Pellino Proteins: Novel Players in TLR and IL-1R Signalling (original) (raw)
Abstract
Members of the Toll-like receptor (TLR) and interleukin-1 receptor (IL-1R) family play important roles in immunity and inflammation. They initiate common intracellular signalling cascades leading to the activation of nuclear factor-κB (NF-κB) and other transcription factors that stimulate the expression of a variety of genes that shape an appropriate immune response. TLR/IL-1R signalling involves multiple protein–protein interactions, but the mechanisms that regulate these interactions are still largely unclear. In this context, Pellino proteins have been suggested to function as evolutionary conserved scaffold proteins in TLR/IL-1R signalling. However, recently Pellino proteins were also proposed to function as novel ubiquitin ligases for IL-1R associated kinase 1 (IRAK-1). Here we review our current knowledge on the expression, biological role and mechanism of action of Pellino proteins in TLR/IL-1R-induced signalling.
Keywords: Pellino, Toll-like receptor, interleukin-1, NF-κB, MAPK ubiquitination, IRAK-1
Introduction
The signal transduction pathways activated by Toll-like receptors (TLRs) have been the focus of much attention because of the important role that TLRs play in inflammatory diseases (reviewed by [1, 2]). Thirteen TLRs (named simply TLR1 to TLR13) have been identified in humans and mice together, with each TLR recognizing distinct microbial components. For example, TLR2 responds to lipoproteins, TLR3 to viral double-stranded RNA, TLR4 to lipopolysaccharide (LPS), TLR5 to flagellin, TLR7 and TLR8 to viral single stranded RNA, and TLR9 to unmethylated CpG islands of bacterial and viral DNA. Moreover, next to microbial ligands, stimulation of TLR signalling by endogenous ligands has been linked with the development of different diseases such as atherosclerosis [3]. The intracellular domain of TLRs is similar to that of members of the interleukin-1 receptor (IL-1R) family, which activate similar intracellular signalling path-ways. TLR/IL-1R signalling results in the activation of several transcription factors, such as members of the nuclear factor-κB (NF-κB) family and the interferon regulatory factor (IRF) family, which stimulate the expression of multiple proinflammatory genes that help to shape the immune response. The last couple of years, our understanding of the molecular mechanisms of TLR/IL-1R signalling to NF-κB and IRFs has increased considerably. All TLR/IL-1R family members contain a similar intracellular domain, known as the TIR domain, which binds specific adaptor proteins through homotypic TIR–TIR interactions. Four different adaptor proteins have been described: MyD88, Mal (TIRAP), TRIF (TICAM-1) and TRAM (TICAM-2) (reviewed by [4]). With the exception of TLR3, all TLRs as well as the IL-1R recruit the adaptor protein MyD88 (reviewed by [5]). In the case of TLR2 and TLR4, MyD88 recruitment requires Mal [6], whereas the other TLRs and the IL-1R signal independently of Mal. TLR4 can also initiate a Mal/MyD88-independent signalling pathway through the recruitment of TRAM and TRIF. Whereas the Mal/MyD88 pathway signals to NF-κB, the TRAM/TRIF pathway mainly activates members of the IRF family [7–9]. Because Pellino proteins have not yet been implicated in the activation of IRFs, we will focus in this review on the Mal/MyD88 pathway. The major steps in the MyD88-dependent signalling cascade initiated by TLR4 are depicted in Figure 1. Binding of MyD88 to TLR4 allows the recruitment of IL-1R associated kinase (IRAK)-1 and IRAK-4 through the death domains (DD) of both kinases and the DD and intermediate domain of MyD88 (Fig. 1, step 1). Although final proof is still missing, it has been suggested that close proximity in the receptor complex (Complex I, as described by [10]) enables IRAK-4 to phosphorylate IRAK-1, leading to its activation and IRAK–1 autophosphorylation ([11–13], Fig. 1, steps 2 and 3). Active IRAK–1 also phosphorylates Tollip, supposedly a silencer for quiescent IRAK-1 [14,15], which subsequently leaves the receptor complex (Fig. 1, step 4). IRAK-1 in turn interacts with the signalling molecule TRAF6 and TIFA, which is an adaptor protein that has been suggested to promote the oligomerization and ubiquitination of TRAF6 [16]. The IRAK-1/TRAF6/TIFA complex then leaves the receptor complex but remains at the membrane where it interacts with a preformed (but inactive) complex consisting of the mitogen activated protein kinase (MAPK) kinase kinase TAK1 [17], and the adaptor molecules TAB1 [18] and TAB2 [19] or TAB3 [20] (Fig. 1, steps 5–7), thus forming Complex II [10]. After phosphorylation of TAK1 and TAB2, the newly formed TRAF6-TAB2/3-TAK1-TAB1 complex migrates to the cytosol (Fig. 1, step 8; now called Complex III [10]), while IRAK-1 has been suggested to remain at the membrane and to disappear by proteasomal degradation (Fig. 1, step 9). Interestingly, a redistribution of IRAK-1 from the receptor complex to the nucleus has also been shown upon receptor triggering, but the function of nuclear IRAK-1 remains speculative [21]. Cytosolic TRAF6 is modified via a K63-linked polyubiquitin chain, which is a prerequisite for TAK1 activation ([22], Fig. 1, step 10). Active TAK1 then leads to the downstream activation of IκB kinases (IKK) and JNK or p38 MAPKs (Fig. 1, step 11) [22, 23]. IKKs eventually activate NF-κB by phosphorylating the NF-κB inhibitory protein IκBα, leading to its ubiquitination and proteasome-dependent degradation, whereas JNK and p38 further contribute to the innate immune response by phosphorylating and activating several other transcription factors.
1.
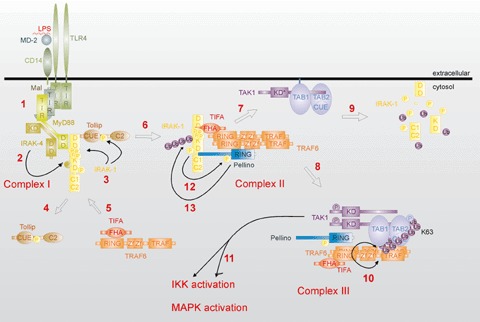
Schematic overview of LPS-induced MyD88-dependent signalling leading to IKK and MAPK activation. Upon LPS-binding (mediated by MD-2 and CD14) to TLR4 and subsequent TLR4 clustering, Mal, MyD88, IRAK-1, Tollip and IRAK-4 are recruited to the cytoplasmic part of the receptor via TLR4/MyD88 TIR–TIR interactions (1). IRAK-4 phosphorylates IRAK-1 (2), leading to IRAK-1 activity and autophosphorylation (3). Tollip is also phosphorylated by IRAK-1 and leaves the receptor complex (Complex I) (4). IRAK-1 interacts with TRAF6 and the TRAF6 adaptor protein TIFA (5). IRAK-1/TRAF6/TIFA leave the receptor complex (6) and associate with the preformed (but inactive) TAK1/TAB1/TAB2 complex (7), resulting in the formation of Complex II. There, TAK1 and TAB1 are phosphorylated after which TRAF6/TAK1/TAB1/TAB2 migrate back to the cytosol (8), while IRAK-1 remains at the membrane and is most likely degraded by the proteasome (9). TIFA promotes the oligomerization and activation of TRAF6, leading to TRAF6 K63-linked polyubiquitination and subsequent TAK1 activation (10). TAK1 is then responsible for the downstream activation of IKK as well as MAPK pathways (11). Pellino proteins associate with IRAK-1, TRAF6 and TAK1 in Complex II and III, and become phosphorylated by IRAK-1 (12). Pellino-1 and -2 also induce IRAK-1 polyubiquitination (13). DD = Death Domain, KD = active kinase domain, KD*= inactive kinase domain, RING = RING domain, FHA = Forkhead Associated domain, Ub = Ubiquitin, Zf = Zinc-finger, TRAF = TRAF domain and P = phosphorylation. For more information, see text.
It should be clear from the above described signalling pathways that protein–protein interactions play crucial roles in TLR/IL-1R signalling. However, whereas the role of specific domains (e.g. TIR and DD) in mediating these protein–protein interactions is generally accepted, it is still largely unknown how these interactions are regulated. An important role for scaffold proteins in regulating signalling is now emerging, as is exemplified by the ERK/MAPK pathway [24]. Some scaffolds augment the signal flux, but also mediate ‘cross–talk’ with other path-ways; certain adaptors may target signalling complexes to other subcellular locations; others provide regulated inhibition. Recently, members of a novel family of proteins, consisting of Pellino-1, Pellino-2 and-3 in humans, have been proposed to function as scaffold proteins in the TLR/IL-1 signalling path-way, where they bind TRAF6, TAK1, IRAK-1 and IRAK-4. Moreover, we recently showed that Pellino proteins are also putative E3 ubiquitin ligases, capable of inducing IRAK-1 polyubiquitination [25]. However, although Pellino proteins are evolutionary extremely conserved both in vertebrates and invertebrates, making it reasonable to assume they do serve important functions, their physiological function remains largely obscure.
Pellino proteins are extremely well conserved during evolution
The signalling pathways initiated by members of the mammalian TLR/IL-1R family are evolutionary conserved with the Drosophila Toll protein-mediated pathway [26]. Toll initiates its signal through the adapter molecule Tube and the Ser/Thr kinase Pelle, which is highly homologous to IRAKs in mammals. Pellino was originally identified as a novel Pelle-interacting protein in Drosophila [27]. Soon after its discovery in the fruitfly, several Pellino-related sequences were found in other animals, including mammals. Today more than 40 Pellino-related sequences are included in the NCBI–Gene database. In mammals, the Pellino family consists of three members (Pellino-1/-2/-3) that are highly related. Pellino-1 and -2 are most similar and are almost identical in size, while Pellino-3 has an extra N-terminal stretch of 27 amino acids. In insects and C. elegans only one Pellino gene is present. In amphibians the situation is less clear as the Pellino sequence found in Xenopus laevis (AAH44117) is closely related to mammalian Pellino-1, while Pellino from Xenopus tropicalis (NP_989171) is the likely homologue of mammalian Pellino-2. A Xenopus sequence related to Pellino-3 is currently not present in the NCBI-databases, so it is not clear how many Pellino genes are conserved in amphibians. Sequence alignment of Pellino proteins from different species immediately shows that Pellino family members are strongly conserved during evolution. For example, human Pellino-1 (NP_065702) and C. elegans Pellino (NP_504501) share more than 40% identical amino acids, while the degree of conservation between all known mammalian Pellino-1 and -2 forms is above 80%.
Genomic organization and protein structure of Pellino
The genes encoding human Pellino-1, Pellino-2 and Pellino-3 have been mapped to chromosome 2, chro-mosome 14, and chromosome 11, respectively [28–29]. At least two splice variants of Pellino-3 exist in humans [29]. The longest form (Pellino-3a) consists of seven coding exons, but a shorter form (Pellino-3b) is missing exon-2, leading to an in frame deletion of 24 amino acids. Surprisingly, in dogs (XM_860735) a third Pellino-3 specific transcript is predicted in which not only exon-2, but also parts of exon-1 and -3 are lacking. A murine Pellino-3a transcript was not found in the public gene databases; therefore we blasted both the human (NM_145065) and rat (XM_219692) Pellino-3a sequences against the mouse genome, but surprisingly no putative mouse Pellino-3a exon-2 was found. Both Pellino-1 and -2 consist of only six coding exons (Fig. 2), which are homologous to the exons of Pellino-3b. For all species of which the genomic sequences are available, we found that intron–exon boundaries are also conserved, except in Drosophila where there are eight coding exons: the equivalent of human Pellino-3b exons-2 and -3 is Drosophila Pellino exon-2, while the equivalent of human Pellino-3b exon-6 is split into four separate exons (Fig. 2).
2.
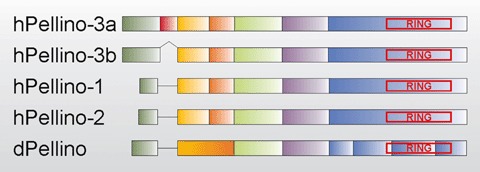
Schematic alignment of some representative Pellino proteins. Different exons of a particular Pellino are marked in a different colour, and homologous exons in various Pellino sequences are indicated in the same colour. The RING-like domain in the C-terminal part of every Pellino is boxed in red. (h) = human; (d) =Drosophila. For more information, see text.
So far, the information that is available on the expression pattern of mammalian Pellino proteins is mostly limited to their expression at the mRNA level. Interestingly, while EST sequence analysis demonstrates that all mammalian Pellino mRNAs are almost ubiquitously expressed in most organs, their relative expression levels in different organs differ remarkably, suggesting tissue-specific functions for distinct Pellino proteins [29–31]. In the adult mouse, Pellino-2 transcripts could be detected predominantly in the liver, testis and skin, with only low amounts in other tissues. Intriguingly, Pellino-2 transcripts were either not detected (spleen) or were barely detectable (thymus) in organs that predominate in the adaptive immune response, but were more abundant in organs (liver and skin) that predominate in the innate immune response [31]. Human Pellino-3 mRNA was detected at high levels in brain, heart and testis, and at lower levels in kidney, liver, lung, placenta, small intestine, spleen and stomach. Pellino-3 mRNA was only marginally expressed in colon or muscle tissue [29]. Apart from the tissue-dependent expression of different Pellino proteins, their expression might be further regulated upon stimulation of cells with specific agents. For example, mouse Pellino-1 was upregulated after 3 hours stimulation of bone-marrow-derived macrophages with lipid A (a major constituent of bacterial LPS) [32].
A domain search of Pellino protein sequences with different algorithms did not reveal any significant matches for the more variable N-terminal part of Pellino proteins. However, we and others noted a C3HC4 RING-like motif in the C-terminal part of Pellino proteins [25, 30, 33]. RING domains mediate protein–protein and protein–DNA interactions in a diverse group of proteins (reviewed by [34, 35]) and are best known for their occurrence in RING E3 ubiquitin ligases [34]. The name C3HC4 denotes the sequence order of the 8 Cys and His residues that coordinate two Znatoms, resulting in a ‘cross-brace’ protein motif [36]. Two invariant Cys-Pro-X-Cys motifs (amino acids 333–336 and 395–398 in human Pellino-1) and two absolutely conserved Cys-Gly-His triplets (amino acids 367–369 and 311–313 in human Pellino-1) are present in all Pellino proteins. Alignment of a consensus mammalian Pellino RING sequence with a consensus C3HC4 RING domain sequence revealed that the order of Cys and His residues in both RING domains differs and therefore we proposed to call the Pellino-type RING domain CHC2CHC2 ([25, 37], Fig. 3).
3.
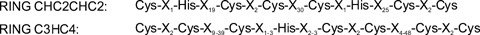
Alignment of the mammalian Pellino-RING (CHC2CHC2) consensus sequence with the C3HC4 RING consensus sequence, which was adopted from [37].
Pellino proteins interact with key mediators in TLR/IL-1R-induced signalling pathways
Similar to the originally described interaction of Drosophila Pellino with Pelle, all mammalian Pellino proteins were shown to interact with IRAK-1 and IRAK-4 (the latter was not yet reported to interact with Pellino-3) [25, 29–31, 38–40]. Moreover, all three mammalian Pellino proteins also interact with TRAF6 and TAK1 [29, 30, 39–41]. Based on the observation that Pellino-3 can simultaneously interact with endogenous IRAK-1, TRAF6 and TAK1 upon IL-1 stimulation, it was hypothesized that Pellino proteins might act as scaffold proteins that regulate signalling branch-points in TLR/IL-1R signalling to NF-κB and MAPKs [29]. Pellino-3 was also shown to bind the Ser/Thr kinase NIK [29]. The latter kinase is known to be involved in the activation of the alternative NF-κ_B_ pathway by specific members of the TNF receptor superfamily, but so far this pathway has not yet been shown to be activated by TLRs or IL-1R. It will also be interesting to analyse whether the Pellino-3/NIK interaction has any role in the activation of NF-κB by stimuli such as lymphotoxin β, which are known to activate the alternative NF-κB pathway. Pellino-1 was recently found to interact with Smad6, an anti-inflammatory protein induced by TGF-β[39]. This Pellino-1/Smad6 interaction completely abrogat-ed the formation of the Pellino-1/IRAK-1/TRAF6 signalling complex induced by IL-1 [39]. In contrast to others [29, 41], the same group also reported an interaction between Pellino-1 and the TLR/IL-1R adaptor protein MyD88 [39], but it is currently unclear if this is a direct interaction or mediated via binding of Pellino-1 to IRAK-1. Finally, Pellino-2 was also picked up via phage display as a Bcl-10 interacting protein and shown to interact with Bcl-10 in RAW264.7 macrophages treated with LPS [42]. The observation that knockdown of Pellino-2 had no effect on the recruitment of Bcl-10 to the TLR4 signalling complex suggested that Pellino-2 is an adaptor downstream of Bcl-10 in the LPS signalling pathway to NF-κB. It has to be mentioned that many of the reported protein–protein interactions were only described upon overexpression of Pellino proteins in HEK293 cells or RAW264.7 cells and still need to be confirmed for the endogenously expressed proteins. A summary of all known interactions of Pellino proteins with intermediates in TLR/IL-1R signalling is given in Table 1. Binding studies with different mutants of human Pellino-3 demonstrated that Y44 is crucial for binding of IRAK-1 [40]. Although this residue is conserved in Pellino-1 (Y17 in human Pellino-1) and -2 (Y19 in human Pellino-2), it is dispensable for IRAK-1 bind-ing of these proteins [40]. Pellino mutants truncated at their C-terminus (e.g. human Pellino-1 (1–332)) are strongly impaired in IRAK-1 binding, suggesting that also the C-terminal part of Pellino contributes to IRAK-1 binding [25]. Pellino-3 binding with TRAF6 and TAK1 occurs through amino acids downstream of position 135 [40], and binding of mouse Pellino-2 to Bcl-10 is mediated by a region spanning amino acids 169–233 [42]. As mentioned before, the more C-terminal part (downstream of residue 300) of Pellino proteins contains a novel CHC2CHC2 RING domain. Whereas efficient binding of Pellino proteins to IRAK-1 requires their C-terminal region, binding is independent of a structurally intact RING domain [25]. Drosophila Pellino/Pelle as well as all human Pellino/IRAK-1 interactions are dependent on an intact IRAK-1 kinase domain [25, 27, 29]. Furthermore, co-expression of IRAK-1 and Pellino-1, -2, or -3 in HEK293 cells leads to Pellino phosphorylation (Fig. 1, step 12) [25, 38], suggesting that phosphorylation of Pellino proteins by IRAK-1 might promote or stabilize Pellino/IRAK-1 binding. Alternatively, the observation that Pellino proteins only interact with kinase active IRAK-1 might also reflect a dependency of this interaction on IRAK-1 hyperphosphorylation. Recently, we found that co-expression of Pellino-1 or -2 with IRAK-1 also results in IRAK-1 polyubiquitination, which was dependent on an intact Pellino RING motif (Fig. 1, step 13) [25]. Although the biological role of IRAK-1 ubiquitination by Pellino proteins still needs to be demonstrated, these data strongly indicate that Pellino proteins are not only scaffold proteins, but can also function as genuine E3 ubiquitin ligases in TLR/IL-1R signalling.
1.
Binding partners of mammalian Pellino proteins. Key mediators of TLR/IL-1R-induced signalling pathways are indicated on top of the table. Binding = (+), no binding = (−), contrasting reports = (+/−); binding to an endogenous signalling protein is indicated by a grey shaded cell in the table (Pellino proteins were always overexpressed)
| MyD88 | IRAK1 | TRAF6 | TAK1 | NIK | IRAK4 | Bcl10 | Smad6 |
|---|---|---|---|---|---|---|---|
| Pellino-1 | +/− | + | + | +/− | + | + | [25, 30, 39, 41] |
| Pellino-2 | − | + | + | + | + | + | [25, 31, 38, 41, 42] |
| Pellino-3 | − | + | + | + | + | [25, 29, 40] |
Role of Pellino proteins in TLR/IL-1R signalling to NF-κB and MAPK
Although Pellino proteins are known to bind different TLR/IL-1R signalling proteins, their specific biological function in TLR/IL-1R signalling is still largely unclear and sometimes controversial (summarized in Table 2). RNAi and overexpression experiments indicated that Pellino-1 is involved in NF-κB activation in response to IL-1 [30, 39], but has no role in the IL-1-induced activation of different MAPKs [40, 41]. For Pellino-2, the situation is less clear. Reporter gene assays initially suggested a role for Pellino-2 in NF-κB, Elk1 and c-Jun/AP-1 activation in response to IL-1 or LPS [31, 41]. However, others could not confirm this and hypothesized that Pellino-2 links TLRs to basic cellular processes that regulate the activity of specific promoters, seemingly independent of the available transcription factor binding sites [38]. The reason for these paradoxical results are unclear, but most likely reflect the use of different expression plasmids and promoters, different species of Pellino proteins, a different cellular context and different cellular assays that were used to measure NF-κB or MAPK activation (e.g. different reporter assays, gelretardation assays, phosphoMAPK-specific western blotting). Finally, overexpression of Pellino-3 has no effect on NF-κB activation [29], but activates ERK, JNK and p38 MAPKs, leading to the activation of Elk-1, c-Jun, CREB and CHOP dependent gene expression [29, 40]. It should be mentioned that the above mentioned data are almost all obtained upon transient overexpression of different Pellino family members in HEK293T cells. In a similar setup, we were unable to observe any effect (positive or negative) of overexpression of different Pellino proteins in NF-κB-dependent reporter assays with HEK293T cells (Schauvliege et al., unpublished observations). It is generally known that such overexpression experiments are prone to artefacts due to high protein expression and outtitration of crucial signalling components. RNAi experiments are therefore a better alternative, but unfortunately this has only been reported in studies investigating the role of Pellino-1 and -2 in IL-1-induced NF-κB activation [30, 31, 38, 39]. It is clear that a lot of work still needs to be done in order to understand the real role of the Pellino proteins in TLR/IL-1R signalling.
2.
Role of Pellino proteins in TLR/IL-1R-induced activation of NF-κB and MAPKs. Evidence supporting a role is indicated by (+); evidence against a role is indicated by (−); when contrasting reports are available a (+/−) is used
| NF-κB | ERK | p38 | JNK | ||
|---|---|---|---|---|---|
| Pellino-1 | + | − | − | − | [30, 39–41] |
| Pellino-2 | +/− | +/− | − | +/− | [31, 38, 41] |
| Pellino-3 | − | + | + | + | [29, 40] |
Perspectives
The identification of many signalling proteins that mediate TLR/IL-1R-induced responses has greatly expanded our knowledge and appreciation of the complexity of the innate immune response. The data summarized in this review indicate an important function for the strongly conserved family of Pellino proteins. As scaffolds for multiple signalling proteins they could modulate the magnitude and specificity of TLR/IL-1R signalling, and thereby regulate the wide variety of signalling outputs. On the other hand, the recent finding that Pellino proteins can also mediate IRAK-1 ubiquitination suggests a novel function as E3 ubiquitin ligases.
There are as yet many outstanding questions that need to be addressed. It will be important to clarify why mammals have three different Pellino proteins that are very similar to each other. Do they have different functions or do they function in a cell-type or tissue-specific manner? Is their function dependent on their relative expression levels and can their expression be modulated by specific stimuli? So far, Pellino proteins have been studied in the context of TLR/IL-1R signalling, because of their interaction with IRAK-1, TRAF6 and TAK1. However, with more Pellino-binding proteins being identified, it is not unlikely that these will reveal also a function for Pellino proteins in signalling pathways initiated by other receptors. Also the observation that Pellino proteins can function as E3 ubiquitin ligases opens interesting perspectives. In this context, it will be interesting to analyse the type and functional consequences of IRAK-1 ubiquitination by Pellino, as well as to reveal whether other TLR/IL-1R signalling intermediates are substrates for Pellino-induced ubiquitination. Importantly, mechanisms that regulate the function of Pellino proteins as scaffolds or ubiquitin ligases need to be clarified. Maybe the already described Pellino phosphorylation by IRAK-1 plays an important regulatory function. Clearly, much more biochemical studies as well as the generation of Pellino-deficient mice will be of utmost importance to disentangle the regulation and function of distinct Pellino proteins in innate immunity.
Acknowledgments
SJ is a post-doctoral research associate with the ‘Fonds voor Wetenschappelijk Onderzoek-Vlaanderen’ (FWO-Vlaanderen). Research in the author's lab is in part financed by grants from the ‘Interuniversitaire Attractiepolen’ (IAP6/18), the FWO-Vlaanderen (grant 3G010505), and the ‘Geconcerteerde Onderzoeksacties’ (GOA grant 01G06B6) of the Ghent University.
References
- 1.Dunne A, O'Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflam-mation and host defense. Sci STKE. 2003;171:re3. doi: 10.1126/stke.2003.171.re3. [DOI] [PubMed] [Google Scholar]
- 2.Beutler B, Jiang Z, Georgel P, Crozat K, Croker B, Rutschmann S, Du X, Hoebe K. Genetic analysis of host resistance: toll-like receptor signaling and immunity at large. Annu Rev Immunol. 2006;24:353–89. doi: 10.1146/annurev.immunol.24.021605.090552. [DOI] [PubMed] [Google Scholar]
- 3.Li H, Sun B. Toll-like receptor 4 in atherosclerosis. J Cell Mol Med. 2007;11:88–95. doi: 10.1111/j.1582-4934.2007.00011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunne A, O'Neill LA. Adaptor usage and Toll-like receptor signaling specificity. FEBS Lett. 2005;579:3330–5. doi: 10.1016/j.febslet.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 5.Janssens S, Beyaert R. A universal role for MyD88 in TLR/IL-1R-mediated signaling. Trends Biochem Sci. 2002;27:474–82. doi: 10.1016/s0968-0004(02)02145-x. [DOI] [PubMed] [Google Scholar]
- 6.Kagan JC, Medzhitov R. Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell. 2006;125:943–55. doi: 10.1016/j.cell.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto M, Sato S, Hemmi H, Uematsu S, Hoshino K, Kaisho T, Takeuchi O, Takeda K, Akira S. TRAM is specifically involved in the Toll-like receptor 4 mediated MyD88-independent signaling pathway. Nat Immunol. 2003;4:1144–50. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald KA, Rowe DC, Barnes BJ, Caffrey DR, Visintin A, Latz E, Monks B, Pitha PM, Golenbock DT. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J Exp Med. 2003;198:1043–55. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oshiumi H, Sasai M, Shida K, Fujita T, Matsumoto M, Seya T. TIR-containing adapter molecule (TICAM)-2, a bridging adapter recruiting to toll-like receptor 4 TICAM-1 that induces interferon-beta. J Biol Chem. 2003;278:49751–62. doi: 10.1074/jbc.M305820200. [DOI] [PubMed] [Google Scholar]
- 10.Jiang Z, Ninomiya-Tsuji J, Qian Y, Matsumoto K, Li X. Interleukin-1 (IL-1) receptor-associated kinase-dependent IL-1-induced signaling complexes phos-phorylate TAK1 and TAB2 at the plasma membrane and activate TAK1 in the cytosol. Mol Cell Biol. 2002;22:7158–67. doi: 10.1128/MCB.22.20.7158-7167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S, Strelow A, Fontana EJ, Wesche H. IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase. Proc Natl Acad Sci USA. 2002;99:5567–72. doi: 10.1073/pnas.082100399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin J, Jiang Z, Qian Y, Casanova JL, Li X. IRAK4 kinase activity is redundant for interleukin-1 (IL-1) receptor-associated kinase phosphorylation and IL-1 responsiveness. J Biol Chem. 2004;279:26748–53. doi: 10.1074/jbc.M400785200. [DOI] [PubMed] [Google Scholar]
- 13.Kollewe C, Mackensen AC, Neumann D, Knop J, Cao P, Li S, Wesche H, Martin MU. Sequential autophosphorylation steps in the interleukin-1 recep-tor-associated kinase-1 regulate its availability as an adapter in interleukin-1 signaling. J Biol Chem. 2004;279:5227–36. doi: 10.1074/jbc.M309251200. [DOI] [PubMed] [Google Scholar]
- 14.Burns K, Clatworthy J, Martin L, Martinon F, Plumpton C, Maschera B, Lewis A, Ray K, Tschopp J, Volpe F. Tollip, a new component of the IL-1RI pathway, links IRAK to the IL-1 receptor. Nat Cell Biol. 2000;2:346–51. doi: 10.1038/35014038. [DOI] [PubMed] [Google Scholar]
- 15.Zhang G, Ghosh S. Negative regulation of toll-like receptor-mediated signaling by Tollip. J Biol Chem. 2002;277:7059–65. doi: 10.1074/jbc.M109537200. [DOI] [PubMed] [Google Scholar]
- 16.Ea CK, Sun L, Inoue J, Chen ZJ. TIFA activates IκB kinase (IKK) by promoting oligomerization and ubiq-uitination of TRAF6. Proc Natl Acad Sci USA. 2004;101:15318–23. doi: 10.1073/pnas.0404132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondo M, Osada H, Uchida K, Yanagisawa K, Masuda A, Takagi K, Takahashi T, Takahashi T. Molecular cloning of human TAK1 and its mutational analysis in human lung cancer. Int J Cancer. 1998;75:559–63. doi: 10.1002/(sici)1097-0215(19980209)75:4<559::aid-ijc11>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 18.Shibuya H, Yamaguchi K, Shirakabe K, Tonegawa A, Gotoh Y, Ueno N, Irie K, Nishida E, Matsumoto K. TAB1: an activator of the TAK1 MAPKKK in TGF-beta signal transduction. Science. 1996;272:1179–82. doi: 10.1126/science.272.5265.1179. [DOI] [PubMed] [Google Scholar]
- 19.Takaesu G, Surabhi RM, Park KJ, Ninomiya-Tsuji J, Matsumoto K, Gaynor RB. TAK1 is critical for IkappaB kinase-mediated activation of the NF-kappaB pathway. J Mol Biol. 2003;326:105–15. doi: 10.1016/s0022-2836(02)01404-3. [DOI] [PubMed] [Google Scholar]
- 20.Cheung PC, Nebreda AR, Cohen P. TAB3, a new binding partner of the protein kinase TAK1. Biochem J. 2004;378:27–34. doi: 10.1042/BJ20031794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Böl GF, Jurrmann N, Brigelius-Flohe R. Cellular trafficking of the IL-1RI-associated kinase-1 requires intact kinase activity. Biochem Biophys Res Commun. 2005;332:279–87. doi: 10.1016/j.bbrc.2005.04.121. [DOI] [PubMed] [Google Scholar]
- 22.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–61. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 23.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–51. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 24.Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol. 2005;6:827–37. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- 25.Schauvliege R, Janssens S, Beyaert R. Pellino proteins are more than scaffold proteins in TLR/IL-1R signalling: a role as novel RING E3-ubiquitinligases. FEBS Lett. 2006;580:4697–702. doi: 10.1016/j.febslet.2006.07.046. [DOI] [PubMed] [Google Scholar]
- 26.Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci USA. 1997;94:14614–9. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grosshans J, Schnorrer F, Nusslein-Volhard C. Oligomerisation of Tube and Pelle leads to nuclear localisation of dorsal. Mech Dev. 1999;81:127–38. doi: 10.1016/s0925-4773(98)00236-6. [DOI] [PubMed] [Google Scholar]
- 28.Resch K, Jockusch H, Schmitt-John T. Assignment of homologous genes, Peli1/PELI1 and Peli2/PELI2, for the Pelle adaptor protein Pellino to mouse chromosomes 11 and 14 and human chromosomes 2p13.3 and 14q21, respectively, by physical and radiation hybrid mapping. Cytogenet Cell Genet. 2001;92:172–4. doi: 10.1159/000056895. [DOI] [PubMed] [Google Scholar]
- 29.Jensen LE, Whitehead AS. Pellino3, a novel member of the Pellino protein family, promotes activation of c-Jun and Elk-1 and may act as a scaffolding protein. J Immunol. 2003;171:1500–6. doi: 10.4049/jimmunol.171.3.1500. [DOI] [PubMed] [Google Scholar]
- 30.Jiang Z, Johnson HJ, Nie H, Qin J, Bird TA, Li X. Pellino 1 is required for interleukin-1 (IL-1)-mediated signaling through its interaction with the IL-1 receptor-associated kinase 4 (IRAK4)-IRAK-tumor necrosis factor receptor-associated factor 6 (TRAF6) complex. J Biol Chem. 2003;278:10952–6. doi: 10.1074/jbc.M212112200. [DOI] [PubMed] [Google Scholar]
- 31.Yu KY, Kwon HJ, Norman DA, Vig E, Goebl MG, Harrington MA. Cutting edge: mouse pellino-2 mod-ulates IL-1 and lipopolysaccharide signaling. J Immunol. 2002;169:4075–8. doi: 10.4049/jimmunol.169.8.4075. [DOI] [PubMed] [Google Scholar]
- 32.Weighardt H, Jusek G, Mages J, Lang R, Hoebe K, Beutler B, Holzmann B. Identification of a TLR4-and TRIF-dependent activation program of dendritic cells. Eur J Immunol. 2004;34:558–64. doi: 10.1002/eji.200324714. [DOI] [PubMed] [Google Scholar]
- 33.Rich T, Allen RL, Lucas AM, Stewart A, Trowsdale J. Pellino-related sequences from Caenorhabditis elegans and Homo sapiens. Immunogenetics. 2000;52:145–9. doi: 10.1007/s002510000249. [DOI] [PubMed] [Google Scholar]
- 34.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 35.Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–87. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 36.Hanzawa H, De Ruwe MJ, Albert TK, Van Der Vliet PC, Timmers HT, Boelens R. The structure of the C4C4 ring finger of human NOT4 reveals features distinct from those of C3HC4 RING fingers. J Biol Chem. 2001;276:10185–90. doi: 10.1074/jbc.M009298200. [DOI] [PubMed] [Google Scholar]
- 37.Freemont PS. The RING finger: a novel protein sequence motif related to the zinc finger. Ann NY Acad Sci. 1993;684:174–92. doi: 10.1111/j.1749-6632.1993.tb32280.x. [DOI] [PubMed] [Google Scholar]
- 38.Strelow A, Kollewe C, Wesche H. Characterization of Pellino2, a substrate of IRAK1 and IRAK4. FEBS Lett. 2003;547:157–61. doi: 10.1016/s0014-5793(03)00697-5. [DOI] [PubMed] [Google Scholar]
- 39.Choi KC, Lee YS, Lim S, Choi HK, Lee CH, Lee EK, Hong S, Kim IH, Kim SJ, Park SH. Smad6 negatively regulates interleukin 1-receptor-Toll-like receptor signaling through direct interaction with the adaptor Pellino-1. Nat Immunol. 2006;7:1057–65. doi: 10.1038/ni1383. [DOI] [PubMed] [Google Scholar]
- 40.Butler MP, Hanly JA, Moynagh PN. Pellino3 is a novel upstream regulator of p38 MAPK and activates CREB in a p38-dependent manner. J Biol Chem. 2005;280:27759–68. doi: 10.1074/jbc.M500756200. [DOI] [PubMed] [Google Scholar]
- 41.Jensen LE, Whitehead AS. Pellino2 activates the mitogen activated protein kinase pathway. FEBS Lett. 2003;545:199–202. doi: 10.1016/s0014-5793(03)00533-7. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Dong W, Chen L, Xiang R, Xiao H, De G, Wang Z, Qi Y. BCL10 mediates lipopolysaccharide/toll-like receptor-4 signaling through interaction with Pellino2. J Biol Chem. 2004;279:37436–44. doi: 10.1074/jbc.M400241200. [DOI] [PubMed] [Google Scholar]