NeuroMorpho.Org: A Central Resource for Neuronal Morphologies (original) (raw)
The structure of dendrites and axons plays fundamental roles in synaptic integration and network connectivity. Synergistic advances in neurobiology (e.g., intracellular injections, fluorescent protein expression), microscopy (e.g., multiphoton laser scanning, computer controllers), and imaging software (e.g., Neurolucida tracing, blind deconvolution) are rapidly transforming the three-dimensional (3D) reconstruction of neuronal morphology into a mainstream technique. In the course of electrophysiological, pharmacological, or histological studies, neuritic arborizations are accurately traced into digital files representing location, orientation, and branching details as a parsimonious set of interconnected cylinders. The process is still considerably labor intensive (typically 1 person-week per neuron), but it enables nearly unlimited analytical power, including, e.g., quantitative morphological and stereological characterizations. Digital neuronal reconstructions also constitute primary sources for comparative anatomical investigations, computational models of biophysics, and the statistical assessment of potential synaptic contacts. These scientific applications are often independent of, and unrelated to, the research projects originating the datasets. Thus neuromorphological files constitute an ideal case of value added by data sharing.
Here we present NeuroMorpho.Org, a web-accessible archive of digital reconstructions of neuronal morphology (neuromorpho.org). The goal is to illustrate the value of this resource rather than describe it exhaustively. Readers are encouraged to complement the present text by navigating the website to explore specific issues. NeuroMorpho.Org also contains additional information, including an introduction page, a QuickStart guide, and answers to frequently asked questions. The two main sections of this article introduce NeuroMorpho.Org's essential functionality and content, respectively. The former offers several examples of the principal capabilities enabled by this resource. The latter overviews the data in terms of raw file availability, type of accompanying information, and potential applications. Emphasizing the user's, rather than developer's, perspective, this ToolBox does not cover the technical aspects of database structure and underlying back-end engine. Instead, the closing remarks focus on ongoing and planned developments, comparison with and interoperation with other similar or related resources, and mechanisms to contribute data.
NeuroMorpho.Org functionality
NeuroMorpho.Org is publicly available, free of charge, and accessible through any modern web browser (e.g., Explorer, Firefox, or Safari) and operating system (including Windows, Mac, and Linux), without login/registration. The entire repository of neuronal morphologies can be browsed by cell type (e.g., pyramidal vs basket), brain region (hippocampus, cerebellum, etc.), animal species, or by the contributing laboratory. These complementary options correspond to intuitive elements immediately associated with every study (which neuron, where in the nervous system, what organism, and who did it). The four browse modes are each further organized based on the most common subcriteria and can be seamlessly intertoggled with single clicks. Browsing provides for easy navigation and a comprehensive outlook of the database. Users can select (and deselect) sets of neurons for additional exploration.
Neurons can also be searched by a variety of criteria, including the categories described for browsing, additional metadata (animal age or developmental stage, experimental conditions, etc.), and morphometric measures (such as soma volume, number of branches, average bifurcation angles). Searching by metadata is enabled by a series of user-friendly drop-down menus (Fig. 1). Morphometric searches require the additional selection of a relationship (equal to, greater/smaller than) and a value to find (e.g., all cells with “>200 branches”). An option is offered to show a set of values for the chosen morphometric parameter extracted from a sample of neurons, providing an idea of the range and order of magnitude for the given measure (Fig. 2). The available metadata types and morphometry parameters are detailed in the next section. Users can preview the number of neurons their criteria will return, and may refine the search by combining multiple filters, as in a query for, e.g., layer 4 stellate cells reconstructed in vitro at 100× from the somatosensory cortex of Wistar rats. Search results are displayed in a new window or tab (thus retaining the specified criteria for subsequent use), either as a summary of thumbnail images and selected metadata or as a list of cells organized in one of the browse modes (Figs. 1, 2).
Figure 1.
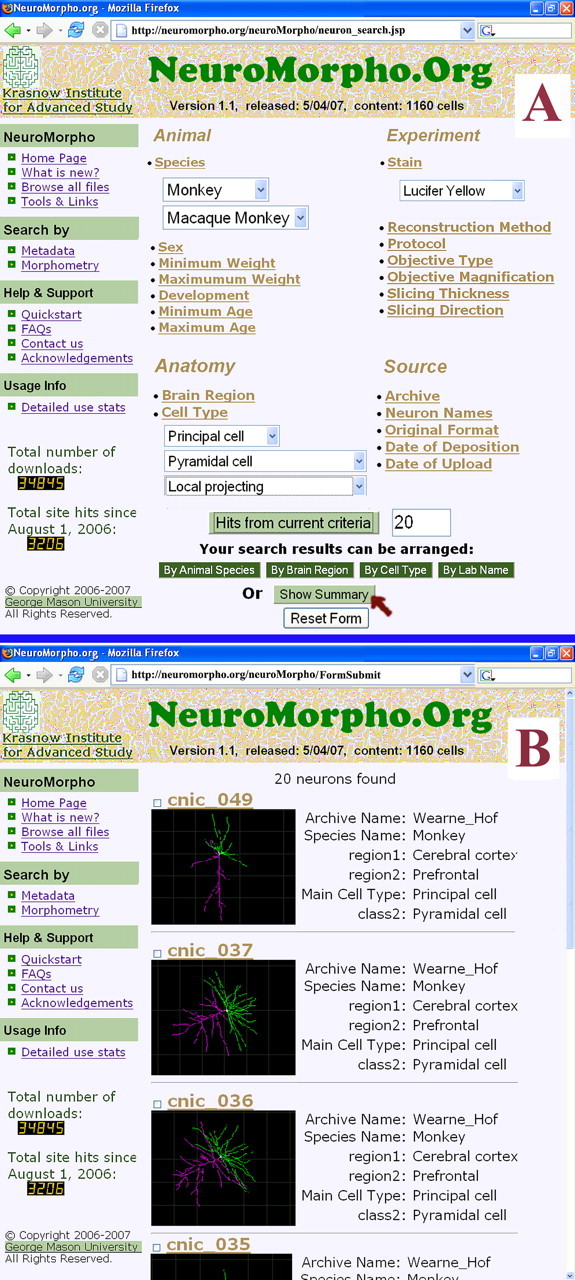
Reconstruction search and retrieval by metadata. A, Screenshot of the drop-down menus illustrating a query based on species and strain, morphological class and subclasses, and a histological detail, matched by 20 cells. B, Organization of the pop-up results requested as a “summary” with images and selected information (individual cells are hyperlinked and can be selected/deselected with toggle clicks).
Figure 2.
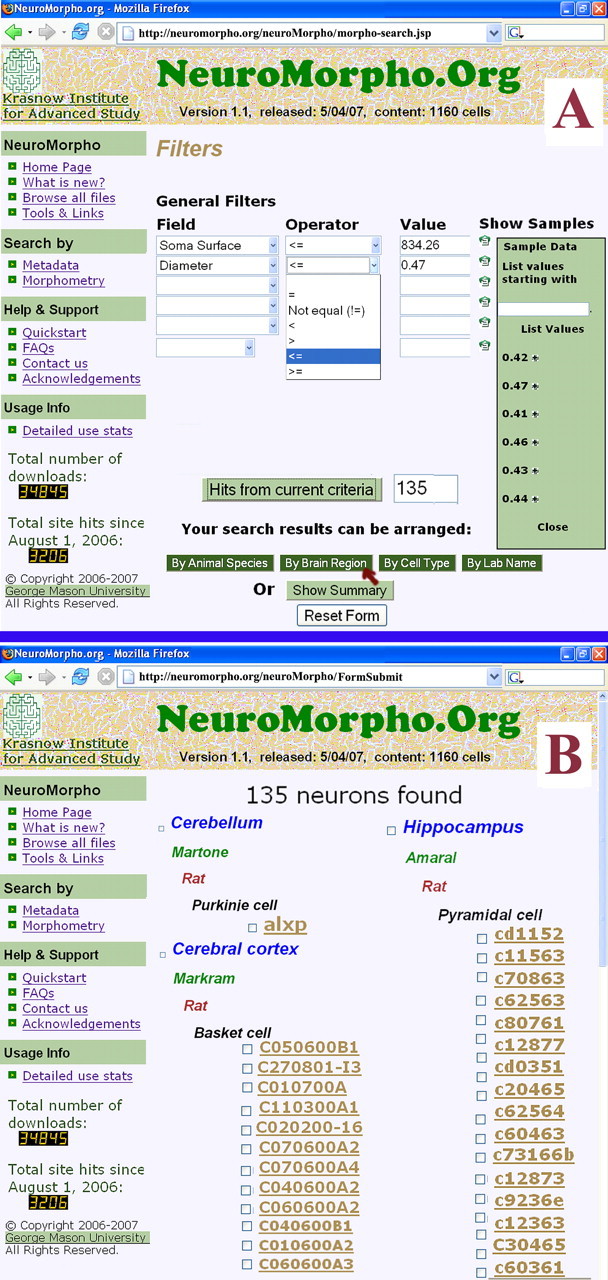
Morphometric content-based queries. A, In a search by morphometry, available geometrical parameters are selected along with appropriate numerical ranges, facilitated by an optional dynamical overlay of representative values. B, In this example, the user chose to return results organized by brain region.
Neurons selected through browsing or a search can be bundled in a single compressed folder for download or further inspected in dedicated individual windows. For each cell, downloadable material includes the digital reconstructions and auxiliary text. Reconstructions consist of the original shared files and a standardized version checked/edited for integrity and converted in a uniform, nonproprietary ASCII format. The text specifies all the standardization edits and logs any residual anomaly. In addition to linking to these files, the detailed cell pages show a 2D projection image, a 3D dynamical rendering of the growing and revolving morphology, and an assortment of metadata and morphometric features (Fig. 3). Moreover, a virtual reality display allows interactive zooming, panning, and multidirectional turning of the neuron, as well as conversion to computational model formats for the popular NEURON and GENESIS simulation environments.
Figure 3.
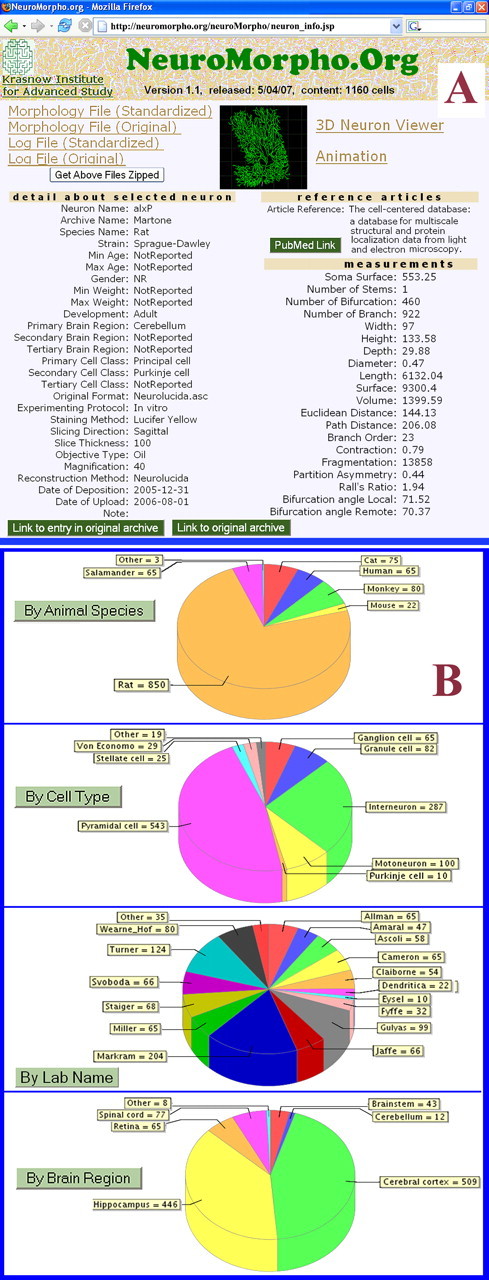
Available data in NeuroMorpho.Org. A, An individual neuron page, including download links of the raw and processed data files and information logs, images and options for dynamic displays, and experimental, literature, and morphometric details. B, Distribution of the 1160 reconstructions available in version 1.1 by major “browse” features.
Each neuron is hyperlinked to the PubMed reference of the article describing it. The same PubMed abstract, in turn, closes the loop with a “linkout” call to NeuroMorpho.Org via the SfN Neuroscience Database Gateway, thus establishing a reciprocal correspondence between the peer-reviewed literature and the underlying scientific data. NeuroMorpho.Org also connects to a number of resources and tools for the reconstruction, visualization, analysis, and modeling of neuronal morphology, as well as to the contributing laboratories and databases. Usage statistics are summarized based on web visits (nonconsecutive hits), users (unique internet identifiers), and downloads (by cell type, brain region, animal species, and contributing laboratory).
NeuroMorpho.Org data content
The Spring 2007 release (version 1.1) contains 1160 neurons from 23 contributing laboratories, 7 species (from salamander to human), 9 main brain regions, and >30 cell types. The distribution of data reflects both research trends and different subcommunity emphases on digital technology and data sharing (Fig. 3). In particular, the vast majority (>75%) of cells comes from rats, >40% from the hippocampus (with the neocortex close second), and nearly half consist of pyramidal neurons. An additional ∼600 reconstructions have been collected and are in the processing pipeline for inclusion in the database. In particular, >250 neurons from the Drosophila olfactory system will be included in the Summer 2007 release (version 1.2).
For every neuron, a rich set of relevant information and annotations (metadata) are stored in addition to the morphological files. These include the source (archive or provider, date of deposition and upload, file format, and publication reference), the animal (species, strain, sex, and age, developmental stage, or weight), and additional notes from the data owner and/or curator. Experimental details are also recorded, including on the histology (injection protocol, slicing thickness and direction, and stain), microscopy (objective type and magnification), and reconstruction hardware or software. A three-level hierarchy is used to specify brain region (e.g., cerebral cortex, somatosensory, layer 5) and cell type (e.g., interneuron, CCK+, VIP−).
Finally, several morphometric characteristics are recorded from every cell. These include measures of size both at the local (soma surface area, average branch diameter) and whole neuron level (width, height, and depth; number of trees and of bifurcations; total length and volume; maximum Euclidian distance, path to terminals, and branch order). Spatial orientation is captured by bifurcation angles, either between local tracing points or between remote nodes. Finally, topological and functional properties are reflected in parameters such as tortuosity, number of points per branch, partition asymmetry, and Rall's power ratio. A large number of additional morphometrics can be extracted with the companion software L-Measure (Scorcioni and Ascoli, 2001).
The main usage and potential applications of NeuroMorpho.Org include comparative morphological and stereological analysis, compartmental simulations of neuronal electrophysiology, computational models of structure and development, scientific education and illustration, large-scale data mining, and anatomically realistic neural networks (Stepanyants and Chklovskii, 2005; Samsonovich and Ascoli, 2006; Gleeson et al., 2007). Examples of “success stories” in the re-use of neuronal reconstructions are particularly abundant in computational neuroscience, where biophysical studies are increasingly based on accurate morphological data (Migliore et al., 2003). Here we provide a recent use case from our own experience for a different problem solved using the database.
In the course of efforts to describe dendritic structure quantitatively, we noticed a tendency of pyramidal cells with larger basal tree to have smaller apical trees, and vice versa. This observation was qualitatively consistent with published illustrations and tabulated data, but access to the complete reconstructions in digital format enabled its quantitative and statistical demonstration (Samsonovich and Ascoli, 2006). Moreover, analysis of the raw data files revealed similar trends at different scales, e.g., among individual trees of the basal arborizations, and even between subtrees stemming from a bifurcation, as well as with different metrics (number of branches, length, surface area, etc.). Thus, dendritic dimension exhibits significantly higher stability than if individual trees and their parts developed independently. Such systematic propensity of neurons to compensate internally for random structural fluctuations is contrary to what would be expected by genetic and/or environmental regulation (e.g., people with big hands tend to have big feet). Instead, it suggests the action of direct intracellular competition for resources. The availability of multiple data sets allowed the extension of these findings to different species (rat and monkey), brain regions (hippocampus and prefrontal cortex), and cell classes (pyramidal and granule) but excluded experimental bias of specific methods (robust across in vitro and in vivo histology over multiple laboratories).
Concluding remarks
First publicly released in Fall 2006, NeuroMorpho.Org underwent thorough and robust testing from active researchers (>3800 hits at the time of this manuscript submission), and nearly 35 times the entire volume content has been downloaded already. We attribute this success to the effective fulfillment of a community need, the mature functionality of the site, the density of data content (Ascoli, 2006), and the sheer richness of available information. A Google/PubMed-like search bar is under development for future release, with automated expansion of query terms to their synonyms (e.g., six-layer cortex vs neocortex) and inclusive relationships (e.g., extension of “cortex” to cerebellar, hippocampal, and cerebral cortices). User requests and feedback are also continuously addressed in the regular version updates.
NeuroMorpho.Org is related to several modern neuroinformatics resources. With respect to other databases of digital morphologies, NeuroMorpho.Org is distinct as a dynamic, centrally curated, umbrella inventory as opposed to single laboratory sources (Cannon et al., 1998). As such, it constitutes to date the largest collection of publicly accessible 3D reconstructions and associated metadata. At the same time, the content of NeuroMorpho.Org is highly specialized and emphasizes dense coverage of available data (aiming to inventory most, if not all, shared morphological files) rather than integration with other dimensions, such as electrophysiological or molecular (e.g., Markram, 2006). Nevertheless, a broad format and tool interoperability ensures the functional integration of NeuroMorpho.Org with complementary resources, such as the computational neuroscience ModelDB archive (Migliore et al., 2003). Neuroscientists can engineer novel biophysically and anatomically detailed simulations in a matter of minutes by combining any of the structures from NeuroMorpho.Org with one of many physiological models from ModelDB.
Contributing neuronal reconstructions to NeuroMorpho.Org is simple, with the only conditions being the researchers' willingness to share their data and for the morphologies to be in digital format (e.g., Neurolucida or any of several freeware alternatives) (Brown et al., 2005). The database administrators (authors of this article), contacted through the web or by e-mail, handle the file transfer (FTP or CD/DVD), conversion, annotation, and curation, with minimal burden on the data owners, who are given the opportunity to preview their uploads before public release (the processing pipeline is detailed online). Only reconstructions associated with a published study are posted, to ensure a “data quality” criterion for acceptance of data. However, owners are welcome to submit their data at any time (before submission, at time of acceptance, etc.) to speed up processing.
The results of research projects developed at the time of the database first release may have not been published yet. Thus, it is still too early to quantitatively evaluate how this data enhanced the users' studies. However, in addition to benefiting science, data sharing augments visibility and impact of both the original results and their authors (Piwowar et al., 2007). The existence of NeuroMorpho.Org also allows publications such as The Journal of Neuroscience and The Journal of Comparative Neurology to instruct contributors to deposit neuronal reconstructions upon manuscript acceptance.
Footnotes
Editor's Note: Toolboxes are intended to briefly highlight a new method or a resource of general use in neuroscience or to critically analyze existing approaches or methods. For more information, see www.jneurosci.org/misc/itoa.shtml.
NeuroMorpho.Org is part of the “Neuroscience Information Framework,” a neuroinformatics consortium led by Cornell University, endorsed by the Society for Neuroscience, and sponsored by the Blueprint Initiative of the National Institutes of Health under National Institute of Drug Abuse Contract HHSN271200577531C. Additional support was provided by National Institute of Neurological Disorders and Stroke–National Institute of Mental Health (NIMH)–National Science Foundation Grant R01 NS39600 and NIMH Grant R01 MH64417, both under the Human Brain Project. We are indebted to the MITRE Corporation for permission to use their technology in developing this project, and in particular to Dr. Ken Smith, Jeffrey Hoyt, and Gail Hamilton for their expert advice and collaboration. The described data and tools were generously shared by many different investigators, credited at http://neuromorpho.org.
References
- Ascoli GA. Mobilizing the base of neuroscience data: the case of neuronal morphologies. Nat Rev Neurosci. 2006;7:318–324. doi: 10.1038/nrn1885. [DOI] [PubMed] [Google Scholar]
- Brown KM, Donohue DE, D'Alessandro G, Ascoli GA. A cross-platform freeware tool for digital reconstruction of neuronal arborizations from image stacks. Neuroinformatics. 2005;3:343–360. doi: 10.1385/NI:3:4:343. [DOI] [PubMed] [Google Scholar]
- Cannon RC, Turner DA, Pyapali GK, Wheal HV. An on-line archive of reconstructed hippocampal neurons. J Neurosci Methods. 1998;84:49–54. doi: 10.1016/s0165-0270(98)00091-0. [DOI] [PubMed] [Google Scholar]
- Gleeson P, Steuber V, Silver RA. NeuroConstruct: a tool for modeling networks of neurons in 3D space. Neuron. 2007;54:219–235. doi: 10.1016/j.neuron.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram H. The blue brain project. Nat Rev Neurosci. 2006;7:153–160. doi: 10.1038/nrn1848. [DOI] [PubMed] [Google Scholar]
- Migliore M, Morse TM, Davison AP, Marenco L, Shepherd GM, Hines ML. ModelDB: making models publicly accessible to support computational neuroscience. Neuroinformatics. 2003;1:135–139. doi: 10.1385/NI:1:1:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwowar HA, Day RS, Fridsma DB. Sharing detailed research data is associated with increased citation rate. PLoS ONE. 2007;2:e308. doi: 10.1371/journal.pone.0000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsonovich AV, Ascoli GA. Morphological homeostasis in cortical dendrites. Proc Natl Acad Sci USA. 2006;103:1569–1574. doi: 10.1073/pnas.0510057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorcioni R, Ascoli GA. Algorithmic extraction of morphological statistics from electronic archives of neuroanatomy. Lect Notes Comp Sci. 2001;2084:30–37. [Google Scholar]
- Stepanyants A, Chklovskii DB. Neurogeometry and potential synaptic connectivity. Trends Neurosci. 2005;28:387–394. doi: 10.1016/j.tins.2005.05.006. [DOI] [PubMed] [Google Scholar]