Human tankyrases are aberrantly expressed in colon tumors and contain multiple epitopes that induce humoral and cellular immune responses in cancer patients (original) (raw)
Abstract
Purpose
Tankyrases 1 and 2 are telomere-associated poly(ADP-ribose) polymerases (PARP) that can positively regulate telomere elongation and interact with multiple cellular proteins. Recent reports implicated tankyrases as tumor antigens and potential targets of anticancer treatment. We examined expression of tankyrases in colon tumors and immune response to these enzymes in patients with different types of cancer.
Methods
mRNA and protein expression was evaluated by quantitative real-time RT-PCR and Western blotting, respectively. Humoral immune response to recombinant tankyrases was investigated by modified enzyme-linked immunoassays. Cellular immune response was analysed by ELISPOT and 51Cr release assays.
Results
We found that both mRNA and protein levels of tankyrase 2 (TNKL) are upregulated in colon tumors. In contrast, protein level of tankyrase 1 (TNKS) is downregulated, while mRNA level shows variable changes. More than a quarter of colon cancer patients develop humoral immune response to at least one of the two tankyrases. In this study we mapped common and unique B-cell epitopes located in different domains of the two proteins. Additionally, we present evidence for T-cell responses both to epitopes that are unique for TNKL and to those shared between TNKL and TNKS.
Conclusion
Our study favors a biomarker usage of antibody response to tankyrases. Spontaneous CD8+ T-cell responses to these enzymes are rare and further investigation is needed to evaluate tankyrases as potential targets for cancer immunotherapy.
Keywords: SEREX, Telomerase-interacting proteins, Epitope mapping
Introduction
Human telomeres are regions of repetitive DNA at the chromosome ends, containing up to 15 kb of tandem repeats of the hexanucleotide TTAGGG [33]. Telomere shortening by an average of 50–150 base pairs with each cell division limits the number of divisions for normal cells [19]. To compensate for the loss of telomeres, cells with high proliferative capacity express a two-component ribonucleoprotein enzyme, telomerase, which is active in the majority of cancer cells [46]. Telomere length control in telomerase-positive cells is provided by TTAGGG repeat binding factor 1 (TRF1), a small dimeric protein, which inhibits the action of telomerase at the ends of individual telomeres [54]. The binding of human TRF1 to telomeres can be inhibited as a result of ADP-ribosylation of TRF1 by tankyrases 1 (TNKS) and 2 (TNKL), two highly homologous telomeric poly(ADP-ribose) polymerases (PARP) [6, 52]. PARP catalytic activity of tankyrases is also essential for correct separation of sister chromatids during mitosis [2, 13].
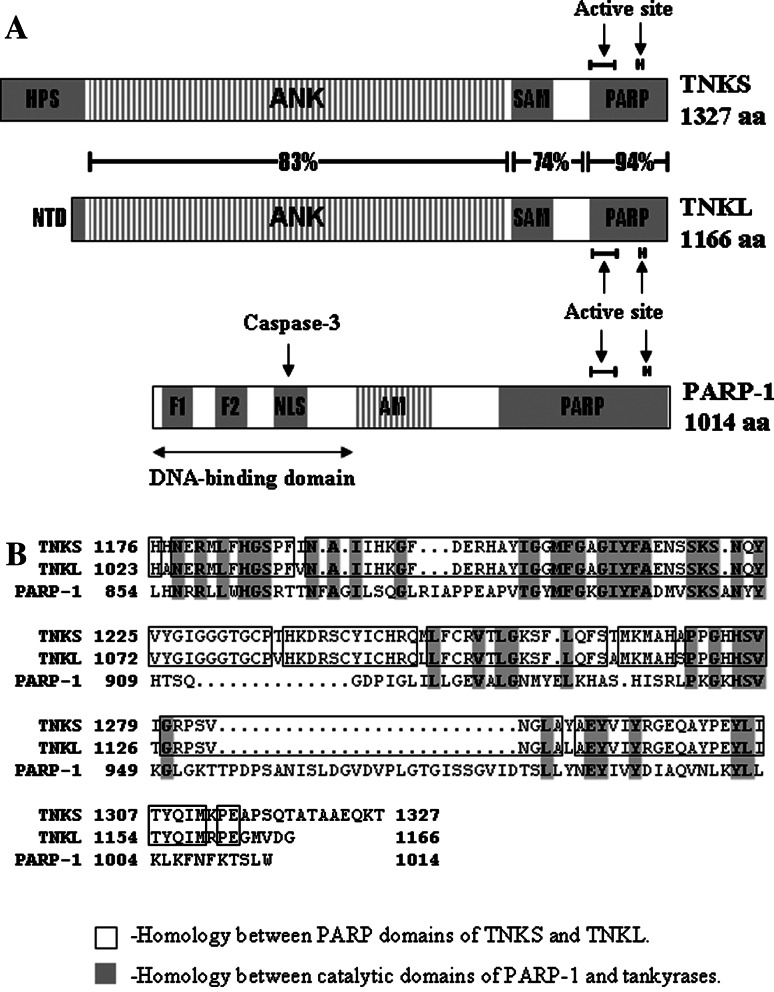
Each tankyrase consists of four distinct domains, three of which are highly homologous between the two proteins (Fig. 1). The long ankyrin (ANK) domain consisting of 24 consecutive ANK repeats is responsible for interactions with various partners and target proteins, including TRF1, Epstein Barr nuclear antigen 1 (EBNA1), nuclear/mitotic apparatus protein (NuMA) and myeloid cell leukemia-1 (Mcl-1) proteins in the nucleus, the insulin-responsive aminopeptidase IRAP in the Golgi, the adaptor protein Grb14 and the formin-binding protein 17 (FBP17) in the cytoplasm, and a 182-kDa tankyrase-binding protein (TAB182) which localizes to both the nucleus and the cytoplasm [1, 3, 11, 15, 22, 28, 40, 41, 45, 50, 52]. Some of the tankyrase partners bind to ANK repeats and share common tankyrase binding motif RXXPDG (IRAP, TAB182 and NuMA) while TRF1 contains a variant motif RGCADG with decreased affinity to tankyrases [40]. The sterile α motif (SAM) also mediates protein–protein interactions and is responsible for homo- and hetero-multimerization of tankyrases [7, 8]. The C-terminal part of each tankyrase contains the PARP domain responsible for poly(ADP-ribosyl)ation of tankyrases themselves and the numerous tankyrase-interacting proteins [7].
Fig. 1.
Immunogenic human poly(ADP-ribose)polymerases. a Domain structure of tankyrases and PARP-1. Tankyrases: HPS histidine-proline-serine rich domain of TNKS, NTD N-terminal domain of TNKL, ANK ankyrin-repeat domain, SAM sterile α motif, PARP poly(ADP-ribose) polymerase domain [6]. PARP-1 F1 and F2 zinc fingers, NLS nuclear localization signal motif, AM auto modification domain, PARP poly(ADP-ribose)polymerase domain [10]. b Alignment of catalytic domains of human tankyrases and PARP-1
The possible link between tankyrases and cancer has not been clearly established. It was reported that TNKS overexpression has a pro-survival effect and provides protection against genotoxin-induced cell death [60]. Overexpressed TNKS appears to inhibit PARP-1 [60], a ubiquitously expressed member of PARP family, responsible for many cellular processes involved in suppression of tumorigenesis, including DNA repair and genomic stability [53]. Knock-down of TNKS by siRNA brings tumor cells to mitotic arrest [2, 13] while combined chemical inhibition of tankyrases (affecting their PARP activity) and telomerase caused telomere shortening and apoptosis in telomerase-positive target cells [43, 44]. Even in the case of shortened telomeres and acquired resistance to telomerase inhibitors, administration of PARP inhibitors affecting tankyrases could restore telomerase sensitivity to inhibition and result in further loss of telomeres [44]. Although tankyrase expression is not restricted to tumor cells and differs among malignant tumors, telomerase activity is observed predominantly in cancer cells [16, 23, 29–31, 56, 57]. Therefore, simultaneous targeting of tankyrases and telomerase may be effective and specific for cancer cells [43, 44].
TNKL was identified as a tumor antigen by serological screening of recombinant tumor-derived libraries (SEREX method [36,39]) in independent studies on breast cancer [25], meningioma [32] and acute lymphoblastic T-cell leukemia [12]. Additionally, humoral immune response against TNKS in meningioma patients was recently reported [5]. PARP-1 (see Fig. 1) was repeatedly identified as an autoantigen without a cancer-related serological profile [9, 10, 24, 34, 42, 58, 59]. In this study, we investigated expression patterns of tankyrases in colon tumors and characterized humoral and cellular immune responses to TNKL and TNKS in cancer patients.
Materials and methods
Specimens
Tissue and sera samples from cancer patients who had undergone surgical resection of the colon were obtained from Blokhin Cancer Research Center (Moscow, Russia), Municipal Hospital # 24 (Moscow, Russia) and Krankenhaus Nordwest (Frankfurt, Germany). Normal sera obtained during routine diagnostic procedures were provided by the outpatient clinic of the Russian Ministry of Economics and Krankenhaus Nordwest. Tissue samples were either snap-frozen in liquid nitrogen and stored at −80°C or immediately stabilized in a tissue storage reagent (RNAlater, Ambion, Inc., Austin, TX) and stored at −20°C. Sera were aliquoted and stored at −80°C.
The study protocols were approved by the local ethics committees of the participating clinical centers. The research activities at Moscow State University were designated exempt from Institutional Review Board (IRB) reviews.
Real-time RT-PCR
Total RNA was extracted from tissue samples by TRI reagent (Sigma, Saint Louis, MO), converted to cDNA with random nanomers by ImProm-II Reverse Transcriptase (Promega, Madison, WI) and amplified in LightCycler 2.0 (Roche Applied Science, Mannheim, Germany) using LightCyclet FastStart DNA Master SYBR Green I pre-made PCR mix (Roche Applied Science). Primers used for PCR were the following: TNKS forward GCAGTACCACCAGCACAATC, TNKS reverse AGGGGAGGATGGAGAGGAAG, TNKL forward ATCTGCTCTGCCCTCTTGTTACAA, TNKL reverse GCTAAAATCTACTCCTGGAACCTC, β-actin forward GCTACGAGCTGCCTGACGG, β-actin reverse GATGGAGTTGAAGGTAGTTTCG. Program for PCR was following: denaturation for 10 min at 95°C and 50 cycles: 95°C for 10 s, 60°C for 10 s and 72°C for 30 s, acquisition at 82°C.
Affinity precipitation of TNKL
Bacterial vectors pGEX-4T1 (Amersham, Little Chalfont, Buckinghamshire, UK) encoding GST and GST-IRAP (aa 86–101) were expressed in E.coli DH5 α and recombinant proteins were purified on GST Bind Resins (Novagen, San Diego, CA) as described [14] except that sarkosyl was not used and that Triton X-100 (Sigma) concentration was limited to 1%. Tissue samples were homogenized in TBS with 1% Triton X-100, 0.05% NP40, 1 mM phenylmethylsulfonyl fluoride (Sigma) and a protease inhibitor cocktail (Roche Applied Science). Homogenates were sedimented by centrifugation and cleared lysates were incubated overnight at 4°C with GST Bind Resin preincubated with GST or GST-IRAP. GST Bind Resin was washed five times with 20 volumes of TBS. The lysates after affinity-precipitation were subjected to Western blot analysis. Normalization was performed by silver staining of lysates separated in 8% SDS-PAGE.
Western blotting
Proteins were separated in 8% SDS-PAGE, transferred to a Hybond-C Extra nitrocellulose membrane (Amersham) and incubated overnight with one of following antibodies: polyclonal rabbit anti-TNKS and goat anti-TNKL antibodies purchased from Santa Cruz Biotechnology (Santa Cruz, CA), polyclonal rabbit anti-TNKL antibodies raised against a GST-fused fragment of human TNKL aa 813–881. Then blots were washed 3–4 times, incubated for 1 h with appropriate HRP-conjugated antibodies (goat-anti-rabbit from Jackson ImmunoResearch (West Grove, PA; 1:20 000) or mouse-anti-goat antibodies (Santa Cruz; 1:20 000)) and developed using an enhanced chemoluminescence kit (ECL) (Amersham). Staining of the gels was performed using the Silver staining kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol.
Proteins
(His)6-tagged proteins were cloned in pET-vectors from Novagen (pET-17b/TNKL and pET-28b/TNKS) and pBK-CMV vector from Stratagene, La Jolla, CA (both tankyrases and their fragments, Fig. 4). Transduced E.coli strains Rosetta(DE3)pLys (for pET-vectors) and XLOLR (for pBK-CMV) were grown in LB/Mg media in the presence of 50 μg/ml kanamycin (pET-28b and pBK-CMV) or ampicillin (pET-17b) to OD600 of 0.35. Protein expression was induced with 1 mM (pET-28b and pBK-CMV) or 0.4 mM (pET-17b) IPTG for 4 h at 37°C prior to harvesting the bacteria by centrifugation. Recombinant (His)6-tagged proteins were batch-purified on Ni-NTA Agarose (Qiagen, Valencia, CA, USA) in denaturing urea buffer according to the manufacturer’s recommendations. BSA was purchased from Sigma.
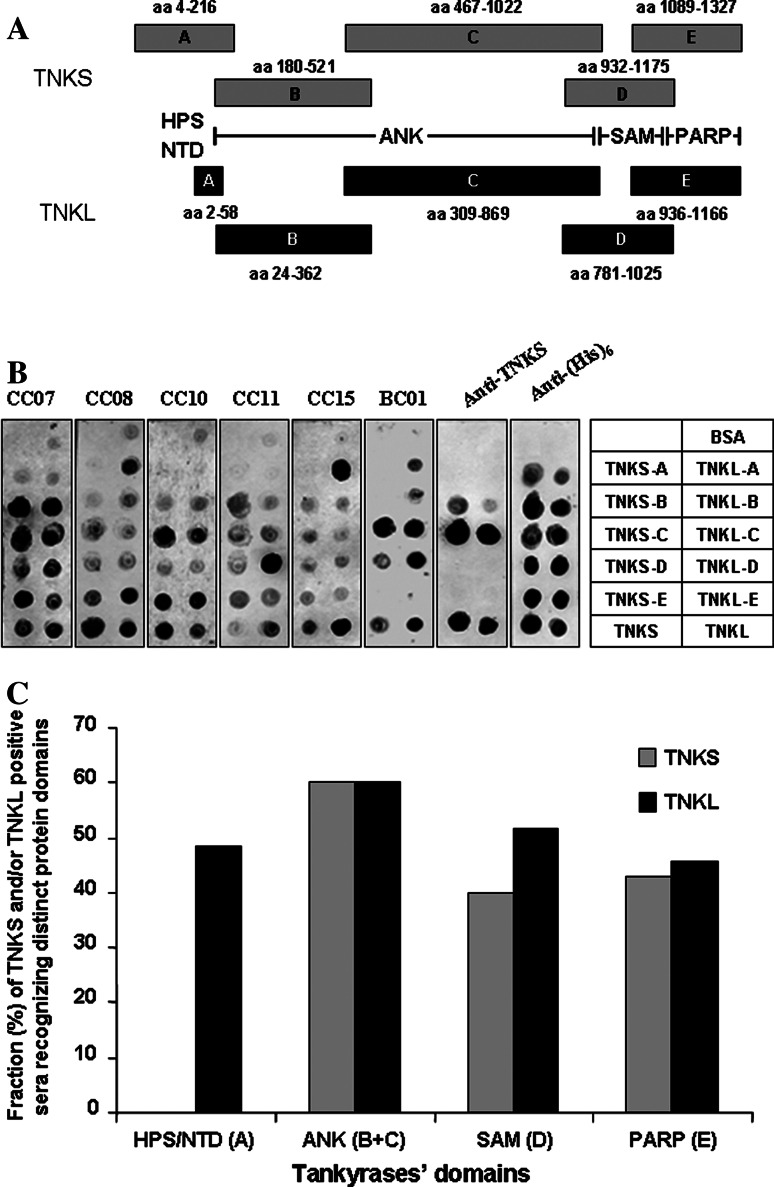
Fig. 4.
Sera of cancer patients recognize various protein domains of tankyrases. a Tankyrase fragments tested for serological reactivity. HPS histidine–proline–serine rich domain of TNKS, NTD N-terminal domain of TNKL, ANK ankyrin-repeat domain, SAM sterile α motif, PARP poly(ADP-ribose) polymerase domain [6]. TNKS-A to -E and TNKL-A to -E: tested tankyrases fragments. b Mini-array of recombinant protein domains and full-length tankyrases. Loading BSA (bovine serum albumin) −negative control, CC colon cancer patients, BC01 breast cancer patient. c Frequencies of humoral immune response to different tankyrases domains. Total of 38 pre-selected TNKL and/or TNKS positive sera tested (colon cancer19 patients, breast cancer 5 patients, ovarian cancer and head and neck cancer: in groups of 3 patients, lung cancer, renal cancer and melanoma: in groups of 2 patients, cervical cancer and prostate cancer: one patient)
Protein mini-arrays
Protein arrays were prepared as described earlier [48]. Briefly, 0.5 μg aliquots of recombinant proteins were applied to Hybond-C Extra nitrocellulose membrane in 1 μl of elution buffer (8 M urea; 0.1 M NaH2PO4; 0.01 M Tris·Cl; pH 4.5). Ten nanograms of total human IgG and 10 ng of rabbit anti-human polyclonal antibodies were applied to the same membrane in 1 μl of 50% glycerol in TBS as positive controls. One microgram of BSA in 1 μl of elution buffer was applied as negative control. Arrays were dried, washed two times in TBST, blocked by 5% NFDM/TBST for 1 h at room temperature, incubated with pre-treated patients’ sera and with alkaline phosphatase-conjugated goat anti-human Fcγ secondary antibodies (Jackson Immunoresearch; 1:2000), and developed by nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) (Sigma). Loading was normalized by mouse monoclonal antibodies against (His)6 (Novagen; 0.2 μg/ml) and TNKS (BD Pharmingen, San Jose, CA; 0.5 μg/ml).
Sera pre-treatment and analysis of serological mini-arrays of recombinant tumor antigens (SMARTA) were performed as described earlier [26, 47].
In vitro sensitization of effector CD8+ T cells with peptides
Peripheral blood mononuclear cells were separated from heparinized blood over Ficoll density gradient. CD8+ T lymphocytes were selected by magnetic cell sorting using the miniMACS system (Miltenyi Biotec, Bergisch Gladbach, Germany). For in vitro presensitization, CD8-/CD4- PBMC were irradiated (3,000 rad), pulsed with HLA-A2-restricted TNKL peptides (10 μg/ml, manufactured by Academisch Ziekenhuis Leiden, Netherlands) for 1 h in serum-free medium, washed and used as antigen presenting cells (APC) to stimulate CD8+ T lymphocytes. The APC/CD8+ cell ratio was 2.5–3:1. Effector cells were restimulated with peptide (1 μg/ml) weekly. Cells were cultured in RPMI 1640 medium supplemented with 10 mM HEPES buffer, l-arginine (242 mg/l), l-asparagine (50 mg/l), l-glutamine (300 mg/l), penicillin (10 IU/ml), streptomycin (100 μg/ml), 1% non-essential amino acids, 10% human serum and IL-2 (10 ng/ml), further referred to as TCM.
ELISPOT assay
Flat-bottomed 96-well nitrocellulose plates were incubated overnight at 4°C with 100 μl of anti-interferon-gamma antibody (5 μg/ml; Hölzel Diagnostic, Köln, Germany), washed with PBS and blocked with 10% equine serum for 1 h at 37°C. Presensitized CD8+ T cells (2.5 × 104) and target cells (5 × 104) were added to each well and incubated for 16 h at 37°C in TCM without IL-2. Plates were then washed 6 times with 0.05% Tween 20/PBS, and biotinylated anti-interferon-gamma detection antibody was added at a concentration of 0.5 μg/ml. After incubation for 2 h at 37°C, plates were washed and incubated with ABC-AP Vectastain (Vector, Burlingame, CA) for 1 h. Plates were washed and developed with NBT/BCIP.
51Cr release assay
Specific cytotoxicity against peptide-pulsed T2 cells was determined in standard 51Cr release assays as described [21]. Briefly, target cells were labeled with 100 μCi Na2 51CrO4 for 2 h and pulsed with peptide (10 μg/ml) for 1 h. Target cells (1 × 103/well) were incubated in V-bottomed 96-well plates with effector cells at various E/T ratios for 4 h at 37°C. T-cells were preincubated with unlabeled K562 cells at a concentration of 40:1 to block non-specific effector reactions. 51Cr release into the supernatant was measured in a gamma counter.
Quantification and statistical analysis
Densitometry analysis was performed by ImageQuant Version 5.2 software (Molecular Dynamics, Sunnyvale, CA). Fisher’s Exact Test was performed by online tool “Consultancy for Research and Statistics” (http://home.clara.net/sisa/fisher.htm), the Mann–Whitney _U_-test (Wilcoxon rank sum test) was performed by Leon Avery’s online tool “Elementary Statistics for Biologists” (http://elegans.swmed.edu/~leon/stats/utest.html). Student’s t test was calculated using Microsoft Excel program.
Results
TNKL mRNA levels are elevated in tumors of early stage colon cancer patients
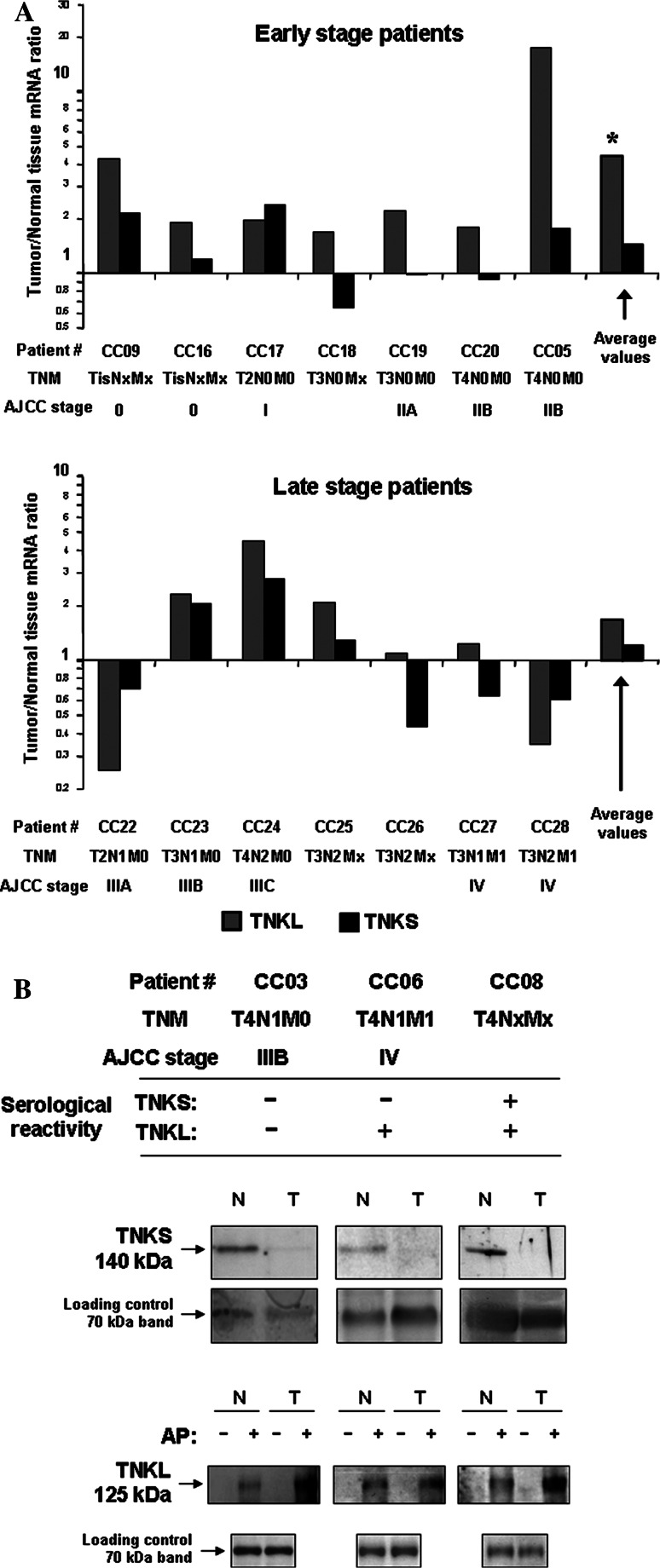
In order to explore the expression patterns of tankyrases in colon cancer patients, we evaluated both TNKS and TNKL protein and mRNA levels in tumor samples and in adjacent normal tissues. We analyzed paired RNA samples derived from tissues of 14 patients with colorectal carcinoma by quantitative real-time PCR and found that TNKL was upregulated (threshold 1.5-fold change) in tumors of 71% of patients tested for colon cancer, while TNKS did not show a consistent pattern and was either upregulated or downregulated with comparable frequencies (36 and 29%, respectively). The average ratio of mRNA expression of tankyrases in paired samples (tumor vs. normal colon tissue) was higher for TNKL (3.04 ± 4.28) than for TNKS (1.31 ± 0.75). The expression levels of TNKL mRNA might be related to the stage of disease. Particularly, we observed more frequent overexpression of TNKL in colon tumors from patients with earlier stages of disease (0–II, 100% cases) compared to later stages (III–IV, 43% cases). In samples from patients with earlier stages of colon cancer, the overexpression of TNKL in tumors was statistically significant according to Student’s test for paired samples (P < 0.005).
Colon tumors contain less TNKS but more TNKL as compared to normal colon tissues
Proteins derived from tissue samples of 19 patients with colorectal cancer (including 11 paired samples analyzed for mRNA expression) were further analyzed by Western blotting. Results representing samples from patients with different serological status are shown in Fig. 2b).
Fig. 2.
Expression levels of human tankyrases in colon tumors. a Quantitative real-time RT-PCR. TNKS and TNKL mRNA levels ratios (tumor vs. normal tissue) in patients with different stages of colorectal cancer are shown. *P < 0.005 according Student’s t test for paired samples. b Western blotting. Upper rows Immunoblotting with anti-TNKS and anti-TNKL antibodies, N normal colon, T tumor tissue. AP (only for TNKL) affinity precipitation. −GST (negative control), _+_GST-IRAP. Lower rows Normalization of loading by silver staining (representative major protein bands of approx. molecular weight 70 kDa are shown). The results of one of two representative experiments are shown
TNKS protein levels were decreased in about half (10 from 19, 52.6%) of tested tumor samples as compared to normal colon tissues (Fig. 2b). In seven cases (36.8%) TNKS protein level was not changed (including two cases when TNKS was undetectable in both tumor and normal tissue) and only in two cases (10.5%) TNKS was upregulated in tumors. Overall, TNKS was more often dowregulated, than upregulated in colon tumors (P < 0.02 according exact Fisher’s test).
TNKL protein levels in tissues were poorly detectable by conventional Western blotting due to a lower sensitivity of available TNKL-specific immunoreagents (data not shown). Therefore, we performed affinity-precipitation of TNKL from homogenized normal and tumor tissues from nine colon cancer patients using GST-fused IRAP fragment (aa 86–101) which contained tankyrase binding motif RXXPDG [3, 41]. We found that TNKL protein levels in tumors were significantly higher (on average 2.39 ± 0.81-fold, P < 0.001) than in adjacent normal tissues in contrast to those of TNKS (Fig. 2b). Somewhat surprisingly, protein expression levels of TNKS and TNKL did not correlate with the stage of disease or with the serological status of patients.
Tankyrases are serologically defined cancer-associated antigens for colon and other types of cancer
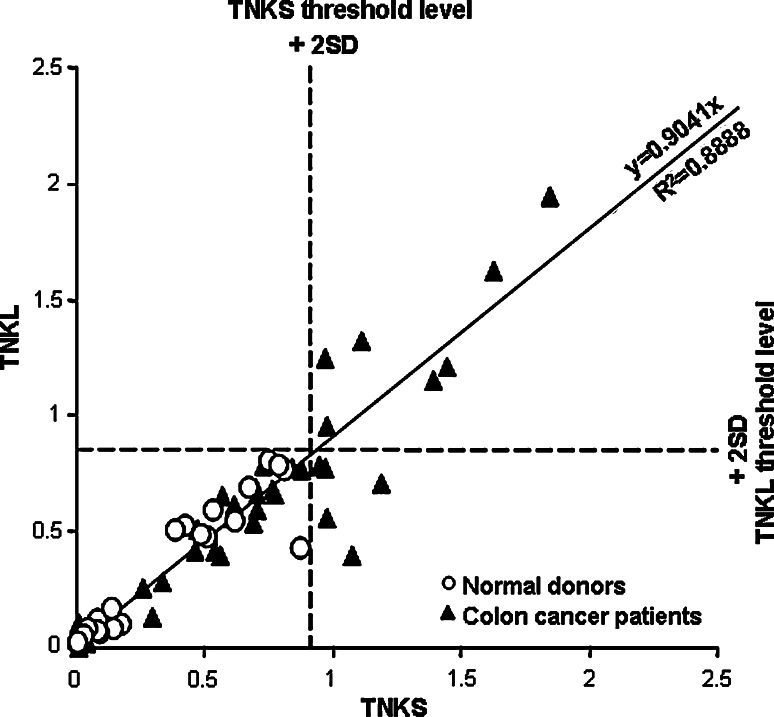
Initial characterization of the SEREX-derived truncated TNKL clone MO-BC-203 [25] (lacking first 1,150 bp/257 aa) by SMARTA arrays [26] demonstrated significantly more frequent serological response in patients with breast (P < 0.05) and colon cancer (P < 0.0001) compared to normal donors (Table 1). There was no difference in frequencies of humoral immune response to truncated TNKL in patients with different stages of colon cancer (data not shown). For more robust serological analysis, we cloned and expressed full-length tankyrases and performed all subsequent analyses of reactivity against recombinant purified proteins in a mini-array format [48]. In such assay, both tankyrases demonstrated frequent and intensive immune reaction with sera of colon cancer patients (Table 2; Fig. 3). Scatter plot of intensities of serological reactions to TNKS and TNKL demonstrated very strong linear correlation (linear regression trend y = 0.9041x, R 2 = 0.8886) suggesting cross-reactivity between TNKS and TNKL (Fig. 3).
Table 1.
Serological responses to truncated human TNKL (clone MO-BC-203) in patients with different types of cancer assayed in SEREX-based SMARTA format
| Positive/total (%) | |
|---|---|
| Normal donors | 6/228 (2.6) |
| Breast cancer | 6/60* (10.0) |
| Colon cancer | 17/116** (14.7) |
| Head and neck cancer | 2/31 (6.5) |
| Melanoma | 3/45 (6.7) |
| Ovarian cancer | 3/75 (4.0) |
| Renal cancer | 2/51 (3.9) |
| Cervical cancer | 1/23 (4.3) |
Table 2.
Serological responses to full-length purified human tankyrases
| TNKS | TNKL | |||
|---|---|---|---|---|
| Positive/total | % | Positive/total | % | |
| Threshold level defined as: average value for sera from normal donors + 2 standard deviations (SD) | ||||
| Normal donors | 0/26 | 0.0% | 0/26 | 0.0% |
| Colon cancer patients | 12/42** # | 28.6% | 7/42* # | 16.7% |
| Threshold level defined as: average value for sera from normal donors + 3 standard deviations (SD) | ||||
| Normal donors | 0/26 | 0.0% | 0/26 | 0.0% |
| Colon cancer patients | 4/42 # | 9.5% | 6/42 # | 14.3% |
Fig. 3.
Serological cross-reactivity between TNKS and TNKL. Axes: relative (normalized to positive controls, see Table 2) intensities of serological reactions of tankyrases. Serological reactions to TNKS and TNKL demonstrate correlation designated as linear regression trend. Threshold levels (designated by dashed lines) separating serologically positive and negative samples: average value for sera from normal donors + 2 standard deviations
Sera of cancer patients recognize multiple domains of tankyrases
Tankyrases consist of four distinct functional domains, three of which share a high level of homology (Fig. 1). More than 60% of TNKS and 70% of TNKL protein sequences are comprised of ankyrin repeats (Fig. 1). To dissect the serological reactivity in patients against distinct tankyrase domains, we subcloned and expressed the individual domains in the form of (His)6-tagged polypeptides (Fig. 4a) and tested their reactivity in protein mini-array format with 38 pre-selected sera of TNKL and/or TNKS seropositive patients with different malignant tumors (listed in legend to Fig. 4c, representative results are shown in Fig. 4b). We found that all tested TNKL fragments were seroreactive with comparable frequencies (Fig. 4c). On the other hand, only TNKS domains sharing high sequence homology with TNKL were reactive with sera of cancer patients. Of interest, a short unique N-terminal domain of TNKL, representing less than 5% of the entire protein, reacted with nearly half (48.6%) of the tested sera, while the much longer N-terminal domain of TNKS failed to react with any sera (Fig. 4b, c).
CD8+ T-cell immune response against tankyrases
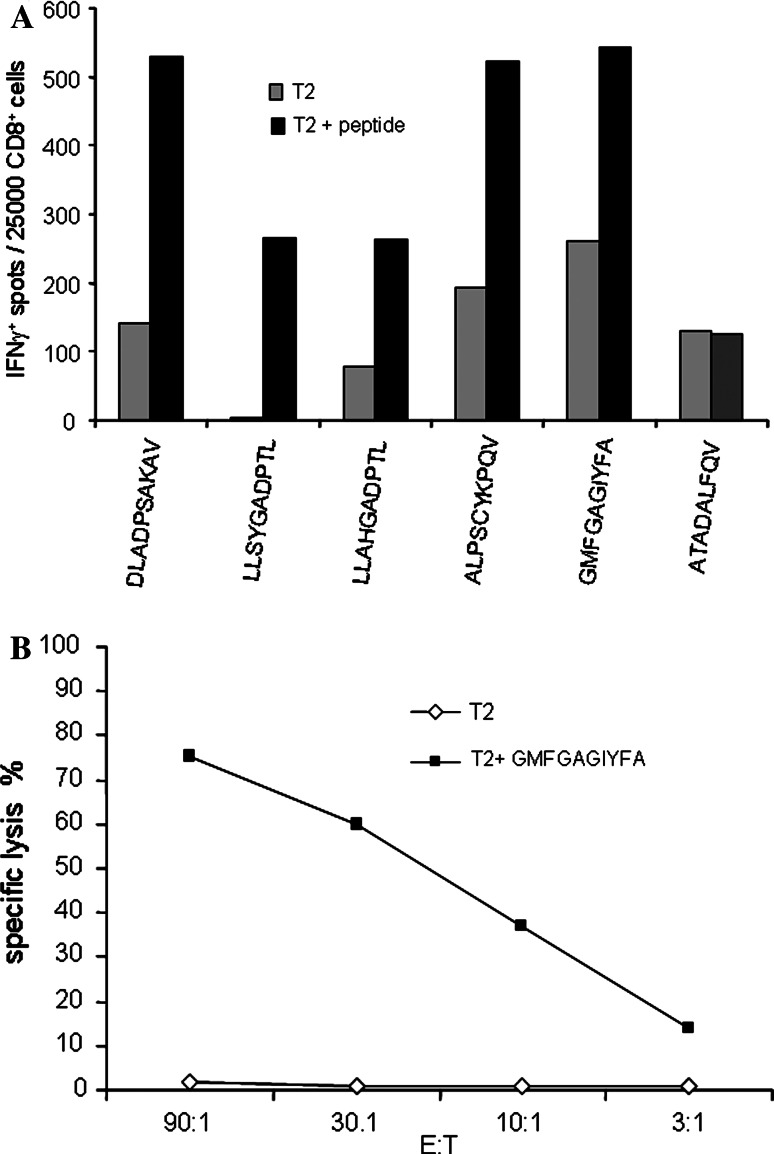
In order to evaluate possible T-cell response against tankyrases in seropositive cancer patients, a set of 33 HLA-A2-restricted peptides was designed based on TNKL amino acid sequence (Table 3). These peptides were tested in ELISPOT assay with primary peripheral blood mononuclear cells (PBMC) from TNKL/TNKS-seropositive, HLA-A2-positive colon and breast cancer patients. None of the three colon cancer patients tested were found positive, but in one breast cancer patient five peptides, DLADPSAKAV (P2, aa 162–170), LLSYGADPTL (P8, aa 296–305), LLAHGADPTL (P23, aa 764–773), ALPSCYKPQV (P24, aa 802–811), and GMFGAGIYFA (P31, aa 1053–1062), were clearly recognized by specific CD8+ T-cells from primary PBMC. This result was confirmed using pre-stimulated effector cells (Fig. 5a), and one of the five peptides (peptide P31) was also recognized in a cytotoxicity test (51Cr release assay) (Fig. 5b). All these peptides belong to conserved tankyrase domains: P2, P8, P23 and P24 are located within the ankyrin domain; P31 is located in the PARP domain. Peptides P2 and P31 are shared by both TNKL and TNKS (Table 3). Four additional patients (all HLA A2-positive and TNKL antibody-positive) failed to show T cell response to any of selected peptides. Therefore, although we have a clear evidence for spontaneous specific CD8+ T-cell response against tankyrases in patients, such responses appear to be rare.
Table 3.
Mapping T-cell epitopes of human tankyrases: TNKL-derived and control peptides with binding motifs to HLA-A2
| Peptide # | Peptide sequence | Position TNKL | Position TNKS |
|---|---|---|---|
| P1 | VLLQHGAEPT | 142–151 | – |
| P2 | DLADPSAKAV a | 162–170 | 320–329 |
| P3 | KMMALLTPL | 192–200 | – |
| P4 | MMALLTPLNV | 193–202 | – |
| P5 | LLLQHGADV | 229–237 | 387–395 |
| P6 | LLVKHGACV | 262–270 | – |
| P7 | LLLSYGADPT | 295–304 | – |
| P8 | LLSYGADPTL a | 296–305 | – |
| P9 | LLQAAREADV | 335–344 | – |
| P10 | NINEKTKEFL | 392–401 | – |
| P11 | ALQMGNENV | 469–477 | – |
| P12 | QMGNENVQQL | 471–480 | – |
| P13 | QLLQEGISL | 479–487 | – |
| P14 | YLLQHGADV | 544–552 | – |
| P15 | LLVKHGAVV | 577–585 | – |
| P16 | NVADLWKFT | 586–594 | 744–752 |
| P17 | LLLQHGADPT | 610–619 | – |
| P18 | ALLDAAKKGC | 648–657 | 806–815 |
| P19 | YLLQHGADV | 697–705 | – |
| P20 | ALLIKYNACV | 729–738 | – |
| P21 | LLIKYNACV | 730–738 | – |
| P22 | LLLAHGADPT | 763–772 | 921–930 |
| P23 | LLAHGADPTL a | 764–773 | – |
| P24 | ALPSCYKPQV a | 802–811 | – |
| P25 | NLGLEHLMDI | 884–893 | – |
| P26 | VLVEMGHKEL | 903–912 | – |
| P27 | KLIKGVERL | 925–934 | 1078–1087 |
| P28 | RLISGQQGL | 932–940 | – |
| P29 | MLFHGSPFV | 1028–1036 | – |
| P30 | YIGGMFGAGI | 1050–1059 | 1203–1212 |
| P31 | GMFGAGIYFA b | 1053–1062 | 1206–1215 |
| P32 | YICHRQLLFC | 1090–1099 | – |
| P33 | QLLFCRVTL | 1094–1102 | – |
| P34 | FLQFSAMKMA | 1107–1116 | – |
| C1 | ATADALFQV | Control | |
| C2 | ALQMGNENV | Control | |
| C3 | FLQPAVLTTI | Control | |
| C4 | LLLKKGSNI | Control | |
| C5 | WEFSELSSV | Control |
Fig. 5.
Cellular immune response to TNKL. a CD8+ reactivity to HLA-A2-restricted peptides in ELISPOT IFN-γ assay. TNKL peptides DLADPSAKAV (P2, aa 162–170), LLSYGADPTL (P8, aa 296–305), LLAHGADPTL (P23, aa 764–773), ALPSCYKPQV (P24, aa 802–811) and GMFGAGIYFA (P31, aa 1053–1062). ATADALFQV (C1): control irrelevant peptide. b Cytotoxicity test with CD8+ T cells from HLA-A2+ patient. Cytotoxicity was measured by 51Cr release assay
Discussion
Tankyrases are multifunctional proteins with a complex pattern of subcellular localization and multiple interacting partners in vitro. Overexpression of tankyrases may lead to telomere elongation in cancer cells [6, 20, 51], while TNKS knockdown by siRNA causes mitotic arrest due to impaired chromosomal segregation [2, 13]. Recent reports describing phenotypes of TNKL-deficient mice [4] and mice with deleted PAPR domain of TNKL [20] failed to unequivocally demonstrate the telomere-related functions of tankyrases in vivo. These mice showed significant decrease in body weight, but normal telomere length. Such phenotype may be explained by disrupted interactions with tankyrase partners, such as IRAP [3, 41] and Grb14 [28], involved in insulin-mediated signaling. The mild phenotype of TNKL-deficient mice might also suggest that TNKL and TNKS are functionally redundant, and thus the deficient phenotype cannot be fully revealed in a single-gene knockout mouse. Of note, investigation of telomere-related tankyrase functions in mice is complicated by the fact that, unlike its human counterpart, mouse TRF1 lacks the peptide motif necessary for interaction with tankyrases [35, 40].
Based on their effects on the telomerase complex in vitro, their role in mitosis and the interplay with PARP-1 activity [2, 13, 16, 44, 53, 60], tankyrases are considered as positive regulators of mitogenesis and carcinogenesis and as possible targets for anti-cancer interventions. The depletion of TNKS by transfection of small interfering RNA (siRNA) led to mitotic arrest of the cancer cell line HeLaI.2.11, which could be rescued by co-transfection of wild-type (but not of PARP-dead) TNKS [13]. Additionally, combined inhibition of telomerase and of PARP-modifying activities of tankyrases causes apoptosis of cancer cells, implying a possibility of development of new cancer therapeutics [43, 44].
TNKS mRNA overexpression was reported in bladder and colon cancers, multiple myeloma and plasma cell leukemia [17, 18, 56] In breast cancer, upregulation of both mRNA and protein levels of TNKS were observed [16]. In non-Hodgkin lymphomas or gastric cancer TNKS overexpression was not found [23, 29–31, 57]. Analysis of TNKL expression in meningioma and in tissues of breast cancer patients, demonstrated the presence of TNKL mRNA in both tumor and normal tissues without obvious overexpression [25, 32], but in about 11% of breast cancer patients TNKL protein was overexpressed in tumors [49]. Our findings presented here indicated that TNKL mRNA and protein are upregulated in colonic tumors. In contrast, TNKS mRNA level is not significantly changed while protein concentration of TNKS is actually downregulated probably in part due to modifications (including poly-ADP-ribosylation (PARsylation) and ubiquitination) and proteasome-mediated degradation [37, 38, 61]. Manifestation of tankyrase’s PARP activity (beneficial for cancer cells [44]) causes TNKS automodification followed by ubiquitination and proteasome-mediated degradation [61]. The N-terminus of TNKS harbors a histidine–proline–serine (HPS) rich domain that has a potential PEST motif (aa 79-184) (found by PESTfind Analysis Webtool, https://emb1.bcc.univie.ac.at/toolbox/pestfind/pestfind-analysis-webtool.htm) which often serves to target proteins for degradation in the ubiquitin-proteasome pathway [37, 38] and could be involved in the negative regulation of intracellular TNKS levels. The enzymatically active TNKS has a half-live of about 2 hours [61] and we hypothesize that elevated TNKS turnover might result in the decrease of the protein level in colon tumors. In contrast to TNKS the N-terminus of TNKL does not provide a potential site for ubiquitination and the protein might be accumulated in the cell.
Serological analysis of full-length tankyrases and subsequent epitope mapping demonstrate significant serological cross-reactivity between TNKL and TNKS. Common T-cell epitopes are also found during analysis of CD8+ T-cell response to tankyrases. Our study clearly demonstrates that due to immunological cross-reactivity between tankyrases both TNKS and TNKL can be recognized by humoral and cellular arms of the immune system. Highly conserved PARP domains of TNKS and TNKL were frequently recognized by sera of different colon cancer patients (Figure 4b). PARP-1 was repeatedly identified by SEREX in expression libraries derived from breast, colon, renal and lung cancer, but all PARP-1-related clones also frequently reacted with sera of patients with autoimmune diseases and of normal donors [24, 27, 42], making PARP-1 unsuitable as a serological biomarker for cancer. The absence of cross-reactivity between tankyrases and PARP-1 was experimentally tested (data not shown) and can be explained by a modest degree of homology in their catalytic domains, not exceeding 30% [52]. Another member of telomerase complex and the target of tankyrase PARP activity, TRF1 was also identified by SEREX as putative serological cancer antigen in gastric cancer (SEREX clone NGO-St-2, https://www2.licr.org/CancerImmunomeDB/). Finally, the protein moiety of human telomerase (TERT) was previously defined as tumor-associated antigen inducing CD8+ T-cell responses, implicating TERT as target in anti-cancer therapy [55]. In consideration of a possible use of tankyrases as targets of T cell immunity in cancer patients, we demonstrated spontaneous CD8+ T-cell response to several HLA-A2-restricted peptides (Table 3; Fig. 5), but we also found that such responses are rare. We thus favor a biomarker usage of antibody response to tankyrases in which case TNKS and TNKL can be utilized in array format along with other serologically defined cancer antigens and biomarkers with modest frequencies.
Acknowledgments
This study was supported by the Ludwig Institute for Cancer Research (New York), by the Cancer Antigen Discovery Collaborative of the Cancer Research Institute, Cancer Research Institute, by Deutsche Forschungsgemeinschaft Grant SFB 633, by grants from the Russian Academy of Sciences (Molecular and Cellular Biology; Medicine) and by Russian Foundation for Basic Research (grant 05-04-49075-а). We are greatly indebted to H.-G. Rammensee and M. Lagarkova for advise and to L.J. Old and A.M. Malyguine for stimulating discussions. We thank O.V. Gurova, A.A. Meshcheryakov and M.R. Lichinitser for providing clinical samples, D. Liepinsh, P. Lemansky and S. Durum for critical reading and valuable comments on the manuscript and T. Kadachigova and R. Kazaryan for assistance.
References
- 1.Bae J, Donigian JR, Hsueh AJ. Tankyrase 1 interacts with Mcl-1 proteins and inhibits their regulation of apoptosis. J Biol Chem. 2003;278:5195–5204. doi: 10.1074/jbc.M201988200. [DOI] [PubMed] [Google Scholar]
- 2.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 3.Chi NW, Lodish HF. Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J Biol Chem. 2000;275:38437–38444. doi: 10.1074/jbc.M007635200. [DOI] [PubMed] [Google Scholar]
- 4.Chiang YJ, Nguyen ML, Gurunathan S, Kaminker P, Tessarollo L, Campisi J, Hodes RJ. Generation and characterization of telomere length maintenance in tankyrase 2-deficient mice. Mol Cell Biol. 2006;26:2037–2043. doi: 10.1128/MCB.26.6.2037-2043.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comtesse N, Zippel A, Walle S, Monz D, Backes C, Fischer U, Mayer J, Ludwig N, Hildebrandt A, Keller A, Steudel WI, Lenhof HP, Meese E. Complex humoral immune response against a benign tumor: Frequent antibody response against specific antigens as diagnostic targets. Proc Natl Acad Sci USA. 2005;102:9601–9606. doi: 10.1073/pnas.0500404102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook BD, Dynek JN, Chang W, Shostak G, Smith S. Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol Cell Biol. 2002;22:332–342. doi: 10.1128/MCB.22.1.332-342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Rycker M, Price CM. Tankyrase polymerization is controlled by its sterile alpha motif and poly(ADP-ribose) polymerase domains. Mol Cell Biol. 2004;24:9802–9812. doi: 10.1128/MCB.24.22.9802-9812.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Rycker M, Venkatesan RN, Wei C, Price CM. Vertebrate tankyrase domain structure and sterile alpha motif (SAM)-mediated multimerization. Biochem J. 2003;372:87–96. doi: 10.1042/BJ20021450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decker P, Briand JP, de Murcia G, Pero RW, Isenberg DA, Muller S. Zinc is an essential cofactor for recognition of the DNA binding domain of poly(ADP-ribose) polymerase by antibodies in autoimmune rheumatic and bowel diseases. Arthritis Rheum. 1998;41:918–926. doi: 10.1002/1529-0131(199805)41:5<918::AID-ART20>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 10.Decker P, Isenberg D, Muller S. Inhibition of caspase-3-mediated poly(ADP-ribose) polymerase (PARP) apoptotic cleavage by human PARP autoantibodies and effect on cells undergoing apoptosis. J Biol Chem. 2000;275:9043–9046. doi: 10.1074/jbc.275.12.9043. [DOI] [PubMed] [Google Scholar]
- 11.Deng Z, Atanasiu C, Zhao K, Marmorstein R, Sbodio JI, Chi NW, Lieberman PM. Inhibition of Epstein-Barr virus OriP function by tankyrase, a telomere-associated poly-ADP ribose polymerase that binds and modifies EBNA1. J Virol. 2005;79:4640–4650. doi: 10.1128/JVI.79.8.4640-4650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dohnal AM, Inthal A, Felzmann T, Glatt S, Sommergruber W, Mann G, Gadner H, Panzer-Grumayer ER. Leukemia-associated antigenic isoforms induce a specific immune response in children with T-ALL. Int J Cancer. 2006;119:2870–2877. doi: 10.1002/ijc.22224. [DOI] [PubMed] [Google Scholar]
- 13.Dynek JN, Smith S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304:97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- 14.Frangioni JV, Neel BG. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- 15.Fuchs U, Rehkamp GF, Slany R, Follo M, Borkhardt A. The formin-binding protein 17, FBP17, binds via a TNKS binding motif to tankyrase, a protein involved in telomere maintenance. FEBS Lett. 2003;554:10–16. doi: 10.1016/S0014-5793(03)01063-9. [DOI] [PubMed] [Google Scholar]
- 16.Gelmini S, Poggesi M, Distante V, Bianchi S, Simi L, Luconi M, Casini RC, Cataliotti L, Pazzagli M, Orlando C. Tankyrase, a positive regulator of telomere elongation, is over expressed in human breast cancer. Cancer Lett. 2004;216:81–87. doi: 10.1016/j.canlet.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 17.Gelmini S, Poggesi M, Pinzani P, Mannurita SC, Cianchi F, Valanzano R, Orlando C. Distribution of Tankyrase-1 mRNA expression in colon cancer and its prospective correlation with progression stage. Oncol Rep. 2006;16:1261–1266. [PubMed] [Google Scholar]
- 18.Gelmini S, Quattrone S, Malentacchi F, Villari D, Travaglini F, Giannarini G, Della MA, Pazzagli M, Nicita G, Selli C, Orlando C. Tankyrase-1 mRNA expression in bladder cancer and paired urine sediment:preliminary experience. Clin Chem Lab Med. 2007;45:862–866. doi: 10.1515/CCLM.2007.133. [DOI] [PubMed] [Google Scholar]
- 19.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 20.Hsiao SJ, Poitras MF, Cook BD, Liu Y, Smith S. Tankyrase 2 poly(ADP-Ribose) polymerase domain-deleted mice exhibit growth defects but have normal telomere length and capping. Mol Cell Biol. 2006;26:2044–2054. doi: 10.1128/MCB.26.6.2044-2054.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jager E, Chen YT, Drijfhout JW, Karbach J, Ringhoffer M, Jager D, Arand M, Wada H, Noguchi Y, Stockert E, Old LJ, Knuth A. Simultaneous humoral and cellular immune response against cancer-testis antigen NY-ESO-1: definition of human histocompatibility leukocyte antigen (HLA)-A2-binding peptide epitopes. J Exp Med. 1998;187:265–270. doi: 10.1084/jem.187.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaminker PG, Kim SH, Taylor RD, Zebarjadian Y, Funk WD, Morin GB, Yaswen P, Campisi J. TANK2, a new TRF1-associated poly(ADP-ribose) polymerase, causes rapid induction of cell death upon overexpression. J Biol Chem. 2001;276:35891–35899. doi: 10.1074/jbc.M105968200. [DOI] [PubMed] [Google Scholar]
- 23.Klapper W, Krams M, Qian W, Janssen D, Parwaresch R. Telomerase activity in B-cell non-Hodgkin lymphomas is regulated by hTERT transcription and correlated with telomere-binding protein expression but uncoupled from proliferation. Br J Cancer. 2003;89:713–719. doi: 10.1038/sj.bjc.6601112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koroleva EP, Lagarkova MA, Mesheryakov AA, Scanlan MJ, Old LJ, Nedospasov SA, Kuprash DV. Serological identification of antigens associated with renal cell carcinoma. Russ J Immunol. 2002;7:229–238. [PubMed] [Google Scholar]
- 25.Kuimov AN, Kuprash DV, Petrov VN, Vdovichenko KK, Scanlan MJ, Jongeneel CV, Lagarkova MA, Nedospasov SA. Cloning and characterization of TNKL, a member of tankyrase gene family. Genes Immun. 2001;2:52–55. doi: 10.1038/sj.gene.6363722. [DOI] [PubMed] [Google Scholar]
- 26.Lagarkova MA, Koroleva EP, Kuprash DV, Boitchenko VE, Kashkarova UA, Nedospasov SA, Shebzukhov YV. Evaluation of humoral response to tumor antigens using recombinant expression-based serological mini-arrays (SMARTA) Immunol Lett. 2003;85:71–74. doi: 10.1016/S0165-2478(02)00209-2. [DOI] [PubMed] [Google Scholar]
- 27.Lim Y, Lee DY, Lee S, Park SY, Kim J, Cho B, Lee H, Kim HY, Lee E, Song YW, Jeoung DI. Identification of autoantibodies associated with systemic lupus erythematosus. Biochem Biophys Res Commun. 2002;295:119–124. doi: 10.1016/S0006-291X(02)00637-X. [DOI] [PubMed] [Google Scholar]
- 28.Lyons RJ, Deane R, Lynch DK, Ye ZS, Sanderson GM, Eyre HJ, Sutherland GR, Daly RJ. Identification of a novel human tankyrase through its interaction with the adaptor protein Grb14. J Biol Chem. 2001;276:17172–17180. doi: 10.1074/jbc.M009756200. [DOI] [PubMed] [Google Scholar]
- 29.MacNamara B, Wang W, Chen Z, Hou M, Mazur J, Gruber A, Porwit-MacDonald A. Telomerase activity in relation to pro- and anti-apoptotic protein expression in high grade non-Hodgkin’s lymphomas. Haematologica. 2001;86:386–393. [PubMed] [Google Scholar]
- 30.Matsutani N, Yokozaki H, Tahara E, Tahara H, Kuniyasu H, Haruma K, Chayama K, Yasui W. Expression of telomeric repeat binding factor 1 and 2 and TRF1-interacting nuclear protein 2 in human gastric carcinomas. Int J Oncol. 2001;19:507–512. [PubMed] [Google Scholar]
- 31.Matsutani N, Yokozaki H, Tahara E, Tahara H, Kuniyasu H, Kitadai Y, Haruma K, Chayama K, Tahara E, Yasui W. Expression of MRE11 complex (MRE11, RAD50, NBS1) and hRap1 and its relation with telomere regulation, telomerase activity in human gastric carcinomas. Pathobiology. 2001;69:219–224. doi: 10.1159/000055946. [DOI] [PubMed] [Google Scholar]
- 32.Monz D, Munnia A, Comtesse N, Fischer U, Steudel WI, Feiden W, Glass B, Meese EU. Novel tankyrase-related gene detected with meningioma-specific sera. Clin Cancer Res. 2001;7:113–119. [PubMed] [Google Scholar]
- 33.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller S, Briand JP, Barakat S, Lagueux J, Poirier GG, de Murcia G, Isenberg DA. Autoantibodies reacting with poly(ADP-ribose) and with a zinc-finger functional domain of poly(ADP-ribose) polymerase involved in the recognition of damaged DNA. Clin Immunol Immunopathol. 1994;73:187–196. doi: 10.1006/clin.1994.1187. [DOI] [PubMed] [Google Scholar]
- 35.Muramatsu Y, Ohishi T, Sakamoto M, Tsuruo T, Seimiya H. Cross-species difference in telomeric function of tankyrase 1. Cancer Sci. 2007;98:850–857. doi: 10.1111/j.1349-7006.2007.00462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preuss KD, Zwick C, Bormann C, Neumann F, Pfreundschuh M. Analysis of the B-cell repertoire against antigens expressed by human neoplasms. Immunol Rev. 2002;188:43–50. doi: 10.1034/j.1600-065X.2002.18805.x. [DOI] [PubMed] [Google Scholar]
- 37.Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. doi: 10.1016/0968-0004(96)10031-1. [DOI] [PubMed] [Google Scholar]
- 38.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 39.Sahin U, Tureci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc Natl Acad Sci USA. 1995;92:11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sbodio JI, Chi NW. Identification of a tankyrase-binding motif shared by IRAP, TAB182, and human TRF1 but not mouse TRF1. NuMA contains this RXXPDG motif and is a novel tankyrase partner. J Biol Chem. 2002;277:31887–31892. doi: 10.1074/jbc.M203916200. [DOI] [PubMed] [Google Scholar]
- 41.Sbodio JI, Lodish HF, Chi NW. Tankyrase-2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1) and IRAP (insulin-responsive aminopeptidase) Biochem J. 2002;361:451–459. doi: 10.1042/0264-6021:3610451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scanlan MJ, Gout I, Gordon CM, Williamson B, Stockert E, Gure AO, Jager D, Chen Y-T, Mackay A, O’Hare MJ, Old LJ. Humoral immunity to human breast cancer: antigen definition and quantitative analysis of mRNA expression. Cancer Immun. 2001;1:4. [PubMed] [Google Scholar]
- 43.Seimiya H. The telomeric PARP, tankyrases, as targets for cancer therapy. Br J Cancer. 2006;94:341–345. doi: 10.1038/sj.bjc.6602951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seimiya H, Muramatsu Y, Ohishi T, Tsuruo T. Tankyrase 1 as a target for telomere-directed molecular cancer therapeutics. Cancer Cell. 2005;7:25–37. doi: 10.1016/j.ccr.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 45.Seimiya H, Smith S. The telomeric poly(ADP-ribose) polymerase, tankyrase 1, contains multiple binding sites for telomeric repeat binding factor 1 (TRF1) and a novel acceptor, 182-kDa tankyrase-binding protein (TAB182) J Biol Chem. 2002;277:14116–14126. doi: 10.1074/jbc.M112266200. [DOI] [PubMed] [Google Scholar]
- 46.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 47.Shebzukhov YV, Koroleva EP, Khlgatian SV, Belousov PV, Kuz’mina KE, Radko BV, Longpre F, Lagarkova MA, Kadachigova TS, Gurova OV, Meshcheryakov AA, Lichinitser MR, Knuth A, Jager E, Kuprash DV, Nedospasov SA. Antibody response to a non-conserved C-terminal part of human histone deacetylase 3 in colon cancer patients. Int J Cancer. 2005;117:800–806. doi: 10.1002/ijc.21240. [DOI] [PubMed] [Google Scholar]
- 48.Shebzukhov YV, Koroleva EP, Khlgatian SV, Lagarkova MA, Meshcheryakov AA, Lichinitser MR, Karbach J, Jager E, Kuprash DV, Nedospasov SA. Humoral immune response to thymidylate synthase in colon cancer patients after 5-FU chemotherapy. Immunol Lett. 2005;100:88–93. doi: 10.1016/j.imlet.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 49.Sidorova N, Zavalishina L, Kurchashova S, Korsakova N, Nazhimov V, Frank G, Kuimov A. Immunohistochemical detection of tankyrase 2 in human breast tumors and normal renal tissue. Cell Tissue Res. 2006;323:137–145. doi: 10.1007/s00441-005-0053-8. [DOI] [PubMed] [Google Scholar]
- 50.Smith S, de Lange T. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J Cell Sci. 1999;112(Pt 21):649–3656. doi: 10.1242/jcs.112.21.3649. [DOI] [PubMed] [Google Scholar]
- 51.Smith S, de Lange T. Tankyrase promotes telomere elongation in human cells. Curr Biol. 2000;10:1299–1302. doi: 10.1016/S0960-9822(00)00752-1. [DOI] [PubMed] [Google Scholar]
- 52.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 53.Tong WM, Cortes U, Wang ZQ. Poly(ADP-ribose) polymerase: a guardian angel protecting the genome and suppressing tumorigenesis. Biochim Biophys Acta. 2001;1552:27–37. doi: 10.1016/s0304-419x(01)00035-x. [DOI] [PubMed] [Google Scholar]
- 54.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 55.Vonderheide RH. Telomerase as a universal tumor-associated antigen for cancer immunotherapy. Oncogene. 2002;21:674–679. doi: 10.1038/sj.onc.1205074. [DOI] [PubMed] [Google Scholar]
- 56.Xu D, Zheng C, Bergenbrant S, Holm G, Bjorkholm M, Yi Q, Gruber A. Telomerase activity in plasma cell dyscrasias. Br J Cancer. 2001;84:621–625. doi: 10.1054/bjoc.2000.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamada M, Tsuji N, Nakamura M, Moriai R, Kobayashi D, Yagihashi A, Watanabe N. Down-regulation of TRF1, TRF2 and TIN2 genes is important to maintain telomeric DNA for gastric cancers. Anticancer Res. 2002;22:3303–3307. [PubMed] [Google Scholar]
- 58.Yamanaka H, Willis EH, Carson DA. Human autoantibodies to poly(adenosine diphosphate-ribose) polymerase recognize cross-reactive epitopes associated with the catalytic site of the enzyme. J Clin Invest. 1989;83:180–186. doi: 10.1172/JCI113856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamanaka H, Willis EH, Penning CA, Peebles CL, Tan EM, Carson DA. Human autoantibodies to poly(adenosine diphosphate-ribose) polymerase. J Clin Invest. 1987;80:900–904. doi: 10.1172/JCI113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yeh T-YJ, Sbodio JI, Nguyen MTA, Meyer TN, Lee RM, Chi NW. Tankyrase-1 overexpression reduces genotoxin-induced cell death by inhibiting PARP1. Mol Cell Biochem. 2005;276:183–192. doi: 10.1007/s11010-005-4059-z. [DOI] [PubMed] [Google Scholar]
- 61.Yeh TY, Meyer TN, Schwesinger C, Tsun ZY, Lee RM, Chi NW. Tankyrase recruitment to the lateral membrane in polarized epithelial cells. Regulation by cell-cell contact and protein PARsylation. Biochem J. 2006;399:415–425. doi: 10.1042/BJ20060713. [DOI] [PMC free article] [PubMed] [Google Scholar]