Characterization of Integrative and Conjugative Element ICEKp1-Associated Genomic Heterogeneity in a Klebsiella pneumoniae Strain Isolated from a Primary Liver Abscess (original) (raw)
Abstract
Genomic heterogeneity has been shown to be associated with Klebsiella pneumoniae strains causing pyogenic liver abscesses (PLA) and metastatic infections. In order to explore the mechanism responsible for genomic heterogeneity in K. pneumoniae, we compared the complete genomic sequences of strains NTUH-K2044 and MGH78578. An ∼76-kbp DNA fragment located adjacent to an asparagine (asn) tRNA gene was present in NTUH-K2044 but not in MGH78578. This fragment could be divided into three regions with different functions, and structurally it resembled a functional integrative and conjugative element (ICE), ICE_Ec1_, in Escherichia coli. The 5′ region of this fragment contained genes similar to a high-pathogenicity island (HPI) of Yersinia pestis and Yersinia pseudotuberculosis. The middle region was similar to part of a large plasmid in K. pneumoniae, and the 3′ region contained genes responsible for DNA conjugative transfer. Therefore, this DNA fragment was designated ICE_Kp1_. Precise excision and extrachromosomal circularization of ICE_Kp1_ were detected in K. pneumoniae wild-type strain NTUH-K2044. ICE_Kp1_ could integrate into the asn tRNA loci of the chromosome of another K. pneumoniae isolate. The prevalence of ICE_Kp1_ was higher in PLA strains (38 of 42 strains) than in non-tissue-invasive strains (5 of 32 strains). Therefore, ICE_Kp1_ may contribute to the transmission of the HPI and result in K. pneumoniae PLA infection-associated genomic heterogeneity.
Klebsiella pneumoniae, a gram-negative enteric bacterium, is a common pathogen that causes hospital-acquired urinary tract infections, septicemia, and pneumonia, as well as community-acquired pneumonia (1). Recently, community-acquired pyogenic liver abscesses (PLA) caused by K. pneumoniae complicated with metastatic meningitis and endophthalmitis have been found globally (13, 15, 19, 21, 25, 27, 37). Capsular serotypes have been reported to play a vital role in the pathogenicity of this organism (18, 20). Using transposon mutagenesis, the magA virulence gene has been shown to be associated with mucoviscosity, resistance to serum, and phagocyte killing and virulence in mice (18). Sequencing of the magA flanking region (∼25 kb) revealed a capsular polysaccharide synthesis (cps) region. The region containing magA has been shown to be responsible for capsular serotype K1 (17). Genomic heterogeneity is also important in the virulence of K. pneumoniae (16, 29). Therefore, both genomic heterogeneity and capsular serotypes play important roles in the pathogenesis of K. pneumoniae strains causing liver abscesses.
Horizontal gene transfer contributes substantially to genomic heterogeneity among bacteria. The exchange of DNA plays a critical role in the evolution of bacteria and facilitates the rapid adaptation of bacteria to environmental alterations (24). Horizontal gene transfer is known to be mediated by three mechanisms: transformation, transduction, and conjugation. Pathogenicity islands (PAIs) are defined as 10- to 200-kb DNA fragments that contain gene clusters associated with virulence and are closely related to pathogenic strains. PAIs have G+C contents that are different from the G+C contents of the rest of the whole genome and mobile elements or insertion sequences. Therefore, PAIs are believed to be acquired by horizontal gene transfer. The acquisition of PAIs allows bacteria to grow in and colonize existing niches. However, the mechanism of transmission of PAIs has not been clearly demonstrated yet. Several conjugative and self-transmissible elements that integrate into the bacterial chromosome have been discovered recently (11, 12). These elements had features of plasmids and phages; they could be transferred via conjugation (plasmidlike), and they could integrate into and replicate with the host chromosome (phagelike). Therefore, they were classified as integrative and conjugative elements (ICEs). Because the PAIs were proposed to be transmitted horizontally but no longer appear to be mobile, the progenitors of PAIs might be ICEs.
The high-pathogenicity island (HPI) of Yersinia species that carries the yersiniabactin siderophore system is essential for the virulence of Yersinia (7, 9, 10). HPI is widely distributed in the family Enterobacteriaceae (4, 34, 36), but its mechanism of transmission has not been demonstrated yet. In a recent study, a novel ICE (ICE_Ec1_) of Escherichia coli strain ECOR31 was suggested to be a mobilizable progenitor of the HPI (35).
To explore the mechanism of genomic heterogeneity in K. pneumoniae strains causing PLA, we compared the complete genome sequences of K. pneumoniae strains NTUH-K2044 and MGH78578. A putative ICE containing an HPI was identified in NTUH-K2044 but not in MGH78578, and it was designated ICE_Kp1_. Self-transmission and chromosomal integration were demonstrated in this study.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. The 74 clinical isolates were collected from National Taiwan University Hospital from 1997 to 2003 (18, 29). Forty-two of these strains were PLA strains isolated from the blood of patients with PLA with or without meningitis or endophthalmitis complications, and 32 were non-tissue-invasive strains isolated from patients with sepsis but without PLA or other metastatic infections in any tissue. K. pneumoniae and E. coli were cultured in LB medium supplemented with appropriate antibiotics, including 50 μg/ml kanamycin, 100 μg/ml chloramphenicol, 100 μg/ml streptomycin, and 1,000 μg/ml rifampin.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain(s) or plasmid | Description | Reference or source |
|---|---|---|
| K. pneumoniae strains | ||
| Clinical isolates | 74 isolates collected from National Taiwan University Hospital from 1997 to 2003 | 18 |
| NTUH-K2044 | Clinical strain causing PLA and meningitis | 18 |
| MGH78578 | ATCC 700721, used for determination of the complete genome sequence at Washington University | ATCC |
| NTUH-K2044 int mutant | NTUH-K2044 isogenic int mutant (insertion mutation) | This study |
| NTUH-K2044 irp2 mutant | NTUH-K2044 isogenic irp2 mutant (irp2 replaced by a kanamycin resistance gene) | This study |
| NTUH-K2044 rmpA mutant | NTUH-K2044 isogenic rmpA mutant (mutant 3-20, transposon insertion mutant) | 18 |
| NTUH-K2044 virB1 mutant | NTUH-K2044 isogenic virB1 mutant (insertion mutation) | This study |
| NTUH-K2044 mobB mutant | NTUH-K2044 isogenic mobB mutant (insertion mutation) | This study |
| N4252 Rifr | Non-tissue-invasive strain, spontaneous rifampin-resistant mutant | This study |
| E. coli strains | ||
| HB101 | supE44 hsdS20(rB− mB−) _recA13 ara_-_14 proA2 lacY1 galK2 rpsL20 xyl_-_5 mtl_-_1 leuB6 thi_-1 | Takara |
| CC118λ_pir_ | Δ(ara-leu) araD Δ_lacX74 galE galK phoA20 thi-1 rpsE rpoB argE_(Am) recA1 λ_pir_ phage lysogen | 23 |
| S17-1λ_pir_ | hsdR recA pro RP4-2 (Tc::Mu; Km::Tn_7_)(λ_pir_) | 31 |
| Plasmids | ||
| pACYC184 | Cloning vector, P15A replicon, Cmr | New England Biolabs |
| pUT-km | pUTKm1-derived plasmid with mini-Tn_5_ excised by EcoRI, tnp excised by SalI, and bla removed by ApaLI and with insertion of a kanamycin resistance cassette from pUC4K into a PstI site | 17 |
Sequence analysis.
The complete genome sequence of K. pneumoniae MGH78578 was determined at Washington University (http://genome.wustl.edu/projects/bacterial/kpneumoniae/). The complete genome sequence of K. pneumoniae NTUH-K2044 was determined by collaboration between National Taiwan University College of Medicine and Yangming University in Taiwan (http://genome.nhri.org.tw/kp/index.php). Sequence analysis was performed with Sequencher software (Gene Codes, Wisconsin) and ClustalW (http://www.ebi.ac.uk/Tools/clustalw/index.html). Analysis of homology was performed by comparing sequences with the NCBI BLAST DNA and protein databases.
Construction of K. pneumoniae mutant strains.
int, virB1, and mobB insertion mutants of K. pneumoniae NTUH-K2044 were constructed as previously described (17). Primer pairs (Table 2) were designed to amplify partial regions containing the int (int-101F and int-490R), virB1 (virB1-60F and virB1-359R), and mobB (mobB-101F and mobB-500R) genes for mutant construction. The irp2 deletion mutant was constructed by replacing the irp2 gene with a kanamycin cassette as previously described (29). The NTUH-K2044 rmpA mutant (mutant 3-20) was identified by inverse PCR and sequencing after screening for decreased mucoviscosity by a string test using a mutant library in a previous study (18).
TABLE 2.
Primers used in this study
| Primer | Sequence | Purpose | Reference |
|---|---|---|---|
| oriT-F | 5′-CAAATAAAATGACAGTCATC-3′ | oriT cloning | This study |
| oriT-R | 5′-GGCATCGCCCCATCAATT-3′ | oriT cloning | This study |
| int-101F | 5′-TGGCTCCCGCCACTGGTATC-3′ | int mutant construction | This study |
| int-490R | 5′-CGGGACGCAACTTCCAGCAG-3′ | int mutant construction | This study |
| virB1-60F | 5′-GTCCACGGCATTCGACGT-3′ | virB1 mutant construction | This study |
| virB1-359R | 5′-CGGTAGCAGTCTCTGAG-3′ | virB1 mutant construction | This study |
| mobB-101F | 5′-TGCAACAGAAACAACTGGTG-3′ | mobB mutant construction | This study |
| mobB-500R | 5′-TAATGCAGTGTTTGCGGCAG-3′ | mobB mutant construction | This study |
| 1 | 5′-GCACACAGGCATTTTCCAGCAG-3′ | Detection of excision | This study |
| 2 | 5′-AGATGGCTGGATAGGCAC-3′ | Detection of excision | This study |
| 3 | 5′-GACCCAGATACAGATAGC-3′ | Detection of excision | This study |
| 4 | 5′-GGCTGACGTTGCCGACGATAAG-3′ | Detection of excision | This study |
| 1′ | 5′-GGTGACGTTCAAGAGAGACC-3′ | Detection of excision | This study |
| 2′ | 5′-AGAAGGCTTGAGGGTGCGGATT-3′ | Detection of excision | This study |
| 3′ | 5′-GACTCATACCGCTCATGGGCAC-3′ | Detection of excision | This study |
| 4′ | 5′-GTGAATTCATCCTACTGGC-3′ | Detection of excision | This study |
| 1166F | 5′-GGTGCTCTTTACATCATTGC-3′ | magA detection | 29 |
| 500R | 5′-GTTCACAACAGCTGCCTGAC-3′ | magA detection | 29 |
| 7C4-T71 | 5′-ATCCTGGAAGCTCACCGTTC-3′ | kfu/PTS region detection | 29 |
| 26D6-T32 | 5′-ATTTCCACGCGGATACCGTC-3′ | kfu/PTS region detection | 29 |
| 10E4-2-5F | 5′-AGTCGGCCTGGGGTTTAAGG-3′ | allS region detection | 29 |
| 10E4-2-475R | 5′-CAGTCAACGTGGCGATTCGC-3′ | allS region detection | 29 |
| asn1-F | 5′-GTAGACGTGAGAATGGCCTG-3′ | ICE integration site analysis | This study |
| asn1-R | 5′-GACAGGCTGTTTTCAGCATC-3′ | ICE integration site analysis | This study |
| asn2-F | 5′-CTGGGAGTATGATGCACTGG-3′ | ICE integration site analysis | This study |
| asn2-R | 5′-GGTTAATGATCGGCCATTGC-3′ | ICE integration site analysis | This study |
| asn4-F | 5′-GTCGCTGTCGTGGACTTTAC-3′ | ICE integration site analysis | This study |
| asn4-R | 5′-AGTGATCCTGATGTCGCTGG-3′ | ICE integration site analysis | This study |
| asn3-F | 5′-AAGATACGACTCCACACG-3′ | ICE integration site analysis | This study |
| ybtS-R inverse | 5′-CGCCCTATTTAATGGTGTAG-3′ | ICE integration site analysis | This study |
| fyuA-F | 5′-ATGAAAATGACACGGCT-3′ | 5′ region prevalence analysis | This study |
| fyuA-R | 5′-TCAGAAGAAATCAATTC-3′ | 5′ region prevalence analysis | This study |
| virB1-F | 5′-ATGCTTTCCACCACAGC-3′ | 3′ region prevalence analysis | This study |
| virB1-R | 5′-TTATTCCTCCTCCTCAC-3′ | 3′ region prevalence analysis | This study |
| iroN-F | 5′-GTCCGGCGGTAACTTCAGCC-3′ | Middle region prevalence analysis | This study |
| iroN-R | 5′-TCAGAATGATGCGGTGACAC-3′ | Middle region prevalence analysis | This study |
| HPI 3′-F | 5′-CTGCGGTAAATAAATGACG-3′ | Connection of 5′ and 3′ regions | This study |
| virB1-F inverse | 5′-GCTGTGGTGGAAAGCAT-3′ | Connection of 5′ and 3′ regions | This study |
| orf3-R | 5′-CTTGATGAAAATCTGGTG-3′ | Left junction of middle region | This study |
| orf16-F | 5′-AATGTTCTCTGCGCATGG-3′ | Right junction of middle region | This study |
| asn probe-F | 5′-CGTTCACACGATTCCTC-3′ | asn tRNA probe | This study |
| asn probe-R | 5′-GGCTTCTTAAATTTGGCTC-3′ | asn tRNA probe | This study |
Construction of plasmid pACYC184-oriT.
Sequences containing oriT (nucleotides 65013 to 66772 in the accession number AB298504 sequence after virB11 and before mobB) were amplified from K. pneumoniae NTUH-K2044 genomic DNA by PCR using primers oriT-F and oriT-R (Table 2) and cloned into the HincII site of plasmid pACYC184 (Table 1).
Bacterial conjugation.
Donors (1 × 108 CFU) and recipients (1 × 108 CFU) grown overnight in 2 ml of 10 mM MgSO4 were mated on a 0.22-μm membrane filter (Advantec, Tokyo, Japan). The membrane was then transferred to Columbia agar plates containing 5% sheep blood and incubated at 37°C overnight. Next, the bacteria were plated on an LB plate supplemented with appropriate antibiotics.
Southern blotting.
Approximately 5-μg portions of genomic DNA from various K. pneumoniae strains were digested by EcoRV and subjected to Southern hybridization according to manufacturer's instructions (Roche Molecular Biochemicals, Mannheim, Germany). Primers (asn-F and asn-R primers [Table 2]) were designed to generate the digoxigenin-labeled asn tRNA gene probe by PCR. A digoxigenin-labeled int DNA probe was generated by PCR using primers int-101F and int-490R (Table 2).
Nucleotide sequence accession number.
The nucleotide sequence reported in this study has been deposited in the GenBank database under accession number AB298504.
RESULTS
Comparison of the complete genome sequences of K. pneumoniae strains.
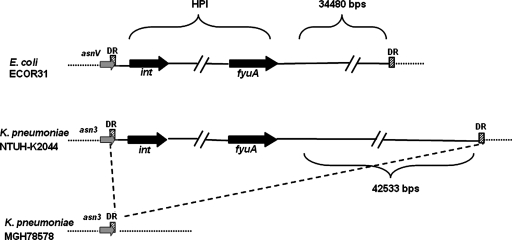
Comparison of the complete genome sequences of K. pneumoniae strains NTUH-K2044 and MGH78578 (Table 1) revealed an ∼76-kb insertion (accession no. AB298504) in strain NTUH-K2044 (Fig. 1). This DNA fragment was located adjacent to an asparagine (asn) tRNA gene and was flanked by 17-bp direct repeats. The average G+C content of the NTUH-K2044 whole genome was ∼58%, while the G+C content of this fragment was ∼52%. These results suggested that this chromosomal fragment might have been acquired by horizontal transfer. Nucleotide sequence analysis of the 76-kb fragment divided it into three functional regions, and this chromosomal fragment was structurally similar to ICE_Ec1_ of E. coli strain ECOR31 (35). The 5′ region was similar to the HPI of Yersinia pestis and Yersinia pseudotuberculosis, which is responsible for synthesis of the yersiniabactin siderophore and is closely related to bacterial virulence (7, 9, 10). The production of yersiniabactin in NTUH-K2044 was demonstrated by cross-feeding assays which showed that culture supernatants of NTUH-K2044 promoted the growth of an indicator strain (Yersinia enterocolitica strain 5030) under iron-deficient conditions, whereas culture supernatants of the irp2 mutant did not (22) (data not shown). The middle region was similar to part of the large plasmid in K. pneumoniae. The 3′ region resembled genes encoding both a functional mating pair formation system and a DNA-processing region for DNA mobilization in ICE_Ec1_ (35). Therefore, this chromosomal fragment was designated ICE_Kp1_.
FIG. 1.
Seventy-six-kilobase pair insertion in K. pneumoniae strain NTUH-K2044. The dashed line indicates that that there was an insertion in K. pneumoniae strain NTUH-K2044 compared with strain MGH78578. The insertion was near the asparagine tRNA gene (the asn tRNA genes are indicated by gray arrows [asnV tRNA gene in E. coli ECOR31 and asn3 tRNA gene in K. pneumoniae NTUH-K2044 and MGH78578]). The cross-hatched box indicates the attachment site (attO) composed of a 17-bp direct repeat (DR). The black arrows indicate the int and fyuA genes, which were located in the 5′ and 3′ ends of the HPI. In contrast to the 34,480-bp fragment located adjacent to the HPI core in E. coli strain ECOR31, a 42,533-bp fragment was located in the right border of HPI in K. pneumoniae strain NTUH-K2044.
Sequence analysis of the middle and 3′ regions of ICE_Kp1_.
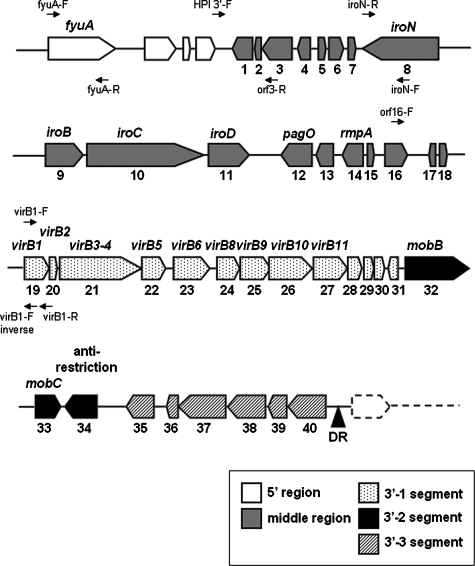
Forty open reading frames (ORFs) larger than 150 nucleotides were identified in the middle and 3′ regions of ICE_Kp1_ (Fig. 2 and Table 3).
FIG. 2.
Genetic alignment of the middle and 3′ regions of ICE_Kp1_. The black triangle indicates the 17-bp direct repeat (DR) and the end of ICE_Kp1_. The large arrows indicate the locations and orientations of ORFs. The ORF numbers and the designations of ORFs are indicated below and above the arrows, respectively. The middle region exhibited similarity to part of large plasmid pLVPK in K. pneumoniae strain CG43. The 3′ region contained three functionally distinct segments (segments 3′-1, 3′-2, and 3′-3). The small arrows indicate the locations and orientations of primers used to study the prevalence of ICE_Kp1_.
TABLE 3.
ORFs in the middle and 3′ regions of ICE_Kp1_
| ORF | Product size (amino acids) | Locationa | Homologous protein(s) | Source | Amino acid identityb | Accession no. | |||
|---|---|---|---|---|---|---|---|---|---|
| No. | Designation | Start site | Stop site | No. of matching amino acids | Total no. of amino acids | ||||
| 1 | ORF1 | 203 | 36256 | 35645 | Transposase and inactivated derivatives | Y. pestis | 179 | 188 | ZP_00797222 |
| 2 | ORF2 | 63 | 36371 | 36180 | IS_100_ ORF1 | Shigella dysenteriae | 60 | 63 | ZP_406030 |
| 3 | ORF3 | 300 | 37368 | 36466 | Transposase and inactivated derivatives | Y. pestis | 297 | 300 | ZP_01175356.1 |
| 4 | ORF4 | 126 | 37936 | 37556 | Hypothetical protein | ||||
| 5 | ORF5 | 76 | 38379 | 38609 | VagC | Salmonella enterica | 71 | 75 | ZP_271744.1 |
| 6 | ORF6 | 138 | 38606 | 39022 | VagD | K. pneumoniae CG43 | 135 | 138 | NP_943352.1 |
| 7 | ORF7 | 87 | 39040 | 39303 | Hypothetical protein LV173 | K. pneumoniae CG43 | 51 | 74 | NP_943463.1 |
| 8 | iroN | 724 | 41632 | 39458 | IroN | E. coli UTI89 | 686 | 725 | ZP_540133.1 |
| 9 | iroB | 371 | 42504 | 43619 | IroB | K. pneumoniae CG43 | 362 | 371 | NP_943394.1 |
| 10 | iroC | 1231 | 43706 | 47401 | IroC | K. pneumoniae CG43 | 1198 | 1214 | NP_943393.1 |
| 11 | iroD | 409 | 47507 | 48736 | IroD | K. pneumoniae CG43 | 404 | 409 | NP_943392.1 |
| 12 | pagO | 304 | 50655 | 49741 | PagO | K. pneumoniae CG43 | 275 | 300 | NP_943390.1 |
| 13 | ORF13 | 183 | 51279 | 50728 | Hypothetical protein | ||||
| 14 | rmpA | 210 | 52286 | 51654 | Regulator of mucoid phenotype | K. pneumoniae CG43 | 183 | 210 | AAL25252.1 |
| 15 | ORF15 | 54 | 52300 | 52464 | Hypothetical protein | ||||
| 16 | ORF16 | 217 | 52828 | 53481 | Hypothetical protein LV256 | K. pneumoniae CG43 | 175 | 179 | NP_943388.1 |
| 17 | ORF17 | 61 | 54304 | 54119 | Transposase and inactivated derivatives | Y. pestis | 61 | 61 | ZP_00797222.1 |
| 18 | ORF18 | 77 | 54789 | 55022 | Putative ATP/GTP-binding protein remnant | Y. pestis | 46 | 66 | NP_995420 |
| 19 | virB1 | 236 | 55095 | 55805 | Pilx1/VirB1-like protein | E. coli ECOR31 | 232 | 236 | AAP70283.1 |
| 20 | virB2 | 97 | 55805 | 56098 | VirB2-like protein | E. coli ECOR31 | 97 | 97 | AAP70284.1 |
| 21 | virB3-virB 4 | 912 | 56111 | 58849 | Pilx3-4/VirB3-4-like protein | E. coli ECOR31 | 897 | 912 | AAP70302.1 |
| 22 | virB5 | 234 | 58867 | 59571 | Pilx5/VirB5-like protein | E. coli ECOR31 | 224 | 229 | AAP70285.1 |
| 23 | virB6 | 359 | 59820 | 60899 | Pilx6/VirB6-like protein | E. coli ECOR31 | 296 | 354 | AAP70286.1 |
| 24 | virB8 | 227 | 61115 | 61798 | Type IV secretion system VirB8 component | Y. pestis | 226 | 227 | NP_995427.1 |
| 25 | virB9 | 302 | 61795 | 62703 | Type IV secretory pathway VirB9 component | Y. pestis | 293 | 301 | NP_995428.1 |
| 26 | virB10 | 416 | 62747 | 63997 | Pilx10/VirB10-like protein | E. coli ECOR31 | 402 | 416 | AAP70288.1 |
| 27 | virB11 | 341 | 63987 | 65012 | Type IV secretory pathway VirB11 component | Y. pestis | 335 | 341 | NP_995430.1 |
| 28 | ORF28 | 132 | 65009 | 65407 | Hypothetical protein YP_pCRY17 | Yersinia pestis | 118 | 132 | NP_995431.1 |
| 29 | ORF29 | 100 | 65443 | 65745 | YggA-like protein | E. coli ECOR31 | 69 | 101 | AAP70290.1 |
| 30 | ORF30 | 101 | 65788 | 66093 | Unknown | E. coli ECOR31 | 83 | 101 | AAP70307.1 |
| 31 | ORF31 | 100 | 66204 | 66506 | Unknown | E. coli ECOR31 | 99 | 100 | AAP70308.1 |
| 32 | mobB | 629 | 66773 | 68662 | MobB-like protein | E. coli ECOR31 | 607 | 629 | AAP70291.1 |
| 33 | mobC | 248 | 68672 | 69418 | MobC-like protein | E. coli ECOR31 | 212 | 245 | AAP70292.1 |
| 34 | ardC | 315 | 70561 | 69614 | Antirestriction protein | E. coli ECOR31 | 290 | 315 | AAP70295.1 |
| 35 | ORF35 | 271 | 72361 | 71546 | Hypothetical protein | ||||
| 36 | ORF36 | 115 | 73009 | 72662 | Hypothetical protein Nham_1368 | N. hamburgensis | 50 | 93 | ZP_576652.1 |
| 37 | ORF37 | 464 | 74403 | 73009 | UBA/THIF-type NAD/flavin adenine dinucleotide binding fold | N. hamburgensis | 235 | 459 | ZP_576653.1 |
| 38 | ORF38 | 375 | 75503 | 74376 | Hypothetical protein Nham_1370 | N. hamburgensis | 176 | 353 | ZP_576654.1 |
| 39 | ORF39 | 182 | 76146 | 75598 | Conserved hypothetical protein | Gram-negative bacterium | 93 | 175 | AAQ10297.1 |
| 40 | ORF40 | 367 | 77292 | 76189 | Hypothetical protein Shewmr7DRAFT_1704 | Shewanella sp. strain MR-7 | 147 | 365 | ZP_00855252.1 |
The middle region (ORF1 to ORF18) contained 12 ORFs (ORF5 to ORF16) that exhibited similarity to part of large plasmid pLVPK in K. pneumoniae strain CG43 (14). The vagC-vagD operon that was associated with virulence (32) and the _iroN-iroB_-_iroC_-iroD operon that was responsible for the uptake of a catecholate-type siderophore were also found (5, 6). The function of IroN (the receptor of siderophore) in the middle region was proven by iroN transfer to E. coli strain H5058, which allowed bacteria to uptake salmochelin siderophores (33) (data not shown). A regulator of the mucoid phenotype (rmpA) was also identified in this region (30). The ORFs located in the middle region were related to bacterial virulence, including iron acquisition and mucoviscosity.
The 3′ region contained 22 ORFs (ORF19 to ORF40) that exhibited similarity to the genes responsible for conjugative transfer in ICE_Ec1_ of E. coli strain ECOR31. Similar to ICE_Ec1_ of E. coli strain ECOR31, this region also contained three functional distinct segments (segments 3′-1, 3′-2, and 3′-3).
Segment 3′-1 (ORF19 to ORF31) contained nine ORFs (ORF19 to ORF27; virB1 to virB11) that were highly similar to region I of ICE_Ec1_. These ORFs were all in the same orientation and might comprise an operon encoding a putative mating pair formation system.
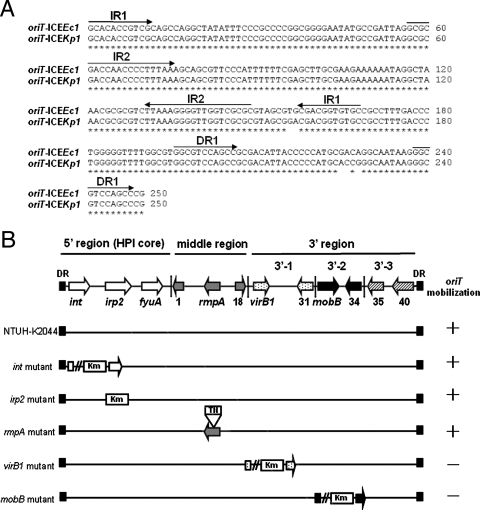
Segment 3′-2 contained three ORFs (ORF32 to ORF34) that were highly similar to region II of ICE_Ec1_. ORF32 to ORF34 (similar to mobB, mobC, and the anti-restriction protein gene) were associated with mobilization of the conjugative plasmid. Unlike ICE_Ec1_, a putative helicase gene (ORF17) was not found in this segment. A putative origin of transfer (oriT), which was located ∼140 bp upstream of the mobB gene, was also identified. The sequences of oriT in ICE_Kp1_ had five nucleotide substitutions compared with the sequences in ICE_Ec1_ (Fig. 3A). Like the oriT sequences of ICE_Ec1_, two inverted repeats and one direct repeat were present in the sequences of ICE_Kp1_. The putative duplicated nic site was also found in the second inverted repeat sequence. The ORFs in segment 3′-2 might encode the ability to process the oriT sequences and cleave the nic site for DNA mobilization.
FIG. 3.
Functional analysis of ICE_Kp1_. (A) Comparison of oriT sequences of ICE_Kp1_ and ICE_Ec1_. Asterisks indicate identical nucleotides in the oriT sequences of ICE_Kp1_ and ICE_Ec1_. The arrows above the sequences indicate the orientations and locations of two inverted repeats (IR1 and IR2) and one direct repeat (DR1). (B) DNA mobilization of ICE_Kp1_. The arrows in the diagram at the top indicate the simplified structure of ICE_Kp1_, which contains three regions. K. pneumoniae wild-type strain NTUH-K2044 and mutant strains harboring plasmid pACYC184-oriT were mated with E. coli strain HB101. A plus sign indicates that the pACYC184-oriT plasmid was mobilized to the recipient. A minus sign indicates that mobilization of plasmid pACYC184-oriT did not occur. DR, direct repeat.
Segment 3′-3 contained six ORFs (ORF 35 to ORF40) that exhibited similarity to hypothetical genes found in Nitrobacter hamburgensis. The sequences in this segment were very different from those in ICE_Ec1_.
Mating pair formation and DNA mobilization of ICE_Kp1_.
Five ORFs (int, irp2, rmpA, virB1, and mobB) were mutated to study the function of ICE_Kp1_. Plasmid pACYC184 carrying cloned oriT (plasmid pACYC184-oriT) was used to analyze the DNA mobilization ability. K. pneumoniae wild-type strain NTUH-K2044 (donor) transformed with plasmid pACYC184 or pACYC184-oriT was mated with E. coli strain HB101 (recipient). Transconjugants were selected with streptomycin and chloramphenicol to monitor the transfer of plasmid pACYC184 to the recipient. K. pneumoniae wild-type strains could mediate mobilization of the pACYC184-oriT plasmid at a frequency of 4.8 × 10−6 but not mobilization of the pACYC184 plasmid (<1 × 10−8). int, irp2, and rmpA mutant strains were able to transfer plasmid pACYC184-oriT (Fig. 3B). virB1 and mobB mutants were not able to mobilize plasmid pACYC184-oriT (<1 × 10−8). These results indicated that ICE_Kp1_ contains the virB1 and mobB genes responsible for DNA mobilization.
Precise excision and extrachromosomal circularization of ICE_Kp1_.
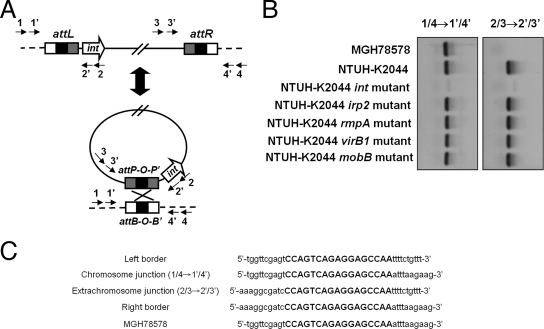
Generation of circular extrachromosomal intermediates by recombination between the direct repeats at the left and right ICE_Kp1_-chromosome junctions (attL and attR) was required for dissemination of ICEs (11, 12). A nested PCR assay using primers outside attL and attR was used to detect whether an extrachromosomal circular ICE_Kp1_ was formed (Table 2 and Fig. 4A). Excision and extrachromosomal circularization of ICE_Kp1_ in NTUH-K2044 were detected by PCR (Fig. 4B). Sequencing of the PCR products which represented the extrachromosomal and chromosomal junctions confirmed the precise excision and recircularization of ICE_Kp1_ in NTUH-K2044 (Fig. 4C). The 17-bp direct repeat identical to the ICE_Ec1_ sequence which was the attachment site (attO) for recombination was detected in the sequences of both extrachromosomal and chromosomal junctions. Formation of a circular extrachromosomal intermediate was not detected in the NTUH-K2044 int mutant (Fig. 4B). Knockout of the other four genes (irp2, rmpA, virB1, and mobB) in ICE_Kp1_ did not affect the recircularization (Fig. 4B). Therefore, the int gene encoding the integrase which catalyzes the recombination reaction was shown to be essential for recircularization of ICE_Kp1_.
FIG. 4.
Precise excision and extrachromosomal circularization of ICE_Kp1_. (A) Integration and excision model of ICE_Kp1_. The precise excision and extrachromosomal circularization of ICE_Kp1_ were mediated by recombination of attO sequences (indicated by a black box). Arrows 1, 1′, 2, 2′, 3, 3′, 4, and 4′ represent the primers outside the attL and attR sequences used to detect the precise excision and extrachromosomal circularization of ICE_Kp1_. (B) Extrachromosomal circular form of ICE_Kp1_ detected by nested PCR. PCR using primers 1 and 4 followed by nested PCR using primers 1′ and 4′ detected the attB site after excision (left gel). PCR using primers 2 and 3 followed by nested PCR using primers 2′ and 3′ detected the attP site in the extrachromosomal circular form (right gel). (C) Sequences of the chromosome and extrachromosomal junctions. Sequences of the chromosomal (1/4→1′/4′) and extrachromosomal (2/3→2′/3′) junctions were compared and aligned with the sequences of the left and right borders of ICE_Kp1_ and the sequences without an insertion in MGH78578. The 17-bp direct repeat is indicated by uppercase letters.
Self-transmission of ICE_Kp1_.
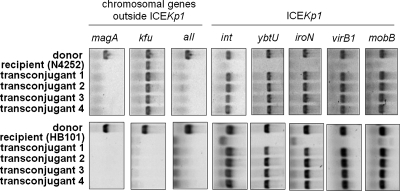
After the circular extrachromosomal intermediate was detected, whether ICE_Kp1_ in K. pneumoniae could be transmitted by conjugation was examined. ICE_Kp1_-negative (ICE_Kp1_−) K. pneumoniae strain N4252 was used to study the transmission of ICE_Kp1_ between K. pneumoniae strains. An NTUH-K2044 rmpA mutant strain (donor) in which the mobilization of oriT and excision of ICE_Kp1_ were not affected was mated with spontaneous rifampin-resistant K. pneumoniae mutant strain N4252 (recipient). Transconjugants were selected with rifampin and kanamycin to monitor the transfer of ICE_Kp1_ to the recipient. The transfer of ICE_Kp1_ to the recipient was confirmed by PCR (Fig. 5). Donor strain NTUH-K2044 was positive for the magA, kfu, and all genes and ICE_Kp1_. Recipient strain N4252 was negative for the magA and all genes and ICE_Kp1_ but positive for the kfu gene (29). The presence of the magA, kfu, and all genes, which were outside ICE_Kp1_, in four randomly selected transconjugants was the same as that in recipient strain N4252. The genes in ICE_Kp1_ (such as int, ybtU, iroN, virB1, and mobB) of the four transconjugants were found to be active for transfer. To further confirm that the transmission of ICE_Kp1_ was recA independent, recA mutant E. coli strain HB101 was used as the recipient. rmpA mutant strain NTUH-K2044 (donor) was mated with E. coli strain HB101 (recipient). Transconjugants were selected with streptomycin and kanamycin and then confirmed by PCR (Fig. 5). The absence of the magA, kfu, and all genes, which were outside ICE_Kp1_, and the presence of genes in the ICE_Kp1_ of four randomly selected transconjugants revealed that ICE_Kp1_ was transferred to E. coli strain HB101. The frequencies of ICE_Kp1_ transfer from K. pneumoniae strain NTUH-K2044 to K. pneumoniae strain N4252 or to E. coli strain HB101 were similar (∼4 × 10−6). In addition, NTUH-K2044 int, virB1, and mobB mutants all failed to transfer ICE_Kp1_ to strain N4252 (<1 × 10−8). These results confirmed that ICE_Kp1_ was transferred to the recipient and that the transfer of ICE_Kp1_ was recA independent. int, virB1, and mobB located in ICE_Kp1_ were essential for conjugative transfer of ICE_Kp1_.
FIG. 5.
Self-transmission of ICE_Kp1_. The transfer of ICE_Kp1_ from the NTUH-K2044 rmpA mutant (donor) to K. pneumoniae strain N4252 or E. coli strain HB101 (recipient) in four randomly selected transconjugants was analyzed by PCR. Chromosomal genes (magA, kfu, and all) outside ICE_Kp1_ were used to differentiate the donor and the recipient. Genes (int, ybtU, iroN, virB1, and mobB) located in ICE_Kp1_ were used to examine the presence of ICE_Kp1_.
Integration site of ICE_Kp1_.
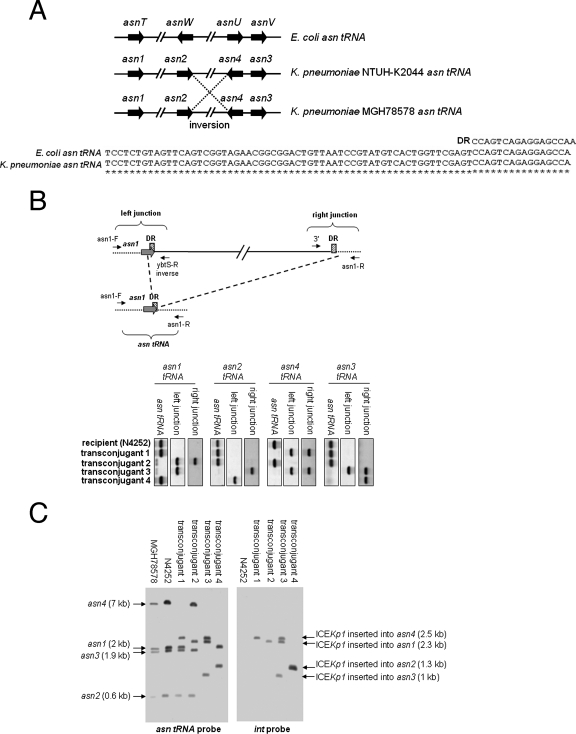
ICE_Kp1_ was located adjacent to the asn tRNA gene. The sequences of the asn tRNA gene in K. pneumoniae were the same as those in E. coli, and the genes have the same attachment site (Fig. 6A). An alignment of four asn tRNA genes of E. coli and K. pneumoniae strains NTUH-K2044 and MGH78578 is shown in Fig. 6A. Compared to K. pneumoniae strain NTUH-K2044, an inversion between the asn2 and asn4 tRNA genes was found in strain MGH78578. The insertion site of ICE_Kp1_ in the four N4252 transconjugants was examined further. Primers flanking the four asn tRNA genes and primers derived from the left and right ends of ICE_Kp1_ were designed to detect the insertion site of ICE_Kp1_ (Table 2 and Fig. 6B). If a 76-kb ICE_Kp1_ sequence was inserted into an asn tRNA gene, primer pairs flanking the asn tRNA gene should not amplify the asn tRNA gene; however, the primers flanking the asn tRNA gene combined with primers at the left and right ends of ICE_Kp1_ should detect the left and right junctions for insertion of ICE_Kp1_. By contrast, if ICE_Kp1_ was not inserted into an asn tRNA gene, the PCR for detecting the asn tRNA gene should remain positive, whereas the PCR for detecting the left and right junctions for the insertion of ICE_Kp1_ should be negative. The PCR results showed that ICE_Kp1_ was inserted into the asn4 and asn1 tRNA genes of transconjugants 1 and 2, respectively (Fig. 6B). Two insertions in the asn4 and asn3 tRNA genes of transconjugant 3 were detected. The left junction of another insertion in transconjugant 3 was detected in the asn1 tRNA gene, whereas the right junction of this insertion was detected in the asn2 tRNA gene. This insertion might have resulted in a deletion between the asn1 and asn2 tRNA genes. In transconjugant 4, the left junction of the ICE_Kp1_ insertion was detected in the asn2 tRNA gene, whereas the right junction was detected in the asn3 tRNA gene. Similar to what occurred in transconjugant 3, this insertion might have resulted in a deletion between the asn2 and asn3 tRNA genes. PCR to determine the genes in the predicted deletion regions in transconjugants 3 and 4 confirmed the occurrence of deletions during the insertion of ICE_Kp1_ (data not shown). The integration site of ICE_Kp1_ was also confirmed by Southern blotting by hybridization with asn tRNA and int gene probes (Fig. 6C). The sizes of four asn tRNA gene fragments in strain N4252 were same as those in strain MGH78578. The asn4 tRNA gene fragment (7 kb) was replaced by a 2.5-kb fragment which corresponded to the fragment that encompassed the left border (int) of ICE_Kp1_ in transconjugant 1. The asn1 tRNA gene fragment (2 kb) was replaced by a 2.3-kb fragment in transconjugant 2; therefore, the ICE_Kp1_ of transconjugant 2 was inserted in the asn1 tRNA gene. In transconjugant 3, three ICE_Kp1_ sequences were inserted in the asn1, asn4, and asn3 tRNA genes, and the asn2 tRNA gene fragment was not present. In transconjugant 4, ICE_Kp1_ was inserted in the asn2 tRNA gene, and the asn4 and asn3 tRNA gene fragments were not present. The insertion sites of ICE_Kp1_ in four transconjugants analyzed by PCR and Southern blotting were consistent. These results showed that ICE_Kp1_ could integrate into all four of the asn tRNA gene loci present on the chromosome. There could be more than one ICE_Kp1_ insertion into the chromosome, and insertion might result in a deletion between the asn tRNA genes.
FIG. 6.
Integration of ICE_Kp1_. (A) (Top) Alignment of four asn tRNA genes of E. coli and K. pneumoniae strains NTUH-K2044 and MGH78578. The arrows indicate the locations and orientations of asn tRNA genes. An inversion between the asn_2 and asn_4 tRNA genes was found in strain MGH78578 (indicated by dashed lines). (Bottom) Alignment of sequences of the 17-bp direct repeat (DR) and asn tRNA genes of E. coli and K. pneumoniae. Asterisks indicate identical nucleotides in the E. coli and K. pneumoniae asn tRNA genes. (B) (Top) Integration of ICE_Kp1 adjacent to asn tRNA genes (asn1 tRNA, for example). The asn tRNA is indicated by gray arrows, and the cross-hatched box indicates the 17-bp direct repeat (DR). The small arrows indicate the orientations of primers. The asn tRNA gene was detected by primers flanking the asn1 tRNA gene (asn1-F and asn1-R). The left junction of the ICE_Kp1 insertion was detected by a primer flanking the asn1 tRNA gene combined with a primer in the left end of ICE_Kp1_ (asn1-F and ybtS-R inverse). The right junction of the ICE_Kp1_ insertion was detected by a primer flanking the asn1 tRNA gene combined with a primer in the right end of ICE_Kp1_ (asn1-R and 3′). (Bottom) PCR analysis of the integration site of ICE_Kp1_ in the four N4252 transconjugants. (C) Southern hybridization of EcoRV-digested DNA from various strains with asn tRNA gene (left gel) and int (right gel) probes. The arrows indicate the positions of asn tRNA gene fragments with or without an ICE_Kp1_ insertion.
Mobility of ICE_Kp1_ in NTUH-K2044.
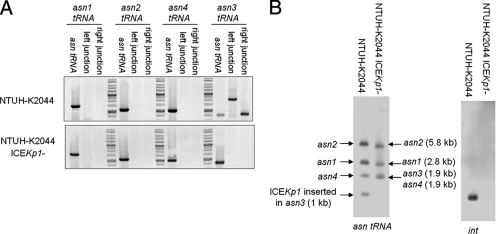
For the stock culture of strain NTUH-K2044 kept at room temperature, spontaneous loss of the entire ICE_Kp1_ sequence was observed in 1 of 300 colonies (NTUH-K2044 ICE_Kp1_−) (Fig. 7A). PCR using primers flanking the four asn tRNA genes combined with primers derived from the left and right ends of ICE_Kp1_ revealed that the ICE_Kp1_ sequence in strain NTUH-K2044 was located adjacent to the asn3 tRNA gene, and precise excision of ICE_Kp1_ was detected (Fig. 7A). A Southern blot hybridized with asn tRNA and int gene probes revealed that the asn3 tRNA gene fragment (1.9 kb) was replaced by a 1-kb fragment corresponding to the fragment that encompasses the left border of ICE_Kp1_ (Fig. 7B). The results of PCR and Southern blotting confirmed that ICE_Kp1_ of strain NTUH-K2044 was inserted into only the asn3 tRNA and did not mobilize to different asn tRNA loci.
FIG. 7.
Mobility of ICE_Kp1_ in strain NTUH-K2044. (A) PCR analysis of the location of ICE_Kp1_ in strains NTUH-K2044 and NTUH-K2044 ICE_Kp1_−. (B) Southern hybridization of EcoRV-digested DNA from strains NTUH-K2044 and NTUH-K2044 ICE_Kp1_− with asn tRNA gene (left gel) and int (right gel) probes. The arrows indicate the positions of asn tRNA gene fragments with or without ICE_Kp1_ insertion.
Prevalence of ICE_Kp1_in PLA and non-tissue-invasive K. pneumoniae strains.
The prevalence of ICE_Kp1_ in the 42 strains from patients with PLA and the 32 non-tissue-invasive strains was analyzed by PCR using primers for the fyuA (5′ region), iroN (middle region), and virB1 (3′ region) genes (Table 2 and Fig. 2). Primers outside the middle region (HPI 3′-F and virB1-F inverse) and primers derived from the left (orf3-R) and right (orf16-F) parts of the middle region were also used (Table 2 and Fig. 2). Seven strains isolated from patients with PLA were positive for fyuA, iroN, and virB1. The presence of the middle region was confirmed by PCR using primers outside the middle region combined with primers derived from the left and right parts of the middle region. Thirty-one PLA strains and five non-tissue-invasive strains were positive for fyuA and virB1 but not for iroN. PCR using primers outside the middle region confirmed the absence of the middle region and the connection of the 5′ and 3′ regions in these strains. Therefore, these strains positive for fyuA and virB1 were suggested to contain ICE_Kp1_ but not the middle region. Two non-tissue-invasive strains which were positive for fyuA but negative for iroN and virB1 were suggested to contain a small HPI. Therefore, the prevalence of ICE_Kp1_ (with or without the middle region) in PLA strains (38 of 42 strains) was higher than the prevalence in the non-tissue-invasive strains (5 of 32 strains). Previous results indicated that 35 of 42 strains from patients with PLA were serotype K1 (magA+), whereas 1 of 32 non-tissue-invasive strains was serotype K1 (17). All 36 serotype K1 strains contained this region. Moreover, the nine strains causing PLA with metastatic complications, such as meningitis or endophthalmitis, contained not only the magA, allS, and kfu/PTS (phosphotransferase) regions (29) but also the ICE_Kp1_.
DISCUSSION
Genomic heterogeneity and capsular serotypes both play important roles in the pathogenesis of K. pneumoniae causing liver abscesses (16, 18, 29). This study explored the mechanism of genomic heterogeneity by comparing the complete genome sequences of K. pneumoniae strains NTUH-K2044 and MGH78578. An approximately 76-kb DNA insertion adjacent to an asn tRNA gene was identified in K. pneumoniae strain NTUH-K2044. A novel ICE_Ec1_ of E. coli strain ECOR31 was recently shown to encode a functional mating pair formation system and a DNA-processing system (35). After induction of the integrase, the ICE_Ec1_ was shown to excise precisely from the chromosome and recircularize. Based on the similarity of sequences and the genetic structure of ICE_Ec1_, the insertion in K. pneumoniae strain NTUH-K2044 was suggested to be an ICE and was designated ICE_Kp1_. Mobilization of oriT sequences by ICE_Kp1_ and precise excision of ICE_Kp1_ were demonstrated. Furthermore, transfer of ICE_Kp1_ to another strain was directly demonstrated.
int in ICE_Kp1_ was proven to be essential for excision and integration of ICE_Kp1_. Recent studies showed that the HPI excision factor (Hef) and excisionase (XisHPI) of HPI were also required for the excision and integration of HPI (3, 28). A hypothetical protein composed of 61 amino acids (nucleotides 34091 to 34276 of the accession number AB298504 sequence of ICE_Kp1_) was identical to Hef in Y. pestis and Y. pseudotuberculosis and was considered to be a Hef homologue that might have similar functions. However, the actual function of this putative Hef protein encoded in ICE_Kp1_ needs to be investigated further.
Based on a comparison of sequences with the NCBI BLAST DNA database, the genome of K. pneumoniae strain MGH78578 did not have sequences homologous to sequences in ICE_Kp1_. Strain NTUH-K2044 did not have a homologue outside the ICE_Kp1_ region either. Integration of ICE_Kp1_ resulted from recombination of attO sequences present in asn tRNA loci of K. pneumoniae and not from recombination of other homologous sequences in ICE_Kp1_. ICE_Kp1_ contained the HPI (5′ region) and segments (segments 3′-1 and 3′-2) for the formation of the conjugative apparatus and for DNA mobilization which were highly similar to those of ICE_Ec1_. The middle region of ICE_Kp1_, which exhibited similarity to part of large plasmid pLVPK in K. pneumoniae strain CG43, was absent in ICE_Ec1_. There were similar 284-bp DNA sequences in the left (nucleotides 35579 to 35862 of the accession number AB298504 sequence) and right (nucleotides 54053 to 54336 of the accession number AB298504 sequence) parts of the middle region (Fig. 8). The 284-bp DNA sequences were also found to be similar to nucleotides 193069 to 193352 of the pLVPK plasmid. The large plasmid of strain NTUH-K2044 also contained these 284-bp DNA sequences (data not shown). These 284-bp DNA sequences flanking the middle region and on the large plasmid might be repeat structures for integration of the middle region from the large plasmid. However, we did not obtain evidence that ICE_Kp1_ is a larger progenitor of ICE_Ec1_. Putative virulence genes, such as the vagC-vagD and iroN-iroB_-iroC_-iroD operons and rmpA, were found in the middle region of ICE_Kp1 (5, 6, 30, 32). However, the middle region of ICE_Kp1 was detected in only 7 of the 42 strains isolated from patients with PLA. The middle region of ICE_Kp1_ was associated with iron acquisition and capsule regulation and could enhance virulence, but it seemed to be not essential for infection. Another difference between ICE_Kp1_ and ICE_Ec1_ was segment 3′-3. Region III of ICE_Ec1_ contained genes either with low homology to chromosomal genes found in Vibrio cholerae or with no significant similarity to sequences in the GenBank database. Segment 3′-3 of ICE_Kp1_ carried genes which exhibited similarity to the hypothetical genes found in N. hamburgensis. These results suggested that these two closely related ICEs might have become diverse during the evolution of bacteria.
FIG. 8.
Comparison of sequences from the left and right parts of the middle region and the large plasmid. Asterisks indicate identical nucleotides in nucleotides 35579 to 35862 and 54053 to 54336 in the accession number AB298504 sequence and nucleotides 193069 to 193352 in the pLVPK plasmid sequence.
The HPI of Yersinia species that carries the yersiniabactin siderophore system was essential for the virulence of bacteria (7, 9, 10). The HPI is widely distributed in the family Enterobacteriaceae (4, 34, 36), but the mechanism of HPI transfer remains unknown. A recent study suggested that a novel ICE_Ec1_ of E. coli strain ECOR31 was a mobilizable progenitor of the HPI (35). However, the HPI carried in the ICE was restricted to a single E. coli strain, and conjugative transfer of this ICE has not been demonstrated. Another study reported a possible mechanism of HPI dissemination based on site-specific recombination of the excised HPI with the attB_-presenting RP4 conjugative shuttle plasmid (2). The authors hypothesized that the general model for “trapping” the PAI on a conjugative plasmid bearing an attB site could also explain the dissemination of other PAIs. Here we report that an ICE_Kp1 carrying HPI was widely present in K. pneumoniae strains causing PLA. The conjugative transfer of ICE_Kp1_ to another strain was directly demonstrated in our study, suggesting that it was responsible for the dissemination of HPI in K. pneumoniae.
After conjugative transfer of the excised extrachromosomal ICE_Kp1_ circular form, the ICE_Kp1_ could integrate into any of the four asn tRNA loci on the chromosome. As observed for the HPI of Y. pseudotuberculosis, the excised form could be found to be inserted in any of the three asn tRNA loci present on the chromosome (8). However, ICE_Kp1_ in strain NTUH-K2044 did not mobilize to another asn tRNA locus, and ICE_Kp1_ was more often located adjacent to the asn3 tRNA gene in our strains (data not shown). PCR and Southern blot analysis of transconjugants revealed that more than one ICE_Kp1_ could be inserted into the chromosome (such as transconjugant 3) and that the insertion might result in a deletion between the asn tRNA genes (such as transconjugants 3 and 4). These findings implied that the recombination during the integration of ICEs might also facilitate genetic diversity. Most ICEs are thought to be transferred as single-stranded DNA (11, 12). ICEs would be nicked, and single strands of ICEs would be transferred to recipients. After replication, the donor and recipient would each contain a copy of the ICE. Therefore, a copy of ICE_Kp1_ was thought to be present in the donor after conjugation. However, we frequently observed spontaneous loss of the entire ICE_Kp1_ sequence in our strain. It was difficult to determine whether the donor strain lost or maintained the entire element in the chromosome during transfer of ICE_Kp1_.
In a previous study, 18% of clinical K. pneumoniae isolates contained the HPI region (26). ICE_Kp1_ carrying HPI was present in many of our clinical K. pneumoniae isolates (58%). ICE_Kp1_ was more prevalent in PLA strains than in non-tissue-invasive strains. The HPI and iroN-iroB_-iroC_-iroD operon responsible for iron acquisition may increase bacterial growth during infection (5-7). The rmpA gene has been shown to be involved in mucoviscosity, which plays an important role in the pathogenesis of bacteria (30). The acquisition of ICE_Kp1 should contribute the pathogenesis of K. pneumoniae causing PLA. Serotype K1 has been shown to be the most common serotype in K. pneumoniae strains causing PLA (17, 20). A previous pulsed-field gel electrophoresis typing study showed that all of our K1 strains had different clonal origins (13). Our 36 serotype K1 strains all contained the ICE_Kp1 region. Moreover, the nine strains causing PLA with metastatic complications contained not only three virulence-associated regions (magA, allS, and kfu/PTS), as previously described (29), but also the ICE_Kp1_ region. The clustering of virulence-associated regions in our strains might indicate that together these regions play an important role in the pathogenesis of this invasive K. pneumoniae infection.
Acknowledgments
This study was supported by grants from the National Science Council, by postdoctoral fellowship PD9503 from the National Health Research Institute, and by a grant from the Liver Disease Prevention and Treatment Research Foundation, Taiwan.
Footnotes
▿
Published ahead of print on 2 November 2007.
REFERENCES
- 1.Abbott, S. 2003. Klebsiella, Enterobacter, Citrobacter, Serratia, Plesiomonas, and other Enterobacteriaceae, p. 684-700. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology Press, Washington, DC.
- 2.Antonenka, U., C. Nolting, J. Heesemann, and A. Rakin. 2005. Horizontal transfer of Yersinia high-pathogenicity island by the conjugative RP4 attB target-presenting shuttle plasmid. Mol. Microbiol. 57727-734. [DOI] [PubMed] [Google Scholar]
- 3.Antonenka, U., C. Nolting, J. Heesemann, and A. Rakin. 2006. Independent acquisition of site-specific recombination factors by asn tRNA gene-targeting genomic islands. Int. J. Med. Microbiol. 296341-352. [DOI] [PubMed] [Google Scholar]
- 4.Bach, S., A. de Almeida, and E. Carniel. 2000. The Yersinia high-pathogenicity island is present in different members of the family Enterobacteriaceae. FEMS Microbiol. Lett. 183289-294. [DOI] [PubMed] [Google Scholar]
- 5.Baumler, A. J., T. L. Norris, T. Lasco, W. Voight, R. Reissbrodt, W. Rabsch, and F. Heffron. 1998. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J. Bacteriol. 1801446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumler, A. J., R. M. Tsolis, A. W. van der Velden, I. Stojiljkovic, S. Anic, and F. Heffron. 1996. Identification of a new iron regulated locus of Salmonella typhi. Gene 183207-213. [DOI] [PubMed] [Google Scholar]
- 7.Bearden, S. W., J. D. Fetherston, and R. D. Perry. 1997. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect. Immun. 651659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchrieser, C., R. Brosch, S. Bach, A. Guiyoule, and E. Carniel. 1998. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol. Microbiol. 30965-978. [DOI] [PubMed] [Google Scholar]
- 9.Buchrieser, C., M. Prentice, and E. Carniel. 1998. The 102-kilobase unstable region of Yersinia pestis comprises a high-pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J. Bacteriol. 1802321-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchrieser, C., C. Rusniok, L. Frangeul, E. Couve, A. Billault, F. Kunst, E. Carniel, and P. Glaser. 1999. The 102-kilobase pgm locus of Yersinia pestis: sequence analysis and comparison of selected regions among different Yersinia pestis and Yersinia pseudotuberculosis strains. Infect. Immun. 674851-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46601-610. [DOI] [PubMed] [Google Scholar]
- 12.Burrus, V., and M. K. Waldor. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155376-386. [DOI] [PubMed] [Google Scholar]
- 13.Chang, S. C., C. T. Fang, P. R. Hsueh, Y. C. Chen, and K. T. Luh. 2000. Klebsiella pneumoniae isolates causing liver abscess in Taiwan. Diagn. Microbiol. Infect. Dis. 37279-284. [DOI] [PubMed] [Google Scholar]
- 14.Chen, Y. T., H. Y. Chang, Y. C. Lai, C. C. Pan, S. F. Tsai, and H. L. Peng. 2004. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 337189-198. [DOI] [PubMed] [Google Scholar]
- 15.Chiu, C. T., D. Y. Lin, and Y. F. Liaw. 1988. Metastatic septic endophthalmitis in pyogenic liver abscess. J. Clin. Gastroenterol. 10524-527. [DOI] [PubMed] [Google Scholar]
- 16.Chou, H. C., C. Z. Lee, L. C. Ma, C. T. Fang, S. C. Chang, and J. T. Wang. 2004. Isolation of a chromosomal region of Klebsiella pneumoniae associated with allantoin metabolism and liver infection. Infect. Immun. 723783-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chuang, Y. P., C. T. Fang, S. Y. Lai, S. C. Chang, and J. T. Wang. 2006. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 193645-654. [DOI] [PubMed] [Google Scholar]
- 18.Fang, C. T., Y. P. Chuang, C. T. Shun, S. C. Chang, and J. T. Wang. 2004. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 199697-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang, F. C., N. Sandler, and S. J. Libby. 2005. Liver abscess caused by magA+ Klebsiella pneumoniae in North America. J. Clin. Microbiol. 43991-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fung, C. P., F. Y. Chang, S. C. Lee, B. S. Hu, B. I. Kuo, C. Y. Liu, M. Ho, and L. K. Siu. 2002. A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut 50420-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golia, P., and M. Sadler. 2006. Pyogenic liver abscess: Klebsiella as an emerging pathogen. Emerg. Radiol. 1387-88. [DOI] [PubMed] [Google Scholar]
- 22.Haag, H., K. Hantke, H. Drechsel, I. Stojiljkovic, G. Jung, and H. Zahner. 1993. Purification of yersiniabactin: a siderophore and possible virulence factor of Yersinia enterocolitica. J. Gen. Microbiol. 1392159-2165. [DOI] [PubMed] [Google Scholar]
- 23.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1726557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain, R., M. C. Rivera, J. E. Moore, and J. A. Lake. 2002. Horizontal gene transfer in microbial genome evolution. Theor. Popul. Biol. 61489-495. [DOI] [PubMed] [Google Scholar]
- 25.Ko, W. C., D. L. Paterson, A. J. Sagnimeni, D. S. Hansen, A. Von Gottberg, S. Mohapatra, J. M. Casellas, H. Goossens, L. Mulazimoglu, G. Trenholme, K. P. Klugman, J. G. McCormack, and V. L. Yu. 2002. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg. Infect. Dis. 8160-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koczura, R., and A. Kaznowski. 2003. Occurrence of the Yersinia high-pathogenicity island and iron uptake systems in clinical isolates of Klebsiella pneumoniae. Microb. Pathog. 35197-202. [DOI] [PubMed] [Google Scholar]
- 27.Lederman, E. R., and N. F. Crum. 2005. Pyogenic liver abscess with a focus on Klebsiella pneumoniae as a primary pathogen: an emerging disease with unique clinical characteristics. Am. J. Gastroenterol. 100322-331. [DOI] [PubMed] [Google Scholar]
- 28.Lesic, B., S. Bach, J. M. Ghigo, U. Dobrindt, J. Hacker, and E. Carniel. 2004. Excision of the high-pathogenicity island of Yersinia pseudotuberculosis requires the combined actions of its cognate integrase and Hef, a new recombination directionality factor. Mol. Microbiol. 521337-1348. [DOI] [PubMed] [Google Scholar]
- 29.Ma, L. C., C. T. Fang, C. Z. Lee, C. T. Shun, and J. T. Wang. 2005. Genomic heterogeneity in Klebsiella pneumoniae strains is associated with primary pyogenic liver abscess and metastatic infection. J. Infect. Dis. 192117-128. [DOI] [PubMed] [Google Scholar]
- 30.Nassif, X., N. Honore, T. Vasselon, S. T. Cole, and P. J. Sansonetti. 1989. Positive control of colanic acid synthesis in Escherichia coli by rmpA and rmpB, two virulence-plasmid genes of Klebsiella pneumoniae. Mol. Microbiol. 31349-1359. [DOI] [PubMed] [Google Scholar]
- 31.Parke, D. 1990. Construction of mobilizable vectors derived from plasmids RP4, pUC18 and pUC19. Gene 93135-137. [DOI] [PubMed] [Google Scholar]
- 32.Pullinger, G. D., and A. J. Lax. 1992. A Salmonella dublin virulence plasmid locus that affects bacterial growth under nutrient-limited conditions. Mol. Microbiol. 61631-1643. [DOI] [PubMed] [Google Scholar]
- 33.Reissbrodt, R., and W. Rabsch. 1988. Further differentiation of Enterobacteriaceae by means of siderophore-pattern analysis. Zentralbl. Bakteriol. Mikrobiol. Hyg. Ser. A 268306-317. [DOI] [PubMed] [Google Scholar]
- 34.Schubert, S., S. Cuenca, D. Fischer, and J. Heesemann. 2000. High-pathogenicity island of Yersinia pestis in Enterobacteriaceae isolated from blood cultures and urine samples: prevalence and functional expression. J. Infect. Dis. 1821268-1271. [DOI] [PubMed] [Google Scholar]
- 35.Schubert, S., S. Dufke, J. Sorsa, and J. Heesemann. 2004. A novel integrative and conjugative element (ICE) of Escherichia coli: the putative progenitor of the Yersinia high-pathogenicity island. Mol. Microbiol. 51837-848. [DOI] [PubMed] [Google Scholar]
- 36.Schubert, S., A. Rakin, H. Karch, E. Carniel, and J. Heesemann. 1998. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect. Immun. 66480-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, J. H., Y. C. Liu, S. S. Lee, M. Y. Yen, Y. S. Chen, J. H. Wang, S. R. Wann, and H. H. Lin. 1998. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 261434-1438. [DOI] [PubMed] [Google Scholar]