Activated Renal Macrophages Are Markers of Disease Onset and Disease Remission in Lupus Nephritis (original) (raw)
. Author manuscript; available in PMC: 2009 Feb 1.
Abstract
Costimulatory blockade with CTLA4Ig and anti-CD40L along with a single dose of cyclophosphamide induces remission of systemic lupus erythematosus nephritis in NZB/W F1 mice. To understand the mechanisms for remission and for impending relapse, we examined the expression profiles of 61 inflammatory molecules in the perfused kidneys of treated mice and untreated mice at different stages of disease. Further studies using flow cytometry and immunohistochemistry allowed us to determine the cellular origins of several key markers. We show that only a limited set of inflammatory mediators is expressed in the kidney following glomerular immune complex deposition but before the onset of proteinuria. Formation of a lymphoid aggregate in the renal pelvis precedes the invasion of the kidney by inflammatory cells. Regulatory molecules are expressed early in the disease process and during remission but do not prevent the inevitable progression of active inflammation. Onset of proliferative glomerulonephritis and proteinuria is associated with activation of the renal endothelium, expression of chemokines that mediate glomerular cell infiltration, and infiltration by activated dendritic cells and macrophages that migrate to different topographical areas of the kidney but express a similar profile of inflammatory cytokines. Increasing interstitial infiltration by macrophages and progressive tubular damage, manifested by production of lipocalin-2, occur later in the disease process. Studies of treated mice identify a type II (M2b)-activated macrophage as a marker of remission induction and impending relapse and suggest that therapy for systemic lupus erythematosus nephritis should include strategies that prevent both activation of monocytes and their migration to the kidney.
Systemic lupus erythematosus (SLE)5 nephritis is characterized by immune complex-mediated glomerular and tubulo-interstitial inflammation, leading to chronicrenal insufficiency in up to 30% of affected patients. Maintenance of disease remission after treatment of a renal flare remains a challenging clinical problem (1). Recently. it has become possible to study the mechanisms involved in induction of complete remission of nephritis in NZB/W mice. The combination of a single dose of cyclophosphamide administered along with six doses of CTLA4Ig and six doses of anti-CD154 (triple therapy) induces prompt reversal of proteinuria in NZB/W mice with established nephritis (2). Although immune complexes and complement persist in the glomeruli, histologic changes in the glomeruli reverse and there is a decrease in expression of several chemokines with efflux or death of renal inflammatory cells (2).
To further understand how inflammatory cells migrate to and from the inflamed NZB/W kidney during active disease and remission, we undertook targeted real-time PCR analysis of 61 inflammatory molecules in the kidneys of NZB/W F1 at various disease stages. Our results show that expression of inflammatory mediators follows the deposition of immune complexes in the glomeruli but that distinct subsets of genes are up-regulated at sequential stages of disease. Our findings yield insight into the progressive inflammatory process in SLE nephritis and identify an activated type II macrophage population as a key marker of proteinuria onset and disease remission.
Materials and Methods
Animals
NZB/NZW F1 females were purchased from The Jackson Laboratory. Urine was tested weekly for proteinuria by dipstick (Multistick; Fisher Scientific). Once fixed proteinuria of >300 mg/dl on two occasions 24-h apart appeared, a single dose of 50 mg/kg cyclophosphamide and six doses of 100 _μ_g of CTLA4Ig and 250 _μ_g of anti-CD154 were administered as previously described (2). Sixteen treated mice that entered remission (≤30 mg/dl proteinuria on at least two occasions 3 days apart) were sacrificed at intervals starting 3 wk after treatment. Proteinuria status was confirmed by overnight collection of urine in metabolic cages before sacrifice. Control groups of untreated NZB/W F1 mice were sacrificed at the following ages: 6 wk (5 mice), 16 wk (5 mice), 23 wk without any proteinuria (23w NP: 10 mice, 5 with and 5 without high-titer IgG anti-DNA Abs), 23 wk with new onset proteinuria >300 mg/dl (23w P: 6 mice), and 36 – 40 wk with established proteinuria >300 mg/dl (36 – 40w: 12 mice).
Quantitative real-time PCR (qPCR)
After perfusion with 60 ml of cold sterile saline, kidneys were immediately removed into TRIzol (Invitrogen Life Technologies) and total RNA was prepared. Quality of the RNA was verified on a NanoRNA chip (Agilent). Five micrograms of RNA was reverse-transcribed with SuperScript II reverse transcriptase (Invitrogen Life Technologies). Amplification was performed in triplicate using SYBR Green PCR Master Mix (Roche Diagnostics) and specific primers (Table I) in a LightCycler480 (Roche Diagnostics). Dissociation curve analysis confirmed amplification of one specific product per primer pair.
Table I.
List of primers used for ANOVA analysis
| Molecule | Forward Sequence (5′–3′) | Reverse Sequence (5′–3′) |
|---|---|---|
| APRIL | CCG TGC CTA CAA TAG CTG C | CCG TGG AAT TTT GAC AGT GAT |
| β-Actin | ACC GTG AAA AGA TGA CCC AG | AGC CTG GAT GGC TAC GTA CA |
| BAFF | AAT ATG CCC AAA ACA CTG CC | CAT CTC CTT CTT CCA GCC TC |
| CCL11 | PPM02967A-200 | Superarray (Frederick, MD) |
| CCL19 | AGG AGC CCT GTG TCT TGA GT | GAG GCC TGG TCC TCT CTT CT |
| CCL2 | AAG AGG ATC ACC AGC AGC AG | GGT CAG CAC AGA CCT CTC TCT T |
| CCL20 | GGT ACT GCT GGC TCA CCT CT | TGT ACG AGA GGC AAC AGT CG |
| CCL21 | CTT TCT TAC CTG GCG GAG C | TCC CTG GGA GAC ACT CTT TC |
| CCL3 | CAA GTC TTC TCA GCG CCA TA | GGA ATC TTC CGG CTG TAG G |
| CCL4 | CTC CAA GCC AGC TGT GGT AT | GGC TCA CTG GGG TTA GCA C |
| CCL5 | GCA AGT GCT CCA ATC TTG C | CTT CTT CTC TGG GTT GGC AC |
| CCL8 | TTC TTT GCC TGC TGC TCA TA | AGC AGG TGA CTG GAG CCT TA |
| CCL9 | GAT TTA TAG GTT ATT TTC CCA CCA | GAA CCC CCT CTT GCT GAT AA |
| CCR1 | PPM0317A-200 | Superarray (Frederick, MD) |
| CCR10 | CAA GCC CAC AGA GCA GGT | GCC ACC ATC AGG GAG ACA |
| CCR2 | PPM03176A-200 | Superarray (Frederick, MD) |
| CCR5 | GAG ACA TCC GTT CCC CCT AC | GTC GGA ACT GAC CCT TGA AA |
| CCR6 | CCC TGG AAA GCT GGG TAA AG | GAA TGT GAG GAC TAT TAG AAC TGT GAA |
| CCR7 | GGT GGC TCT CCT TGT CAT TT | TCA TCG GTG ACC TCA TCT TG |
| CCR8 | AGA AGA AAG GCT CGC TCA GA | GGC TCC ATC GTG TAA TCC AT |
| CD138 | CCC CTC CTT TGA CTT CTG CCT | GCA GTC GGG TCC CCT TTC T |
| CD40 | PPM03426A-200 | Superarray (Frederick, MD) |
| CHST4 | GGC AAG CAG AAG GGT TAG G | CTG GGA ACC CAG GAA CAT C |
| CX3CL1 | PPM02959A-200 | Superarray (Frederick, MD) |
| CXCL10 | GCT GCC GTC ATT TTC TGC | TCT CAC TGG CCC GTC ATC |
| CXCL11 | AGT AAC GGC TGC GAC AAA GT | GCA CCT TTG TCG TTT ATG AGC |
| CXCL12 | CCA ACG TCA AGC ATC TGA AA | CTG TTG TTG TTC TTC AGC CG |
| CXCL13 | TCT GGA CCA AGA TGA AGA AAG TT | GCT TGG GGA GTT GAA GAC AG |
| CXCL16 | GGC CTA TGT GCT GTG CAA C | GGA GTG TAC CAG AGC TGC AA |
| CXCL9 | GAA GTC CGC TGT TCT TTT CCT | GCA TCG TGC ATT CCT TAT CA |
| CXCR3 | GCT CTA TGC CTT TGT GGG AG | GCG TGA ACA ACA TCC ACA TT |
| CXCR4 | AAC CCC ATC CTC TAT GCC TT | TGG GCA GAG CTT TTG AAC TT |
| CXCR5 | TGG GCT CCA TCA CAT ACA AT | GGG AAT CTC CGT GCT GTT AC |
| CXCR6 | TGG AAC AAA GCT ACT GGG CT | ACT CTT GAT GCC CAT CAT CC |
| E-Selectin | TCC TCT GGA GAG TGG AGT GC | GGT GGG TCA AAG CTT CAC AT |
| Foxp3 | AGA AGC TGG GAG CTA TGC AG | GCT ACG ATG CAG CAA GAG C |
| ICAM-1 | GGG AAT GTC ACC AGG AAT GT | GCA CCA GAA TGA TTA TAG TCC AGT TA |
| ICOS | GCA GCT TTC GTT GTG GTA CTC | TGC ACA CTG GAT CCG TAT TT |
| ICOS-L | GCT CTC CAC CAG AAC ATC ACT | GGT TTC CTG TGG GTT CTT TG |
| IFN-γ | GCA TTC ATG AGT ATT GCC AAG | GGT GGA CCA CTC GGA TGA |
| IL-10 | GTG GAG CAG GTG AAG AGT GA | TTC ATG GCC TTG TAG ACA CCT |
| IL-12A | PPM03019A-200 | Superarray (Frederick, MD) |
| IL-17 | PPM03023A-200 | Superarray (Frederick, MD) |
| IL-1β | GAT CCA CAC TCT CCA GCT GCA | CAA CCA ACA AGT GAT ATT CTC CAT G |
| IL-2 | TGA GAA TTT CAT CAG CAA TAT CA | TCG AAT TGG CAC TCA AAT GT |
| IL-21 | PPM01684A-200 | Superarray (Frederick, MD) |
| IL-4 | CAT CGG CAT TTT GAA CGA G | ACG TTT GGC ACA TCC ATC TC |
| IL-6 | PPM03015A-200 | Superarray (Frederick, MD) |
| LCN-2 | CCA TCT ATG AGC TAC AAG AGA ACA AT | TCT GAT CCA GTA GCG ACA GC |
| LFA-1 | TCC TTC CGG AAA GTG GAG AT | GAG CTC CTC GCA GCT CAC |
| LIGHT | CGA TCT CAC CAG GCC AAC | TCC ACC AAT ACC TAT CAA GCT G |
| L-selectin | GGT CAT CTC CAG AGC CAA TC | GGG GGT TGT AGT CAC CTT CTT |
| Lymphotoxin α | AAA CCT GCT GCT CAC CTT GT | AGA GCA GTG AGT TCT GCT TGC |
| MADCAM | GAC CCC GAA TTC CTC CTC | GCA AGG AAG ACC AGT GCA A |
| Ox-40L | GAC CCT CCA ATC CAA AGA CTC | TCA TTC TTG TAT GAG CTG ATG AAT |
| PD-1 | GGG CAC TTG ACT CTC CCT ACT | ATA TCC CCC TTT CCT TCC CT |
| PDL1 | ATC CTG TCT CAA AGG AAA GAC G | TGA ACA GGT AAC AGA GGG GG |
| PDL2 | CTT TCC GCA GGG AGA CAT | CGG AGG AAA TGA GGG CTA T |
| P-selectin | PPM03205A-200 | Superarray (Frederick, MD) |
| TGF-β | TGG AGC AAC ATG TGG AAC TC | GTC AGC AGC CGG TTA CCA |
| TNF-α | TGG GAG TAG ACA AGG TAC AAC CC | CAT CTT CTC AAA ATT CGA GTG ACA A |
| VCAM-1 | AAG ACT GAA GTT GGC TCA CAA TTA | GGC GAA AAA TAG TTC TTG TTA TGT T |
Flow cytometry and cell sorting
Kidneys from five proteinuric mice were treated for 30 min with collagenase (Worthington Biochemical), followed by progressive sieving to obtain single-cell suspensions. Whole kidney preparations, spleen cells, and PBMC from the same mice were stained with Abs to CD11c, CD11b, CD19, IgM, IgD, CD5, CD21, CD23, CD4, CD69, CD44, CD62L (BD Pharmingen), and F/80 (Caltag Laboratories and Invitrogen Life Technologies) as previously described (3). Ox-40L staining was detected using murine Ox-40Ig followed by FITC-labeled anti-IgG2a. Kidney preparations from three 8-wk- old mice were used as negative controls.
To analyze gene expression in the various cell subsets, renal lymphoid and nonlymphoid cells were separated from five nephritic mice using pooled Abs to CD4, CD8, F4/80, CD11b, CD11c, CD5, CD49b, and B220. CD19-positive B cells (n = 14), CD4-positive T cells (n = 10) CD11b/CD11chigh dendritic cells (n = 4), and CD11b/CD11clow or CD11b/F4/80high macrophages (n = 10) were then isolated from kidneys of nephritic mice using a FACSAria (BD Biosciences). Isolated cells were >90% pure. RNA was synthesized from the isolated cell populations using a picopure RNA isolation kit (Arcturus Molecular Device) and qPCR was performed using primers specific for IL-1, IL-10, BAFF, TNF-α, CXCL13, and CXCR5 as above.
Histologic and immunohistochemical analysis of kidneys
H&E sections of kidneys were scored by a single observer (M.M.) blinded to the treatment group as previously described (4). In brief, glomerular and interstitial disease were scored separately for each kidney using a semi-quantitative scale from 0 to 1 (absent) to 3 to 4 (severe). Immunohistochemistry was performed using Abs for IgG (Southern Biotechnology Associates), CD4, CD8, B220, CD62L, CD11c, CD138, F4/80, IL-17 (all from BD Pharmingen), and Foxp3 (eBioscience). Slides were counterstained with 4′,6-diamidino-2-phenylindole (Molecular Probes and Invitrogen Life Technologies) and images were captured using a digital charge-coupled device camera system connected to a Zeiss microscope.
Statistical analysis
The TIGR Multi Experiment Viewer (TMEV) software package was used for statistical analysis of qPCR data. The average of the raw data for each sample (Ct value) was normalized to the internal control (housekeeping gene _β_-actin). Normalized expression data were log 2-transformed and scaled to the expression value for a single 6-wk-old mouse given an arbitrary value of 1 (0 by log scale). Cluster analyses show these values and are scaled from −2 or −3 (green) to 3 (red). Statistical analysis between experimental groups was performed using specialized implementations of ANOVA with the t test statistic and permutations available in the TMEV package. Unsupervised hierarchical clustering with bootstrap procedures of samples based on gene expression data was performed using Euclidean metrics with average or complete linkage and visualized using TMEV. To graphically display the results of the statistical analyses, data were scaled to the mean of the 10 young control mice. Data shown in the cluster analyses are for all significant genes. For simplicity, data shown in the graphs represent genes that were significantly different both by statistical analysis of microarray (SAM) and t test analysis and that had >2-fold increased expression over the mean of young controls.
Results
Histologic analysis of kidneys
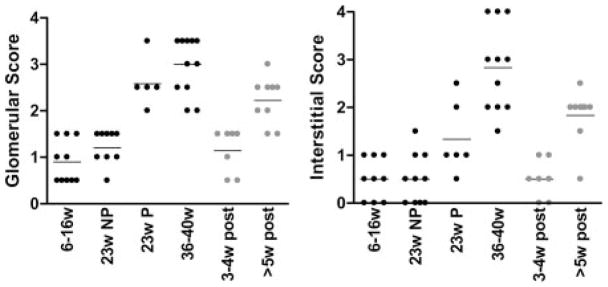
Six groups of mice were used in this study: 1) 10 mice 6 –16 wk old (6 –16w); 2) 10 mice 23 wk old without proteinuria (23w NP); 3) 6 mice 23 wk old with new onset proteinuria >300 mg/dl (23w P); 4) 10 mice 36 – 40 wk old with established proteinuria >2 wk (36 – 40w); 5) 7 mice sacrificed 3– 4 wk after beginning remission induction therapy and with complete resolution of proteinuria (3– 4w post); and 6) 9 mice sacrificed ≥5 wk after remission induction therapy and with complete resolution of proteinuria confirmed on multiple occasions (≥5w post). Kidneys were scored separately for glomerulonephritis and interstitial inflammation using a semiquantitative score from 0 to 4 rated from early or focal to severe as previously described (4). Glomerular inflammation was absent or mild in mice younger than 16 wk of age and in 23-wk nonproteinuric mice. Proliferative glomerulonephritis appeared at the onset of proteinuria ( p < 0.001 23w P vs 23w NP; Fig. 1_A_). Mild interstitial inflammation also appeared at the onset of proteinuria ( p < 0.04 23w P vs 6 –16w) and increased in severity with disease progression ( p < 0.01 23w P vs 36 – 40w; Fig. 1_B_) Thus, proteinuria onset is associated with glomerular inflammation but severe interstitial inflammation and tubular necrosis are a feature of late-stage renal disease in this mouse model.
FIGURE 1.
Histologic scores of H&E sections of kidneys examined by light microscopy. The six groups of mice used in this study are described in the text. Untreated mice are in black and treated mice in gray. A, Glomerular scores and interstitial scores (B). Each symbol indicates an individual mouse.
All mice sacrificed 3– 4 wk after remission induction had complete histologic and clinical remission defined as a glomerular score <2 and an interstitial score ≤1. Mice sacrificed 5–14 wk after treatment had no proteinuria but were histologically similar to 23-wk-old mice with new onset proteinuria, indicating that they had impending disease relapse (Fig. 1).
Early expression of inflammatory markers in the kidneys of prenephritic NZB/W mice
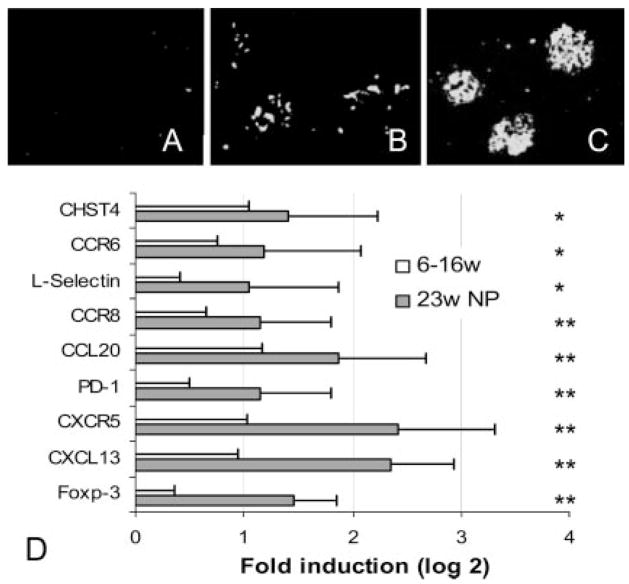
There was no glomerular Ab deposition (Fig. 2_A_) and no difference in the expression of any of the inflammatory markers in the kidneys between 6- and 16-wk-old mice and these two groups were pooled and used as controls for all subsequent analyses (6 –16 wk). Although the kidneys of 23- wk-old nonproteinuric mice appeared histologically normal, faint staining of the glomeruli with anti-IgG was observed in all of the mice in this group regardless of levels of anti-dsDNA Abs in the serum (Fig. 2_B_). Expression of a limited set of inflammatory markers was increased in the kidneys of these mice compared with mice ≤16 wk of age (Fig. 2_D_). These early markers included the chemokine CXCL13 (BLC) and its receptor CXCR5, CCL20 (MIP-3α) and its receptor CCR6, and CCR8 whose ligand CCL1 is expressed by monocytes and T cells. L-selectin (CD62L) was also increased, indicating migration of L-selectin-positive lymphocytes into the kidney, as was GlcNAc2 (CHST4), a marker for high endothelial venules. The negative regulatory molecules PD-1 and Foxp3 were also increased in the prenephritic kidneys. H&E staining of the kidneys of prenephritic mice revealed small collections of lymphocytes in the perihilar region but no cellular infiltration of the renal parenchyma and no glomerular or interstitial damage (see Fig. 1). Foxp3 (see Fig. 7) and L-selectin staining (data not shown) were confined to this perihilar region.
FIGURE 2.
Early expression of inflammatory markers in NZB/W kidneys: upper panels, fluorescence for IgG at 16 wk (A), 23 wk before proteinuria onset (B), and 23 wk at onset of proteinuria (C). Data are representative of five mice examined in each group. D, Significantly up-regulated (>2-fold) genes by SAM and t test (mean + 1 SD) in the kidneys of 23-wk nonproteinuric mice (23w NP). Values are normalized to the mean of 6- to 16-wk mice given a value of 1. Data are shown as log 2. Values of p for the up-regulated genes are p < 0.01 (*) or p < 0.001 (**) as shown.
FIGURE 7.
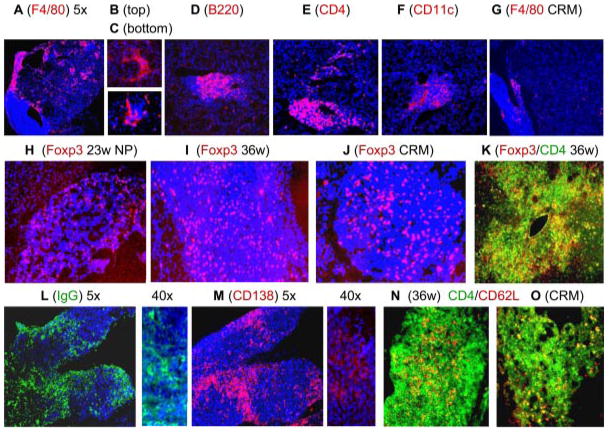
Immunohistochemical analysis of kidneys. A–F, Stained with PE-conjugated Abs as indicated and counterstained with 4′,6-diamidino-2-phenylindole (blue). A–C, F4/80high macrophages are scattered throughout the renal parenchyma and are found surrounding glomeruli and form a cap around perivascular and perihilar lymphoid aggregates. B cells and CD11chigh dendritic cells are found in the lymphoid aggregates, whereas CD4 T cells are found in aggregates and around glomeruli (D–F). Note the virtual disappearance of macrophages from the kidneys of mice in remission (G). H–K, Foxp3 staining of lymphoid aggregates from mice at different disease stages as marked. Foxp3-positive cells appeared in the hilar aggregates early in disease and did not disappear upon remission induction. L and M, Large numbers of plasma cells localized mainly in the hilar aggregates of a 36-wk proteinuric mouse. N and O, Double staining for CD4 and L-selectin (CD62L). L-selectin-positive T cells remained in the renal hilar aggregate after remission induction. Images are ×10 unless indicated otherwise. Images are representative of at least three mice examined in each group.
Expression of inflammatory markers associated with proteinuria onset and disease progression
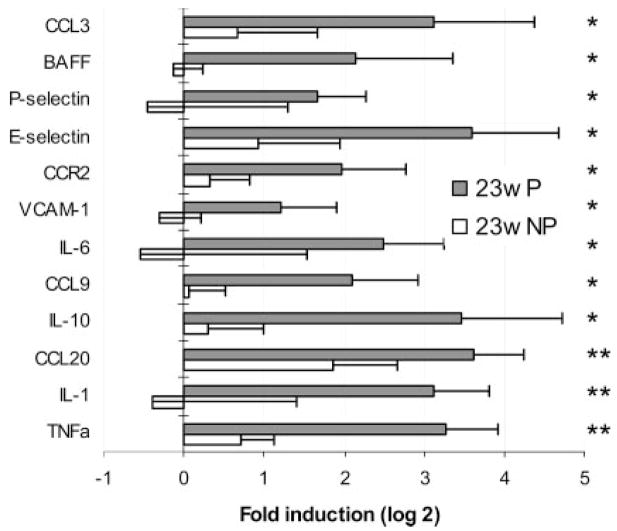
Bright staining with anti-IgG was observed in the glomeruli of proteinuric 23-wk-old mice (Fig. 2_C_). Of 61 markers, 15 were up-regulated in the kidneys of these mice compared with young or nonproteinuric controls. Several cytokines and chemokines were up-regulated in the kidneys of these mice compared with age-matched nonproteinuric mice (23w NP). The five cytokines were IL-1, TNF-α, IL-6, BAFF, and IL-10. The up-regulated chemokines were CCL3 (MIP-1α), and CCL9 (MIP-1γ) as well as the CCR2 receptor. In addition, up-regulation of endothelial activation markers VCAM-1, E-selectin, and P-selectin was noted (Fig. 3). When mice with new onset proteinuria (23w P) were compared with 6- to 16-wk-old mice, CCL2 (MCP-1), CCL5 (RANTES), CCR5, and CXCR3 were also found to be up-regulated (data not shown).
FIGURE 3.
Genes up-regulated at the onset of proteinuria. Gene expression for age-matched mice with (23w P) and without (23w NP) proteinuria was normalized to the mean of 6- to 16-wk controls given a value of 1. Data are shown as log 2. Genes significantly up- or down-regulated (>2-fold) by SAM and t test (mean + 1 SD) are shown. Values of p for the up-regulated genes are p < 0.01 (*) or p < 0.001 (**) as shown.
We next determined which inflammatory markers were increased over the entire course of disease compared with young mice. A comparison of all four untreated groups using ANOVA showed that expression of 52 of 61 inflammatory markers analyzed increased over time as disease progressed to the terminal stages (Table I). Only nine genes (IL-4, IL-12, IL-17, PDL1, CX3CL1, CCL11, CCL21, and CXCL12) were not significantly up-regulated across the four untreated groups during the disease course. Some of these genes (IL-4, PDL1, CCL21, CXCL12) were not expressed at any time during the disease course, whereas others were variably expressed in individual mice (e.g., IL-12 and IL-17 were expressed at high levels in three of eight 36- to 40-wk mice tested but were not expressed in any mice at earlier disease stages).
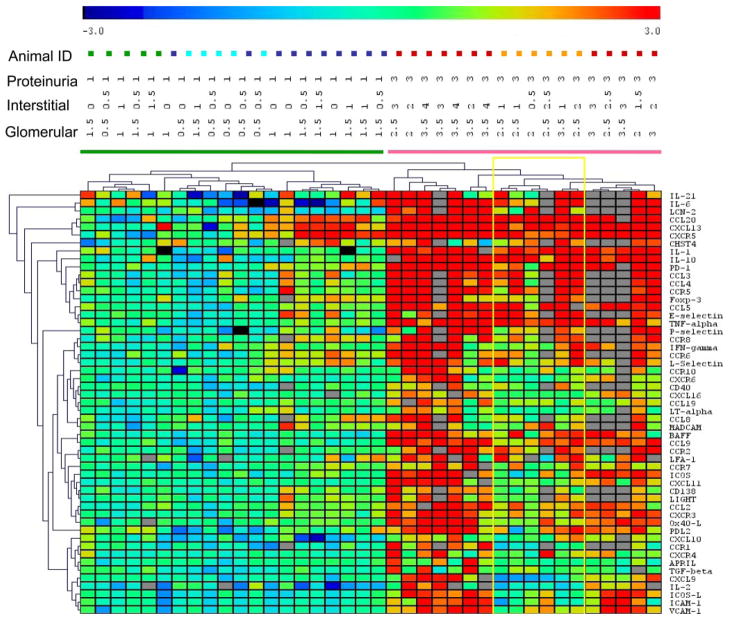
To further determine whether the expression of inflammatory markers in the kidneys corresponded with disease stage, we performed two-dimensional cluster analysis of the data from the four untreated groups. This analysis showed that the inflammatory kidney phenotype corresponded with the presence or absence of proteinuria (Fig. 4) and with the glomerular (but not interstitial) score. Two clearly distinguishable clusters accurately identified mice with and without proteinuria; all of the proteinuric mice had glomerular scores of ≥2, whereas all of the nonproteinuric mice had glomerular scores ≤1.5. Furthermore, within the nonproteinuric group, older mice segregated from younger mice and within the proteinuric group all mice with new onset proteinuria clustered together (Fig. 4).
FIGURE 4.
Two-way hierarchical cluster analysis of genes with significantly altered expression by ANOVA in the four groups of untreated mice color coded as follows: 6w, green; 16w, light blue; 23w NP, dark blue; 23w P, orange; and 36 – 40w, red. Proteinuria and renal glomerular and interstitial scores are shown for each mouse. Gray squares indicate assay not done. Nonproteinuric and proteinuric mice cluster separately. Within the nonproteinuric group, the older mice almost all cluster together and within the proteinuric group the mice with new onset proteinuria cluster together (yellow box).
Change in expression of inflammatory markers upon remission induction
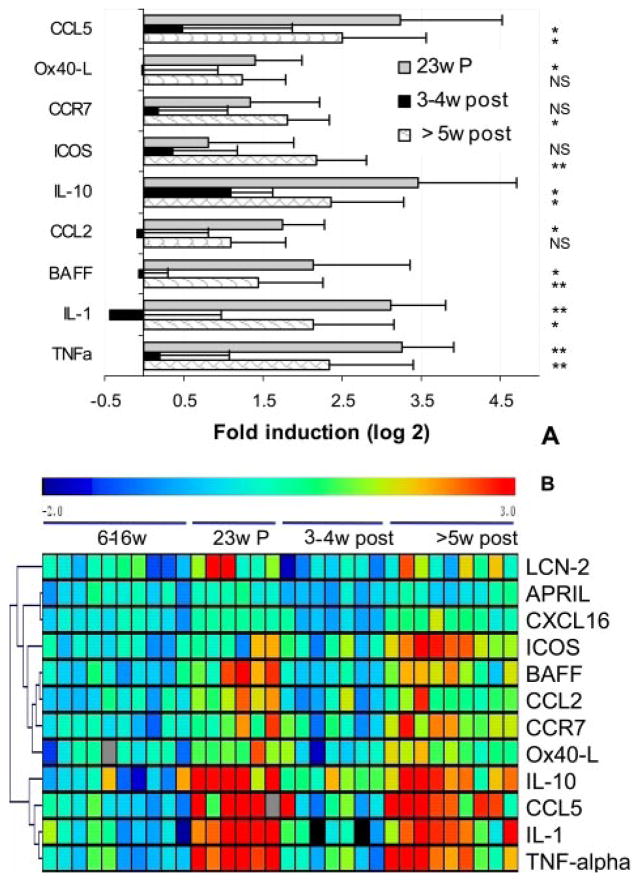
Because remission induction therapy was begun immediately upon development of proteinuria, comparisons were performed between 23-wk-old mice with new onset proteinuria and mice entering remission. Significant decreases were observed in expression of TNF-α, BAFF, IL-1, IL-10, CCL2, and CCL5 upon remission induction. When kidneys from mice sacrificed ≥5 wk after remission induction, most of which had histologic evidence of impending relapse, were compared with those from mice that had just entered remission expression of BAFF, IL-1, IL-10, TNF-α, and CCL5 could again be detected along with increases in the T cell-expressed molecules CCR7 and ICOS (Fig. 5_A_). Results of ANOVA analysis for the three groups are shown in Fig. 5_B_.
FIGURE 5.
Altered renal gene expression after remission induction. A, The genes that were significantly altered 3– 4 wk after remission induction compared with mice with new onset proteinuria (23w P) and genes that changed between early remission (3– 4w post) and the later stages of remission in which relapse is impending (≥5w post). Values were normalized to the mean of young 6- to 16-wk mice given a value of 1. Data are shown as log 2. Genes significantly up- or down-regulated (>2-fold) by SAM and t test (mean + 1 SD) are shown. Values of p for the up-regulated genes are NS, p < 0.01 (*), or p < 0.001 (**) as shown. B, The individual results for genes significantly different by ANOVA analysis in the three groups are shown in the upper panel. Young mice are shown for comparison but were not included in the ANOVA analysis.
Another marker weakly associated with disease remission was lipocalin-2 neutrophil gelatinase-associated lipocalin, a marker of renal damage that is produced by tubular cells and excreted in the urine (5, 6). Lipocalin-2 was not expressed in the NZB/W kidneys before the onset of proteinuria but increased 6-fold in mice with new onset proteinuria and >100-fold in kidneys of mice with established proteinuria. Lipocalin-2 expression completely normalized upon remission induction and although it began to increase again upon histologic relapse, there was significant mouse to mouse variability and average values were only 2-fold over those of young mice (Fig. 5_B_ and Table I). These data show that lipocalin-2 is a marker for interstitial rather than glomerular disease. Urinary NAG (7) may therefore be a useful candidate marker for identifying patients with interstitial and tubular disease that carries a poorer prognosis and for following responses to therapy in such patients.
Almost no differences were noted between the 3- to 4-wk post-treatment kidneys and the 23-wk prenephritic kidneys; the only significant differences were in expression of CXCL13 and ICOS-L (both higher in the remission kidneys). Thus, remission induction returns the kidneys to a late prenephritic state.
Flow cytometry and analysis of gene expression in sorted lymphocyte populations
The number of lymphoid cells in the kidneys of nephritic mice was significantly higher than in young mice. Young kidneys contained fewer resident CD4 T cells (0.5 ± 0.1% in 6 –16 wk vs 2.6 ± 2.2% in 36 – 40 wk; p < 0.02) and macrophages (1.4 ± 0.3% in 6 –16 wk vs 3.8 ± 1.5% in 36 – 40 wk; p < 0.02) than old kidneys and had almost no B cells (0.07 ± 0.02% in 6 –16 wk vs 0.8 ± 0.3% in 36 – 40 wk; p < 0.04) or dendritic cells (0.15 ± 0.02% in 6 –16 wk vs 1.4 ± 0.2% in 36 – 40 wk; p < 0.02).
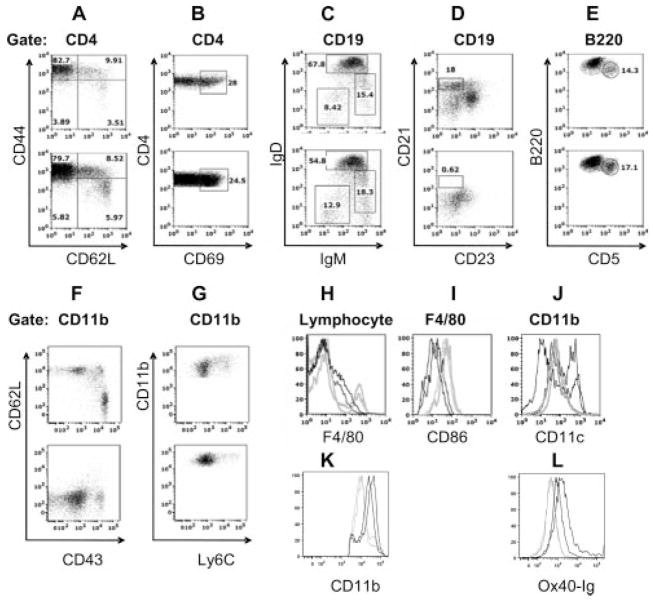
The cell surface phenotype of single-cell preparations from perfused kidneys of nephritic mice was compared with that of spleen cells from the same mice using flow cytometry. The spleen cell phenotype of young and nephritic NZB/W mice has previously been described in detail (3, 8). Nearly all lymphocyte populations were represented in the kidneys (Table II and Fig. 6) in proportions similar to those in matched spleens from the same mice. Both CD4 and CD8 T cells were present in a 2:1 ratio (data not shown); the CD4 T cells, like those in the spleens, were predominantly of the activated/memory (CD44highCD62Llow) phenotype and expressed the early activation marker CD69 (Fig. 6, A and B). B cells were heterogeneous including mature B cells (IgDhighIgMint), class-switched cells (IgD−IgM−) (Fig. 6_C_), and plasma cells (Fig. 7). There was a notable absence of marginal zone cells (Fig. 6_D_) but large numbers of B1 cells were present (Fig. 6_E_).
Table II.
Mean and SD of values of inflammatory markers for all groups analyzed by qPCR
| Average | SD | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6–16w | 23w | 23w P | 36–40w | CRM | PRM | Adjusted pa | 6–16w | 23w | 23w P | 36–40w | CRM | PRM | |
| APRIL | 1.00 | 1.22 | 1.25 | 2.48 | 0.75 | 1.11 | 0.008 | 0.36 | 0.23 | 0.23 | 2.01 | 0.18 | 0.29 |
| BAFF | 1.00 | 0.89 | 5.50 | 8.20 | 0.93 | 2.87 | 9.17E-09 | 0.41 | 0.23 | 4.45 | 6.51 | 0.25 | 1.22 |
| CCL11 | 1.00 | 0.92 | 1.68 | 0.71 | 0.80 | 0.19 | NS | 0.53 | 0.7 | 1.12 | 0.66 | 0.79 | 0.24 |
| CCL19 | 1.00 | 1.33 | 1.98 | 3.21 | 1.32 | 1.83 | 3.59E-06 | 0.37 | 0.36 | 0.52 | 1.58 | 0.36 | 0.74 |
| CCL2 | 1.00 | 1.98 | 3.38 | 6.70 | 1.09 | 2.33 | 2.48E-10 | 0.35 | 0.56 | 1.25 | 3.99 | 0.9 | 1.6 |
| CCL20 | 1.00 | 3.06 | 9.87 | 34.47 | 2.98 | 7.99 | 3.70E-12 | 0.8 | 1.34 | 4.19 | 22.57 | 2.69 | 7.23 |
| CCL21 | 1.00 | 1.03 | 1.32 | 1.08 | ND | ND | NS | 0.33 | 0.37 | 0.39 | 0.34 | ND | ND |
| CCL3 | 1.00 | 1.69 | 9.45 | 37.27 | 6.15 | 10.69 | 4.74E-08 | 0.69 | 1.34 | 6.59 | 59.04 | 7.01 | 7.16 |
| CCL4 | 1.00 | 1.48 | 7.37 | 33.34 | 5.14 | 8.36 | 6.13E-07 | 0.69 | 0.89 | 5.76 | 58.04 | 5.68 | 5.43 |
| CCL5 | 1.00 | 1.69 | 11.05 | 12.59 | 2.06 | 6.33 | 3.46E-11 | 0.48 | 0.51 | 6.9 | 9.32 | 2.6 | 3.52 |
| CCL8 | 1.00 | 1.95 | 1.86 | 8.19 | 1.85 | 2.42 | 9.58E-04 | 0.77 | 1 | 0.63 | 9.44 | 0.9 | 0.94 |
| CCL9 | 1.00 | 0.99 | 4.51 | 8.58 | 1.95 | 1.86 | 1.65E-12 | 0.45 | 0.33 | 2.82 | 6.15 | 0.73 | 1.09 |
| CCR1 | 1.00 | 0.79 | 0.84 | 4.14 | 1.60 | 1.12 | 1.66E-05 | 0.48 | 0.4 | 0.32 | 2.71 | 1.71 | 1.21 |
| CCR10 | 1.00 | 1.52 | 1.68 | 3.55 | 1.87 | 1.77 | 0.002 | 0.42 | 0.61 | 0.72 | 2.85 | 0.74 | 0.38 |
| CCR2 | 1.00 | 1.20 | 4.53 | 7.82 | 3.49 | 3.70 | 3.21E-08 | 0.41 | 0.35 | 1.74 | 5.52 | 2.64 | 1.95 |
| CCR5 | 1.00 | 2.21 | 6.21 | 20.51 | 5.16 | 7.10 | 1.43E-07 | 0.47 | 2.85 | 4.09 | 26.88 | 3.48 | 5.56 |
| CCR6 | 1.00 | 2.31 | 3.95 | 11.00 | 3.47 | 9.34 | 3.82E-06 | 0.56 | 1.14 | 2.42 | 9.86 | 1.64 | 6.15 |
| CCR7 | 1.00 | 1.55 | 2.87 | 4.31 | 1.25 | 3.60 | 7.85E-06 | 0.28 | 0.64 | 1.91 | 3.21 | 0.7 | 1.5 |
| CCR8 | 1.00 | 2.21 | 3.85 | 12.44 | 6.58 | 7.48 | 2.04E-06 | 0.49 | 0.94 | 1.57 | 11.75 | 6.27 | 4.91 |
| CD138 | 1.00 | 1.70 | 1.78 | 4.54 | 5.54 | 1.99 | 1.60E-06 | 0.34 | 0.75 | 0.63 | 3.15 | 6.02 | 0.54 |
| CD40 | 1.00 | 1.05 | 2.40 | 2.74 | 1.15 | 1.88 | 9.97E-06 | 0.36 | 0.35 | 1.2 | 1.22 | 0.54 | 1.18 |
| CHST4 | 1.00 | 2.37 | 3.78 | 3.63 | 5.66 | 2.63 | 0.004 | 0.82 | 1.33 | 2.24 | 3.12 | 6.49 | 1.58 |
| CX3CL1 | 1.00 | 1.93 | 1.85 | 2.42 | 2.75 | 1.28 | NS | 0.51 | 3.2 | 1.11 | 1.33 | 2.42 | 0.36 |
| CXCL10 | 1.00 | 0.62 | 1.91 | 2.31 | 0.98 | 1.43 | 1.33E-05 | 0.56 | 0.5 | 1.3 | 1.05 | 0.49 | 1.09 |
| CXCL11 | 1.00 | 1.09 | 1.96 | 6.78 | 1.00 | 1.73 | 1.75E-09 | 0.31 | 0.17 | 0.53 | 4.59 | 0.41 | 1.19 |
| CXCL12 | 1.00 | 1.38 | 0.99 | 0.72 | 1.08 | 1.01 | NS | 0.28 | 0.86 | 0.52 | 0.47 | 0.35 | 0.52 |
| CXCL13 | 1.00 | 4.76 | 9.41 | 52.62 | 10.88 | 10.53 | 1.00E-13 | 0.44 | 1.9 | 5.42 | 18.19 | 4.78 | 5.26 |
| CXCL16 | 1.00 | 1.27 | 1.47 | 3.39 | 0.84 | 1.76 | 1.03E-11 | 0.22 | 0.35 | 0.04 | 1.03 | 0.31 | 0.59 |
| CXCL9 | 1.00 | 0.98 | 0.67 | 5.12 | 1.43 | 2.01 | 7.04E-09 | 0.23 | 0.26 | 0.53 | 4.12 | 0.99 | 1.59 |
| CXCR3 | 1.00 | 1.69 | 3.29 | 7.49 | 1.74 | 2.22 | 5.32E-11 | 0.27 | 0.53 | 1.52 | 5.29 | 0.59 | 0.96 |
| CXCR4 | 1.00 | 0.94 | 2.47 | 3.70 | 0.86 | 1.17 | 1.47E-04 | 0.58 | 0.15 | 2.2 | 3.41 | 0.28 | 1.03 |
| CXCR5 | 1.00 | 5.03 | 6.20 | 17.50 | 6.85 | 14.94 | 2.27E-11 | 0.69 | 2.61 | 2.38 | 10.26 | 4.41 | 11.4 |
| CXCR6 | 1.00 | 0.91 | 1.26 | 3.77 | 1.23 | 1.21 | 1.63E-05 | 0.49 | 0.22 | 0.49 | 3.12 | 0.58 | 0.7 |
| E-selectin | 1.00 | 2.28 | 13.99 | 8.40 | 5.03 | 7.63 | 3.55E-08 | 0.35 | 1.77 | 9.55 | 5.99 | 4.33 | 4.84 |
| Foxp3 | 1.00 | 2.76 | 3.82 | 17.75 | 8.56 | 7.03 | 1.14E-07 | 0.26 | 0.64 | 1.73 | 24.66 | 10.12 | 4.31 |
| ICAM-1 | 1.00 | 0.83 | 1.51 | 5.87 | 2.19 | 1.79 | 1.89E-07 | 0.28 | 0.31 | 1.32 | 3.94 | 2.73 | 0.62 |
| ICOS | 1.00 | 1.38 | 2.08 | 8.80 | 1.39 | 4.70 | 6.55E-12 | 0.3 | 0.47 | 1.47 | 3.56 | 0.74 | 2.02 |
| ICOS-L | 1.00 | 0.74 | 1.51 | 7.25 | 2.58 | 1.45 | 1.36E-10 | 0.3 | 0.3 | 0.85 | 7.45 | 2.21 | 0.69 |
| IFN-γ | 1.00 | 1.51 | 2.83 | 10.43 | 2.17 | 4.13 | 4.38E-06 | 0.66 | 0.63 | 1.95 | 10.42 | 1.45 | 3.48 |
| IL-1β | 1.00 | 0.92 | 7.76 | 13.19 | 0.84 | 4.29 | 3.79E-09 | 0.66 | 0.5 | 3.63 | 8.7 | 0.6 | 2.32 |
| IL-10 | 1.00 | 0.94 | 10.25 | 14.59 | 1.57 | 4.21 | 6.63E-11 | 1.01 | 0.37 | 8.08 | 11.21 | 0.65 | 2.53 |
| IL-12A | 1.00 | 1.18 | 1.15 | 9.97 | 0.65 | 1.45 | NS | 0.61 | 1.57 | 0.72 | 17.55 | 0.57 | 2.18 |
| IL-17 | 1.00 | 1.88 | 1.97 | 18.10 | 0.96 | 4.76 | NS | 0.92 | 2.54 | 2.1 | 30.76 | 1.01 | 13.21 |
| IL-2 | 1.00 | 0.45 | 0.63 | 3.86 | 0.99 | 1.21 | 2.91E-07 | 0.6 | 0.38 | 0.16 | 3.2 | 0.56 | 0.7 |
| IL-21 | 1.00 | 2.17 | 5.81 | 43.26 | 5.85 | 5.80 | 0.001 | 1.06 | 2.74 | 5.14 | 42.24 | 6.89 | 4.93 |
| IL-4 | 1.00 | 3.85 | 1.22 | 6.63 | 2.98 | 2.44 | NS | 0.42 | 9.09 | 0.56 | 6.11 | 3.14 | 3.91 |
| IL-6 | 1.00 | 0.97 | 3.91 | 47.19 | 1.67 | 3.90 | 3.26E-07 | 0.89 | 1.16 | 1.97 | 53.59 | 1.98 | 4.31 |
| LCN-2 | 1.00 | 0.79 | 5.87 | 138.12 | 0.71 | 2.14 | 4.22E-10 | 0.52 | 0.76 | 6.53 | 156.34 | 0.32 | 1.51 |
| LFA-1 | 1.00 | 1.71 | 4.76 | 5.44 | 1.12 | 2.15 | 0.002 | 0.41 | 1.18 | 3.41 | 4.58 | 0.51 | 1.98 |
| LIGHT | 1.00 | 1.98 | 2.44 | 6.11 | 6.08 | 2.97 | 5.00E-06 | 0.27 | 1.1 | 1.12 | 6.31 | 6.46 | 1.1 |
| L-selectin | 1.00 | 2.29 | 5.15 | 9.02 | 4.88 | 7.87 | 2.12E-06 | 0.3 | 1.21 | 3.84 | 11.34 | 3.11 | 7.31 |
| Lymphotoxinα | 1.00 | 1.24 | 1.14 | 2.54 | 2.10 | 2.33 | 5.08E-04 | 0.47 | 0.68 | 0.22 | 1.35 | 1.57 | 1.08 |
| MADCAM | 1.00 | 2.13 | 3.03 | 5.11 | 2.03 | 3.71 | 0.002 | 0.41 | 1.45 | 1.73 | 5.85 | 1.01 | 5.18 |
| Ox-40L | 1.00 | 1.53 | 2.61 | 8.42 | 1.06 | 2.29 | 3.12E-10 | 0.39 | 0.24 | 1.36 | 5.23 | 0.71 | 0.84 |
| PD-1 | 1.00 | 2.28 | 9.63 | 35.26 | 4.57 | 10.99 | 4.43E-13 | 0.41 | 0.96 | 9.02 | 28.57 | 3.05 | 7.76 |
| PDL1 | 1.00 | 0.94 | 1.54 | 1.24 | 0.88 | 1.26 | NS | 0.62 | 0.25 | 0.65 | 0.47 | 0.48 | 0.56 |
| PDL2 | 1.00 | 1.12 | 2.80 | 8.24 | 1.98 | 1.25 | 2.33E-05 | 0.73 | 0.63 | 2.04 | 8.94 | 1.69 | 1.33 |
| P-selectin | 1.00 | 1.04 | 3.02 | 14.61 | 1.46 | 2.20 | 1.46E-05 | 0.52 | 0.98 | 1.26 | 22.01 | 1.62 | 0.98 |
| TGF-β | 1.00 | 0.93 | 1.67 | 2.64 | 2.21 | 1.02 | 0.006 | 0.47 | 0.3 | 0.48 | 2.36 | 2.86 | 0.72 |
| TNF-α | 1.00 | 1.51 | 9.31 | 10.21 | 1.20 | 5.68 | 2.74E-12 | 0.57 | 0.44 | 3.99 | 6.43 | 0.73 | 3.9 |
| VCAM-1 | 1.00 | 0.79 | 2.34 | 7.10 | 2.34 | 1.83 | 2.62E-11 | 0.35 | 0.25 | 1.04 | 4.26 | 2.96 | 0.72 |
FIGURE 6.
Flow cytometry analysis of renal-infiltrating cells: A–G, The upper plot shows spleen (A–E) or PBMC (F and G) and the lower plot shows kidney cells from the same mouse. Cells were gated on live cells and lymphocytes with further gates as shown above each set of plots. Renal CD4 T cells were activated and were similar in phenotype to those found in the spleen (A and B). Renal B cells included follicular (IgMintIgDhigh), immature (IgMhigh IgDlow), and class-switched (IgMlowIgDlow) cells (C), as well as B1cells (B220int CD5high) (D). Marginal zone B cells (CD23lowCD21high) were absent (E). Both peripheral blood and kidney CD11b- positive monocytes were predominantly negative for Ly6C, but kidney cells had also down-regulated CD43 (F and G), Kidney (gray histograms) and spleen cells (black histograms) from matched mice are compared in H–J. Kidney CD11b cells consisted of a major population of F4/80high macrophages (H) that had up-regulated CD80 and CD86 (I) and a smaller population of CD11chigh dendritic cells (J). Splenic CD11b-positive cells from the same mice contained a small population of F4/80-positive cells that had not up-regulated CD86 and a large population of CD11chigh dendritic cells. Renal CD11b cells from young (gray histograms) and nephritic mice (black histograms) are compared in K and L. CD11b cells from nephritic mice have higher expression of CD11b (K) and Ox-40L (L). Results are representative of samples from at least three mice examined in each group.
A large population of CD11bhigh cells was present both in the kidneys and in the peripheral blood of sick mice. In the peripheral blood, these cells were predominantly of the Ly6ClowGr-1lowCD62LlowCD43high phenotype typical of M2 monocytes (Fig. 6, F and G). There was a small population of Ly6Chigh/intGr1high cells in the peripheral blood; these were CD62LhighCD43int and CD11bintCD11chigh (data not shown), typical of M1 monocytes. In the kidneys, the CD11b-positive cells were also predominantly Ly6low and Gr1low but they had down-regulated expression of CD43 (Fig. 6, F and G). The renal CD11b-positive cells were mostly F4/80high macrophages but there was also a smaller population of CD11bF4/80intCD11chigh dendritic cells (Table I and Fig. 6_H_). In contrast, analysis of CD11b-positive cells in the spleens showed only a small population of F4/80int cells and a larger population of CD11chigh cells (Fig. 6, H and J). Further analysis of the renal F4/80-positive population showed up-regulation of both CD80 and CD86 compared with F4/80-positive spleen cells (Fig. 6_I_).
We next compared the renal CD11b population from nephritic mice with that of resident CD11b-positive cells from young mice. CD11b-positive cells from nephritic mice had significantly higher expression of F4/80 (Fig. 7), CD11b, and Ox-40L (Fig. 6, K and L). Only very small numbers of CD11b-positive cells were found in the spleens and peripheral blood of young mice. To determine the location of inflammatory cells in the kidney, immunohistochemical staining was performed for B220, CD138, CD4, CD8, F4/80, and CD11c. Large perihilar collections first started to appear at the age of 23 wk and these consisted of B cells, T cells, and CD11c-positive dendritic cells with a “cap” of F4/80-positive macrophages (Fig. 7). With the onset of proteinuria, large numbers of strongly positive F4/80 interstitial macrophages appeared in the renal parenchyma and in and around the glomeruli (Fig. 7, A–C) but not within the lymphoid aggregates (arrow, Fig. 7_A_). With further progression of disease, large disorganized perivascular and periglomerular collections of mixed B cells, T cells, and dendritic cells appeared within the kidney parenchyma, again surrounded by several layers of macrophages (Fig. 7, D–F). Small numbers of CD4 T cells were also observed in the interstitium and around glomeruli (Fig. 7_E_). In contrast, B cells, CD8 T cells, and dendritic cells rarely invaded the interstitium but were confined to mixed inflammatory infiltrates around blood vessels and occasionally around glomeruli (Fig. 7, D and F). Large numbers of plasma cells were also found in the perihilar and periglomerular infiltrates (Fig. 7, L and M). Upon remission induction, parenchymal infiltrates resolved, leaving just the initial perihilar collections of cells (Fig. 7_G_). Although L-selectin and Foxp3 were expressed early in the disease process mRNA expression of these markers did not decrease after remission induction. Immunohistochemistry revealed that both L-selectin (Fig. 7, N and O) and CD4+Foxp3-expressing cells (Fig. 7, H–K) were still present in the perihilar collections of remission kidneys, consistent with the qPCR data. Immunohistochemistry also confirmed expression of IL-17 in lymphocytic aggregates in only a few of the older mice (data not shown).
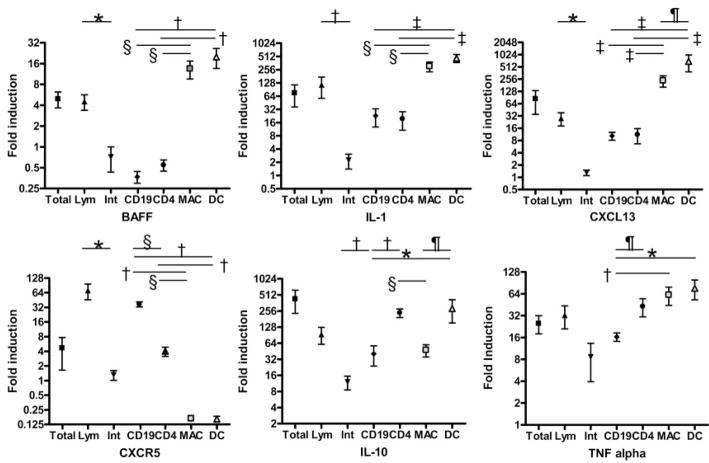
As shown in Fig. 5, remission induction was associated with decreased expression of only a small set of inflammatory mediators whose expression increased again during impending relapse. To determine which cells were making IL-1, BAFF, TNF-α, IL-10, CXCR5, and CXCL13, we performed cell sorting from kidneys of mice with established proteinuria followed by qPCR. We first established that lymphoid cells were making the markers of interest by performing qPCR on separated lymphoid cells and intrinsic renal cells. Fig. 8 shows that BAFF, IL-1, and CXCL13 were made by macrophages and dendritic cells, whereas IL-10 was made by CD4 T cells and dendritic cells with less production by macrophages, CXCR5 was made predominantly by B cells and TNF-α was made by all lymphoid cells except B cells as well as by non-lymphoid cells. (Fig. 8). We also showed that CCL20 and lipocalin-2 were made by intrinsic renal cells and that CCL5 was made predominantly by lymphoid cells but the exact cellular origins of these three markers were not further examined (data not shown). Our findings identify activated macrophages and dendritic cells as the source of critical markers that herald proteinuria onset, remission, and impending relapse.
FIGURE 8.
Cellular source of key markers: fold induction of the indicated inflammatory markers in sorted cells from the kidneys of nephritic mice shown as mean ± SEM. Total kidney cells and sorted lymphoid (Lym) and intrinsic renal cells (Int) were obtained from one group of five mice and CD4 T cells, CD19 B cells, macrophages, and dendritic cells were sorted from a second group as described in the text. Significant differences between groups are as indicated (*, p < 0.03; †, p < 0.01; ‡, p < 0.02; §, p < 0.001; ¶, p < 0.05).
Discussion
The inflamed kidneys of patients with SLE and NZB/W F1 mice contain many lymphocytes around glomeruli, blood vessels, and in the interstitium (9 –11). Our goal was to understand the mechanisms of lymphocytic invasion of the kidneys in SLE nephritis, resolution of nephritis induced by a combination cyclophosphamide and costimulatory blockade and, relapse following cessation of therapy.
Changes in gene expression in the kidney became apparent before either glomerular or interstitial inflammation could be detected by light microscopy and were associated with early immune complex deposition in the glomeruli and formation of a disorganized collection of lymphocytes in the perihilar region. Of the 17 chemokines tested, only CCL20 and CXCL13 and their monogamous receptors were up-regulated at this time. CCL20, a homeostatic chemokine in gut and skin, is also expressed on inflamed endothelial cells (12). CCL20 has been found in the lymph nodes and spinal cords of mice during the sensitization phase of experimental autoimmune encephalomyelitis and may thus play an important role in the early stages of autoimmune inflammation (13). The CCL20 receptor CCR6 is expressed on memory T cells, monocytes, and follicular and memory B cells (14) all of which were found in the perihilar infiltrates.
CXCL13 is found in ectopic lymphoid tissues in other inflammatory diseases and has been associated with focal B cell infiltrates in some diseases (15, 16) and with accumulation of CD4-positive T cells in others (17). The CXCL13 receptor CXCR5 was expressed predominantly by B cells in the NZB/W kidney. CXCL13 has been reported to derive from CD11b/CD11c myeloid dendritic cells in the NZB/W kidney (18) but our studies show that it is expressed both by CD11b/CD11chigh dendritic cells and CD11b/CD11cint macrophages. Although CXCL13 expression is regulated by lymphotoxin _α_1_β_2 in secondary lymphoid organs (19), our data suggest that this is not the means by which it is up-regulated in the lupus kidney. Up-regulation of CXCL13 by peripheral blood monocytes of NZB/W mice can be induced by exposure to IL-1 and TNF-α (20).
Cells expressing regulatory molecules including Foxp3 also appeared in the kidney early in the course of nephritis. Recently, it was reported that Foxp3-expressing T cells appear in the CNS early in the course of experimental autoimmuune encephalomyelitis and remain in the CNS during convalescence, similar to what we have observed here (21, 22). However, presence of these cells was not sufficient to prevent nephritis onset perhaps because activated effector cells become resistant to suppression by regulatory T cells as a result of exposure to IL-6 and TNF-α (22), both of which are highly expressed in the nephritic lupus kidney. PD-1, a CTLA4-like molecule expressed on activated B and T cells, was also expressed early in the disease course. In autoimmune diabetes, the interaction of PDL1, but not PDL2, on pancreatic cells with PD-1 on lymphocytes mediates inhibition of inflammation in the early phases of disease (23). In the NZB/W kidney however only increased expression of PDL2, a receptor expressed predominantly on activated macrophages and dendritic cells (24) was detected.
Significant increases in the expression of several renal chemokines, endothelial activation markers, and monocyte-derived chemokines were observed concomitant with the onset of proteinuria. Four chemokines, CCL2 (MCP-1), CCL3 (MIP-1_α_), CCL5 (RANTES), and CCL9 (MIP-1_γ_), were overexpressed at the onset of proteinuria. These chemokines have previously been identified as crucial mediators of SLE nephritis in other mouse models and in humans (25, 26) and are produced both by intrinsic renal cells and by infiltrating lymphocytes. CCR5, the receptor for CCL3, and CCL5 and CCR2, the receptors for CCL2, were also up-regulated at this time. These receptors have previously been identified on glomerular-infiltrating cells in both murine and human SLE nephritis (25, 27, 28). Interestingly, CCR1, a receptor that can interact with both CCL3 and CCL9, was up-regulated only in established disease. This is consistent with studies showing that CCR1 is involved mainly in interstitial infiltration that occurs in the later stages of disease (29).
The cytokine expression profile at the onset of proteinuria, namely, IL-1, IL-10, TNF-α, IL-6, and BAFF, was suggestive of infiltration of the kidney with activated macrophages and dendritic cells. Examination of sorted cell populations from nephritic kidneys showed that macrophages and dendritic cells were the main source of BAFF, IL-1, and CXCL13 and that IL-10 was made by both CD4 T cells and dendritic cells. BAFF is induced on myeloid cells by TLR ligation and is a survival factor for most B cells. It has also been suggested that activated macrophages express BAFF receptors and can stimulate their own survival via BAFF in an autocrine fashion (30). This may help explain why BAFF blockade can induce remission in nephritic NZB/W mice (3). IL-1, derived from infiltrating monocytes, is a pathogenic cytokine in experimental immune complex glomerulonephritis and induces TNF-α production by intrinsic renal cells (31). Sorting experiments showed that TNF-α derived from multiple cell types including intrinsic renal cells but was regulated concomitantly with macrophage and dendritic cell-derived cytokines, confirming its relationship with IL-1 expression. Although systemic TNF-α protects against initiation of SLE (32), local TNF-α production can promote nephritis once immune complex deposition has occurred (33). IL-10 is also pathogenic in the NZB/W model (34) perhaps because it loses its anti-inflammatory properties in the setting of chronic inflammation (35). Blockade of IL-1, IL-10, and TNF-α can all ameliorate nephritis in mice and humans (36 –38).
Established proteinuria and increasing renal damage were associated with extensive spreading of the inflammatory response, creating a local chemokine and cytokine “storm.” Interestingly, few T cell-derived cytokines were detected in the kidneys until the late stages of disease. In particular, IL-17 was found in none of the mice with new onset proteinuria and in less than half of the aged mice. This is in contrast to other autoimmune inflammatory diseases such as multiple sclerosis and rheumatoid arthritis in which IL-17 is the predominant inflammatory cytokine made by CD4 T cells (39). Similarly, ICOS, a marker of activated and memory T cells, appeared relatively late in the disease course.
Because there have not previously been murine models of remission of lupus nephritis, little is known about the mechanisms for remission and subsequent relapse of this disease. In experimental immune complex disease, spontaneous remission is associated with complete disappearance of inflammatory markers from the kidney (40); however, in our model there was only partial resolution of the inflammatory phenotype. Disease remission was associated with significant down-regulation of TNF-α, IL-1, BAFF, CCL5, IL-10, and Ox-40L, nearly are all expressed either entirely or in part by activated renal macrophages or dendritic cells. Histologic analysis confirmed disappearance of both F4/80high and CD11chigh cells from the kidneys early in the remission process (2); whether these cells died in situ following therapy or migrated out of the kidney remains to be determined. Histologic disease relapse was associated with reexpression of most of these markers, indicating return of activated monocytes to the kidney. Studies in human SLE have similarly shown that clinical and outcome parameters correlate with the degree of glomerular and tubular macrophage infiltration (41).
In nephritic mice, the renal dendritic cell and macrophage populations were segregated topologically: dendritic cells were confined to foci of inflammatory cells surrounding blood vessels and some glomeruli, whereas macrophages were found surrounding the lymphoid aggregates, throughout the interstitium and surrounding area, and occasionally within glomeruli. We show here that the activated renal macrophages were phenotypically different both from peripheral blood monocytes and from the resident macrophages found in young NZB/W kidneys. Renal macrophages in the inflamed kidneys, like those in the peripheral blood, were Ly6Clow but they had acquired a high-level expression of F4/80 and increased expression of CD11b, Ox-40L, CD80, and CD86, confirming an activated phenotype. Renal macrophages also lost expression of CD43, a transmembrane glycoprotein that may have a role in cell adhesion. Finally, these macrophages produced cytokines that can amplify the inflammatory response, leading to irreversible renal damage. Thus, the renal macrophages in the NZB/W kidney appear to be most like type II-activated macrophages (also known as M2b macrophages) that can be induced by ligation of Fc receptors and by exposure to TLRs (42, 43).
In summary, our studies show that there is an ordered progression of the inflammatory process SLE nephritis that is recapitulated during relapse after remission induction therapy. We suggest that therapy for SLE nephritis may require an initial combination regimen that depletes activated effector cells followed by maintenance therapy that targets renal endothelial activation, activation of monocytes, and their migration to the kidney.
Footnotes
1
This work was supported by grants from the Lupus Research Institute (to A.D.), the Alliance for Lupus Research (to A.D.), the New York Systemic Lupus Erythematosus Foundation (to L.S., R.B., and M.R.), and National Institute of Allergy and Infectious Diseases.
5
Abbreviations used in this paper: SLE, systemic lupus erythematosus; qPCR, quantitative PCR; SAM, statistical analysis of microarray.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Davidson A, Aranow C. Pathogenesis and treatment of systemic lupus erythematosus nephritis. Curr Opin Rheumatol. 2006;18:468 – 475. doi: 10.1097/01.bor.0000240356.45550.13. [DOI] [PubMed] [Google Scholar]
- 2.Schiffer L, Sinha J, Wang X, Huang W, von Gersdorff G, Schiffer M, Madaio MP, Davidson A. Short-term administration of costimulatory blockade and cyclophosphamide induces remission of systemic lupus erythematosus nephritis in NZB/W F1 mice by a mechanism downstream of renal immune complex deposition. J Immunol. 2003;171:489 – 497. doi: 10.4049/jimmunol.171.1.489. [DOI] [PubMed] [Google Scholar]
- 3.Ramanujam M, Wang X, Huang W, Liu Z, Schiffer L, Tao H, Frank D, Rice J, Diamond B, Yu KO, et al. Similarities and differences between selective and nonselective BAFF blockade in murine SLE. J Clin Invest. 2006;116:724 –734. doi: 10.1172/JCI26385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan O, Madaio MP, Shlomchik MJ. The roles of B cells in MRL/lpr murine lupus. Ann NY Acad Sci. 1997;815:75– 87. doi: 10.1111/j.1749-6632.1997.tb52046.x. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt-Ott KM, Mori K, Li JY, Kalandadze A, Cohen DJ, Devarajan P, Barasch J. Dual action of neutrophil gelatinase-associated lipocalin. J Am Soc Nephrol. 2007;18:407– 413. doi: 10.1681/ASN.2006080882. [DOI] [PubMed] [Google Scholar]
- 6.Mori K, Nakao K. Neutrophil gelatinase-associated lipocalin as the real-time indicator of active kidney damage. Kidney Int. 2007;71:967–970. doi: 10.1038/sj.ki.5002165. [DOI] [PubMed] [Google Scholar]
- 7.Brunner HI, Mueller M, Rutherford C, Passo MH, Witte D, Grom A, Mishra J, Devarajan P. Urinary neutrophil gelatinase-associated lipocalin as a biomarker of nephritis in childhood-onset systemic lupus erythematosus. Arthritis Rheum. 2006;54:2577–2584. doi: 10.1002/art.22008. [DOI] [PubMed] [Google Scholar]
- 8.Ramanujam M, Wang X, Huang W, Schiffer L, Grimaldi C, Akkerman A, Diamond B, Madaio MP, Davidson A. Mechanism of action of transmembrane activator and calcium modulator ligand interactor-Ig in murine systemic lupus erythematosus. J Immunol. 2004;173:3524 –3534. doi: 10.4049/jimmunol.173.5.3524. [DOI] [PubMed] [Google Scholar]
- 9.Muehrcke RC, Kark RM, Pirani CL, Pollak VE. Lupus nephritis: a clinical and pathologic study based on renal biopsies. Medicine. 1957;36:1–145. [PubMed] [Google Scholar]
- 10.Kuroiwa T, Lee EG. Cellular interactions in the pathogenesis of lupus nephritis: the role of T cells and macrophages in the amplification of the inflammatory process in the kidney. Lupus. 1998;7:597– 603. doi: 10.1191/096120398678920712. [DOI] [PubMed] [Google Scholar]
- 11.Andrews BS, Eisenberg RA, Theofilopoulos AN, Izui S, Wilson CB, McConahey PJ, Murphy ED, Roths JB, Dixon FJ. Spontaneous murine lupus-like syndromes: clinical and immunopathological manifestations in several strains. J Exp Med. 1978;148:1198 –1215. doi: 10.1084/jem.148.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzhugh DJ, Naik S, Caughman SW, Hwang ST. Cutting edge: C-C chemokine receptor 6 is essential for arrest of a subset of memory T cells on activated dermal microvascular endothelial cells under physiologic flow conditions in vitro. J Immunol. 2000;165:6677– 6681. doi: 10.4049/jimmunol.165.12.6677. [DOI] [PubMed] [Google Scholar]
- 13.Kohler RE, Caon AC, Willenborg DO, Clark-Lewis I, McColl SR. A role for macrophage inflammatory protein-3 α/CC chemokine ligand 20 in immune priming during T cell-mediated inflammation of the central nervous system. J Immunol. 2003;170:6298 – 6306. doi: 10.4049/jimmunol.170.12.6298. [DOI] [PubMed] [Google Scholar]
- 14.Kunkel EJ, Butcher EC. Plasma-cell homing. Nat Rev Immunol. 2003;3:822– 829. doi: 10.1038/nri1203. [DOI] [PubMed] [Google Scholar]
- 15.Manzo A, Paoletti S, Carulli M, Blades MC, Barone F, Yanni G, Fitzgerald O, Bresnihan B, Caporali R, Montecucco C, et al. Systematic microanatomical analysis of CXCL13 and CCL21 in situ production and progressive lymphoid organization in rheumatoid synovitis. Eur J Immunol. 2005;35:1347–1359. doi: 10.1002/eji.200425830. [DOI] [PubMed] [Google Scholar]
- 16.Steinmetz OM, Panzer U, Kneissler U, Harendza S, Lipp M, Helmchen U, Stahl RA. BCA-1/CXCL13 expression is associated with CXCR5-positive B-cell cluster formation in acute renal transplant rejection. Kidney Int. 2005;67:1616 –1621. doi: 10.1111/j.1523-1755.2005.00244.x. [DOI] [PubMed] [Google Scholar]
- 17.Bagaeva LV, Rao P, Powers JM, Segal BM. CXC chemokine ligand 13 plays a role in experimental autoimmune encephalomyelitis. J Immunol. 2006;176:7676 –7685. doi: 10.4049/jimmunol.176.12.7676. [DOI] [PubMed] [Google Scholar]
- 18.Ishikawa S, Sato T, Abe M, Nagai S, Onai N, Yoneyama H, Zhang Y, Suzuki T, Hashimoto S, Shirai T, et al. Aberrant high expression of B lymphocyte chemokine (BLC/CXCL13) by CD11b+CD11c+dendritic cells in murine lupus and preferential chemotaxis of B1 cells toward BLC. J Exp Med. 2001;193:1393–1402. doi: 10.1084/jem.193.12.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider K, Potter KG, Ware CF. Lymphotoxin and LIGHT signaling pathways and target genes. Immunol Rev. 2004;202:49 – 66. doi: 10.1111/j.0105-2896.2004.00206.x. [DOI] [PubMed] [Google Scholar]
- 20.Ishikawa S, Nagai S, Sato T, Akadegawa K, Yoneyama H, Zhang YY, Onai N, Matsushima K. Increased circulating CD11b+CD11c+den-dritic cells (DC) in aged BWF1 mice which can be matured by TNF-α into BLC/CXCL13-producing DC. Eur J Immunol. 2002;32:1881–1887. doi: 10.1002/1521-4141(200207)32:7<1881::AID-IMMU1881>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 21.McGeachy MJ, Stephens LA, Anderton SM. Natural recovery and protection from autoimmune encephalomyelitis: contribution of CD4+CD25+regulatory cells within the central nervous system. J Immunol. 2005;175:3025–3032. doi: 10.4049/jimmunol.175.5.3025. [DOI] [PubMed] [Google Scholar]
- 22.Korn T, Reddy J, Gao W, Bettelli E, Awasthi A, Petersen TR, Backstrom BT, Sobel RA, Wucherpfennig KW, Strom TB, et al. Myelin-specific regulatory T cells accumulate in the CNS but fail to control autoimmune inflammation. Nat Med. 2007;13:423– 431. doi: 10.1038/nm1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fife BT, Guleria I, Gubbels Bupp M, Eagar TN, Tang Q, Bour-Jordan H, Yagita H, Azuma M, Sayegh MH, Bluestone JA. Insulin-induced remission in new-onset NOD mice is maintained by the PD-1-PD-L1 pathway. J Exp Med. 2006;203:2737–2747. doi: 10.1084/jem.20061577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keir ME, Francisco LM, Sharpe AH. PD-1 and its ligands in T-cell immunity. Curr Opin Immunol. 2007;19:309 –314. doi: 10.1016/j.coi.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 25.Vielhauer V, Anders HJ, Schlondorff D. Chemokines and chemokine receptors as therapeutic targets in lupus nephritis. Semin Nephrol. 2007;27:81–97. doi: 10.1016/j.semnephrol.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Peterson KS, Huang JF, Zhu J, D’Agati V, Liu X, Miller N, Erlander MG, Jackson MR, Winchester RJ. Characterization of heterogeneity in the molecular pathogenesis of lupus nephritis from transcriptional profiles of laser-captured glomeruli. J Clin Invest. 2004;113:1722–1733. doi: 10.1172/JCI19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tesch GH, Maifert S, Schwarting A, Rollins BJ, Kelley VR. Monocyte chemoattractant protein 1-dependent leukocytic infiltrates are responsible for autoimmune disease in MRL-Faslpr mice. J Exp Med. 1999;190:1813–1824. doi: 10.1084/jem.190.12.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panzer U, Steinmetz OM, Stahl RA, Wolf G. Kidney diseases and chemokines. Curr Drug Targets. 2006;7:65– 80. doi: 10.2174/138945006775270213. [DOI] [PubMed] [Google Scholar]
- 29.Anders HJ, Belemezova E, Eis V, Segerer S, Vielhauer V, Perez de Lema G, Kretzler M, Cohen CD, Frink M, Horuk R, et al. Late onset of treatment with a chemokine receptor CCR1 antagonist prevents progression of lupus nephritis in MRL-Faslpr mice. J Am Soc Nephrol. 2004;15:1504 –1513. doi: 10.1097/01.asn.0000130082.67775.60. [DOI] [PubMed] [Google Scholar]
- 30.Chang SK, Arendt BK, Darce JR, Wu X, Jelinek DF. A role for BLyS in the activation of innate immune cells. Blood. 2006;108:2687–2694. doi: 10.1182/blood-2005-12-017319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timoshanko JR, Kitching AR, Iwakura Y, Holdsworth SR, Tipping PG. Leukocyte-derived interleukin-1β interacts with renal interleukin-1 receptor I to promote renal tumor necrosis factor and glomerular injury in murine crescentic glomerulonephritis. Am J Pathol. 2004;164:1967–1977. doi: 10.1016/s0002-9440(10)63757-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacob CO, McDevitt HO. Tumour necrosis factor-α in murine autoimmune “lupus” nephritis. Nature. 1988;331:356 –358. doi: 10.1038/331356a0. [DOI] [PubMed] [Google Scholar]
- 33.Brennan DC, Yui MA, Wuthrich RP, Kelley VE. Tumor necrosis factor and IL-1 in New Zealand Black/White mice: enhanced gene expression and acceleration of renal injury. J Immunol. 1989;143:3470 –3475. [PubMed] [Google Scholar]
- 34.Ishida H, Muchamuel T, Sakaguchi S, Andrade S, Menon S, Howard M. Continuous administration of anti-interleukin 10 antibodies delays onset of autoimmunity in NZB/W F1 mice. J Exp Med. 1994;179:305–310. doi: 10.1084/jem.179.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji JD, Tassiulas I, Park-Min KH, Aydin A, Mecklenbrauker I, Tarakhovsky A, Pricop L, Salmon JE, Ivashkiv LB. Inhibition of interleukin 10 signaling after Fc receptor ligation and during rheumatoid arthritis. J Exp Med. 2003;197:1573–1583. doi: 10.1084/jem.20021820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tipping PG, Timoshanko J. Contributions of intrinsic renal cells to crescentic glomerulonephritis. Nephron Exp Nephrol. 2005;101:e173– e178. doi: 10.1159/000088165. [DOI] [PubMed] [Google Scholar]
- 37.Llorente L, Richaud-Patin Y, Garcia-Padilla C, Claret E, Jakez-Ocampo J, Cardiel MH, Alcocer-Varela J, Grangeot-Keros L, Alarcon-Segovia D, Wijdenes J, et al. Clinical and biologic effects of anti-interleukin-10 monoclonal antibody administration in systemic lupus erythematosus. Arthritis Rheum. 2000;43:1790 –1800. doi: 10.1002/1529-0131(200008)43:8<1790::AID-ANR15>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 38.Aringer M, Steiner G, Graninger WB, Hofler E, Steiner CW, Smolen JS. Effects of short-term infliximab therapy on autoantibodies in systemic lupus erythematosus. Arthritis Rheum. 2007;56:274 –279. doi: 10.1002/art.22327. [DOI] [PubMed] [Google Scholar]
- 39.Furuzawa-Carballeda J, Vargas-Rojas MI, Cabral AR. Autoimmune inflammation from the Th17 perspective. Autoimmun Rev. 2007;6:169 –175. doi: 10.1016/j.autrev.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Anders HJ, Vielhauer V, Kretzler M, Cohen CD, Segerer S, Luckow B, Weller L, Grone HJ, Schlondorff D. Chemokine and chemokine receptor expression during initiation and resolution of immune complex glomerulonephritis. J Am Soc Nephrol. 2001;12:919 –931. doi: 10.1681/ASN.V125919. [DOI] [PubMed] [Google Scholar]
- 41.Hill GS, Delahousse M, Nochy D, Remy P, Mignon F, Mery JP, Bariety J. Predictive power of the second renal biopsy in lupus nephritis: significance of macrophages. Kidney Int. 2001;59:304 –316. doi: 10.1046/j.1523-1755.2001.00492.x. [DOI] [PubMed] [Google Scholar]
- 42.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677– 686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 43.Mosser DM. The many faces of macrophage activation. J Leukocyte Biol. 2003;73:209 –212. doi: 10.1189/jlb.0602325. [DOI] [PubMed] [Google Scholar]