Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase (original) (raw)
. Author manuscript; available in PMC: 2008 Jun 18.
Published in final edited form as: Science. 2007 Nov 15;318(5856):1637–1640. doi: 10.1126/science.1150034
Abstract
Cells respond to DNA double-strand breaks (DSBs) by recruiting factors such as the DNA-damage mediator protein MDC1, p53-binding protein 1 (53BP1) and the breast cancer susceptibility protein BRCA1 to sites of damaged DNA. Here, we reveal that the ubiquitin ligase RNF8 mediates ubiquitin conjugation and 53BP1 and BRCA1 focal accumulation at sites of DNA lesions. Moreover, we establish that MDC1 recruits RNF8 via phospho-dependent interactions between the RNF8 forkhead-associated (FHA) domain and motifs in MDC1 that are phosphorylated by the DNA-damage activated protein kinase ATM. We also show that depletion of the E2 enzyme UBC13 impairs 53BP1 recruitment to sites of damage suggesting that it cooperates with RNF8. Finally, we reveal that RNF8 promotes the G2/M DNA damage checkpoint and resistance to ionizing radiation. These results demonstrate how the DNA-damage response is orchestrated by ATM-dependent phosphorylation of MDC1 and RNF8-mediated ubiquitination.
DNA DSBs are highly cytotoxic lesions; and to ensure that they are repaired with minimal impact on genome stability, cells mount a complex DNA-damage response (DDR) that includes the spatial reorganization of DSB repair and signaling proteins into subnuclear structures – ionizing radiation-induced foci (IRIF) – that surround DSB sites (1, 2). Most IRIF formation depends on phosphorylation of the histone variant H2AX (to form γH2AX) by the DNA-PK and ATM protein kinases (3-6). The γH2AX epitope is bound by MDC1 (7-10) that then promotes IRIF formation by other proteins, including 53BP1, Nijmegen-breakage-syndrome protein NBS1 and BRCA1 (11, 12). BRCA1 recruitment to IRIF requires its interaction with the ubiquitin-binding protein RAP80 (13-16) that interacts with lysine 63 (K63)-linked poly-ubiquitinated protein(s) at sites of DNA damage (15). Here, we identify RNF8 as the prime ubiquitin ligase for poly-ubiquitination at DSB sites, define its functional importance in the DDR, and establish how RNF8 is recruited to sites of DNA damage via interactions with MDC1.
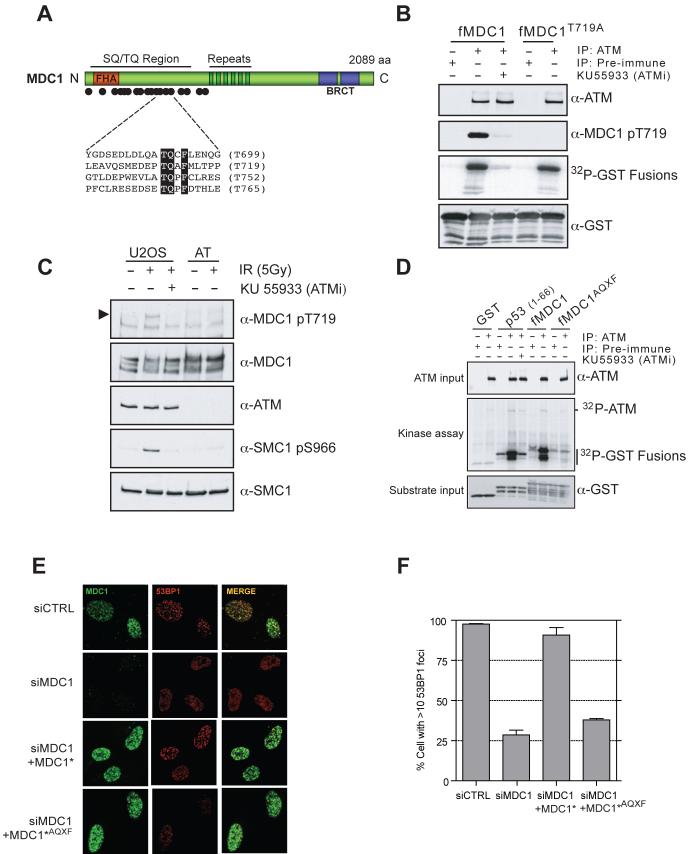
MDC1 is phosphorylated in an ATM-dependent manner in response to ionizing radiation (IR) (11, 12). Potential ATM target sites (consensus S/T-Q) cluster in the MDC1 N-terminus, the most notable being four adjacent motifs conforming to the consensus TQXF (Figs. 1A and S1). Significantly, antibodies raised against a peptide encoding phospho-T719 (Fig. S2A) indicated that it is targeted by ATM in vitro (Fig. 1B), and in vivo (Fig. 1C and S2B). However, in vitro assays with bacterially-expressed MDC1 fragments revealed that T719 was not the only site of ATM modification. Phosphorylation was only abolished when an “AQXF” mutant protein bearing threonine-to-alanine substitutions in all four TQXF motifs was used as substrate (Fig. 1D). These data and the recent identification of another TQXF site (T752) as an ATM target (17) therefore imply that MDC1 TQXF motifs are likely all modified by ATM and may function redundantly with one another.
Fig. 1.
The MDC1 TQXF motifs are ATM targets required for 53BP1 IRIF. (A) Domain architecture of MDC1, with ATM consensus sites (dots). (B) MDC1 T719 is phosphorylated by ATM in vitro. GST-MDC1679-778 (fMDC1) or GST-MDC1679-778-T719A (fMDC1T719A) were incubated with anti-ATM or pre-immune complexes in the presence or absence of the ATM kinase inhibitor KU55933. (C) MDC1 TQXF motifs are phosphorylated by ATM in vivo. Lysates from U2OS or AT22IJE (AT) cells were immunoblotted with the indicated antibodies. The arrowhead points to phospho-MDC1. (D) The MDC1 TQXF motifs are phosphorylated by ATM in vitro. Kinase reactions with anti-ATM or pre-immune complexes and the following substrates: GST, GST-p531-56 (p53), fMDC1 or GST-MDC1679-778-AQXF (fMDC1AQXF) as in (B). (E-F) The TQXF cluster is required for 53BP1 IRIF. U2OS cells expressing siRNA-resistant GFP-MDC1 (MDC1*) or GFP-MDC1AQXF (MDC1*AQXF) were transfected with siRNA against MDC1 (siMDC1) or luciferase (siCTRL) and post-irradiation (5 Gy) were stained for MDC1 and 53BP1 (E) and quantitated (F; N=4 +/-SD).
To address the function of the MDC1 TQXF motifs, we used siRNA to deplete endogenous MDC1 in human U2OS cells stably expressing siRNA-resistant wild-type MDC1 or the MDC1 AQXF mutant. While both wild-type and AQXF mutant proteins formed IRIF and supported IRIF formation by NBS1 (Fig. S3), only wild-type MDC1 promoted effective IRIF formation by 53BP1 (Figs. 1E-F), revealing that ATM-mediated phosphorylation of MDC1 facilitates 53BP1 focus formation. Indeed, although 53BP1 IRIF formation can occur in ATM-deficient AT cells (18, 19), we found that pharmacological inhibition of ATM impaired 53BP1 focus formation (Fig. S4). While these results suggested that 53BP1 might directly bind the phosphorylated TQXF motifs, we were unable to detect such interactions (Fig. 3B), suggesting that MDC1-dependent 53BP1 IRIF formation is likely mediated by an additional factor.
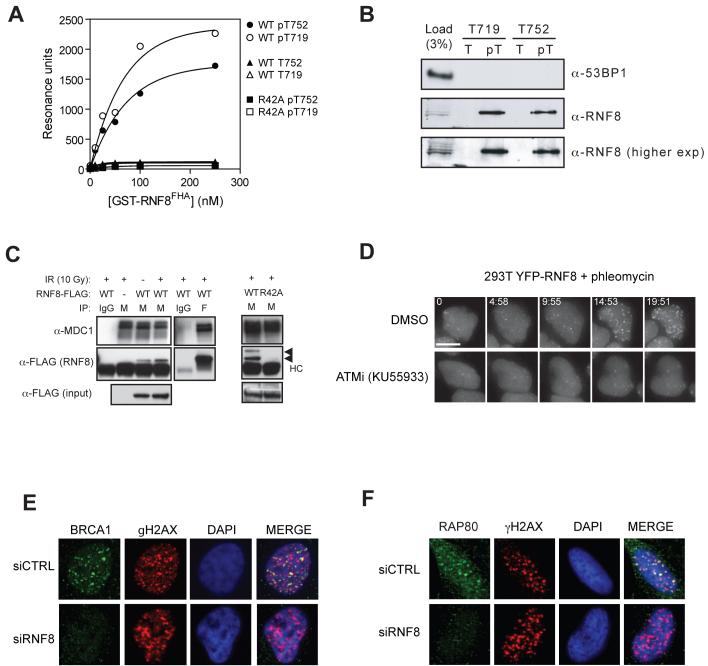
Fig. 3.
RNF8 mediates BRCA1-RAP80 IRIF via a physical interaction with MDC1. (A) Binding curves of GST-RNF8FHA (WT) the R42A mutant obtained by surface plasmon resonance with peptides corresponding to MDC1 epitopes surrounding T719, T752 or their phosphorylated counterparts (pT719 and pT752). (B) Peptide pull-downs of HeLa nuclear extracts with immobilized peptides; phosphorylated (pT) or unphosphorylated (T), encompassing MDC1 T719 or T752 residues. (C) RNF8 interacts with MDC1 in vivo. Extracts from 293T cells mock-transfected (-) or transfected with RNF8-FLAG (WT) or the R42A FHA mutant were immunoprecipitated (IP) with anti-MDC1 (M), anti-FLAG (F) or normal mouse IgG and probed for MDC1 or RNF8-FLAG as indicated. Arrowheads indicate the RNF8-specific signal. Note that RNF8 appears modified when interacting with MDC1. HC, IgG heavy chains. (D) Time-lapse microscopy of 293T cells stably expressing YFP-RNF8 pre-incubated with ATM inhibitor KU55933 or DMSO and treated with the radio-mimetic drug phleomycin (1.5 mg/ml) for the indicated times (min:sec). Three-dimensional image datasets were computationally deconvolved and shown as two-dimensional projections. Scale bar = 10μm. (E-F) Irradiated (10 Gy) HeLa cells transfected with the indicated siRNAs were stained with anti-γH2AX, anti-BRCA1 (D) or anti-RAP80 (E) antibodies 1hr post-IR.
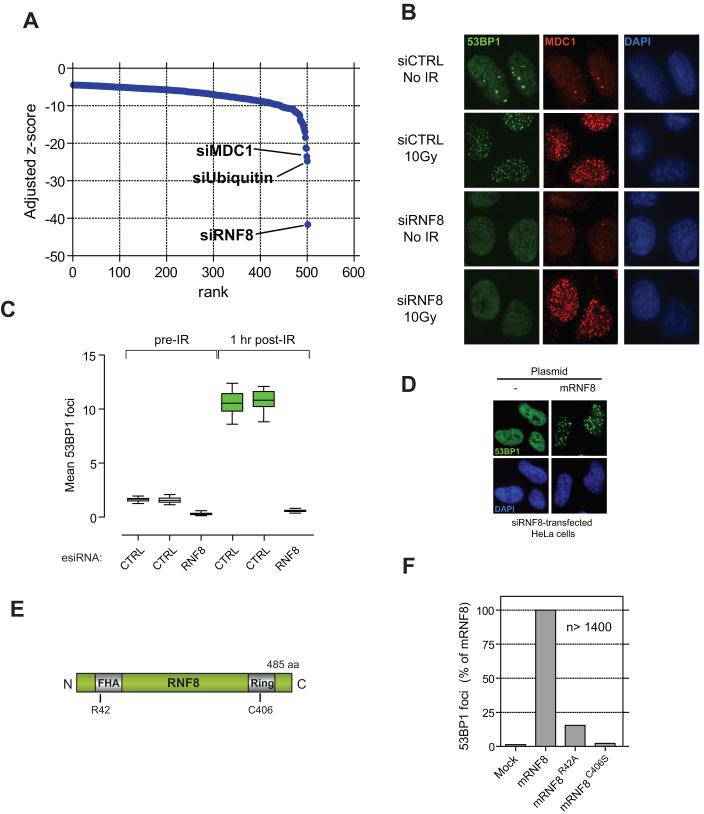
To uncover proteins that regulate 53BP1 focus formation, we mined an ongoing RNAi screen that employs 53BP1 focus formation as a readout (Figs. 2A and S5A-C). Strikingly, the three siRNAs that most potently impaired 53BP1 focus formation targeted transcripts encoding MDC1, ubiquitin, and the RNF8 E3 ubiquitin ligase that was recently shown to control mitosis (20) (Fig. 2A). Notably, we found that depletion of RNF8 by siRNA or enzyme-generated siRNA pools (esiRNAs) (21) abrogated 53BP1 foci while preserving MDC1 IRIF, thus phenocopying the MDC1AQXF mutation (Figs. 2B-C and S6A). Introduction of RNAi-resistant murine RNF8 into cells transfected with RNF8 siRNA restored 53BP1 focus formation (Figs. 2D and S7), indicating the specificity of RNF8 depletion and confirming that RNF8 promotes 53BP1 IRIF formation.
Fig. 2.
RNF8 promotes 53BP1 IRIF assembly. (A) Ranking by z-score of 500 siRNAs giving the least 53BP1 foci from an ongoing siRNA screen examining 53BP1 focus formation. See Fig. S5 for details. (B) 53BP1 and MDC1 immunofluorescence of U2OS cells transfected with control siRNA (siCTRL) or RNF8 siRNA (siRNF8) and fixed prior (no IR) or 1 hr following 10 Gy irradiation. (C) Quantitation of 53BP1 IRIF in HeLa cells transfected with the indicated esiRNAs (CTRL against luciferase). N=16, data displayed using box-and-whisker plots. (D) Transfection of siRNAi-resistant murine RNF8 in HeLa cells restores 53BP1 IRIF formation caused by RNF8 depletion. (E) Domain architecture of RNF8. Numbering refers to murine RNF8. (F) Rescue of RNF8 depletion by murine RNF8 but not the FHA- (R42A) or RING finger-mutated (C406) mutants. 53BP1 foci were quantitated 1 hr after 10 Gy irradiation in siRNF8-treated cells. Data for wild-type RNF8 were set at 100%. Over 1400 cells per condition were counted.
RNF8 possesses an N-terminal FHA domain (22) and a C-terminal RING-finger domain responsible for its ubiquitin ligase activity (Fig. 2E) (23). By complementing the human RNF8 RNAi phenotype with siRNA-resistant murine RNF8, we established that mutations in either the FHA domain (RNF8R42A) or RING-finger domain (RNF8C403S) abrogated the ability of RNF8 to support 53BP1 IRIF formation (Figs. 2E-F and S7). As FHA domains bind phosphothreonine-bearing epitopes, the above data suggested that RNF8 might interact with ATM-phosphorylated MDC1. Indeed, the RNF8 FHA domain, but not an FHA-domain mutant (RNF8R42A), bound specifically and in a phospho-dependent manner to TQXF peptides corresponding to MDC1 Thr719 or Thr752 (Fig. 3A). Furthermore, these phosphorylated TQXF peptides retrieved RNF8 from HeLa nuclear extracts, while the corresponding unphosphorylated peptides did not (Fig. 3B). Notably, epitope-tagged RNF8, but not RNF8R42A, was detected in MDC1 immunoprecipitates in a manner that was enhanced by irradiation (Fig. 3C), suggesting that MDC1 might recruit RNF8 to sites of DNA damage. To test this idea, we generated cell lines stably expressing RNF8 fused to yellow fluorescent protein (YFP). As shown by analysis of of live cells (Fig. 3D) and fixed samples (Fig. S8), addition of the radio-mimetic drug phleomycin or irradiation resulted in the accumulation of YFP-RNF8 into foci that colocalized with γH2AX. In support of such events being brought about by ATM-mediated phosphorylation of MDC1, YFP-RNF8 recruitment was impaired by the selective ATM inhibitor KU55933 (24) (Fig. 3D).
As was the case for MDC1 foci, we found that γH2AX, NBS1 and FANCD2 foci formed efficiently in RNF8-depleted cells (Fig. S8). In contrast, BRCA1 IRIF formation was impaired upon RNF8 depletion (Fig. 3F). BRCA1-interacting protein RAP80 is required for BRCA1 IRIF formation (13-16) and RAP80 itself forms IRIF in a manner that involves interactions between its ubiquitin-interacting motif (UIM) and K63-linked ubiquitin chains at sites of DNA lesions (13-16). Because RNF8 is a ubiquitin ligase required for BRCA1 IRIF, we speculated that RNF8 might mediate IRIF formation by RAP80 and conjugated ubiquitin. Indeed, RNF8 was essential for IRIF formation in both cases (Figs. 3F, 4A and S10A). As RAP80 is not required for 53BP1 focus formation after IR (14) and 53BP1 is not needed for RAP80 IRIF (Fig. S10B), these results indicated that RNF8 acts downstream of MDC1 to promote at least two types of IRIF: those containing 53BP1 and those containing BRCA1. In support of this model, mutation of the TQXF motifs in MDC1 also impaired BRCA1 and conjugated ubiquitin IRIF (Fig. S11).
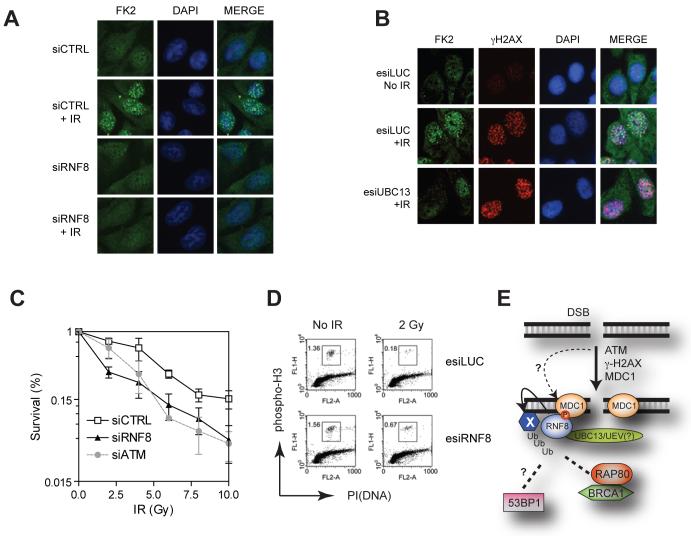
Fig. 4.
RNF8 cooperates with UBC13 to mediate ubiquitin IRIF and a functional DDR. (A-B) Irradiated (10 Gy) U2OS cells transfected with the indicated siRNA (A) or esiRNAs (B) were stained with FK2 anti- conjugated ubiquitin and γH2AX antibodies to assess IRIF. Cells were fixed 1 hr post-IR. (C) Clonogenic survival of HeLa cells transfected with siRNAs against ATM (siATM), RNF8 (siRNF8) or a non-targeting control (siCTRL). N=3 +/- SEM. (D) G2/M checkpoint analysis of U2OS cells transfected with the indicated esiRNAs. Fixed mock-treated (No IR) or irradiated (2 Gy) cells were stained with an anti-phospho-histone H3 antibody and propidium iodide (PI). The percentage of mitotic cells was determined by fluorescence-activated cell sorting. (E) Model of RNF8 action at DSBs. RNF8 is recruited by ATM-phosphorylated MDC1 to DSBs where it ubiquitinates an unknown protein (X), recruiting RAP80-BRCA1 and allowing 53BP1 to recognize methylated histones, as suggested by the RNF8-dependent recruitment of the 53BP1 Tudor domain to DSB sites (Fig. S13). Our results also suggest the presence of a MDC1-independent pathway of RNF8 action (dashed line) that mediate 53BP1 and BRCA1 IRIF formation.
RNF8 can bind to multiple E2-conjugating enzymes to catalyze both K63-linked and K48-linked ubiquitin chains (23, 25). When we screened a panel of thirteen E2 enzymes by RNAi and quantitative microscopy, only UBC13 depletion markedly impaired 53BP1 IRIF formation (Fig. S12). UBC13 depletion also impaired IRIF by conjugated ubiquitin (Fig. 4B). Entirely consistent with our data, genetic ablation of UBC13 in DT40 cells abrogates BRCA1 and ubiquitin IRIFs (26). As UBC13 is the only known E2-conjugating enzyme that catalyzes K63-linked poly-ubiquitination, these findings are consistent with data indicating that ubiquitin IRIF form in part via K63-linked poly-ubiquitination (15, 26). Moreover, as UBC13 physically interacts with RNF8 to catalyze K63-linked ubiquitin chains (27), the available data imply that RNF8 is responsible for formation of K63-linked ubiquitin chains at DSB sites.
Consistent with RNF8 playing an important function in the DDR, we found that RNF8 depletion caused IR hyper-sensitivity in clonogenic cell-survival assays (Fig. 4C). Furthermore, a greater proportion of RNF8-depleted cells progressed into M-phase following irradiation than did cells transfected with a control esiRNA indicating that RNF8 enforces the G2/M DNA-damage checkpoint (Fig. 4D).
In conclusion, our results identify mammalian RNF8 as an important component of the DDR. Specifically, RNF8 binds to ATM-target motifs on MDC1, thus recruiting RNF8 to DSB sites. RNF8 then triggers the formation of poly-ubiquitin chains that promote recruitment of the RAP80-BRCA1 complex and 53BP1 to DSB sites, thereby enhancing DNA-damage checkpoint events and promoting cell survival (Fig. 4E).
Supplementary Material
Supporting Online Material
Acknowledgments
We thank members from the Durocher and Jackson laboratories for input on the manuscript and Milijana Vojvodic for experimental assistance. We also thank AC Gingras for the stable YFP-RNF8 line, KuDOS Pharmaceuticals for providing inhibitors and reagents and Abcam for the MDC1 pT719 antibody. This work was supported by grants from the Canadian Institutes of Health Research (CIHR) to DD and by funding from Cancer Research UK and the European Union (SPJ). NKK is a CIHR post-doctoral fellow and an EIRR21st training program alumnus; FDS holds a Terry-Fox studentship from the National Cancer Institute of Canada; SN is a Gail-Posluns Fellow. DD is a Canada Research Chair (Tier II) in Proteomics, Functional Genomics and Bioinformatics.
References
- 1.Bartek J, Lukas J. Curr Opin Cell Biol. 2007;19:238. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Maser RS, Monsen KJ, Nelms BE, Petrini JH. Mol Cell Biol. 1997;17:6087. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stiff T, et al. Cancer Res. 2004;64:2390. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 4.Paull TT, et al. Curr Biol. 2000;10:886. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 5.Celeste A, et al. Nat Cell Biol. 2003;5:675. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 6.Bekker-Jensen S, et al. J Cell Biol. 2006;173:195. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stucki M, et al. Cell. 2005;123:1213. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 8.Lou Z, et al. Mol Cell. 2006;21:187. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 9.Lee MS, Edwards RA, Thede GL, Glover JN. J Biol Chem. 2005;280:32053. doi: 10.1074/jbc.C500273200. [DOI] [PubMed] [Google Scholar]
- 10.Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J. J Cell Biol. 2005;170:201. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. Nature. 2003;421:961. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 12.Goldberg M, et al. Nature. 2003;421:952. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 13.Yan J, et al. Cancer Res. 2007;67:6647. doi: 10.1158/0008-5472.CAN-07-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim H, Chen J, Yu X. Science. 2007;316:1202. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 15.Sobhian B, et al. Science. 2007;316:1198. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang B, et al. Science. 2007;316:1194. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuoka S, et al. Science. 2007;316:1160. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 18.DiTullio RA, Jr., et al. Nat Cell Biol. 2002;4:998. doi: 10.1038/ncb892. [DOI] [PubMed] [Google Scholar]
- 19.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. J Cell Biol. 2000;151:1381. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plans V, Guerra-Rebollo M, Thomson TM. Oncogene. 2007 doi: 10.1038/sj.onc.1210782. [DOI] [PubMed] [Google Scholar]
- 21.Kittler R, Heninger AK, Franke K, Habermann B, Buchholz F. Nat Methods. 2005;2:779. doi: 10.1038/nmeth1005-779. [DOI] [PubMed] [Google Scholar]
- 22.Durocher D, Henckel J, Fersht AR, Jackson SP. Mol Cell. 1999;4:387. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- 23.Ito K, et al. Eur J Biochem. 2001;268:2725. doi: 10.1046/j.1432-1327.2001.02169.x. [DOI] [PubMed] [Google Scholar]
- 24.Hickson I, et al. Cancer Res. 2004;64:9152. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 25.Bothos J, Summers MK, Venere M, Scolnick DM, Halazonetis TD. Oncogene. 2003;22:7101. doi: 10.1038/sj.onc.1206831. [DOI] [PubMed] [Google Scholar]
- 26.Zhao GY, et al. Mol Cell. 2007;25:663. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 27.Plans V, et al. J Cell Biochem. 2006;97:572. doi: 10.1002/jcb.20587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Online Material