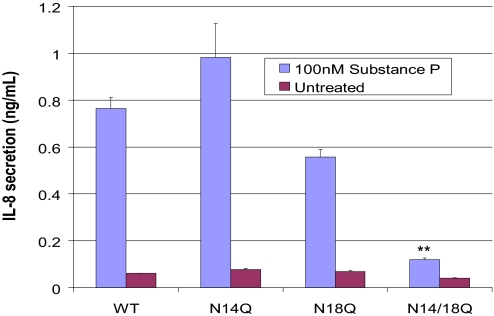
The NK1R has been shown to mediate SP activation of inflammatory pathways, including IL-8 production (15). Therefore, an IL-8 secretion assay to functionally characterize the lentivirally introduced wild-type and mutant NK1R was undertaken. NCM 460 cells transfected with wild-type NK1R showed increased IL-8 production as a function of time after bolus SP treatment (0–8 h) and SP concentration (0.3 nM-30 μM). Four hours of 100 nM SP treatment increased IL-8 secretion, which remained elevated after 8 h. After 4 h of 100 nM SP treatment, NCM 460 cells with wild-type NK1R showed a 13-fold increase in IL-8 secretion as measured by ELISA, compared with untreated transfected cells, indicating that NK1R is functionally active in these cells (Fig. 7). In contrast, the amount of IL-8 produced by unstimulated NCM 460 did not plateau but increased steadily over time, indicating that the IL-8 secreting machinery of unstimulated cells is not saturated. Furthermore, the NK1R is activated by SP specifically, because a NK1R antagonist abrogates this observed IL-8 production (data not shown). In contrast, NCM 460 cells with the double nonglycosylated mutant NK1R demonstrated a pronounced diminution of IL-8 production after 100 nM SP stimulation for 4 h, showing an 85% decrease from IL-8 production (Fig. 7).
Fig. 7.
Transfected NCM 460 cells were stimulated for 4 h with 100 nM SP, demonstrating by ELISA a pronounced diminution of IL-8 production in the N14Q/N18Q mutant cell line lacking both glycosylation sites. This reduction was significant when compared with cells containing the wild-type and single mutant receptors (α = 0.01, P < 0.001).
Discussion
Our evidence shows that glycosylation of the NK1R has functional consequences. We demonstrate here that NK1R glycosylation may not be necessary for ligand binding and downstream MAPK signaling. However, nonglycosylated NK1R has significantly reduced cell surface levels of functional receptor. As well, nonglycosylated NK1Rs elicit reduced IL-8 secretion in response to SP activation.
The presence of N-glycosylation sites in the N-terminal domain is a common characteristic of GPCRs, although oligosaccharide function in GPCR function varies with different systems, and therefore the role of glycosylation must be determined empirically. For example, studies with N-glycosylation of the rat angiotensin II receptor (37) showed that receptors with all three defective N-glycosylation sites are not expressed on the plasma membrane, but instead are accumulated in the endoplasmic reticulum; interestingly, the preservation of Asn-176 in the second extracellular loop enabled surface expression similar to wild-type receptors. Glycosylation-deficient receptors displayed wild-type _K_D values for its ligand sarcosine, whereas levels of inositol phosphate production upon activation were unchanged. In contrast, site-directed mutagenesis of two glycosylation sites at the extracellular domain of the follicle-stimulating hormone receptor showed that one intact site is sufficient for receptor expression at the cell surface and ligand binding with high affinity (38). This group also reported that removal of six N-glycosylation sites on rat lutropin receptor had no effect on binding to human chorionic gonadotropin, although glycosylation may be involved in proper receptor folding (39). Similarly, gonadotropin-releasing hormone (GnRH) receptor mutants in which the consensus N-linked glycosylation sites were ablated showed unchanged ligand affinity and were able to stimulate downstream signaling pathways. However, the glycosylation-defective GnRH receptors displayed decreased _B_max values at 39–46% of wild-type depending on mutation site, implying decreased receptor expression and/or stability at the plasma membrane (40).
In other systems, GPCRs do not require N-glycosylation for membrane targeting, ligand binding, or downstream signaling. For example, glycosylation-defective α1-adrenergic, H2 histamine, and M2-muscarinic receptors do not have altered cell surface expression and/or stability. Further, site-directed mutagenesis at two of three consensus sites of the histamine H2 receptor show that lack of glycosylation does not alter the receptor's ability to bind ligand, nor does it affect cAMP activation and intracellular calcium accumulation. Immunostaining and binding experiments localized the glycosylation-defective receptors to the plasma membrane, implying that N-glycosylation is not required for intracellular targeting of the H2 receptor (41).
The present studies of a glycosylation-defective NK1R may indicate that N-glycosylation is necessary for stable receptor expression in the plasma membrane, because the glycosylation-defective receptors show reduced functional cell surface expression and more rapid internalization upon ligand binding. Interestingly, previous work on the extensively glycosylated EGFR shows that receptor self-dimers have 5-fold more kinase activity than monomeric receptors (independent from ligand binding); this self-dimerization and thus kinase activity highly depends on glycosylation (26). Fernandes et al. (26) suggest that glycosylation confers a “kinase-active” conformation that enables self-dimerization. As in the present studies with NK1R, lack of the oligosaccharides might contribute to receptor destabilization and degradation. The above studies do not, however, rule out the possibility of impaired receptor export, regulation of which requires endoplasmic reticulum chaperones, accessory proteins, and receptor activity modifying proteins, which can dimerize with GPCRs and modulate receptor folding.
Ligand affinities for SP and NKA (data not shown) of glycosylation-defective receptors are similar to that of the wild-type receptor, which suggests that for the NK1R, N-glycosylation is not involved in ligand binding, because the nonglycosylated receptor is able to attain proper folding to bind ligand. As well, glycosylation-deficient receptors are able to activate various MAPKs similarly to wild-type receptors, leading to NF-κB nuclear translocation (data not shown) and IL-8 production. It is somewhat unexpected, however, that we observe a substantial reduction in IL-8 secretion in cells containing the glycosylation-deficient receptors, because proinflammatory cytokine production has been shown to be downstream of and dependent on protein kinase pathways. This finding may reflect the sensitivity of IL-8 production to small changes in the timing of effector phosphorylation, which, in turn, are amplified throughout the activation cascades. For example, it has been previously shown that IL-8 secretion is sensitive to the synergistic effects of the ERKs, JNK, and the p38 MAPK cascades, such that a subtle change in one pathway dramatically affects IL-8 induction (42). It is also possible that the NK1R-induced IL-8 secretion observed here depends on protein kinases outside of the scope of this present study.
It is clear from the study of various GPCRs that glycosylation can have a wide range of effects on receptor function, and further study of the receptor mutants created here will enhance our knowledge on the role of NK1R in inflammation associated with several disease states. As observed here, the decreased expression of glycosylation-deficient NK1R at the plasma membrane may prove to be a critical point of therapeutic manipulation.
Materials and Methods
Creation of Lentiviral Vector with NK1R.
Plasmids containing the wild-type human NK1R were obtained from N. Bunnett (University of California, San Francisco, CA). Using restriction enzyme digest, the NK1R sequence was extracted and ligated into PC3 DNA cloning vector (Stratagene, La Jolla, CA). Then, using splicing by overlap extension (43), single glycosylation mutants of the NK1R were made, changing Asn → Gln-14, and Asn → Gln-18, to create the N14Q and N18Q mutants, respectively. As well, a double N14/18Q glycosylation mutant was created. These three mutant plasmids, and the wild-type NK1R in the PC3 cloning vector, were used as templates for a modified PCR, in which HotStart Ultra Pfu polymerase (Stratagene) and primers with a Topo-directional four-base sequence and a Kozak sequence were used to create Topo-directional fragments. The blunt-end fragments containing the NK1R sequence were incorporated into the Lenti6/V5-D-TOPO vector (Invitrogen, Carlsbad CA). These constructs were verified by sequencing, purified, and transfected into 293FT cells with Lipofectamine 2000 (Invitrogen). The virus-containing supernatant was harvested, titered, and used to create stable NCM 460 cell lines.
Real-Time Quantitative RT-PCR.
Total RNA was isolated from NCM 460 cells with an RNeasy mini kit (Qiagen, Valencia, CA) and reverse-transcribed with the One-Step RT-PCR kit (PerkinElmer, Shelton, CT). The resulting DNA was used for real-time PCRs on a 96-well plate using either NK1R-specific Taqman master mix from Applied Biosystems (Foster City, CA) or a human TATA-box binding protein Taqman master mix for endogenous control and analyzed in a GeneAmp 5700 detection system (ABI/PerkinElmer, Boston, MA) (44).
ELISA.
The secretion of IL-8 in NCM-460 cells was measured by using an ELISA kit developed by R & D Systems (Minneapolis, MN). Cells were grown to 80% confluence in 24-well plates and serum-starved overnight (≈18 h). Media were then replaced with fresh serum-free media containing peptide agonist, antagonist, or both and incubated for the indicated amount of time. The media were then analyzed for IL-8 levels as detailed by the R & D Systems protocol.
Immunoblot.
The cell lysates were run on 4–12% SDS/PAGE gels, transferred, and immobilized on nitrocellulose or PVDF membranes using a modified Laemmli and Tobin method (45, 46). Cell lysates were boiled for 10 min and loaded on NuPAGE gels (Invitrogen) and run at 150 V for 2 h using Mops buffer (50 mM Tris base/50 mM Mops/1 mM EDTA/3.5 mM SDS, pH 7.7). The gels were transferred at 30 V for 4 h in NuPAGE transfer buffer (25 mM Bis-Tris/1 mM EDTA/25 mM Bicine/15% methanol). Membranes were blocked overnight at 4°C in 5% skim milk or 5% BSA and washed with 0.05% Tween-20 in Tris-buffered saline, pH 7.4. The membranes were then incubated with the appropriate primary antibodies. NK1R was visualized by using rabbit anti-NK1R antibodies (H-83; Santa Cruz Biotechnology, Santa Cruz CA) and HRP–labeled secondary antibodies, and detected by SuperSignal Chemiluminescent Substrate (Pierce, Rockford, IL).
MAP kinase activation was assessed by stimulation of NCM 460 cells cultured in complete medium, and when 75% confluent, incubated for 18 h in 0% FBS medium. The cells were stimulated with 10 or 100 nM SP or NKA or 20 ng/ml EGF for the stated times. The cells were processed for immunoblot as described above, using phosphospecific antibodies (0.2 μg/ml) directed against ERK1 or ERK2 to detect MAPK activation, anti-JNK rabbit antibody or anti-p38 rabbit antibody (Cell Signaling Technology, Beverly, MA).
Binding Kinetics.
For competitive binding analysis of the NK1R in the various cell lines, a fixed concentration of 125I-BH-SP (62.5 pM) and a variable concentration of SP (1 pM—1 μM) were incubated with a fixed number of cells (5 × 104) for 2 h at 4°C in Krebs–Ringer's solution–Hepes buffer (20 mM Hepes/1 mM MgCl2/5 mM KCl/120 mM NaCl, pH 7.4) supplemented with 6 mg/ml glucose and 0.6 mg/ml BSA. For saturation analysis experiments, cells were treated with increasing concentrations of 125I-BH-SP with and without 1 μM SP, representing nonspecific and total binding, respectively.
Quantitative Receptor Internalization.
NCM 460 cells plated in 24-well plates were washed three times with 4°C Hanks' buffered saline solution (HBSS) with 0.1% BSA at 80% confluence. The cells were blocked for 1 h with 4°C HBSS plus 0.1% BSA. After blocking, the cells were treated with either 62 pM 125I-BH-SP or 62 pM 125I-BH-SP and 1 μM SP for 1 h at 4°C. After incubation at 37°C for the specified time (0–45 min), the cells were treated with 0.2 M acetic acid/0.5 M NaCl for 10 min, which constituted the acid-wash fraction or surface receptors. The cells were washed three times with chilled PBS and then treated with 0.5 M NaOH for 30 min at room temperature. The base-wash fraction represented primarily the internalized receptor. The acid and base solutions containing the lysed cells were individually read for 10 min in a 1470 gamma counter (Wallac, Gaithersburg, MD). The amounts of ligand in both the acid wash (surface-bound ligand) and alkaline wash (internalized ligand) were then determined by radioactive count; the data are represented as a ratio of internalized to total ligand (total is the sum of acid and alkaline washes). The labeled SP was prepared by using 125I Bolton-Hunter reagent (PerkinElmer, Waltham MA) and SP (Bachem, King of Prussia, PA) in a conjugation reaction (47).
Immunofluorescence.
Cells were treated with 10 nM SP for 0 min to 2 h, fixed with 4% formaldehyde, probed with a polyclonal antibody raised against the H83 epitope of the NK1R and a rhodamine-conjugated secondary antibody, and finally visualized by confocal microscopy.
Statistical Analyses.
Results were analyzed by using Statview for Windows (SAS Institute, Cary, NC) and KaleidaGraph (Synergy, Reading, PA). The data were subjected to a Student's–Newman–Keuls' test to determine significance.
Acknowledgments
We thank Dr. A. Stucchi (Boston University School of Medicine) for NK1R antagonist, C. Song for technical assistance, C.P.'s laboratory for technical assistance and helpful discussion (particularly Dr. D. Zhao), and Dr. N. Bunnett for the wild-type NK1R plasmid. This work was funded in part by National Institutes of Health Grants 5R21NS04322-02 and R0-1 DK 47343 and the Russek Foundation.
Abbreviations
SP
substance P
NK1R
neurokinin 1 receptor
NKA
neurokinin A
GPCR
G protein-coupled receptor
125I-BH-SP
125I Bolton Hunter-SP.
Footnotes
The authors declare no conflict of interest.
References
- 1.Reed KL, Fruin AB, Gower AC, Stucchi AF, Leeman SE, Becker JM. Proc Natl Acad Sci USA. 2004;101:9115–9120. doi: 10.1073/pnas.0403210101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pothoulakis C, Castagliuolo I, Leeman SE, Wang CC, Li H, Hoffman BJ, Mezey E. Am J Physiol. 1998;275:G68–G75. doi: 10.1152/ajpgi.1998.275.1.G68. [DOI] [PubMed] [Google Scholar]
- 3.Mantyh CR, Gates TS, Zimmerman RP, Welton ML, Passaro EP, Vigna SR, Maggio JE, Kruger L, Mantyh PW. Proc Natl Acad Sci USA. 1988;85:3235–3239. doi: 10.1073/pnas.85.9.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhatia M, Saluja AK, Hofbauer B, Frossard JL, Lee HS, Castagliuolo I, Wang CC, Gerard N, Pothoulakis C, Steer ML. Proc Natl Acad Sci USA. 1998;95:4760–4765. doi: 10.1073/pnas.95.8.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lilly CM, Kobzik L, Hall AE, Drazen JM. J Clin Invest. 1994;93:2667–2674. doi: 10.1172/JCI117280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schelfhout V, Louis R, Lenz W, Heyrman R, Pauwels R, Joos G. Pulm Pharmacol Ther. 2005;19:413–418. doi: 10.1016/j.pupt.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Ho WZ, Douglas SD. J Neuroimmunol. 2004;157:48–55. doi: 10.1016/j.jneuroim.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 8.Masu Y, Nakayama K, Tamaki H, Harada Y, Kuno M, Nakanishi S. Nature. 1987;329:836–838. doi: 10.1038/329836a0. [DOI] [PubMed] [Google Scholar]
- 9.Shigemoto R, Yokota Y, Tsuchida K, Nakanishi S. J Biol Chem. 1990;265:623–628. [PubMed] [Google Scholar]
- 10.Yokota Y, Sasai Y, Tanaka K, Fujiwara T, Tsuchida K, Shigemoto R, Kakizuka A, Ohkubo H, Nakanishi S. J Biol Chem. 1989;264:17649–17652. [PubMed] [Google Scholar]
- 11.von Euler US, Gaddum JH. J Physiol (London) 1931;72:74–87. doi: 10.1113/jphysiol.1931.sp002763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang MM, Leeman SE. J Biol Chem. 1970;245:4784–4790. [PubMed] [Google Scholar]
- 13.Tregear GW, Niall HD, Potts JT, Jr, Leeman SE, Chang MM. Nat New Biol. 1971;232:87–89. doi: 10.1038/newbio232087a0. [DOI] [PubMed] [Google Scholar]
- 14.Tansky MF, Leeman S. In: Encyclopedia of Neuroscience. Adelman G, Smith B, editors. Amsterdam: Elsevier; 2004. CD-ROM. [Google Scholar]
- 15.Zhao D, Kuhnt-Moore S, Zeng H, Pan A, Wu JS, Simeonidis S, Moyer MP, Pothoulakis C. Biochem J. 2002;368:665–672. doi: 10.1042/BJ20020950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koon HW, Zhao D, Zhan Y, Simeonidis S, Moyer MP, Pothoulakis C. J Pharmacol Exp Ther. 2005;314:1393–1400. doi: 10.1124/jpet.105.088013. [DOI] [PubMed] [Google Scholar]
- 17.Rollandy I, Dreux C, Imhoff V, Rossignol B. Neuropeptides. 1989;13:175–185. doi: 10.1016/0143-4179(89)90089-9. [DOI] [PubMed] [Google Scholar]
- 18.Pradier L, Heuillet E, Hubert JP, Laville M, Le Guern S, Doble A. J Neurochem. 1993;61:1850–1858. doi: 10.1111/j.1471-4159.1993.tb09826.x. [DOI] [PubMed] [Google Scholar]
- 19.Luo W, Sharif TR, Sharif M. Cancer Res. 1996;56:4983–4991. [PubMed] [Google Scholar]
- 20.Castagliuolo I, Valenick L, Liu J, Pothoulakis C. J Biol Chem. 2000;275:26545–26550. doi: 10.1074/jbc.M003990200. [DOI] [PubMed] [Google Scholar]
- 21.Koon HW, Zhao D, Na X, Moyer MP, Pothoulakis C. J Biol Chem. 2004;279:45519–45527. doi: 10.1074/jbc.M408523200. [DOI] [PubMed] [Google Scholar]
- 22.Sachon E, Girault-Lagrange S, Chassaing G, Lavielle S, Sagan S. J Pept Res. 2002;59:232–240. doi: 10.1034/j.1399-3011.2002.01977.x. [DOI] [PubMed] [Google Scholar]
- 23.Sagan S, Chassaing G, Pradier L, Lavielle S. J Pharmacol Exp Ther. 1996;276:1039–1048. [PubMed] [Google Scholar]
- 24.Sagan S, Karoyan P, Chassaing G, Lavielle S. J Biol Chem. 1999;274:23770–23776. doi: 10.1074/jbc.274.34.23770. [DOI] [PubMed] [Google Scholar]
- 25.Bremer AA, Leeman SE, Boyd ND. FEBS Lett. 2000;486:43–48. doi: 10.1016/s0014-5793(00)02228-6. [DOI] [PubMed] [Google Scholar]
- 26.Fernandes H, Cohen S, Bishayee S. J Biol Chem. 2001;276:5375–5383. doi: 10.1074/jbc.M005599200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang R, Cai H, Fatima N, Buczko E, Dufau ML. J Biol Chem. 1995;270:21722–21728. doi: 10.1074/jbc.270.37.21722. [DOI] [PubMed] [Google Scholar]
- 28.Boudin H, Lazaroff B, Bachelet CM, Pelaprat D, Rostene W, Beaudet A. J Comp Neurol. 2000;425:45–57. [PubMed] [Google Scholar]
- 29.Petaja-Repo UE, Hogue M, Laperriere A, Walker P, Bouvier M. J Biol Chem. 2000;275:13727–13736. doi: 10.1074/jbc.275.18.13727. [DOI] [PubMed] [Google Scholar]
- 30.Nehring RB, Richter D, Meyerhof W. J Physiol (Paris) 2000;94:185–192. doi: 10.1016/s0928-4257(00)00203-5. [DOI] [PubMed] [Google Scholar]
- 31.Zhou AT, Assil I, Abou-Samra AB. Biochemistry. 2000;39:6514–6520. doi: 10.1021/bi992706f. [DOI] [PubMed] [Google Scholar]
- 32.Bremer AA, Leeman SE, Boyd ND. J Biol Chem. 2001;276:22857–22861. doi: 10.1074/jbc.M100824200. [DOI] [PubMed] [Google Scholar]
- 33.Gether U, Nilsson L, Lowe JA, 3rd, Schwartz TW. J Biol Chem. 1994;269:23959–23964. [PubMed] [Google Scholar]
- 34.Huang RR, Huang D, Strader CD, Fong TM. Biochemistry. 1995;34:16467–16472. doi: 10.1021/bi00050a030. [DOI] [PubMed] [Google Scholar]
- 35.Huang RR, Yu H, Strader CD, Fong TM. Biochemistry. 1994;33:3007–3013. doi: 10.1021/bi00176a033. [DOI] [PubMed] [Google Scholar]
- 36.Garland AM, Grady EF, Payan DG, Vigna SR, Bunnett NW. Biochem J. 1994;303:177–186. doi: 10.1042/bj3030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deslauriers B, Ponce C, Lombard C, Larguier R, Bonnafous JC, Marie J. Biochem J. 1999;339:397–405. [PMC free article] [PubMed] [Google Scholar]
- 38.Davis D, Liu X, Segaloff DL. Mol Endocrinol. 1995;9:159–170. doi: 10.1210/mend.9.2.7776966. [DOI] [PubMed] [Google Scholar]
- 39.Davis DP, Rozell TG, Liu X, Segaloff DL. Mol Endocrinol. 1997;11:550–562. doi: 10.1210/mend.11.5.9927. [DOI] [PubMed] [Google Scholar]
- 40.Davidson JS, Flanagan CA, Zhou W, Becker II, Elario R, Emeran W, Sealfon SC, Millar RP. Mol Cell Endocrinol. 1995;107:241–245. doi: 10.1016/0303-7207(94)03449-4. [DOI] [PubMed] [Google Scholar]
- 41.Fukushima Y, Oka Y, Saitoh T, Katagiri H, Asano T, Matsuhashi N, Takata K, van Breda E, Yazaki Y, Sugano K. Biochem J. 1995;310:553–558. doi: 10.1042/bj3100553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holtmann H, Winzen R, Holland P, Eickemeier S, Hoffmann E, Wallach D, Malinin NL, Cooper JA, Resch K, Kracht M. Mol Cell Biol. 1999;19:6742–6753. doi: 10.1128/mcb.19.10.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 44.Holland PM, Abramson RD, Watson R, Gelfand DH. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 46.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gaudriault G, Vincent JP. Peptides. 1992;13:1187–1192. doi: 10.1016/0196-9781(92)90027-z. [DOI] [PubMed] [Google Scholar]