ΔNp63 regulates thymic development through enhanced expression of FgfR2 and Jag2 (original) (raw)
Abstract
p63, a homologue of the tumor suppressor p53, is pivotal for epithelial development, because its loss causes severe epithelial dysgenesis, although no information is so far available on the role of p63 in the thymus. We identified the expression of all p63 isoforms in the developing thymus. The p63−/− thymi show severe abnormalities in size and cellularity, even though the organ expresses normal levels of keratins 5 and 8, indicating a p63-independent differentiation of thymic epithelial cells (TEC). TEC were sufficiently developed to allow a significant degree of education to produce CD4/CD8 single- and double-positive T cells. To study the selective contribution of transactivation-active p63 (TAp63) and amino-deleted p63 (ΔNp63) isoforms to the function of the TEC, we genetically complemented p63−/− mice by crossing p63+/− mice with transgenic mice expressing either TAp63α or ΔNp63α under the control of the keratin 5 promoter. Thymic morphology and cellularity were partially restored by complementation with ΔNp63, but not TAp63, one downstream effector being fibroblast growth factor receptor 2-IIIb (FgfR2-IIIb). Indeed, FgfR2-IIIb is regulated directly by p63, via its interaction with apobec-1-binding protein-1, and its knockout shows thymic defects similar to those observed in p63−/− thymi. In addition, expression of Jag2, a component of the Notch signaling pathway known to be required for thymic development, was enhanced by p63 in vivo genetic complementation. Like Jag2−/− thymi, p63−/− thymi also show reduced γδ cell formation. Therefore, p63, and particularly the ΔNp63 isoform, is essential for thymic development via enhanced expression of FgfR2 and Jag2. The action of ΔNp63 is not due to a direct regulation of TEC differentiation, but it is compatible with maintenance of their “stemness,” the thymic abnormalities resulting from epithelial failure due to loss of stem cells.
Keywords: epithelia, thymus
TP63 is a homologue of the well known TP53 tumor suppressor gene (1). Although p63 can mimic the actions of p53 by responding to DNA damage and triggering cell cycle arrest and apoptosis, it soon became clear that its principal function was developmental (2–4). Molecularly, p63 is more complex than p53. p63 transcription can be initiated from two promoters, giving rise to isoforms containing an N-terminal transactivation domain [transactivation-active p63 (TAp63) isoforms], or lacking the transactivation domain [amino-deleted p63 (ΔNp63) isoforms]. In addition, both TAp63 and ΔNp63 isoforms undergo alternative splicing at the C terminus, and some C-terminal splice variants of ΔNp63 contain a second transactivation domain, and can therefore regulate expression of a distinct subset of genes (5, 6).
Mice deficient in all p63 isoforms have no epidermis or other stratified epithelia, and also show aborted limb development (3, 4). Consequently, they die shortly after birth due to dehydration. Although two independent laboratories generated p63 knockout mice with identical results, their interpretation of the underlying mechanism was different. One group proposed that p63, and particularly the ΔNp63 isoform, is important in the maintenance of “stemness” of all stratified epithelia, and that the phenotype is the result of epithelial failure due to loss of stem cells (3). The second has argued that p63 is required for the very basic steps of commitment and differentiation (4).
The thymus also has an epithelial component, and thymic epithelial cells (TEC) are an absolute requirement for the successful maturation of thymocytes into mature T lymphocytes (7). Impaired TEC function may lead to inappropriate positive and negative selection of maturing thymocytes with consequences such as autoimmunity (8–10). However, although previous work on p63-deficient mice has focused on the epidermal defects (11), there are no data on the role of p63 and of its N-terminal isoforms, in thymic development.
Here, we show that all p63 isoforms are expressed in the normal thymus. The p63−/− thymi show severe abnormalities in size and cellularity, even though TEC were sufficiently developed to allow a significant degree of education to produce CD4/CD8 single- and double-positive T cells. In addition, we have genetically complemented p63−/− mice by crossing p63 heterozygotes with transgenic mice expressing either TAp63α or/and ΔNp63α in TEC to generate mice selectively complemented with TAp63 or ΔNp63 isoforms or with both. The data show that the gross morphological thymic defects in the p63−/− mice are at least partially restored by complementation with ΔNp63, but not with the TAp63 isoform, and that this is mediated through regulation of a particular fibroblast growth factor receptor (FgfR) isoform. Moreover, the defective p63−/− thymus shows defective Jag2 expression, which is restored by selective p63 complementation, in this case operating through a component of the Notch signaling pathway. The data show therefore that ΔNp63 is essential for thymic development via enhanced expression of FgfR2 and Jag2, as downstream effectors of p63. Moreover, p63-null mice, like the Jag2 knockouts, are deficient in γδ cells.
Results and Discussion
Morphological Abnormalities in p63−/− Thymi.
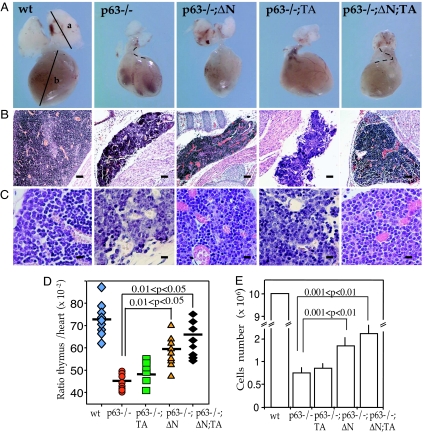
p63 is expressed by epithelial progenitor cells in the embryonic thymus [supporting information (SI) Fig. 5_A_], and p63-positive epithelial cells also remain in thymus into adulthood (data not shown). All p63 isoforms were detected by RT-PCR (SI Fig. 5_B_) in embryonic thymi. p63 knockout mouse embryos at the stage of embryonic day (E)19.5 show a severely hypoplastic thymus (Fig. 1A) in comparison with wild-type or heterozygous littermates, whereas hearts (used here as reference for the thymus) were normal in size (Fig. 1 A–C). In addition, thymi of p63-deficient embryos showed grossly distorted architecture and pronounced hypocellularity (Fig. 1 B and C). Thus, thymocyte numbers in p63-deficient embryos at stage E19.5 were <8% of those found in wild-type mice (Fig. 1E).
Fig. 1.
Fetal thymic development in p63−/− and genetically complemented mice. (A) Relative size of thymi (E19.5 embryos), dissected with the heart attached to provide a frame of reference. Thymi from p63−/− animals are reduced in size and exhibit severe structural anomalies. (B) Thymic histology (E19.5 sections stained with hematoxylin–eosin) in p63−/− mice before and after the reintroduction of TAp63α and/or ΔNp63α. (Scale bars, 300 μm.) (C) Histology of thymic cortical area in p63−/− mice before and after the reintroduction of TAp63α and/or ΔNp63α at bigger magnification. (Scale bars, 10 μm.) (D) The thymus/heart (E19.5 embryos) ratio of p63−/−, and p63−/−;TA mice was markedly reduced compared with wild type (wt), and increased in p63−/−;ΔN and p63−/−;ΔN;TA mice (n = 11; P value is indicated). (E) Thymic cell number (mechanical extraction from thymus from E19.5 embryos) in the different genotypes. Bar indicates standard deviation (n = 11; statistical significance is indicated).
Analysis of the state of thymocyte development in p63−/− embryos reveals a degree of epithelial morphological differentiation, because both wild-type and p63−/− E19.5 thymi showed a similar distribution of K5- and K8-positive epithelial cells (Fig. 2A). This indicates a p63-independent differentiation of TEC. This expression was functional, because transgenes under the control of the K5 promoter were expressed in the complemented mice (see below).
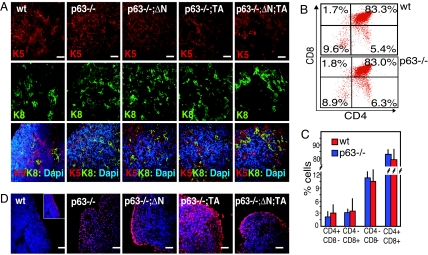
Fig. 2.
Histochemistry of keratins, cell death detection, and T cell counts in complemented mice. (A) Confocal staining of fetal thymus at E19.5 in wild-type (wt); p63−/− mice; and complemented p63−/−;ΔN, p63−/−;TA, and p63−/−;ΔN;TA mice. (Top and Middle) Tissue was stained for K5 (red) (Top) or K8 (green) (Middle). (Bottom) Merged fluorescence with DAPI (blue). The expression of K5 and K8 has a similar pattern in wild-type, p63−/−, and genetically complemented mice. (Scale bars, 60 μm.) (B and C) Staining for CD8 and CD4 in cells extracted from the thymi obtained from seven different mice for each genotype, wild type or p63−/−. Differences between control (wt) and p63−/− (knockout) genotypes for each particular subset were not statistically significant; bar indicates standard deviation. (D) Cell death was evaluated by TUNEL positivity (red); samples are as in A. In the thymi of p63−/− mice, a higher incidence of cell death is detected. The reintroduction of TAp63α results in an increase of cell death. The reintroduction of ΔNp63α results in a slight improvement of both parameters. The experiment was performed three times, and a representative experiment is shown. (Scale bars, 60 μm.)
Moreover, p63−/− thymi displayed an essentially normal profile of CD4+CD8+ double-positive as well as CD4+ and CD8+ single-positive cells at stage E19.5 (Fig. 2B), although with reduced absolute numbers (Fig. 1E), which may reflect the cell death seen in the null thymi (Fig. 2D). TEC were therefore sufficiently developed to allow a significant degree of education to produce CD4/CD8 single- and double-positive T cells.
These three features, K5 expression, the education of mature T cells, and the generation of K5-regulated complemented mice, do not support the hypothesis that p63 controls epithelial differentiation, at least in the thymus (4).
Effect of ΔNp63α and TAp63α Genetic Complementation on Thymic Morphology.
Because p63−/− TEC expressed K5 (Fig. 2A), we used this promoter to reintroduce both p63 isoforms singly and in combination in vivo.
Genetic complementation with ΔNp63α, but not TAp63α, resulted in an increased thymus/heart ratio, with increased cellularity (Fig. 1). Complementation with both TAp63α and ΔNp63α provided only a modest further increase in thymus size and cellularity over single ΔNp63α complementation. Histologically, TAp63α complementation did not improve thymic structure, whereas this was markedly restored by single or double ΔNp63α complementation (Fig. 1 B and C). The expression pattern of K5 and K8 differentiation markers in TEC did not change on reintroduction of either or both p63 isoforms (Fig. 2A), showing that the expression of these genes is not strictly p63-dependent and p63 is not absolutely required for TEC differentiation.
Our data indicate that p63 function in the thymus is predominantly exerted by ΔNp63α.
Defects in FgfR2-IIIb and Jag2 Pathways.
Two genes important for thymic development, FgfR2-IIIb and Jag2, are targets of p63. Specifically, FgfR2-IIIb is expressed in TEC, and knockout mice show a block in thymic growth at developmental stage E12.5 (12). FgfR2 expression is regulated by p63 (13), and the protein interaction between the p63α carboxyl terminus and apobec-1-binding protein-1 leads to a specific shift of FgfR2 alternative splicing toward the FgfR2-IIIb isoform essential for epithelial differentiation (14, 15). However, sufficient epithelial cell differentiation occurs in the FgfR2-IIIb−/− hypoplastic thymi to allow the development of CD4/CD8 double-positive and single-positive thymocytes (12). This phenotype is remarkably similar to that of the p63−/− thymi, as described above. To understand the molecular mechanisms responsible for the partial thymic recovery on reintroduction of ΔNp63α in p63−/− TEC, we therefore investigated the mRNA levels of FgfR2-IIIb on reintroduction of the different p63 isoforms in p63−/− TEC.
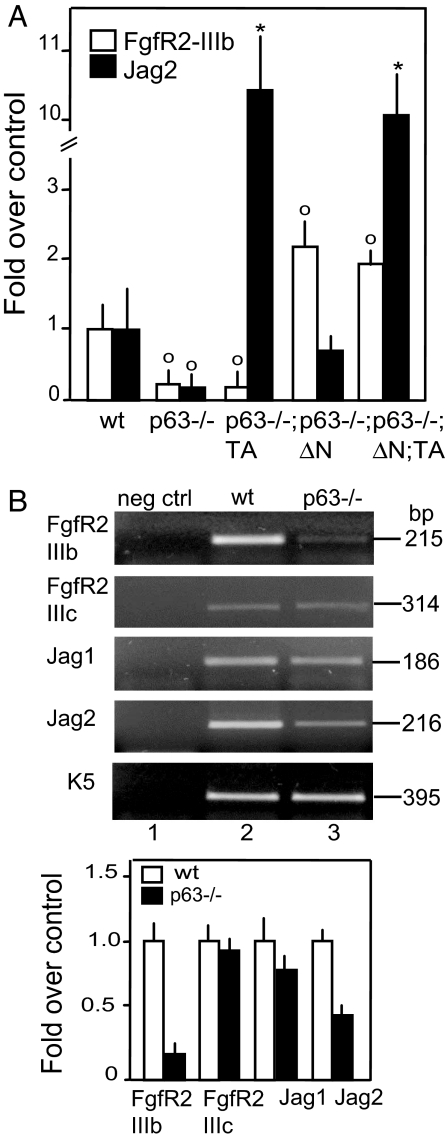
In the p63−/− thymus, the expression of FgfR2-IIIb (Fig. 3), but not the p63-independent FgfR2-IIIc isoform (14) (Fig. 3B), is drastically reduced when compared with wild type. Complementation with ΔNp63α, but not TAp63α, restored FgfR2-IIIb expression (Fig. 3A). Complementation with both TAp63α and ΔNp63α did not further increase FgfR2-IIIb expression level over ΔNp63α alone. This indicates that the known mechanism generating FgfR2-IIIb, downstream of p63, is operational in the thymus, leading to comparable thymic defects, both in p63 (this study) and FgfR2-IIIb (12) knockout mice.
Fig. 3.
Expression of FgfR2 and Jag2 in complemented mice. (A) Quantitative real-time PCR indicates that the p63−/− reduced FgfR2-IIIb expression was restored upon reintroduction of ΔNp63α, but not TAp63α, whereas Jag2 expression was restored upon reintroduction of TAp63α and ΔNp63α [∗, 0.001 < P < 0.01 between wild-type (wt) and genetically complemented mice; ○, 0.01 < P < 0.05 between wt and genetically complemented mice; n = 7]. (B) (Upper) Semiquantitative RT-PCR for FgfR2 splicing variants IIIc and IIIb and for Jag1 and Jag2. The results indicate that only the expression of the IIIb isoform is down-regulated in the p63−/− thymus. Among the Notch ligands, the expression of Jag2 is more markedly reduced in comparison with Jag1. (Lower) Quantification of the semiquantitative RT-PCR (n = 5).
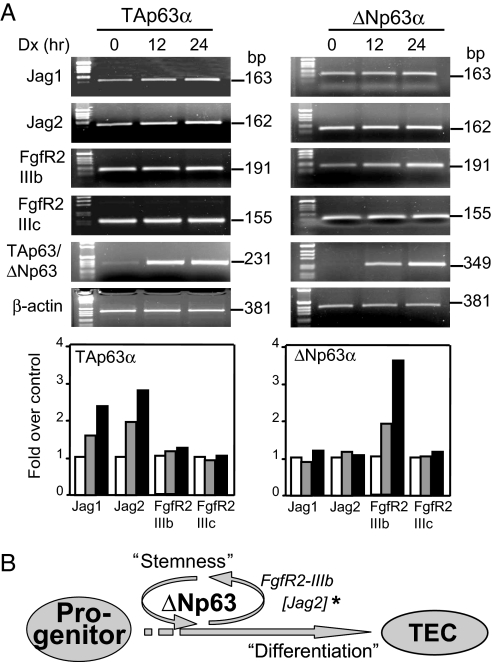
To further elucidate the link between p63 and FgfR2, we investigated the ability of p63 isoforms to induce FgfR2 in vitro by using inducible HA-tagged TAp63α and ΔNp63α Saos-2 cell lines (16). In parallel with the results obtained in vivo by genetic complementation, we found that ΔNp63α, but not TAp63α, significantly increased mRNA levels of FgfR2 spicing variant IIIb (Fig. 4). This increased FgfR2-IIIb mRNA level could be due either to its direct transactivation by ΔNp63α or to interaction of ΔNp63α with apobec-1-binding protein-1 (14), as described above. Apobec-1-binding protein-1, a member of the RNA processing machinery, is indeed expressed in Saos-2 cells (data not shown) and in the thymus, although its expression is not affected by p63 (SI Fig. 6).
Fig. 4.
Expression of FgfR2, Jag1, and Jag2 in TAp63 and ΔNp63 Saos-2 Tet-on inducible cell lines. (A) Semiquantitative RT-PCR in Saos-2 Tet-on inducible cell lines (Dx, 2 μg/ml) overexpressing TAp63α or ΔNp63α isoforms. Actin is shown in the bottom image as a control. FgfR2, Jag1, and Jag2 expression were normalized to actin by densitometry, and results are reported as fold change over zero time (see graphics below actins). We performed three independent experiments, and a representative one is shown. Results obtained from the densitometry are shown as mean from three independent experiments. (B) Scheme of the role of ΔNp63 in “stemness” of thymic development. ∗, Jag2 is also induced by TAp63.
Jag2-null mice also display defects in thymic morphology together with abnormalities in the differentiation of γδ cells (17), and in turn Jag2 is a p63 target gene (18). Thus, FgfR2-IIIb may not be the sole mechanism mediating the role of p63 in the thymus. The prediction would be that, consequent to the TEC–pre-T cell interaction, there will be a reduced number of γδ cells (19). The p63−/− thymi indeed show an impaired γδ cell percentage (Table 1) but the proportion of αβ cells is normal (Table 1), supporting the involvement of Jag2 in the thymus. Interestingly, complementation with p63 isoforms restores Jag2 expression (Fig. 3A). Complementation with TAp63α or both TAp63α and ΔNp63α, increases the proportion of γδ cells in comparison with p63−/− (Table 1). Semiquantitative RT-PCR analysis of Jag1 and Jag2 expression levels in the inducible TAp63α and ΔNp63α Saos-2 cell lines confirms that expression of the TAp63 isoform up-regulates Jag1 and Jag2 mRNA levels after 12 and 24 h of induction with doxocyclin (Dx) (Fig. 4). No change in Jag1 and Jag2 expression was elicited by the ΔNp63α isoform (Fig. 4).
Table 1.
Flow cytometry analysis of fetal thymi (E19.5 embryos) from wild-type, p63−/−, and genetic complemented mice
| Markers | Wild type | p63−/− | p63−/−;TA | p63−/−;ΔN | p63−/−;TA;ΔN |
|---|---|---|---|---|---|
| αβ+ among CD4+CD8+ | 52.1 ± 0.8 (12) | 50.5 ± 1.2 (10) | 50.0 ± 1.2 (2) | 52.7 ± 0.7 (2) | 51.5 ± 1.0 (2) |
| γδ+ among total thymocytes | 5.3 ± 0.8 (12) | 1.5 ± 0.7* (10) | 2.7 ± 0.8 (2) | 1.7 ± 0.8 (2) | 2.3 ± 0.7 (2) |
These data indicate that at least two pathways, FgfR2-IIIb and Jag2, are involved in p63 function in the thymus, in agreement with the thymic defects reported for their individual knockouts.
Conclusions
The data presented indicate that p63 is necessary for thymic development and the severely hypoplastic thymic phenotype of p63−/− mice is markedly restored by complementation with the ΔNp63α isoform. This partial restoration of thymic morphology seems to particularly require FgfR2-IIIb and also Jag2 as downstream effectors.
In addition to describing a novel action of p63 in the thymus, the data presented also indicate that TEC differentiation does not strictly require p63, as previously hypothesized (4), although p63 seems to control organ size and morphology. While this paper was in reevaluation, McKeon and colleagues (20) described a role for p63 in controlling the proliferative capacity of stem cells. These authors show that p63 is not required for lineage commitment and differentiation of epithelial cells, both in developing epidermis and in thymus, but it specifically functions to maintain a high proliferative potential of the stem cells in both the thymus and epidermis. These data indicate that the p63−/− phenotype is the result of epithelial failure due to loss of stem cells (20) as previously hypothesized (3). In the present paper, we confirm McKeon's data on the morphological phenotype of the p63-null thymus and support his conclusion on the importance of p63 in thymic stem cell maintenance. In addition, our results from in vivo genetic complemented mice in both epidermis (8) and thymus (present study) indicate that the ΔNp63α isoform plays a predominant role, even though a minor TAp63 concomitant synergism is suggested. Moreover, we now identify two mechanisms, FgfR2 and Jag2, that mediate this function of p63 in the thymus (Fig. 4B).
Materials and Methods
Cell Cultures.
Saos-2 cells with doxycycline-inducible expression of HA-TAp63α, HA-TAp63β and HA-ΔNp63α, HA-ΔNp63β were generated as described (16). Cells were grown and induction of HA-TAp63α, HA-TAp63β and HA-ΔNp63α, HA-ΔNp63β was obtained by the addition of Dx (2 μg/ml) for 12 and 24 h.
Generation of Transgenic Mice and Maintenance of Mouse Colonies.
Transgenic mice expressing TAp63α or ΔNp63α were generated as described (8). Briefly, the coding sequence for each p63α isoform was placed under the control of the keratin K5 promoter. The cassette containing K5 promoter-TA/ΔNp63α-polyA+ signal was excised and injected directly into pronuclei, which were then implanted in pseudopregnant females. Pups were screened by PCR analysis of genomic DNA. Two founding lines were established and used in successive backcrosses with p63+/− mice to yield p63+/− TA/ΔNp63α Tg; animals that were then used to generate p63−/−;TA, p63−/−;ΔN, and p63−/−;ΔN;TA embryos. Introduction of TAp63α or ΔNp63α did not rescue the lethal aspect of the p63−/− phenotype and the pups died at birth, as with nontransgenic knockout animals. Expression of the transgenes was assessed both by immunohistochemistry of skin sections and by Western analysis on cultured primary keratinocytes. Expression was restricted to tissues where K5 is normally expressed (i.e., epidermal basal layer, thymus, eye, and lung) but not in other tissues including bone, muscle, and liver.
Thymocyte Isolation, Subset Analysis, and Thymus/Heart Ratios.
Both lobes of the thymic glands were dissected with heart attached from E19.5 embryos under a dissecting microscope. Each heart/thymus was examined [Motic (Xiamen, China) stereoscope with Optronics (Muskogee, OK) CCD camera] for thymus/heart ratio determination. Thymi were processed for immunohistochemistry or teased apart with fine forceps to release the thymocytes. Thymocytes were further separated from the glands by squeezing the teased organs through a 40-mm nylon mesh. Recovered thymocytes (1 × 106 cells) were analyzed for CD4 and CD8 surface antigens gating on CD3-Positive cells (FITC-conjugated anti-mouse CD4, phycoerythrin-conjugated anti-mouse CD8a, phosphatidylcholine anti-mouse CD3e; BD Pharmingen, San Diego, CA); 10,000 events were collected by FACSCalibur flow cytometer (BD Biosciences, Mountain View, CA) and analyzed with CellQuest software. Separate aliquots were assessed for the presence of rearranged T cell receptor αβ or γδ complexes with monoclonal antibodies as described previously (17). Data are presented as mean ± SEM of cells expressing the indicated array of differentiation markers.
Immunostaining and Confocal Microscopy.
Mouse embryo tissues were fixed in 4% paraformaldehyde (12–16 h) and embedded in paraffin. Sections (6 mm) were deparaffinized (Histolemon; Carlo Erba, Rodano, Italy) and rehydrated stepwise in alcohol/distilled water. Microwave-assisted antigen retrieval was performed in 0.01 M sodium citrate (pH 6) for three cycles of 5 min (300 W) followed by cooling at 50°C and a final round of microwaving for 5 min, cooling, and a final rinse in PBS. Nonspecific antigens were blocked by incubation in 10% goat serum in PBS for 2 h in a humidified atmosphere at 4°C. Subsequently, sections were incubated overnight with primary antibodies: anti-p63 (Ab4; NeoMarkers, Fremont, CA; 1/500 dilution), anti-MK5 (PRB-160P; Covance, Princeton, NJ; 1/500 dilution), Troma-1 (anti-K8; 1/10,000 dilution; Developmental Studies Hybridoma Bank, Iowa City, IA). Sections were then washed three times with PBS and incubated for 1 h with the secondary antibodies conjugated with FITC or phycoerythrin. After two washes in PBS, the tissue sections were counterstained with DAPI to highlight nuclei. The tissue sections were then mounted by using Prolong Antifade kit, and fluorescence was evaluated by confocal microscopy (Nikon Instruments, Melville, NY; C1 on Eclipse TE200; EZC1 software) fitted with an argon laser (488-nm excitation), He/Ne laser (542-nm excitation), and UV excitation at 405 nm. TUNEL on thymi sections was performed by using In Situ Cell Death Detection kit TMR red (Roche, Basel, Switzerland).
Real-Time PCR Assay.
RNA was extracted from wild-type and p63−/− thymi, by using the RNAeasy kit (Qiagen, Milan, Italy). A total of 500 ng of RNA was used for reverse transcription using the InPromII kit (Promega, Madison, WI), and one-fifth of the reaction was used for PCR. Real-time PCR was performed by using the Platinum SYBR Green qPCR SuperMix UDG with Rox (Invitrogen Life Technologies, Carlsbad, CA; catalog no. 11744-500), with an amplification program as follows: one cycle of 95° for 3 min and 40 cycles of 94° for 15 s and 60° for 1 min. The reaction was followed by a melting curve protocol according the specification of the ABI 7500 instrument (Applied Biosystems, Foster City, CA). Primers were as follows: Jag2 (+, CAA GTT CTG TGA CGA GTG TGT CCC; −, TTG CCC AAG TAG CCA TCT GG; 216 nt), FgfR2-IIIb (+, CCC ATC CTC CAA GCT GGA CTG CCT; −, CAG AAC TGT CAA CCA TGC AGA GTG; 215 nt), FgfR2-IIIc (+, CCC ATC CTC CAA GCT GGA CTG CCT; −, CAG AAC TGT CAA CCA TGC AGA GTG; 314 nt). Mouse β-actin was used as housekeeping gene for quantity normalization (+, TGT CCC TGT ATG CCT CTG GTC G; −, GAA CCG CTC GTT GCC AAT AGTG; 346 nt). Relative quantification of gene expression was calculated according the method of 2−ΔΔCt described in ABI User Bulletin no. 2 (updated October 2001), and the RQ software version 1.3 of Applied Biosystems. For semiquantitative RT-PCR, K5 (+, TCC AGA ACG CCA TTG CTG AAG; −, CCG TAG CCA GAA GAG ACA CTG TTT G; 395 nt) and Jag1 (+, CAA AGT GTG CCT CAA GGA GTA TCA G; −, TCC ACC AGC AAA GTG TAG GAC CTC; 186 nt) were used.
For semiquantitative PCR, RNA was extracted from control and Dx-induced Saos-2 Tet-on HA-tagged TAp63 and -ΔNp63 clones, by using TRIzol (Invitrogen Life Technologies). Primer sequences and PCR condition are available on request.
Supplementary Material
Supporting Information
Acknowledgments
We thank Frank McKeon (Harvard Medical School, Boston, MA) for providing the p63−/− mice and for pivotal discussions and Marco Ranalli and Sek Chow for technical assistance. This work was supported by grants from Cariplo (to R.M.), Telethon, the European Union (Epistem, Active p53), Fondo per gli Investimenti della Ricerca di Base, Ministero dell'Università e della Ricerca, Sanità, and the Medical Research Council (to G.M.).
Abbreviations
TAp63
transactivation-active p63
ΔNp63
amino-deleted p63
TEC
thymic epithelial cell
FgfR
fibroblast growth factor receptor
E_n_
embryonic day n
Dx
doxocyclin.
Footnotes
This article is a PNAS Direct Submission.
The authors declare no conflict of interest.
References
- 1.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 2.Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, et al. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 3.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 4.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 5.Yang A, McKeon F. Nat Rev Mol Cell Biol. 2000;1:199–207. doi: 10.1038/35043127. [DOI] [PubMed] [Google Scholar]
- 6.Duijf PH, Vanmolkot KR, Propping P, Friedl W, Krieger E, McKeon F, Dotsch V, Brunner HG, van Bokhoven H. Hum Mol Genet. 2002;11:799–804. doi: 10.1093/hmg/11.7.799. [DOI] [PubMed] [Google Scholar]
- 7.Blackburn CC, Manley NR. Nat Rev Immunol. 2004;4:278–289. doi: 10.1038/nri1331. [DOI] [PubMed] [Google Scholar]
- 8.Wallace VA, Penninger J, Mak T. Curr Opin Immunol. 1993;5:235–240. doi: 10.1016/0952-7915(93)90010-p. [DOI] [PubMed] [Google Scholar]
- 9.Sebzda E, Wallace VA, Mayer J, Yeung RS, Mak TW, Ohashi PS. Science. 1994;263:1615–1618. doi: 10.1126/science.8128249. [DOI] [PubMed] [Google Scholar]
- 10.Mittrucker HW, Pfeffer K, Schmits R, Mak TW. Immunol Rev. 1995;148:115–150. doi: 10.1111/j.1600-065x.1995.tb00096.x. [DOI] [PubMed] [Google Scholar]
- 11.Candi E, Rufini A, Terrinoni A, Dinsdale D, Ranalli M, Paradisi A, De Laurenzi V, Spagnoli LG, Catani MV, Ramadan S, et al. Cell Death Differ. 2006;13:1037–1047. doi: 10.1038/sj.cdd.4401926. [DOI] [PubMed] [Google Scholar]
- 12.Revest JM, Suniara RK, Kerr K, Owen JJ, Dickson C. J Immunol. 2001;167:1954–1961. doi: 10.4049/jimmunol.167.4.1954. [DOI] [PubMed] [Google Scholar]
- 13.Laurikkala J, Mikkola ML, James M, Tummers M, Mills AA, Thesleff I. Development (Cambridge, UK) 2006;133:1553–1563. doi: 10.1242/dev.02325. [DOI] [PubMed] [Google Scholar]
- 14.Fomenkov A, Huang YP, Topaloglu O, Brechman A, Osada M, Fomenkova T, Yuriditsky E, Trink B, Sidransky D, Ratovitski E. J Biol Chem. 2003;278:23906–23914. doi: 10.1074/jbc.M300746200. [DOI] [PubMed] [Google Scholar]
- 15.Petiot A, Conti FJ, Grose R, Revest JM, Hodivala-Dilke KM, Dickson C. Development (Cambridge, UK) 2003;130:5493–5501. doi: 10.1242/dev.00788. [DOI] [PubMed] [Google Scholar]
- 16.Gressner O, Schilling T, Lorenz K, Schulze Schleithoff E, Koch A, Schulze-Bergkamen H, Lena AM, Candi E, Terrinoni A, Catani MV, et al. EMBO J. 2005;6:2458–2471. doi: 10.1038/sj.emboj.7600708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang R, Lan Y, Chapman HD, Shawber C, Norton CR, Serreze DV, Weinmaster G, Gridley T. Genes Dev. 1998;12:1046–1057. doi: 10.1101/gad.12.7.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki Y, Ishida S, Morimoto I, Yamashita T, Kojima T, Kihara C, Tanaka T, Imai K, Nakamura Y, Tokino T. J Biol Chem. 2001;277:719–724. doi: 10.1074/jbc.M108080200. [DOI] [PubMed] [Google Scholar]
- 19.Cheng SH, Penninger JM, Ferrick DA, Molina TJ, Wallace VA, Mak TW. Crit Rev Immunol. 1991;11:145–166. [PubMed] [Google Scholar]
- 20.Senoo M, Pinto F, Crum CP, McKeon F. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information