TYPE IV PILI: PARADOXES IN FORM AND FUNCTION (original) (raw)
. Author manuscript; available in PMC: 2009 Apr 1.
Published in final edited form as: Curr Opin Struct Biol. 2008 Feb 4;18(2):267–277. doi: 10.1016/j.sbi.2007.12.009
Abstract
Type IV pili are filaments on the surfaces of many Gram-negative bacteria that mediate an extraordinary array of functions, including adhesion, motility, microcolony formation and secretion of proteases and colonization factors. Their prominent display on the surfaces of many bacterial pathogens, their vital role in virulence, and their ability to elicit an immune response make Type IV pilus structures particularly relevant for study as targets for component vaccines and therapies. Structural studies of the pili and components of the pilus assembly apparatus have proven extremely challenging, but new approaches and methods have produced important breakthroughs that are advancing our understanding of pilus functions and their complex assembly mechanism. These structures provide insights into the biology of Type IV pili as well as that of the related bacterial secretion and archaeal flagellar systems. This review will summarize the most recent structural advances on Type IV pili and their assembly components and highlight their significance.
Type IV pili are homopolymers of a 15–20 kDa pilin subunit that emanate from the surfaces of many Gram-negative bacteria and at least one Gram-positive organism. These filaments, which appear smooth and featureless by electron microscopy (EM), are 6–9 nm in diameter and several microns in length (Fig. 1). Beneath their plain façade lies an exquisite helical architecture that provides for strength, flexibility and a multitude of functions, including twitching and gliding motility, adhesion, immune escape, DNA uptake, biofilm formation, microcolony formation, secretion, phage transduction and signal transduction. Unlike other bacterial pili, which use as few as two proteins for assembly [1,2], Type IV pilus biogenesis requires a dozen or more proteins, many of which share sequence conservation among divergent species. Pili are assembled, and in some cases disassembled, rapidly using powerful molecular motors that hydrolyze ATP. The proteins involved in pilus biogenesis form a dynamic but poorly-defined complex that spans both bacterial membranes and the intervening periplasm. Our knowledge of Type IV pili presents several paradoxes: What type of molecular architecture yields such thin flexible filaments that can withstand stresses greater than 100 pN? How does a single filament design provide for such functional diversity? How does the assembly apparatus allow for rapid polymerization and depolymerization at a rate of more than 1000 subunits/second? This review will focus on the latest structural findings, which help to explain these paradoxes and advance our understanding of this remarkable biological machine.
Fig. 1. Neisseria gonorrhoeae GC pili and Vibrio cholerae toxin-coregulated pili imaged by negative-stain electron microscopy.
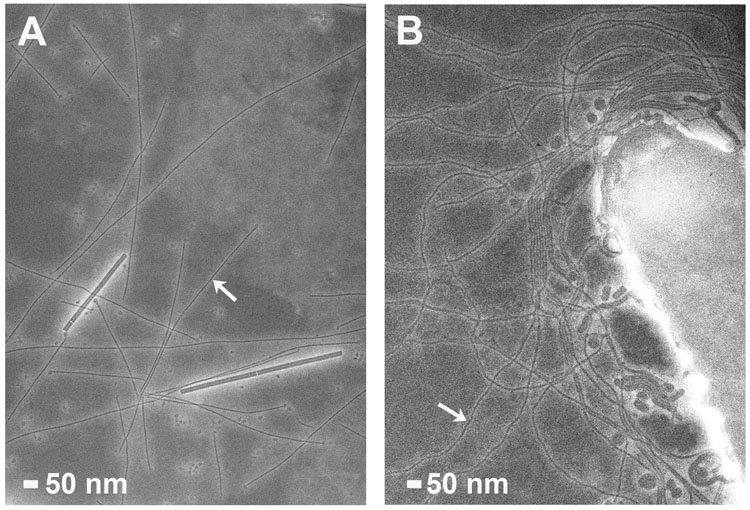
(A) GC pili are indicated by arrows. Tobacco mosaic virus particles (18 Å diameter) are also present. (B) TCP (arrows) emanate from the V. cholerae surface.
Type IV pilus assembly involves 12 or more proteins that in many cases are encoded within the same operon. Several key components are utilized in all Type IV pilus systems and are also found in Type II secretion and archaeal flagellar systems [3–5]. These are: the pilin subunit; an inner membrane prepilin peptidase that cleaves the N-terminal leader peptide; an assembly ATPase that powers pilus assembly; an integral inner-membrane protein that recruits the ATPase from the cytoplasm; and an outer membrane secretin. Many Type IV pilus systems also possess a “retraction” ATPase that drives depolymerization of the pilus filament. The names of the pilus assembly components differ depending on the organism, and are listed in Table 1 for the more well-studied bacteria. In addition, a number of other proteins are required for pilus biogenesis, including pilin-like proteins, which have sequence homology to the pilin subunits in their N-terminal regions.
Table I.
Nomenclature of key Type IV biogenesis components
| Bacteria | Pilin Subunit | Prepilin Peptidase | Assembly ATPase | Retraction ATPase | Inner Membrane Protein | Secretin | Secreted Proteins |
|---|---|---|---|---|---|---|---|
| T4a pili | |||||||
| Pseudomonas aeruginosa | PilA, PilE | PilD | PilB | PilT, PilU | PilC | PilQ | |
| Neisseria gonorrhoeae | PilE | PilD | PilF | PilT | PilG | PilQ | |
| N. meningitidis | PilE | PilD | PilF | PilT | PilG | PilQ | |
| Francisella tularensis | PilE | PilD | PilF | PilT | PilG | PilQ | PepO, Bglx |
| Non-typeable Haemophilus influenzae | PilA | PilD | PilB | PilC | ComE | ||
| Myxococcus xanthus | PilA | PilD | PilT | PilQ | |||
| Clostridium perfringens | PilA1, PilA2 | PilD | PilB | PilT | None | ||
| Dichelobacter nodosus | FimA | FimP | FimN | PilT | FimO | PilQ | Various proteases |
| T4b pili | |||||||
| Vibrio cholerae | TcpA, MshA | TcpJ | TcpT | TcpE | TcpC | TcpF | |
| Enteropathogenic Escherichia coli (EPEC) | BfpA | BfpP | BfpD | BfpF | BfpE | BfpB | |
| Enterotoxigenic E. coli (ETEC) | CofA | CofP | CofH | CofI | CofD | CofJ | |
| Enterotoxigenic E. coli (ETEC) | LngA | LngP | LngH | LngD | |||
| Salmonella Typhi | PilS | PilU | PilR |
The pilin subunit
Type IV pilins are classified based on common features: a homologous and very hydrophobic N-terminal segment (~25 residues), an N-methylated N-terminal residue, and a pair of cysteines in the C-terminal region [6]. To date, two full length Type IV pilin structures have been solved by x-ray crystallography, and structures of a number of truncated pilins, which lack the N-terminal hydrophobic ~28 residues, have been solved by crystallography or nuclear magnetic resonance spectroscopy (NMR) [7]. In spite of limited sequence similarity beyond the first 25 residues, the Type IV pilin subunits all share a common architecture: the N-terminal ~53 residues form an extended α-helix, α1; the N-terminal half of this helix, α1-N, protrudes from the protein and the C-terminal half, α1-C, is embedded in a globular domain and interacts with an anti-parallel four-to five-stranded β-sheet; and the conserved cysteines form a disulfide bond that links the C-terminal segment to the β-sheet (Fig. 2). On either side of this conserved structural scaffold lie two regions that vary substantially from pilin to pilin: the αβ-loop, which is situated between α1 and the β-sheet; and the D-region, encompassed by the conserved cysteines. The Type IV pilins are further classified into two sub-groups, Type IVa and Type IVb, based on the lengths of their signal peptide and mature sequence. The Type IVa pilins are present on a variety of bacteria with broad host ranges, whereas the Type IVb pilins are found almost exclusively on enteric pathogens (Table 1). Although both subtypes share the same overall architecture, the topology of their β-sheets differ, resulting in different protein folds. In the Type IVa pilins, the β-sheet follows the pilin sequence, having N to N+1 nearest neighbor connectivity, as shown for gonococcal (GC) pilin from Neisseria gonorrhoeae (Fig. 2A) [8,9]. In contrast, the β-sheet connectivity for the Type IVb pilins is more complex, with the most C-terminal segment forming the central strand, as shown for Vibrio cholerae TcpA (Fig. 2B) [10]. The most recently-solved pilin structure, an NMR structure of the Type IVb pilin, BfpA, from enteropathogenic E. coli (EPEC), has the general Type IVb pilin architecture, with the C-terminal segment forming the central strand of the β-sheet (Fig. 2C) [11]. However, the β-sheet has 7 β-strands and a different topology and orientation relative to α1-C compared to other Type IVb pilins.
Fig. 2. Structure and schematic representations of Type IV pilin subunits and the pilin-like protein, PilX.
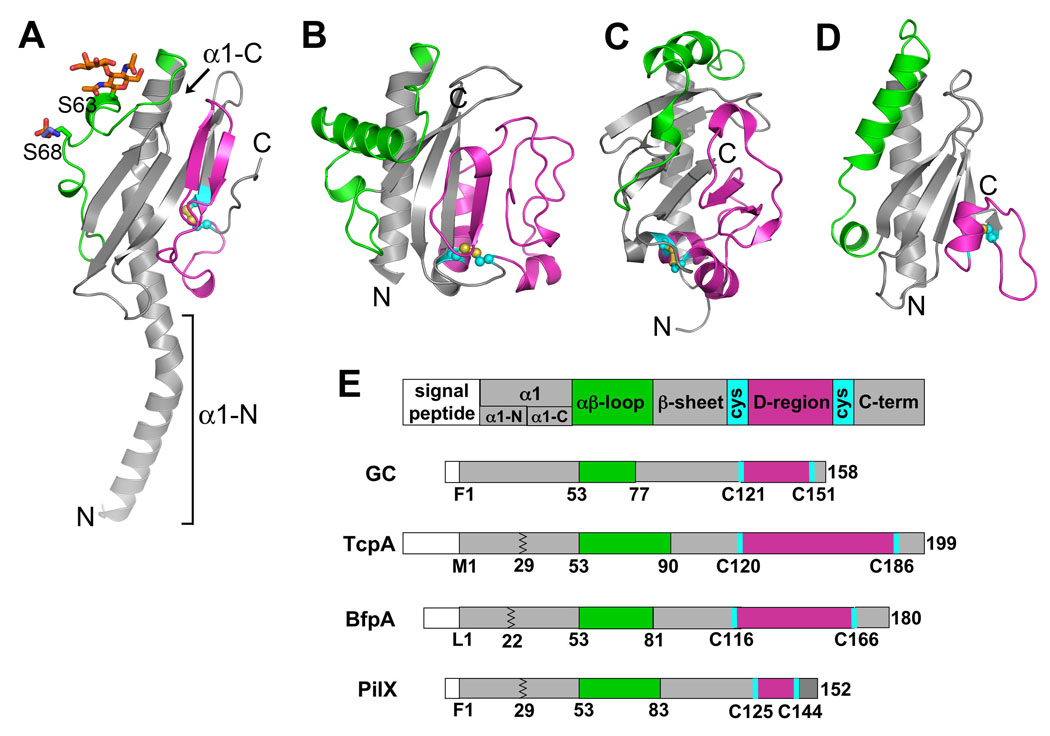
X-ray crystal structures of (A) full length N. gonorrhoeae GC pilin at 2.3 Å resolution [8] and (B) N-terminally truncated V. cholerae TcpA at 1.3 Å resolution [10]. GC pilin has two post-translational modifications: a disaccharide α-D-galactopyranosyl-(1→3)-2,4-diacetamido-2,4-dideoxy-β-D-glucopyranoside covalently attached to Ser63 and a phosphoethanolamine at Ser68. (C) NMR structure of N-terminally truncated EPEC BfpA [11]. (D) X-ray crystal structure of N-terminally truncated N. meningitidis PilX at 2.4 Å resolution [15]. The αβ-loops are colored green and the D-regions are colored magenta. The disulfide-bonded cysteines are shown in yellow and cyan. (E) Schematic representation of the pilins and PilX indicating the relevant regions and residues. The jagged line in TcpA, BfpA and PilX represents the site of truncation for structure determination.
In spite of the different topologies, pilins from many different organisms share the same modular design that allows them to assemble into pilus filaments using the same architectural plan. The conserved structural scaffold holds the subunits together in the filament and the αβ-loop and D-regions define the surface shape and chemistry, and hence functions of the pili. The first Type IV pilus model was proposed based on a single pilin structure, that of N. gonorrhoeae GC pilin [9]. In this model, the hydrophobic α1 helices are twisted in a helical array in the core of the filament, anchoring the globular head domains, which form the outer surface. While new models have been proposed and details have been added or changed, the key features of this early model still hold true. New structural data are providing insights into the mechanism of pilus assembly, the interactions that provide high tensile strength and flexibility, and the molecular strategies used by the pili to accomplish their diverse functions.
The pilus filament
The GC pilus structure, solved to 12.5 Å resolution by cryo-electron microscopy (cryoEM) and iterative helical real space reconstruction (IHRSR), provides the most comprehensive understanding of Type IV pilus structure and assembly to date [8]. The full length pilin subunit structure was computationally docked into the cryoEM reconstruction to produce a “pseudoatomic resolution” structure of the GC pilus (Fig. 3A). The filament is held together by extensive hydrophobic interactions among the N-terminal α-helices in the filament core. The globular domains, on the other hand, are more loosely packed on the filament surface, contacting each other only deep within the filament. This packing results in a highly corrugated filament surface, with grooves running between the globular domains. Some of these grooves are lined with positively charged residues, which may explain the role of GC pili in DNA uptake. DNA could bind in these grooves non-specifically via its negatively charged backbone, and be brought into the cell by pilus retraction. The surface of the globular domains provides additional features relevant to GC pilus functions: the αβ-loop forms a ridge that displays two post-translational modifications, a phosphoethanolamine and a disaccharide, which both undergo phase variation and may also alter their identities; and the D-region forms a second ridge on the subunit surface, which houses the “hypervariable loop”, a region of extreme amino acid sequence variability for both GC pilin and the closely related meningococcal pilin from N. meningitidis. This prominent display of epitopes that continually evolve on the filament surface may explain the ability of the pathogenic Neisseria to evade an effective immune response and establish persistent infections.
Fig. 3. Type IV pilus models.
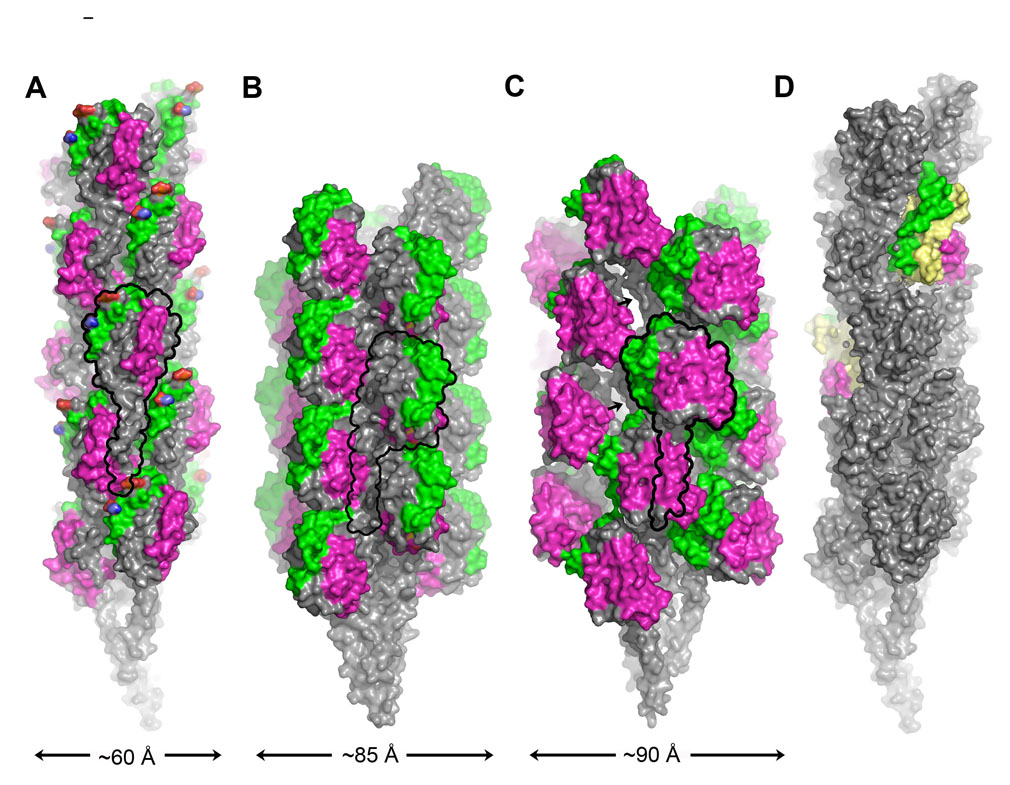
(A) CryoEM reconstruction of the N. gonorrhoeae GC pilus at 12.5 Å resolution colored as in Fig. 2 [8]. (B) EM-based model of EPEC BFP [11]. (C) V. cholerae TCP model based on DXMS analysis and EM-derived structural parameters [12]. Arrows indicate the exposed segment of α1-N. (D) GC pilus model with two subunits replaced by N. meningitidis PilX, colored yellow with the αβ-loop and D-region colored green and magenta, respectively.
A model has also been proposed for the bundle-forming pilus (BFP) from EPEC, based on the BfpA NMR structure and symmetry parameters determined by analysis of negatively-stained filaments [11] (Fig. 3B). A notable feature of BFP is the dominant three-start helix, as indicated by the most visible set of layer lines in the Fourier transforms of the BFP EM images. This feature was also observed for GC pili and V. cholerae toxin-coregulated pili (TCP) [8,10] and has important implications for filament assembly, as discussed below.
Recently, Li et al. used a novel approach, hydrogen/deuterium exchange mass spectrometry (DXMS), to probe the structure of V. cholerae TCP [12]. DXMS exploits the fact that amide hydrogens of proteins and protein complexes exchange with hydrogen in the bulk solvent at a measurable rate that depends on their solvent accessibility. Thus, the relative exposure of a protein can be mapped by incubating it in deuterated buffer for varying amounts of time and measuring deuterium incorporation by mass spectrometry of digested protein fragments. Intact TCP filaments and soluble, monomeric TcpA pilin subunits were analyzed by DXMS to determine the relative surface exposure of different regions of the pilin protein, and hence to identify the subunit:subunit interfaces. The DXMS data were used to refine an earlier computational TCP model, derived from the TcpA crystal structure, crystallographic packing and EM-derived symmetry information [10]. Like GC pili and BFP, TcpA subunits are arranged in a helical array with their N-terminal α-helices oriented toward the filament core (Fig. 3C). In fact, the TcpA subunits are held together almost exclusively by the α1-interactions, while the globular domains make few direct contacts with each other. This architecture produces a more highly variegated surface for TCP than for GC pili: deep pockets or cavities are located between the loosely-packed globular domains, and the D-regions protrude from the filament surface. Surprisingly, these cavities expose a segment of α1-N that was presumed to be buried in TCP and other Type IV pili. The amino acid sequence of this exposed segment is unique to the Type IVb subset of pilins, being glycine-rich and amphipathic. The primary role of TCP in bacterial colonization is to self-aggregate, which holds the bacteria in microcolonies. This new TCP model suggests a mechanism for pilus:pilus interactions, whereby the protruding D-regions of one filament intercalate into the cavities of adjacent filaments, and may even contact the exposed α1-N. In support of this hypothesis, residues shown by mutational analyses of the TcpA subunit to be important for pilus:pilus interactions reside on the exposed D-region [13]. One mutation in particular, Glu158→Leu, did not affect pilus expression levels for the mutant V. cholerae strain, but severely disrupted pilus-mediated cell aggregation and colonization of the infant mouse intestine. The effects of the Glu158→Leu mutation were suppressed by three different mutations in the N-terminal α-helix, two of which converted valines in the exposed segment to glycines. These results imply that pilus:pilus interactions may be mediated in part by a direct interaction between Glu158 in the D-region bulge of one filament and the N-terminal α-helix, which is exposed in the repeating cavities of adjacent filaments. The small side chains corresponding to the Val→Gly suppressors on the exposed α1-N may facilitate this interaction by creating a larger binding pocket to accommodate the bulky uncharged Leu side chain.
In the GC pilus and TCP models, the αβ-loops and the D-regions are optimally exposed and define pilus functions not only by their shape and chemistry, but by the way they come together on the filament surface, creating a complex and repeating landscape on which the pili perform their diverse and essential functions. Furthermore, the Type IV pilus models explain the physical characteristics of these filaments: the extensive interactions among the N-terminal α-helices of the subunits would impart considerable tensile strength, and the presence of cavities and grooves would provide compression spaces to allow the filaments to bend, imparting flexibility. While the GC pilus and TCP structures are held together mostly by interactions among the N-terminal α-helices, it is clear that the globular domains themselves have complementary surfaces that allow them to form helical structures. The N-terminally truncated TcpA subunit crystallized in the P63 space group as a helical filament, held together by complementarity between the αβ-loop and D-region rather than by the extended hydrophobic α-helical interactions present in the native filament [10]. Furthermore, N-terminally truncated pilins from Pseudomonas aeruginosa form helical “nanotubes” when placed in non-polar solvents [14]. These filaments resemble native pili when examined by electron microscopy and have DNA-binding capability. It remains to be seen whether the interactions that hold the nanotubes together also play a prominent role in P. aeruginosa pilus stability, but the current GC and TCP models suggest otherwise. It may be that complementarity between the globular domains facilitates the assembly process but is less important in filament stability, as tight packing among these domains would potentially limit flexibility.
In addition to the functionalities provided by the αβ-loop and D-region, at least one Type IV pilus uses an accessory protein to modify its functions. PilX from N. meningitidis is an 18 kDa pilin-like protein that shares sequence similarity with GC pilin in its N-terminal segment and has a pair of C-terminal cysteines [15]. PilX is not needed for filament assembly but is necessary for pilus:pilus interactions, which are required for N. meningitidis adhesion to host cells. Immunogold labeling showed that PilX associates with meningicoccal pili somewhat randomly along the length of the filaments [15]. To understand the relationship between PilX and the Type IV pili, the N-terminal 28 residues of PilX were deleted to produce a soluble protein, whose crystal structure was determined to 2.4 Å (Fig. 2D). The PilX structure resembles Type IVa pilins, having a conserved structural core comprised of α1-C and an antiparallel 4-stranded β-sheet, with N to N+1 connectivity of the β-strands. PilX also has unique αβ-loop and D-regions. These data led the authors to suggest that PilX is incorporated into the pilus filament during assembly. To visualize this arrangement, GC pilin subunits were replaced with N. meningitidis PilX in the GC pilus filament model (Fig. 3D). This resulted in the αβ-loop and D-region of PilX being exposed on the filament surface as they are for GC pilin. The D-region was shown to mediate pilus:pilus interactions, as deleting this region in whole or in part eliminated N. meningitidis aggregation. Thus, the D-region of PilX may function as it does in V. cholerae TcpA, forming a surface protrusion that interacts with grooves or depressions of adjacent pili to hold cells together. It may be that the hypervariability of the neisserial pilin subunits necessitates the presence of this conserved minor pilin to facilitate pilus:pilus interactions.
Molecular motors driving pilus assembly and disassembly
In contrast to the progress made on the Type IV pilus structure, the mechanism by which these filaments are assembled is still poorly understood. Pilin subunits are synthesized in the cytosol and transported across the inner membrane, most likely via the Sec machinery. These subunits remain anchored in the inner membrane by α1-N, where a dedicated transmembrane prepilin peptidase cleaves off the N-terminal leader sequence on the cytoplasmic side of the subunit, and adds a methyl group to the N-terminal residue [16–18]. The globular domain folds in the periplasm, and disulfide bond formation is catalyzed by an oxidoreductase enzyme [19,20].
However, little is known about the process by which the pilin subunits translocate from this inner membrane reservoir into the growing filament. Polymerization requires ATP hydrolysis by a cytosolic hexameric ATPase, which is recruited to the cytosolic face of the inner membrane by an integral membrane protein [21,22]. For retractile pili, a retraction ATPase is required to rapidly depolymerize the pili, which allows bacteria to move cells along semi-solid surfaces, to transduce phage and transform DNA [23]. Both the assembly and the retraction ATPases belong to the large superfamily of Type II/IV secretion NTPases [24]. Two new crystal structures provide important insights into ATPase-mediated pilus assembly and disassembly.
Until recently, the only structures available for secretion superfamily NTPases were for “traffic ATPases” involved in secretion: the Type II secretion ATPase EpsE from V. cholerae [25], and the Type IV secretion ATPase HP0525 VirB11 from Helicobacter pylori [26,27]. The subunits of these ATPases share a bilobed structure, with an N-terminal domain (NTD) and a C-terminal domain (CTD) connected by a hinge region. Subunits bind nucleotide in the cleft between the two domains via canonical Walker A, Walker B, Asp box and His box ATPase motifs on the CTD, and basic side chains on the NTD. Subunits are arranged in hexameric rings, and are in various conformations ranging from a closed, presumably active conformation where the two domains clamp shut, with nucleotide bound in the cleft, to an open, inactive conformation where the NTD is splayed relative to the CTD. These structures prompted the hypothesis that these ATPase motors function as molecular levers, closing to bind ATP and opening upon ATP hydrolysis to provide a mechanical force that drives secretion [27]. Recently, Forest and co-workers published the first structure of a Type IV pilus retraction motor, PilT, from Aquifex aeolicus [28]. PilT has a bilobed structure and both NTD and CTD are structurally homologous to their corresponding domains in EpsE and HP0525. In each of three structures solved, subunits are arranged in hexameric rings, but only the lowest resolution PilT structure (4.2 Å) possesses active subunit conformations (Fig. 4A–C). In this asymmetric hexamer, the NTD and CTD are brought together in a closed conformation for four of the six subunits, A, C, D and F. Subunit F has clearest density for an ADP in the nucleotide binding site of the CTD, and two arginine fingers in the NTD, Arg95 and Arg110, reach across the binding cleft to potentially stabilize a negatively-charged γ-phosphate leaving group (Fig. 4A). This putative active conformation is thus poised to hydrolyze the bond between the β- and γ-phosphates. The two remaining subunits, B and E, are open, with the NTD twisted ~69° away from the CTD about the linker region, resulting in as much as a 15 Å shift in atom positions. Such a dramatic domain motion, which presumably occurs upon hydrolysis and release of the γ-phosphate, could provide leverage either directly or indirectly, to extract pilin proteins from a pilus filament during retraction. Although the asymmetric structure does not necessarily reflect the native state of the PilT complex, it does imply that subunits can exist in different conformational and active states within a single hexamer, as would be expected for a biological motor.
Fig. 4. Crystal structures of A. aeolicus PilT and A. fulgidus GspE2 ATPases.
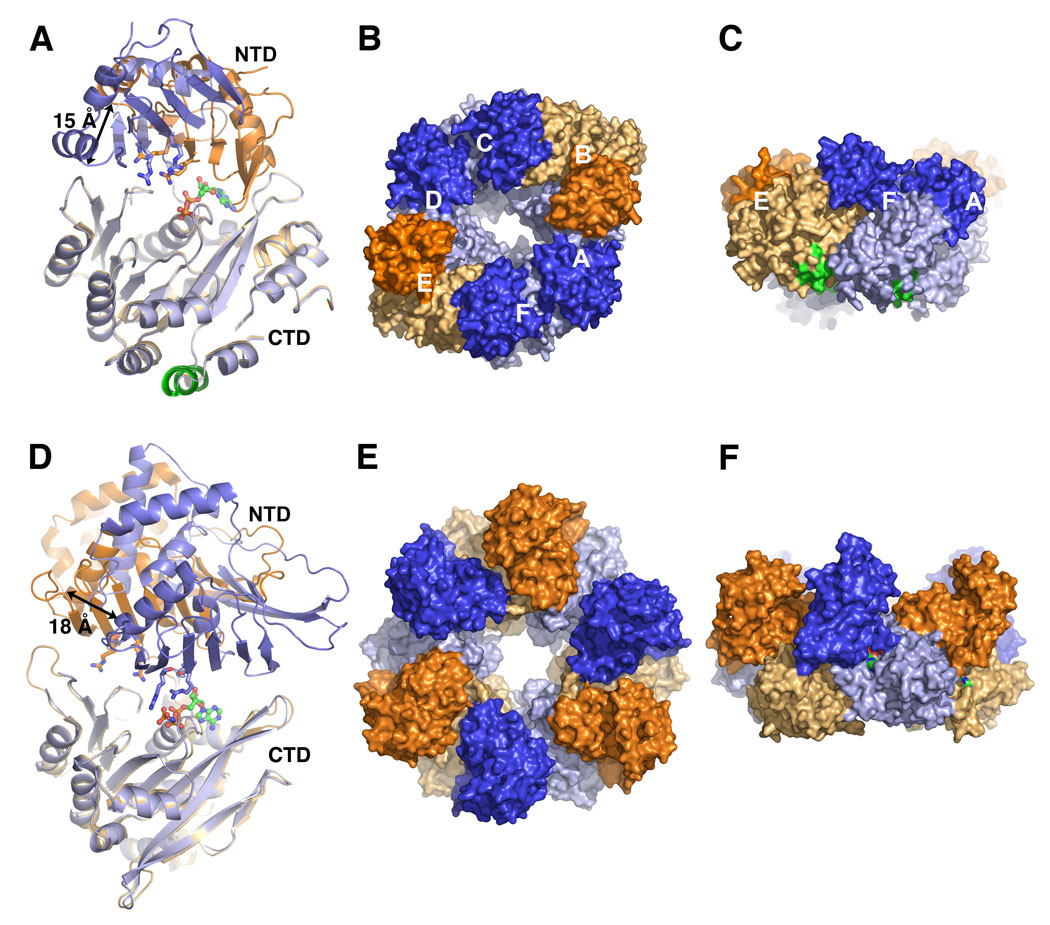
(A) Subunits E (orange) and F (blue) of the 4.2 Å PilT structure [28], representing the open and closed states, respectively, and superimposed via the C-terminal domains (CTD). Dark colors are used for the N-terminal domains (NTD) and light colors are used for the CTD. The NTD arginine fingers are shown as sticks and bound ADP is shown in green and orange ball-and-sticks. The AIRNLIRE motif α-helix (see text) is colored green at the bottom of the CTD. (B) End view of the asymmetric PilT hexamer viewed from the NTD side. (C) Side view of the PilT hexamer. (D) Superposition of the CTDs of the open (orange) and closed (blue) forms of GspE2 bound to AMP-PNP at 2.95 Å resolution [29]. (E) End view and (F) side view of the GspE2 hexamer with alternating open and closed conformations.
Further support for the mechanical lever mechanism for pilus assembly/disassembly comes from a new crystal structure of an archaeal secretion ATPase GspE2 from Archaeoglobus fulgidus, bound to the non-hydrolyzable ATP analog AMP-PNP [29]. Despite minimal sequence similarity, GspE2 is remarkably similar in structure to A. aeolicus PilT, both in overall organization of the NTD and CTD and in the protein fold for each domain (Fig. 4A, D). GspE2 forms a hexameric ring in the crystal lattice, with a bound nucleotide in each subunit, yet the subunits alternate between closed and open forms (Fig. 4D–F). The CTD binds AMP-PNP in both conformations, but the NTD contacts the nucleotide only in the closed conformation via Arg208 and Arg227. Importantly, the presence of a Mg2+ ion, which is necessary for ATP hydrolysis, appears to orient the γ-phosphate of the AMP-PNP such that it can interact with the arginine fingers of the NTD to form the closed and fully active state. GspE2 was further examined in solution by small angle x-ray scattering in the presence of Mg2+ and nucleotide. Interestingly, the scattering profiles for both the ADP- and ATP-bound states fit the profile of the AMP-PNP-bound GspE2 crystal structure with alternating open and closed conformations, whereas AMP-PNP-bound GspE2 produced a scattering curve that best fit a model where all subunits are in a closed position. From these results, it seems that the ATPase can exist with all subunits in a fully active conformation.
In the GspE2 structure, the NTD appears to shift away from the hexameric ring in the open conformation. This domain swing, which presumably occurs upon release of hydrolyzed ATP, would provide a powerstroke that is transmitted across the inner membrane, either directly or via an integral membrane partner, to facilitate extracellular transport and, by analogy, extrusion of the pilus filament [29]. While extracellular secretion and pilus assembly may seem like disparate systems, there is good evidence that the Type II secretion system functions by forming a “pseudopilus” at the inner membrane that spans the periplasm and extrudes toxins and hydrolytic enzymes through an outer membrane secretin [5]. Thus, the mechanisms of the trafficking and pilus assembly ATPases are likely to be similar. The assembly/secretion ATPases have not been shown to interact directly with their corresponding pili or pseudopili, but do interact with inner membrane partners: the EpsE NTD forms a complex with the cytoplasmic N-terminal segment of the integral membrane protein EpsL, a necessary component of Type II secretion in V. cholerae [30]; and the EPEC assembly ATPase BfpD interacts with the N-terminus of BfpE, also an inner membrane protein [31]. It is not known how PilT associates with the inner membrane, but the conserved amino acid sequence AIRNLIRE, which is required for pilus retraction [32], is exposed in an α-helix on the CTD (Fig. 4A, C). In contrast to the domain movements observed for GspE2, it is the CTD that appears to be the mobile elements in PilT. Since PilT functions in depolymerizing pili, its orientation at the inner membrane may differ from those of the assembly/secretion ATPases, which drive polymerization. Obviously, understanding the link between the ATPase motor and the pilus filament is crucial to understanding the pilus assembly mechanism.
The inner membrane protein
New structural data provide a tantalizing glimpse at this putative “missing link”, with a new negative stain EM reconstruction of the inner membrane protein PilG from N. meningitidis [33]. PilG is necessary for pilus assembly [34], but is dispensable when pili are expressed in strains lacking the retraction ATPase, PilT [35]. N. meningitidis PilG was expressed in E. coli with an N-terminal hexa-histidine tag, purified by affinity chromatography, solubilized in detergent and reconstructed using negative stain EM and single particle methods. This ~22 Å structure reveals a missile-shaped molecule with 4-fold symmetry, consistent with a PilG tetramer (Fig. 5). The N-terminus was localized to the cone-shaped bottom in a separate reconstruction using nickel-nitrilotriacetic acid nanogold labeling. The N-terminus of the EPEC PilG ortholog BfpE lies on the cytoplasmic side of the inner membrane [36]. Thus, the narrow “waist” of PilG may represent the transmembrane domains of the PilG subunits, with the upper section, containing four protruding fins, exposed to the periplasm. The PilG architecture provides substantial cytoplasmic and periplasmic domains for interaction with the assembly ATPase and periplasmic proteins, including the pilin subunit.
Fig. 5. EM reconstruction of the N. meningitidis inner membrane protein PilG.
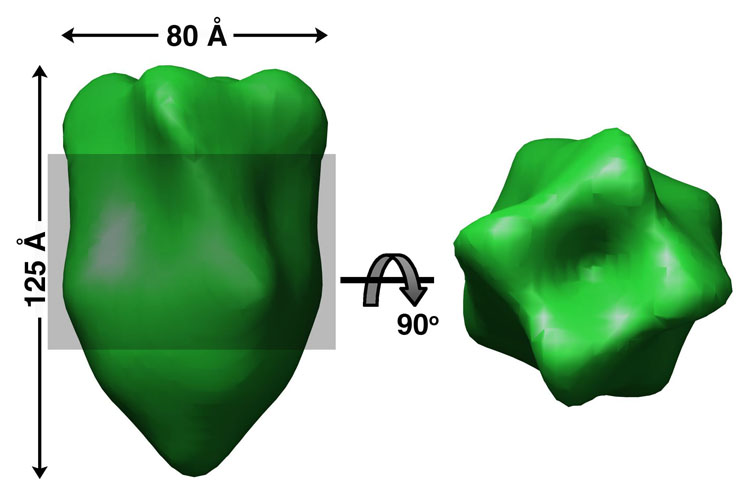
Side and top views of a four-fold symmetrized PilG reconstruction from negatively-stained EM images [33]. The shaded region represents the putative transmembrane “waist”. Nanogold particles bound to the cone-shaped lower domain, indicating the N-terminal region, which is predicted to be located on the cytoplasmic side of the inner membrane.
A model for pilus assembly at the inner membrane
Based on the current state of knowledge, we propose an assembly mechanism whereby pilus filaments assemble from a molecular platform composed minimally of an assembly ATPase and an inner membrane protein (Fig. 6). Pilin subunits are suspended in the inner membrane via their hydrophobic N-terminal α-helices. They are attracted to the growing pilus filament in part due to complementarity between a conserved, negatively-charged Glu5 side chain and the positively-charged N-terminal residue on the terminal subunit, both of which reside in the hydrophobic lipid bilayer and are thus unstable on their own. Additional chemical complementarity between discrete regions of the globular domains, or the globular domain and the N-terminal α-helix, as shown for V. cholerae TCP [12], would help dock the pilin subunits into the polymer. Once a subunit is inserted, the filament is extruded a short distance into the periplasm in response to the mechanical force generated from a single ATP hydrolysis event at one subunit of the assembly ATPase hexamer, located on the cytoplasmic side of the membrane. The ~10 Å upward swing of the GspE2 NTD in going from a closed ATP-bound form to an open ADP-bound form matches that of the rise of the GC pilin subunits in the one-start helix [8,29]. This mechanical force would likely be transmitted through the inner membrane protein (IMP). Subunits would be added to the growing filament one at a time, but at three sites around the base of the filament corresponding to each strand of the 3-start helix. The small outward extrusion of the filament upon addition of one subunit would create a gap at the next strand of the 3-start helix to allow insertion of another subunit, and the filament would subsequently be extruded by ATP hydrolysis at the next active site in the hexameric ATPase. Such a mechanism would allow pilin subunits to be added rapidly and sequentially around the circumference of the filament as each new space opened up. Similarly, pilus retraction would occur by subunits being extracted from the filament, driven by ATP-hydrolysis mediated conformational changes in PilT.
Fig. 6. Model for Type IV pilus assembly.
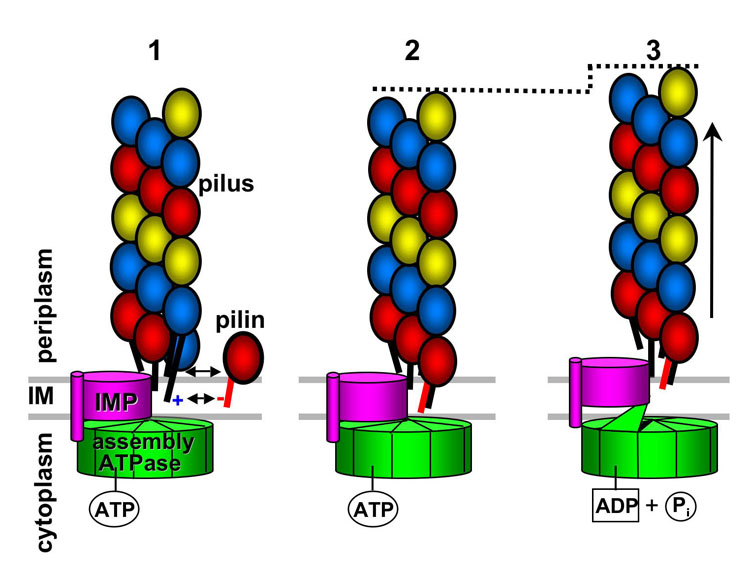
In Step 1, pilin subunits diffuse throughout the inner membrane (IM) and encounter (or are recruited to) the pilus assembly apparatus. The negative charge on Glu5, which makes the subunits somewhat unstable in the lipid bilayer, is attracted to the positively-charged N-terminus of the most terminal pilin subunit in the growing filament. Additional attractive forces between the globular domains allow the subunit to dock into an existing gap at the filament base, thus adding one subunit to the 3-start helical strand colored in red (Step 2). The assembly ATPase is associated with the cytoplasmic side of the inner membrane, possibly via an integral membrane protein (IMP). ATP is bound in the active site cleft of one of the assembly ATPase subunits, causing the N-terminal domain to clamp down on the C-terminal domain. In Step 3, the ATP is hydrolyzed, releasing the N-terminal domain, which twists away from the C-terminal domain. This domain movement induces a conformational change or shift in the associated IMP, which forces the pilus filament out of the membrane by a short distance (~10 Å). This movement results in a new gap at the next strand of the 3-start helix, ready for the addition of a new pilin subunit.
The outer membrane secretin
As the Type IV pilus filament grows, it must pass through the periplasm, including the peptidoglycan layer, and through the outer membrane. To facilitate correct docking of the filament to its outer membrane portal, proteins involved in pilus assembly likely form a large dynamic complex that spans the periplasm, connecting the inner and outer membranes. In support of this model, the BFP assembly complex was isolated by in situ chemical cross-linking and affinity chromatography [37]. This complex contained pilin subunits, integral inner membrane proteins, the assembly and retraction ATPases, the outer membrane secretin, and other proteins involved in pilus assembly. The Type IV pilus secretins are members of a secretin superfamily of complexes that are utilized in pilus assembly, Type II and Type III secretion and filamentous phage release [38]. Secretins are homooligomers of integral membrane proteins with a conserved C-terminal region that is predicted to span the outer membrane and mediate oligomerization. The most well-characterized Type IV pilus secretin is a homododecameric complex of the 82 kDa PilQ protein from N. meningitidis. A 12 Å resolution cryo-negative stain EM reconstruction of this secretin reveals a cage-like structure with four-fold symmetry, consistent with a dodecamer comprised of a tetramer of PilQ trimers (Fig. 7A) [39]. Viewed from the side, the PilQ complex looks like a ring with a “plug” at the bottom and a “cap”, formed by four arms that project from the ring and come together at the top of the complex. The outer diameter of this complex is ~110 Å. Within the complex is a long tapered cavity that is 90 Å in height, and 87 Å in diameter at its broadest point. The topology of the PilQ complex was investigated using insertion epitopes and immunogold labelling [40]. These studies showed that both the N-terminal and C-terminal regions of PilQ localize to the periplasm, including an insert at residue 205, which maps to the arms of the complex. The cavity is large enough to accommodate an assembled GC pilus filament, which is ~60 Å in diameter, but it is obstructed at both ends by the plug and cap structures, as well as by the narrow inner diameter of the ring, and would thus require a substantial conformational change to allow the passage of the filament.
Fig. 7. Structures of the N. meningitidis PilQ secretin complex and PilP lipoprotein.
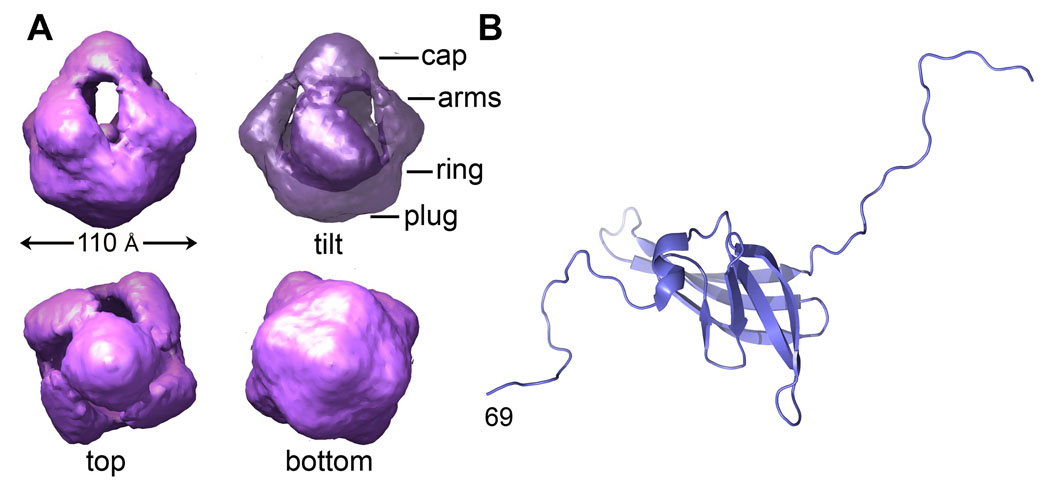
(A) CryoEM reconstruction of the PilQ complex at 12 Å resolution [39]. (B) NMR structure of the PilP lipoprotein fragment [43].
In vitro assays demonstrated a direct interaction between PilQ complexes and one end of purified Type IV pili [41]. A negative-stain reconstruction of the pilus-bound PilQ complex differs from the non-bound complex in that its central cavity was filled and the arms are splayed, thus dissociating the cap. The authors suggest that the observed interaction may represent pili being anchored by and projecting from the extracellular side of the PilQ complex. However, it seems equally plausible given the epitope insertion results that this could represent an interaction on the periplasmic side. The growing pilus filament would insert into the PilQ complex by contacting the cap/arms end, which protrudes into the periplasm, with the ring/plug region spanning the outer membrane. However, substantial changes would still be required for the filament to pass through the ring and plug in the membrane. Additional proteins have been shown to associate with outer membrane secretins and are required for secretin oligomerization and/or pilus assembly. The meningococcal PilP lipoprotein is required for N. meningitidis pilus assembly [35] and interacts directly with the cap region of the PilQ complex, yet also attaches to the inner membrane via a covalently-attached fatty acid [_42_]. An N-terminally truncated PilP structure (residues 69–181) was solved by NMR spectroscopy, revealing a twisted β-sandwich fold and a short α-helix flanked by flexible N- and C-terminal segments [43] (Fig. 7B). Both the N- and the C-terminus of PilP are implicated in PilQ interactions, but the role of PilP in pilus extrusion remains to be elucidated.
Conclusions
New structural studies described here help to explain how a conserved filament architecture for the Type IV pili provides for strength and flexibility, yet displays highly variant filament surfaces, both in terms of their chemistry and molecular landscape, to provide for diverse functionalities. Components of the assembly apparatus form multimeric complexes that must undergo dramatic conformational changes to perform their functions. These changes must be envisioned in the context of an enormous macromolecular machine that physically links the bacterial cytoplasm with the extracellular milieu. We are only beginning to understand how the pilus assembly machinery functions as a coordinated and highly efficient unit. Further progress will require integration of structural results with a broad array of experimental approaches. Parallel studies in bacterial secretion will contribute to this progress and should be especially illuminating with respect to the mechanism of secretion by the Type IV pilus system. The impact of these studies is invaluable for understanding pilus-mediated bacterial functions, and for deriving new strategies to combat and prevent bacterial infections, particularly in the light of rapidly-evolving antibiotic resistances mechanisms.
Acknowledgments
We thank Atsushi Yamagata and Ronald Taylor for insightful discussions, Steve Matthews for the BFP model coordinates and Jeremy Derrick for the PilG and PilQ EM maps. Work in the Craig lab is supported by grants from the Canadian Institutes of Health Research, the National Institute of Allergy and Infectious Diseases and the Natural Sciences and Engineering Research Council of Canada. Figure 2, Figure 3, Figure 4 and Figure 7B were generated using PyMOL (http://www.pymol.org); Figure 5 and Figure 7A were made with Chimera (http://www.cgl.ucsf.edu/chimera).
Abbreviations
ADP
adenosine diphosphate
AMP-PNP
adenylyl imidodiphosphate
ATP
adenosine triphosphate
BFP
bundle-forming pili
CTD
C-terminal domain
DXMS
hydrogen/deuterium exchange mass spectrometry
EM
electron microscopy
EPEC
enteropathogenic E. coli
ETEC
enterotoxigenic E. coli
GC pili
gonococcal pili
NTD
N-terminal domain
pN
piconewtons
TCP
toxin-coregulated pili
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Remaut H, Waksman G. Structural biology of bacterial pathogenesis. Curr Opin Struct Biol. 2004;14:161–170. doi: 10.1016/j.sbi.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Scott JR, Zahner D: Pili with strong attachments: Gram-positive bacteria do it differently. Mol Microbiol. 2006;62:320–330. doi: 10.1111/j.1365-2958.2006.05279.x. [DOI] [PubMed] [Google Scholar]
- 3.Bardy SL, Ng SY, Jarrell KF. Prokaryotic motility structures. Microbiology. 2003;149:295–304. doi: 10.1099/mic.0.25948-0. [DOI] [PubMed] [Google Scholar]
- 4.Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 5.Johnson TL, Abendroth J, Hol WG, Sandkvist M. Type II secretion: from structure to function. FEMS Microbiol Lett. 2006;255:175–186. doi: 10.1111/j.1574-6968.2006.00102.x. [DOI] [PubMed] [Google Scholar]
- 6.Strom MS, Lory S. Structure-function and biogenesis of the Type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 7.Hansen JK, Forest K. Type IV pilin structures: Insights on shared architecture, fiber assembly, receptor binding and Type II secretion. Journal of Molecular Microbiology and Biotechnology. 2006;11:192–207. doi: 10.1159/000094054. [DOI] [PubMed] [Google Scholar]
- 8**.Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Molecular Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004.A new 2.3 Å resolution crystal structure of the full-length GC pilin, showing previously unidentified post-translational modifications, was docked into a cryo-EM reconstruction of the GC pilus. This first high resolution structure of a Type IV pilus has important implications for pilus functions in DNA transport and immune escape and pilus assembly
- 9.Parge HE, Forest KT, Hickey MJ, Christensen DA, Getzoff ED, Tainer JA. Structure of the fibre-forming protein pilin at 2.6 A resolution. Nature. 1995;378:32–38. doi: 10.1038/378032a0. [DOI] [PubMed] [Google Scholar]
- 10.Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, Singh M, Lloyd SJ, Shin DS, Getzoff ED, Yeager M, et al. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell. 2003;11:1139–1150. doi: 10.1016/s1097-2765(03)00170-9. [DOI] [PubMed] [Google Scholar]
- 11.Ramboarina S, Fernandes PJ, Daniell S, Islam S, Simpson P, Frankel G, Booy F, Donnenberg MS, Matthews S. Structure of the bundle-forming pilus from enteropathogenic Escherichia coli. J Biol Chem. 2005;48:40252–40260. doi: 10.1074/jbc.M508099200. [DOI] [PubMed] [Google Scholar]
- 12*.Li J, Lim MS, Li S, Brock M, Pique ME, Woods VLJ, Craig L. Vibrio cholerae toxin-coregulated pilus structure analyzed by hydrogen/deuterium exchange mass spectrometry. Structure. doi: 10.1016/j.str.2007.10.027. in press.This analysis provides a new approach to pilus structure determination and revealed important details about TCP architecture, including a large cavity on the filament surface that exposes the N-terminal α-helix. This new model provides a mechanism for pilus:pilus interactions that may be generalizable to other Type IV pili, and reveals a potential therapeutic target for V. cholerae and other enteric pathogens
- 13.Kirn TJ, Lafferty MJ, Sandoe CM, Taylor RK. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol Microbiol. 2000;35:896–910. doi: 10.1046/j.1365-2958.2000.01764.x. [DOI] [PubMed] [Google Scholar]
- 14*.Audette GF, van Schaik EJ, Hazes B, Irvin RTs. DNA-Binding Protein Nanotubes: Learning from Nature's Nanotech Examples. Nano Lett. 2004;4:1897–1902.A striking observation of solvent-induced filament formation of P. aeruginosa pilins lacking the conserved N-terminal α-helices. While their significance in pilus assembly is not clear, these pilin nanotubes possess pilus functions and have high potential for applications in nanotechnology
- 15.Helaine S, Dyer DH, Nassif X, Pelicic V, Forest KT. 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili. Proc Natl Acad Sci U S A. 2007:15888–15893. doi: 10.1073/pnas.0707581104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaufman MR, Shaw CE, Jones ID, Taylor RK. Biogenesis and regulation of the Vibrio cholerae toxin-coregulated pilus: analogies to other virulence factor secretory systems. Gene. 1993;126:43–49. doi: 10.1016/0378-1119(93)90588-t. [DOI] [PubMed] [Google Scholar]
- 17.Strom MS, Nunn DN, Lory S. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc Natl Acad Sci U S A. 1993;90:2404–2408. doi: 10.1073/pnas.90.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freitag NE, Seifert HS, Koomey M. Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol Microbiol. 1995;16:575–586. doi: 10.1111/j.1365-2958.1995.tb02420.x. [DOI] [PubMed] [Google Scholar]
- 19.Peek JA, Taylor RK. Characterization of a periplasmic thiol:disulfide interchange protein required for the functional maturation of secreted virulence factors of Vibrio cholerae. Proc Natl Acad Sci U S A. 1992;89:6210–6214. doi: 10.1073/pnas.89.13.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang HZ, Donnenberg MS. DsbA is required for stability of the Type IV pilin of enteropathogenic Escherichia coli. Mol Microbiol. 1996;21:787–797. doi: 10.1046/j.1365-2958.1996.431403.x. [DOI] [PubMed] [Google Scholar]
- 21.Crowther LJ, Anantha RP, Donnenberg MS. The inner membrane subassembly of the enteropathogenic Escherichia coli bundle forming pilus. Mol. Microbiology. 2004;52:67–79. doi: 10.1111/j.1365-2958.2003.03963.x. [DOI] [PubMed] [Google Scholar]
- 22.Tripathi SA, Taylor RK. Membrane association and multimerization of TcpT, the cognate ATPase ortholog of the Vibrio cholerae toxin-coregulated-pilus biogenesis apparatus. J Bacteriol. 2007;189:4401–4409. doi: 10.1128/JB.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burrows LL. Weapons of mass retraction. Mol Microbiol. 2005;57:878–888. doi: 10.1111/j.1365-2958.2005.04703.x. [DOI] [PubMed] [Google Scholar]
- 24.Planet PJ, Kachlany SC, DeSalle R, Figurski DH. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc Natl Acad Sci U S A. 2001;98:2503–2508. doi: 10.1073/pnas.051436598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robien MA, Krumm BE, Sandkvist M, Hol WG. Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae. J Mol Biol. 2003;333:657–674. doi: 10.1016/j.jmb.2003.07.015. [DOI] [PubMed] [Google Scholar]
- 26.Yeo HJ, Savvides SN, Herr AB, Lanka E, Waksman G. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori Type IV secretion system. Mol Cell. 2000;6:1461–1472. doi: 10.1016/s1097-2765(00)00142-8. [DOI] [PubMed] [Google Scholar]
- 27.Savvides SN, Yeo HJ, Beck MR, Blaesing F, Lurz R, Lanka E, Buhrdorf R, Fischer W, Haas R, Waksman G. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial Type IV secretion. EMBO J. 2003;22:1969–1980. doi: 10.1093/emboj/cdg223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28**.Satyshur KA, Worzalla GA, Meyer LS, Heiniger EK, Aukema KG, Misic AM, Forest KT. Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility. Structure. 2007;15:363–376. doi: 10.1016/j.str.2007.01.018.This first structure of a pilus retraction ATPase reveals dramatic domain swings and suggests that different binding states can occur in a single PilT hexamer and that conformational changes in one subunit likely affect neighboring subunits. Corresponding residues were mutated in P. aeruginosa and shown to be involved in pilus retraction
- 29**.Yamagata A, Tainer JA. Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. EMBO J. 2007;26:878–890. doi: 10.1038/sj.emboj.7601544.This atomic resolution structure of an archaeal homolog of a Type IV pilus assembly ATPase reveals an active conformational state that depends on the presence of a bound Mg2+ ion. Solution studies support the crystallographic data and imply that all six subunits of the GspE2 hexamer may be in an active conformation in the presence of the non-hydrolyzable ATP analog and Mg2+ but not ADP.
- 30.Abendroth J, Murphy P, Sandkvist M, Bagdasarian M, Hol WG. The X-ray structure of the Type II secretion system complex formed by the N-terminal domain of EpsE and the cytoplasmic domain of EpsL of Vibrio cholerae. J Mol Biol. 2005;348:845–855. doi: 10.1016/j.jmb.2005.02.061. [DOI] [PubMed] [Google Scholar]
- 31.Crowther LJ, Yamagata Y, Craig L, Tainer JA, Donnenberg MS. The ATPase activity of BfpD is greatly enhanced by zinc and allosteric interactions with other Bfp proteins. J. Biol. Chem. 2005;280:24839–24848. doi: 10.1074/jbc.M500253200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Aukema KG, Kron EM, Herdendorf TJ, Forest KT. Functional dissection of a conserved motif within the pilus retraction protein PilT. J Bacteriol. 2005;187:611–618. doi: 10.1128/JB.187.2.611-618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collins RF, Saleem M, Derrick JP. Purification and three-dimensional electron microscopy structure of the Neisseria meningitidis type IV pilus biogenesis protein PilG. J Bacteriol. 2007;189:6389–6396. doi: 10.1128/JB.00648-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tonjum T, Freitag NE, Namork E, Koomey M. Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol Microbiol. 1995;16:451–464. doi: 10.1111/j.1365-2958.1995.tb02410.x. [DOI] [PubMed] [Google Scholar]
- 35.Carbonnelle E, Helaine S, Nassif X, Pelicic V. A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol Microbiol. 2006;61:1510–1522. doi: 10.1111/j.1365-2958.2006.05341.x. [DOI] [PubMed] [Google Scholar]
- 36.Blank TE, Donnenberg MS. Novel topology of BfpE, a cytoplasmic membrane protein required for Type IV fimbrial biogenesis in enteropathogenic Escherichia coli. J Bacteriol. 2001;183:4435–4450. doi: 10.1128/JB.183.15.4435-4450.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hwang J, Bieber D, Ramer SW, Wu CY, Schoolnik GK. Structural and topographical studies of the type IV bundle-forming pilus assembly complex of enteropathogenic Escherichia coli. J Bacteriol. 2003;185:6695–6701. doi: 10.1128/JB.185.22.6695-6701.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bayan N, Guilvout I, Pugsley AP. Secretins take shape. Mol Microbiol. 2006;60:1–4. doi: 10.1111/j.1365-2958.2006.05084.x. [DOI] [PubMed] [Google Scholar]
- 39*.Collins RF, Frye SA, Kitmitto A, Ford RC, Tonjum T, Derrick JP. Structure of the Neisseria meningitidis outer membrane PilQ secretin complex at 12 A resolution. J Biol Chem. 2004;279:39750–39756. doi: 10.1074/jbc.M405971200.This cryoEM reconstruction is the only structure of a pilus secretin. Its closed form implies that the PilQ complex is a gated channel that must undergo a substantial conformational change to allow an intact pilus filament to pass through.
- 40.Frye SA, Assalkhou R, Collins RF, Ford RC, Petersson C, Derrick JP, Tonjum T. Topology of the outer-membrane secretin PilQ from Neisseria meningitidis. Microbiology. 2006;152:3751–3764. doi: 10.1099/mic.0.2006/000315-0. [DOI] [PubMed] [Google Scholar]
- 41.Collins RF, Frye SA, Balasingham S, Ford RC, Tonjum T, Derrick JP. Interaction with Type IV pili induces structural changes in the bacterial outer membrane secretin PilQ. J Biol Chem. 2005;280:18923–18930. doi: 10.1074/jbc.M411603200. [DOI] [PubMed] [Google Scholar]
- 42.Balasingham SV, Collins RF, Assalkhou R, Homberset H, Frye SA, Derrick JP, Tonjum T. Interactions between the lipoprotein PilP and the secretin PilQ in Neisseria meningitidis. J Bacteriol. 2007;189:5716–5727. doi: 10.1128/JB.00060-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Golovanov AP, Balasingham S, Tzitzilonis C, Goult BT, Lian LY, Homberset H, Tonjum T, Derrick JP. The solution structure of a domain from the Neisseria meningitidis lipoprotein PilP reveals a new beta-sandwich fold. J Mol Biol. 2006;364:186–195. doi: 10.1016/j.jmb.2006.08.078. [DOI] [PubMed] [Google Scholar]