A causal role for ERG in neoplastic transformation of prostate epithelium (original) (raw)
Abstract
A significant proportion of human prostate cancers carry a chromosomal rearrangement resulting in the overexpression of the ETS transcription factor, ERG; however, the functional significance of this event is poorly understood. We report here that up-regulation of ERG transcript is sufficient for the initiation of prostate neoplasia. In agreement with measurements of ERG transcripts, we found that ERG protein is expressed in neoplastic human prostate epithelium. Overexpression of ERG in prostate cell lines increased cell invasion. Moreover, targeted expression of this transcript in vivo in luminal prostate epithelial cells of transgenic mice results in initiation of prostate neoplasia observed as the development of focal precancerous prostatic intraepithelial neoplasia (PIN). Similar to human cancers, luminal epithelial cells in these PIN lesions displace diminishing in numbers basal epithelial cells and establish direct contact with the stromal cell compartment. Loss of basal cells is considered to be one of the critical hallmarks of human prostate cancer; however, the mechanisms responsible for this event were unknown. We propose that up-regulation of ERG in human prostate cancer activates cell invasion programs that subsequently displace basal cells by neoplastic epithelium. Our data demonstrate that ERG plays an important causal role in the transformation of prostate epithelium and should be considered as a target for prevention or early therapeutic intervention.
Keywords: mouse models, prostate cancer, prostatic intrepithelial neoplasia (PIN), ETS family
An outlier identification approach for the analysis of transcript profiles recently identified recurrent chromosomal rearrangements in human prostate cancer (1). Fusion events joining the androgen-responsive TMPRSS2 gene and members of the erythroblastosis virus E26 oncogene (ETS) family result in the overexpression of a N-terminally truncated or full-length forms of the ERG transcription factor (ΔN-ERG and ERG). Subsequent studies have confirmed that TMPRSS2-ERG fusion is a common genetic event that occurs early in prostate carcinogenesis at the transition between benign and prostatic intraepithelial neoplasia (PIN) epithelium (2). This rearrangement has been correlated with tumor metastasis and negative patient outcome (3, 4). To determine the role of ERG overexpression in prostate carcinogenesis, we modeled this alteration in prostate epithelial cells in culture and in mouse prostate epithelium in vivo. We report that up-regulation of ERG activates prostate cell invasion and results in the displacement of prostate basal epithelium by the luminal cells and the development of PIN.
Results
ERG Expression Produces an Invasive Phenotype Mediated by Serine Protease Activity.
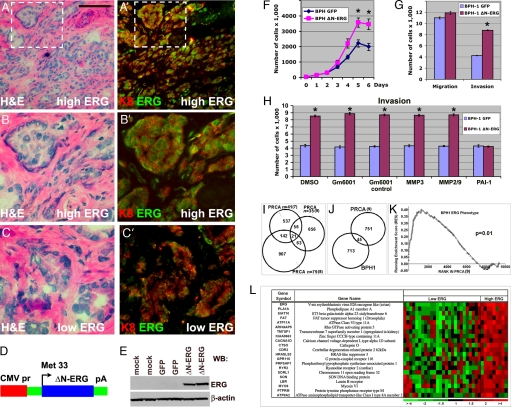
Transcript profiling studies have demonstrated overexpression of the ERG transcription factor in the majority of human primary prostate cancers (5). To determine the presence, cell type specificity of expression, and intracellular localization of ERG protein we performed immunofluorescent stainings with anti-ERG antibodies on human prostate cancers known to express high and low levels of ERG transcripts (n = 4). We found that ERG was confined to the nuclei of malignant prostate epithelial cells coexpressing the luminal cell marker keratin 8 (Fig. 1A–C′). We next modeled the effects of ERG overexpression in cultured prostate epithelial cells. We generated an expression construct encoding ΔN-ERG, identical to the protein predicted to be overexpressed in human prostate cancers after the most frequent TMPRSS2-ERG rearrangement (88% of fusion positive cases), which takes place between the exon 1 of TMPRSS2 and the beginning of ERG exon 4 (6) (Fig. 1 D and E). Overexpression of ΔN-ERG in immortal but nontumorigenic BPH-1 human prostate epithelial cells resulted in increased rates of cell accumulation (Fig. 1F). In addition, Δ-ERG also significantly promoted invasion but had very little effect on migration of BPH-1 cells (Fig. 1G). To differentiate whether increased invasion was driven by serine proteases or matrix metalloproteinases (MMPs), we evaluated the effects of specific inhibitors. Although MMP inhibitors had no effect on invasion, addition of plasminogen activator inhibitor (PAI-1) completely eliminated the differences in invasion between the Δ-ERG expressing and control cells (Fig. 1H). Similar results were obtained with a different transformed but nontumorigenic human prostate cell line RWPE-1 [supporting information (SI) Fig. 5]. These data indicate that Δ-ERG impacts a cell surface serine proteolytic system that promotes invasion of prostate epithelial cells.
Fig. 1.
Cellular and molecular analysis of ERG in primary prostate tumors and prostate cell lines. (A–C′) ERG localizes to the nucleus of luminal epithelial cells in primary human prostate cancer. Serial frozen sections of primary human prostate tumors expressing high (A and B) and low (C and C′) levels of ERG mRNA were stained with H&E (A–C) or anti-ERG (green in A′–C′) and anti-keratin 8 (red in A′–C′, luminal cell marker) antibodies. [Scale bar: 0.11 mm (A and A′) and 50 μm (B–C′).] (D) Schematic representation of ΔN-ERG expression construct. (E) Western blot analysis of cells transfected with GFP or ΔN-ERG construct. Total protein extracts were analyzed with anti-ERG or anti-β-actin antibodies. (F) Changes in the rates of cell accumulation upon overexpression of ΔN-ERG. Growth curves of BPH-1 cells expressing GFP (blue) or ΔN-ERG (pink) are shown (means ± SD, n = 3; *, P < 0.001). (G) Increase in extracellular matrix invasion of BPH-1 cells upon overexpression of ΔN-ERG. Migration and Matrigel invasion of BPH-1 cells overexpressing GFP or ΔN-ERG were analyzed by Transwell migration and invasion assays (means ± SD, n = 3; *, P < 0.001. (H) Increase in ΔN-ERG-induced cell invasion is mediated by cell surface serine proteases. Matrigel invasion of BPH-1 cells overexpressing GFP or ΔN-ERG in the presence of DMSO, GM6001 (broad spectrum MMP inhibitor), GM6001 control, MMP3 inhibitor, MMP2/9 inhibitor, and PAI-1 (cell surface serine protease inhibitor) is shown (means ± SD, n = 3; *, P < 0.001). (I) Transcript alterations associated with high levels of ERG in human prostate tumors (PRCA). Three microarray datasets comprising transcript abundance measurements from human prostate cancers were evaluated. Numbers in Venn diagrams correspond to significant ERG-associated gene expression changes in common between datasets or unique to individual datasets. (J) ERG-associated transcript alterations between immortalized human prostate epithelial cells (BPH-1) overexpressing ERG and human prostate cancers overexpressing ERG. (K) Enrichment plot of BPH ERG gene signature in human tumors. The plot shows the locations of the BPH ERG gene signature in the gene set ranked by the ERG phenotype in human prostate tumors. The running enrichment score (RES) as a function of position in the gene list is shown. The signal-to-noise ranks of all 9,446 genes in the gene set are shown, with low ranks indicating genes up-regulated in high-ERG tumors and high ranks indicating genes down-regulated in high-ERG tumors. BPH-1 ERG signature genes are clearly overrepresented on the left side of the gene list, representing their enrichment in the genes significantly up-regulated in high-ERG tumors (P = 0.01). (L) Genes comprising a signature common between human prostate tumors expressing high ERG levels (7–9). Each heat map column represents a different microdissected human prostate cancer sample, with ratios reflecting transcript abundance relative to the mean.
Transcript Profiles Associated with ERG Expression.
The known functional role of ERG as a transcription factor indicated that overexpression of ΔN-ERG may produce specific changes in gene transcription profiles capable of influencing the development of neoplastic phenotypes. Using oligonucleotide microarrays, we found that high levels of ΔN-ERG altered the expression of 758 genes by at least 1.5-fold in BPH-1 human prostate epithelial cells. To determine whether transcriptional changes caused by overexpression of ΔN-ERG in cultured cells parallel changes associated with ERG expression in human prostate tumors, we identified a signature associated with ERG-expressing primary prostate cancers. We evaluated three different datasets (7, 8) including our own study (9), each comprising at least 35 different cancers, and we partitioned tumors into those with and without high-ERG transcripts. Although a substantial number of genes in each individual dataset correlated with ERG overexpression, the overlap between the datasets numbered only 21 genes (Fig. 1 I and L). The lack of a robust ERG signature is concordant with a prior study delineating ERG coexpression patterns in human prostate cancers that also found minimal overlap and reported only three genes, HDAC1, KCNS3, and MYO6 as being highly significantly associated with ERG-overexpressing tumors across different datasets (10). Further, this result is consistent with the substantial discordance found in the gene expression programs induced by overexpression of ERG fusion proteins (TLS-ERG and EWS-ETS) in different cell lines (11, 12).
The molecular heterogeneity known to be present in human prostate cancers coupled with variables introduced in sample collection and analysis may hamper the ability to discern a reliable ERG signal (13). To address this possibility, we compared the human in vivo ERG signature derived from microdissected prostate cancers with the _in vitro_-derived profiles from BPH-1 cells overexpressing ΔN-ERG (Fig. 1J). Although substantial differences were evident, gene set enrichment analysis (GSEA) determined a significant overlap (P = 0.01) in transcripts associated with elevated ERG expression (Fig. 1 K). Overall, the in vitro studies, coupled with observational data gained from profiling human prostate cancers, suggest that ERG overexpression may differentially influence gene networks depending on the inherent genome state of the cell, and these influences may coevolve with tumor progression.
ΔN-ERG Expression Induces Neoplastic Changes in the Murine Prostate.
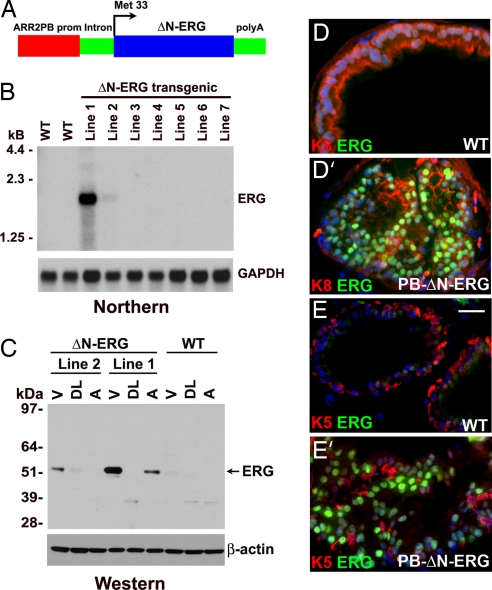
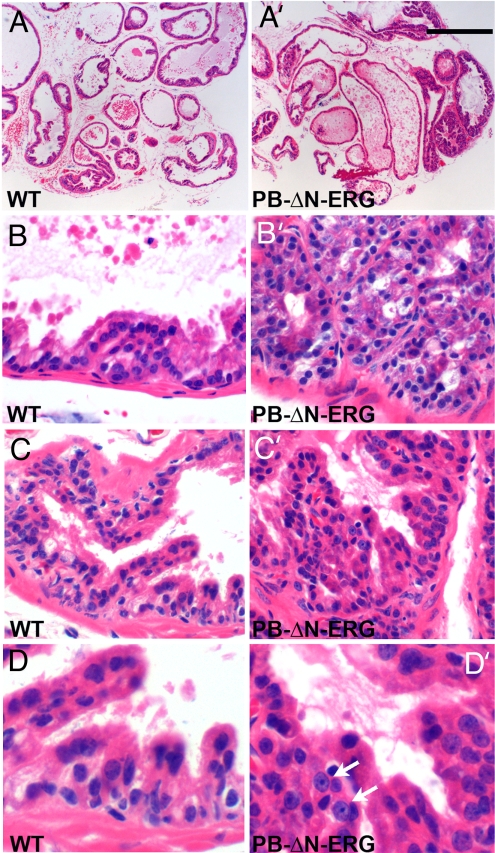
Although suggestive, the in vitro experiments failed to determine whether ΔN-ERG is playing a causal role in prostate neoplasia. To answer this question, we next focused on a direct in vivo approach and developed a mouse model overexpressing ΔN-ERG in prostate epithelial cells. We generated transgenic mice expressing ΔN-ERG protein under the control of modified probasin promoter (ARR2PB) (14, 15) to direct prostate epithelial cell-specific transgenic expression beginning at 10–11 weeks of postnatal development (Fig. 2A). Two transgenic mouse lines expressing ΔN-ERG were generated and analyzed (Fig. 2B). Line 1 expressed significantly higher levels of ERG transcripts than line 2. Western blot analysis of total protein extracts from different lobes of transgenic prostates using anti-ERG antibodies identified the predicted 53-kDa band corresponding to the N-terminally truncated ERG protein (Fig. 2C). The protein was primarily expressed in the ventral prostates with dorsolateral and anterior lobes showing substantially lower levels of expression. Immunofluorescent staining of ventral prostate lobes with anti-ERG antibodies demonstrated epithelial-specific expression of ΔN-ERG protein, which localized to the nuclei of luminal cells (Fig. 2 D and E′). Histologic examination of prostates from 3- to 4-month-old animals revealed no significant abnormalities in the prostate glands of ΔN-ERG transgenic mice (data not shown). However, by 5–6 months of age, focal proliferative and dysplastic PIN lesions were found in the ventral prostates of transgenic line 1 (n = 12, Fig. 3A–D′). Cells in these foci displayed epithelial nuclear pleomorphism and loss of polarization. Both the number of PIN lesions and cellular nuclear pleomorphism increased with the age of the mice. Although only a minority of PIN cells had prominent nucleoli, the percentage of cells with nucleolomegaly increased with advancing age. Overall, the histological features of these lesions were similar to the cribriform and micropapillary patterns of PIN in man (16). Perhaps because of the lower levels of transgene expression in the transgenic line 2, these animals developed PIN later at 10–12 months (n = 3).
Fig. 2.
Generation of transgenic mice overexpressing ΔN-ERG in prostate epithelium. (A) Schematic representation of probasin-ΔN-ERG (PB-ΔN-ERG) transgene. (B) The PB-ΔN-ERG transgenic mouse line 1 expresses highest levels of ΔN-ERG mRNA. Total RNA was extracted from the prostate glands of 13-week-old control (WT) and transgenic (lines 1–7) mice and analyzed by Northern blot hybridization with ERG and GAPDH probes. (C) ΔN-ERG protein is expressed at highest levels in the ventral prostates. Total proteins extracted from ventral (V), dorsolateral (DL), and anterior (A) prostates of 8-month-old transgenic lines 1 and 2 were analyzed by Western blotting with anti-ERG and anti-β-actin antibodies. (D–E′) Immunofluorescent staining of 8-month-old wild-type (WT) and PB-ΔN-ERG ventral mouse prostates with anti-ERG antibodies (green) and anti-keratin 8 (red in D and D′) or anti-keratin 5 (red in E and E′) antibodies is shown. Note nuclear ERG staining in luminal prostate epithelial cells (keratin 8+, keratin 5−) of ΔN-ERG mice. Blue is nuclear DAPI counterstain. [Scale bar: 26 μm (E).]
Fig. 3.
PIN in PB-ΔN-ERG mice. (A–D′) Histologic appearance of ventral prostate lobes from 9-month-old wild-type (WT) and PB-ΔN-ERG animals. Tissue sections were stained with H&E and analyzed with ×4 (A–A′) and ×40 (B–D′) objectives. The PB-ΔN-ERG prostates developed PIN foci of crowded and pseudostratified luminal epithelial cells that formed cribriform (B′) and micropapillary (C′ and D′) architectural patterns. Mutant epithelial cells displayed variable degrees of loss of nuclear polarization and nuclear pleomorphism. Only a minority of PIN cells had prominent nucleoli (arrows in D′). The most cellularized regions of the wild-type glands (B–D) are shown for comparison. [Scale bar: 0.4 mm (A and A′), 40 μm (B–C′), and 20 μm (D and D′).]
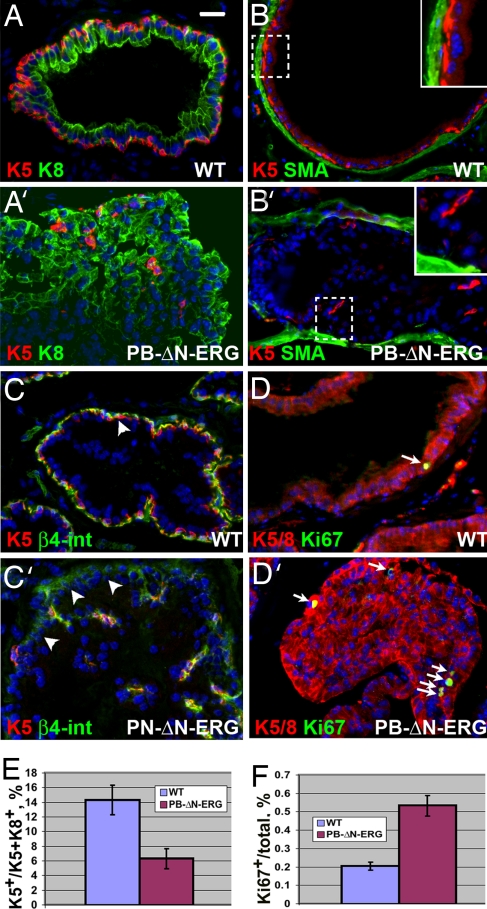
Analyses of prostates from ΔN-ERG transgenic mice with cell type-specific markers revealed substantial changes in glandular cell type composition. In both humans and mice, the normal prostate epithelium consists of two cell layers. The luminal or secretory cell layer contains highly polarized cells facing the prostate lumen, and the basal epithelial layer is located between the luminal cell layer and the basement membrane, a relationship that separates luminal epithelial and stromal cell compartments. One of the critical hallmarks distinguishing the normal human prostate gland from prostate cancer is the loss of basal cells (17), which is paradoxical because almost all epithelial cell proliferation is confined to the basal cell layer in the normal human prostate. Remarkably, both cellular and molecular mechanisms responsible for the loss of basal cells in prostate cancer are completely unknown. As expected, we found that basal (keratin 5+) cells in the control littermates completely encircle the luminal epithelial cell layer and separate luminal cells (keratin 8+) from the stromal cell compartment (smooth muscle actin+ cells) (Fig. 4 A and B and SI Fig. 6_A_). In contrast, basal cells in ΔN-ERG prostates become surrounded by abnormally accumulating luminal cells and are displaced away from the stromal cell compartment (Fig. 4 A′ and B′ and SI Fig. 6_A′_). Examination by electron microscopy confirmed displacement of basal cells and direct contact between luminal cells and the basement membrane in the mutant prostates (SI Fig. 7). Displacement of basal cells was not simply the result of the formation of PIN lesions in the ΔN-ERG prostates because we found that basal cells were not disorganized in PIN lesions from _Nkx3.1_−/−, _PB-Cre4/Pten_−/−, and TRAMP mice (SI Fig. 8). Quantitation revealed a significant decrease in the ratio of basal cells to the total prostate epithelial cells in the ΔN-ERG prostates (Fig. 4E). This finding differs from changes induced by deletion of Pten, which results in increased numbers of basal cells (18). Although in normal prostates the basal epithelial cells use β4-integrin-positive hemidesmosomes to form the contacts with the basement membrane, in ΔN-ERG mice, the luminal epithelial cells start expressing low but detectable levels of β4-integrin as they displace basal cells and directly contact the basement membrane (Fig. 4 C and C′). These changes are likely to have significant consequences because β4-integrin activates multiple survival and proliferation signals in epithelial cells. Analysis of cell proliferation (Ki67+, BrdU+) and apoptotic cell death (active caspase3+, TUNNEL+) revealed very few cycling and apoptotic cells in either wild-type or mutant animals (Fig. 4 D and D′ and data not shown), although the epithelial cell compartment of ΔN-ERG-expressing prostates exhibited a small but significant increase in the proportion of Ki67+ cells (Fig. 4F). These data indicate that similar to human prostate cancer, accumulation of luminal cells in ΔN-ERG mice is a very slow process.
Fig. 4.
Analysis of cell type representation in prostates of PB-ΔN-ERG mice. (A–D′) Immunofluorescent staining of ventral prostate glands from 8-month-old wild-type (WT) and PB-ΔN-ERG transgenic mice with anti-keratin 5 (K5) and anti-keratin 8 (K8) (A and A′), anti-smooth muscle actin (SMA) and anti-keratin 5 (K5) (B and B′), anti-β4-integrin (β4-int) and anti-keratin 5 (K5) (C and C′), and combined anti-keratins 5 and 8 (K5/8) and anti-Ki67 (D and D′) antibodies. Boxed areas in B and B′ are shown at higher magnification in the Insets. Note that although in the wild-type prostate basal cells (K5+) separate stromal (SMO+) and luminal (K8+) cell compartments (A and B), in PB-ΔN-ERG transgenic mice luminal cells displace basal cells and directly contact stromal cell layer (A′ and B′). Note that β4-integrin staining is associated with basal cells (K5+) in the wild-type prostates; however, it is up-regulated in the luminal cells (K5−) in PN-ΔN-ERG prostates (arrowheads in C and C′). (D and D′) Arrows denote cycling cells (Ki67+). (E) Expansion of luminal cells and decrease in proportion of basal cells in 8-month-old PB-ΔN-ERG mice. Ratio of basal cells to the total basal and luminal cell numbers ± SD is shown (n = 3, t test, P = 0.007). (F) Increase in the ratio of proliferating epithelial cells in 8-month-old PB-ΔN-ERG mice. Ratio of Ki67+ cells to the total epithelial cells ± SD is shown (n = 3, t test, P = 0.0006). [Scale bar: 20 μm (A–D′).]
Discussion
Recurrent chromosomal recombination events involving TMPRSS2 and ERG occur commonly in human prostate cancers and often associate with elevated ERG expression. Our studies indicate that overexpression of ERG is likely to be a causal agent in the transformation of prostate epithelial cells. This phenomenon is probably not specific for ERG because similar findings were recently reported in mouse prostates overexpressing ETV1, another ETS family protein overexpressed in a minority of human prostate cancers that results from a less frequent TMPRSS2-ETV1 rearrangement (19). Thus, many of the recombination events that result in overexpression of the ETS family proteins are likely to have tumorigenic effects and play influential roles in human prostate carcinogenesis.
Our analyses of prostate glands from ΔN-ERG mice demonstrate that luminal cells disrupt the basal epithelial layer and establish direct contact with the stromal microenvironment. Mutant luminal cells slowly accumulate and form focal PIN lesions; however, these lesions do not progress to invasive prostate cancer, indicating that additional events are required. These histological features closely parallel early events in human prostate carcinogenesis where disruption of the layer of basal epithelial cells and establishment of a direct contact between luminal cells and the stromal cell compartment is a critical hallmark used by pathologists to differentiate the normal prostate from prostate cancer (17). Remarkably, disorganization and displacement of the basal cells in ΔN-ERG prostates appear to be very specific phenomena because they are not seen in the PIN lesions from other mouse models of prostate neoplasia (SI Fig. 8). Although the exact molecular mechanisms connecting ΔN-ERG to displacement of basal cells by the luminal cells remain to be elucidated, it is likely that a component of these events involves the activation of serine proteases as observed in the in vitro invasion assays. Future studies will be necessary to identify the specific serine proteases responsible for this phenotype and the additional molecular events in ERG expressing neoplastic prostate epithelium that confer the ability to penetrate the basement membrane and extracellular matrix. Together, our data demonstrate that ERG plays an important causal role in prostate cancer initiation and should be considered as a target for prevention or early therapeutic intervention.
Experimental Procedures
Plasmids.
Generation of CMV-driven ΔN-ERG expression construct.
To generate pMAX destination Gateway vector, GFP cDNA in pMAX-GFP vector (Amaxa) was replaced with flanked by attR1-attR2 sequences CmR-ccdB stuffer (Invitrogen). The N-terminally truncated ERG sequence was PCR-amplified from IMAGE clone 6052140 with oligonucleotides 5′-gactagtgagatgaccgcgtcctcctc-3′ and 5′-agcggccgcttagtagtaagtgcccagat-3′ and TA cloned into pCR8/GW/TOPO Gateway entry vector. The PCR-amplified fragment was sequenced and moved by Gateway technology to the pMAX destination vector. The resulting vector expresses N-terminally truncated untagged ERG protein under the control of the CMV promoter.
Generation of PB-ΔN-ERG transgenic construct.
To generate the probasin-TG prostate-specific transgenic construct, the probasin-β-glob-IRES-GFP-poly(A) construct (14) was modified to remove IRES-GFP sequences. The N-terminally truncated ERG sequence was released from pMAX-ΔN-ERG and subcloned into SpeI–NotI sites between the β-globin intron and poly(A) sequences of the probasin-TG construct. Integrity of the transgenic construct was verified by sequencing.
Mice.
The PB-ΔN-ERG-poly(A) fragment was obtained by XhoI–AscI digestion, purified, and injected into fertilized C57BL/6J×CBA mouse eggs. The eggs were transplanted into pseudopregnant females [Fred Hutchinson Cancer Research Center (FHCRC) transgenic facility]. The resulting mice were screened by PCR with primers used for amplification of ERG cDNA (see above). Founder PB-ΔN-ERG mice were bred with 129/Sv animals, and the resulting lines were maintained on the 129/Sv genetic background. For information on TRAMP, _PB-Cre4/Pten_−/−, and _Nkx3.1_−/− mice, see SI Experimental Procedures.
Tissue Dissection, Histology, and Immunofluorescence.
For waxed sections, the ventral, dorsolateral, and anterior prostate lobes were dissected, fixed in 4% formaldehyde in PBS for 30 min, processed, and embedded in paraffin. Sections (5 μm) were stained and examined and photographed with a Nikon TE 200 microscope. For frozen sections, tissue was frozen in OCT and sectioned (7 μm) with a Leica cryostat. For histology, sections were stained with H&E. For immunofluorescent staining, cryosections were subjected to indirect immunostaining and analyzed with the Nikon TE 200 microscope equipped with a COOLSNAP HQ camera. In some cases, tissues were first processed, embedded in paraffin, and sectioned, and the resulting sections were deparaffinized, rehydrated, and processed as described above.
Northern and Western Blotting.
Northern and Western blot analyses were carried out according to standard protocols (20). For Northern analysis, total RNA was extracted by using the TRIzol reagent from Invitrogen. PCR-generated ERG and GAPDH fragments were used as probes.
Microarray Analysis.
Cell line RNA preparation.
Total RNA from BPH-1 cells was isolated by using the RNeasy extraction kit (Qiagen). Samples were treated with DNase by using the Qiagen RNase-free DNase set. RNA was amplified one round by using the Ambion MessageAmp aRNA kit (Applied Biosystems), and sample quality and quantity were assessed by agarose gel electrophoresis and absorbance at _A_260. Cells electroporated with ΔN-ERG or vector controls were harvested at days 3 and 6 for microarray analysis.
Oligonucleotide microarray hybridization.
Labeling with Cy3 and Cy5 fluorescent dyes and hybridization to whole-human genome expression oligonucleotide microarray slides (Agilent Technologies) were performed by following the manufacturer's suggested protocols. Fluorescence array images were collected for both Cy3 and Cy5 by using an Agilent fluorescent scanner, and Feature Extraction software (Agilent Technologies) was used to grid, extract image intensities, and normalize data. Spots of poor quality, as determined by the software, were removed from further analysis.
Generation of ERG signatures.
We had generated a dataset of gene expression from laser capture microdissected human prostate samples (9), and we used it for comparison with the in vitro datasets and other published prostate cancer databases (7, 8). Affimetrix microarray data (8) were normalized by using R (www.r-project.org) and the RMA procedure in the Bioconductor (www.bioconductor.org) package affy. Spotted cDNA microarray data were normalized by fitting a print-tip specific Lowess curve to the log-intensity vs. log-ratio plot. The True et al. data (9) were further normalized with R and the VSN procedure in the Bioconductor package affy to use signal values of tumor rather than tumor vs. matched normal ratios. Samples from the human prostate cancer datasets were determined to be either high-ERG expressers if they achieved 1.4-fold or higher ratios from the mean. Otherwise, they were included in the low-ERG expresser category. Ratios were filtered to include only clones whose expression was measurable in at least 67% of the samples of both groups tested.
We used the Statistical Analysis of Microarray (SAM) program (wwwstat.stanford.edu/∼tibs/SAM) to analyze differences in ratios between high- and low-ERG expressers. Unpaired, two-sample t tests were calculated for each transcript, and genes differentially expressed were identified by using various false discovery rates. Gene expression differences with a false discovery rate of <1% were considered significant. Data from the multiple probes representing the same gene were reduced to unique genes by retaining spots with the highest significance value.
To determine whether phenotypic changes observed in high-ERG tumors were enriched for the genes differentially expressed in BPH-1 cells overexpressing ERG, cDNA array results were subjected to GSEA (www.broad.mit.edu/gsea) (21). To assess the statistical significance of the enrichment score observed in the dataset for the three gene sets, we used permutation testing (1,000) of phenotype labels (e.g., high ERG vs. low ERG), generating a nominal P value.
Antibodies and Apoptosis Staining.
Antibodies used were anti-ERG (Santa Cruz Biotechnology), anti-β-actin and anti-smooth muscle actin (Sigma), anti-β4-integrin (William Carter, FHCRC), anti-Ki67 (Novacastra Laboratories), anti-keratin 8 and anti-vimentin (Hybridoma Bank), anti-activated caspase 3 (Cell Signaling), and anti-keratin 5 (22). Relevant FITC- or Texas red-conjugated donkey or goat antibodies (Jackson Laboratories) were used for detection of primary antibodies. In addition to anti-caspase 3 staining, apoptosis was determined by using FragEL kit from Oncogene Research.
Cells.
BPH-1 cells were provided by Simon Hayward (Vanderbilt University, Nashville, TN) and propagated as described in ref. 23. RWPE-1 cells were from American Type Culture Collection (ATCC) and propagated as suggested by ATCC protocols.
Expression of ΔN-ERG and growth curve analysis.
Cells were nucleoporated with pMAX-ΔN-ERG and pMAX-GFP plasmids by using T buffer and a Amaxa nucleofector device. Efficiency of electroporation varied from 90 to 95%. Immediately after nucleofection, cells were plated at concentration of 25,000 cells per well on 6-well plates and cultured in normal media conditions. Cells were trypsinized every day for 6 consecutive days, and total cell number was determined with a Coulter Counter (Beckman–Coulter).
Motility and invasion analysis.
For motility assays, cells were electroporated with ΔN-ERG or GFP control and allowed to recover for 48 h, and 50,000 cells were placed in serum-free medium on the top of Transwell inserts with 8-μm pore size (BD Biosciences). Ten percent FBS was added to the lower chamber, and 24 h later cells migrating through the membrane were counted. For invasion assays, the upper chambers were first coated with Matrigel, and assays were performed in accordance with published protocols (24). Each experiment was repeated in triplicate. Inhibitors GM6001 (10 μM), GM6001 control (10 μM), MMP3 inhibitor (10 μM), MMP2/9 inhibitor (10 μM), and PAI-1 (500 nM) were from EMD Biosciences. Cells were incubated with inhibitors throughout the experiment starting immediately after electroporation.
Statistical Analysis.
To quantify the ratio of basal (keratin 5+) cells versus total epithelial cells (keratins 5+ and 8+), the prostate sections were stained with antibodies against keratins 5 (K5) and 8 (K8), random ×40 images were taken, and the number of nuclei positive for K5 and K8 per image were counted. To quantify Ki67+ cells, sections were stained with anti-Ki67 and anti-keratin 5 and 8 antibodies, random ×10 images were taken, and the number of K5/8+ and Ki67+ nuclei vs. total number of K5/8+ nuclei was counted. The statistical significance of differences in cell type ratios, proportion of proliferating cells and cell invasion, and proliferation data were determined with a two-tailed Student's t test.
Supplementary Material
Supporting Information
ACKNOWLEDGMENTS.
We thank Dr. William Carter (FHCRC) and Developmental Studies Hybridoma Bank for generous gift of antibodies; Drs. Pradip Roy-Burman (University of Southern California, Los Angeles) Norman Greenberg, Hong Wu, Michael Shen, and Cory Abate Shen (both from the University of Medicine and Dentistry of New Jersey) for providing mouse strains; Nanyan Jiang (FHCRC) for DNA microinjections; Vi Nguyen for help with mouse genotyping; Simon Hayward (FHCRC) for providing cells; and Robert Matusik (Vanderbilt University) for the ARR2B promoter construct. This work was supported by the National Cancer Institute/National Institutes of Health Grants 5R01-CA102365, P01CA85859, and U01CA84294; Department of Defense Grant PC041158; and by the Pacific Northwest Prostate Cancer Special Program of Research Excellence.
Footnotes
The authors declare no conflict of interest.
References
- 1.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 2.Perner S, et al. TMPRSS2-ERG fusion prostate cancer: An early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 3.Perner S, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 4.Demichelis F, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 5.Petrovics G, et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24:3847–3852. doi: 10.1038/sj.onc.1208518. [DOI] [PubMed] [Google Scholar]
- 6.Tu JJ, et al. Gene fusions between TMPRSS2 and ETS family genes in prostate cancer: Frequency and transcript variant analysis by RT-PCR and FISH on paraffin-embedded tissues. Mod Pathol. 2007;20:921–928. doi: 10.1038/modpathol.3800903. [DOI] [PubMed] [Google Scholar]
- 7.Lapointe J, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113:913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.True L, et al. A molecular correlate to the Gleason grading system for prostate adenocarcinoma. Proc Natl Acad Sci USA. 2006;103:10991–10996. doi: 10.1073/pnas.0603678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iljin K, et al. TMPRSS2 fusions with oncogenic ETS factors in prostate cancer involve unbalanced genomic rearrangements and are associated with HDAC1 and epigenetic reprogramming. Cancer Res. 2006;66:10242–10246. doi: 10.1158/0008-5472.CAN-06-1986. [DOI] [PubMed] [Google Scholar]
- 11.Zou J, et al. The oncogenic TLS-ERG fusion protein exerts different effects in hematopoietic cells and fibroblasts. Mol Cell Biol. 2005;25:6235–6246. doi: 10.1128/MCB.25.14.6235-6246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deneen B, et al. PIM3 proto-oncogene kinase is a common transcriptional target of divergent EWS/ETS oncoproteins. Mol Cell Biol. 2003;23:3897–3908. doi: 10.1128/MCB.23.11.3897-3908.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin DW, et al. Influence of surgical manipulation on prostate gene expression: Implications for molecular correlates of treatment effects and disease prognosis. J Clin Oncol. 2006;24:3763–3770. doi: 10.1200/JCO.2005.05.1458. [DOI] [PubMed] [Google Scholar]
- 14.Klezovitch O, et al. Hepsin promotes prostate cancer progression and metastasis. Cancer Cell. 2004;6:185–195. doi: 10.1016/j.ccr.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Thomas TZ, Kasper S, Matusik RJ. A small composite probasin promoter confers high levels of prostate-specific gene expression through regulation by androgens and glucocorticoids in vitro and in vivo. Endocrinology. 2000;141:4698–4710. doi: 10.1210/endo.141.12.7837. [DOI] [PubMed] [Google Scholar]
- 16.Bostwick DG, Brawer MK. Prostatic intraepithelial neoplasia and early invasion in prostate cancer. Cancer. 1987;59:788–794. doi: 10.1002/1097-0142(19870215)59:4<788::aid-cncr2820590421>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 17.Ro JY, Amin MB, Sahin AA, Ayala AG. In: Diagnostic Histopathology of Tumours. Fletcher CDM, editor. Edinburgh: Churchill Livingstone; 1995. pp. 755–761. [Google Scholar]
- 18.Wang S, et al. Pten deletion leads to the expansion of a prostatic stem/progenitor cell subpopulation and tumor initiation. Proc Natl Acad Sci USA. 2006;103:1480–1485. doi: 10.1073/pnas.0510652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomlins SA, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. pp. 7.42–7.44. [Google Scholar]
- 21.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasioukhin V, Bauer C, Degenstein L, Wise B, Fuchs E. Hyperproliferation and defects in epithelial polarity upon conditional ablation of α-catenin in skin. Cell. 2001;104:605–617. doi: 10.1016/s0092-8674(01)00246-x. [DOI] [PubMed] [Google Scholar]
- 23.Hayward SW, et al. Establishment and characterization of an immortalized but nontransformed human prostate epithelial cell line: BPH-1. In Vitro Cell Dev Biol. 1995;31:14–24. doi: 10.1007/BF02631333. [DOI] [PubMed] [Google Scholar]
- 24.Miwa S, et al. The bisphosphonate YM529 inhibits osteolytic and osteoblastic changes and CXCR-4-induced invasion in prostate cancer. Cancer Res. 2005;65:8818–8825. doi: 10.1158/0008-5472.CAN-05-0540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information