Arp2 Links Autophagic Machinery with the Actin Cytoskeleton (original) (raw)
Abstract
Macroautophagy involves lysosomal/vacuolar elimination of long-lived proteins and entire organelles from the cytosol. The process begins with formation of a double-membrane vesicle that sequesters bulk cytoplasm, or a specific cargo destined for lysosomal/vacuolar delivery. The completed vesicle fuses with the lysosome/vacuole limiting membrane, releasing its content into the organelle lumen for subsequent degradation and recycling of the resulting macromolecules. A majority of the autophagy-related (Atg) proteins are required at the step of vesicle formation. The integral membrane protein Atg9 cycles between certain intracellular compartments and the vesicle nucleation site, presumably to supply membranes necessary for macroautophagic vesicle formation. In this study we have tracked the movement of Atg9 over time in living cells by using real-time fluorescence microscopy. Our results reveal that an actin-related protein, Arp2, briefly colocalizes with Atg9 and directly regulates the dynamics of Atg9 movement. We propose that proteins of the Arp2/3 complex regulate Atg9 transport for specific types of autophagy.
INTRODUCTION
Eukaryotic cell homeostasis requires distinct compartmentalization coupled with dynamic exchanges between various endomembrane compartments. To meet changing environmental demands, cellular compartments continually undergo certain morphological adaptations. For example, to mobilize intracellular resources during starvation, cells need to recycle their components, a process that requires degradation and reutilization of the cytoplasm (Klionsky, 2005). Intracellular degradation is achieved mainly by two routes: proteasomal and lysosomal. Proteasomal degradation is limited to single, unfolded proteins. In contrast, macroautophagy (hereafter referred to as autophagy), which is part of the lysosomal degradation system, has a tremendous capacity to degrade large protein complexes and entire organelles (Hamasaki et al., 2005; Mizushima and Klionsky, 2007; Rubinsztein et al., 2007).
Autophagy starts when the intended cargo is surrounded by a double-membrane vesicle, an autophagosome. In starvation conditions, the cargo is bulk cytoplasm. Next, the autophagosome fuses with the lysosome, in mammalian cells, or the vacuole, in yeast. As a result, the autophagosome cargo is released into the organelle lumen and degraded by lysosomal/vacuolar resident hydrolases; the resulting products are subsequently recycled by release back into the cytosol (Shintani and Klionsky, 2004a; Yang et al., 2006).
Analysis in the yeast Saccharomyces cerevisiae has revealed the presence of selective autophagic pathways that target specific cargos, in addition to nonspecific bulk autophagy. These selective pathways morphologically resemble bulk autophagy. An example of a selective autophagy-related process is the cytoplasm-to-vacuole targeting (Cvt) pathway (Baba et al., 1997; Scott et al., 1997). In contrast to starvation-induced autophagy, this is a biosynthetic route, and it occurs constitutively in growing conditions. The Cvt pathway is used by the cell to deliver the precursor form of aminopeptidase I (prApe1), and a second resident hydrolase, α-mannosidase, to the vacuole. These two enzymes are transported to the vacuole in a double-membrane vesicle called a Cvt vesicle. The Cvt vesicle is ∼150 nm in diameter, which is much smaller than the size of autophagosomes (300–900 nm in diameter). Similar to the process of starvation-induced autophagy, the vesicle fuses with the vacuole to release the cargo. Once inside the vacuolar lumen, prApe1 is processed to the mature, fully active hydrolase. Many other examples of selective autophagy have also been described, including specific organelle degradation and the elimination of pathogenic microbes (Dunn et al., 2005; Birmingham and Brumell, 2006; Colombo et al., 2006; Kissova et al., 2007; Zhang et al., 2007).
Approximately 30 _a_u_t_opha_g_y-related (ATG) genes have been identified, predominantly in S. cerevisiae; however, the orthologues of many of the yeast Atg proteins have been found in higher eukaryotes (Klionsky et al., 2003; Levine and Klionsky, 2004). The general process of autophagic degradation has been explored through genetic, cell biological, and biochemical studies. Nevertheless, many aspects of this process still have to be unraveled. For example, the molecular mechanism underlying nucleation of the autophagic sequestering vesicle remains largely unknown.
Many processes involving membrane rearrangement and movement, such as endocytosis, organelle inheritance or membrane ruffling, require the cytoskeleton. Similarly, pharmacological studies have suggested a function for cytoskeletal elements in mammalian autophagy (Aplin et al., 1992; Seglen et al., 1996). Recently, we showed that actin is needed for the selective Cvt pathway (Reggiori et al., 2005a). In conditional actin mutants, or in cells treated with the actin depolymerizing drug latrunculin A, Atg9 cycling is defective. An intriguing question is how actin, and associated factors, regulates the anterograde transport of Atg9 from peripheral sites in the cell to the site of vesicle formation, the phagophore assembly site (PAS). In this study, we discovered that actin-related proteins, comprising the Arp2/3 complex, are required for proper Atg9 function.
The Arp2/3 complex proteins function as nucleators of branched actin filaments, and these proteins are highly conserved through all eukaryotes, including humans (Pinyol et al., 2007; Pollard, 2007). Actin nucleation by the Arp2/3 complex is critical for initial steps of newly formed endocytic vesicle movement. In addition, this complex is involved in intracellular organelle (e.g., mitochondria) transport (Boldogh et al., 2003, Fehrenbacher et al., 2004) as well as being used as a force generator for the movement of invading bacteria inside the host cell (e.g., Listeria monocytogenes and Shigella flexneri) (Boldogh et al., 2001; Welch et al., 1998). The Arp2/3 complex consists of seven subunits: Arp2, Arp3, Arc15/p15, Arc18/p18/p21, Arc19/p19, Arc35/p35, and Arc40/p40. Arp2 and Arp3 are highly similar to the primary actin protein Act1, ∼46 and 39%, respectively, and they are proposed to directly nucleate actin polymerization in the same way that Act1 monomers are assembled into F-actin polymers (actin filaments). The primary logic behind this assumption is the sequence and structural similarity of Arp2 and Arp3 to conventional actin, especially at the barbed end-forming nucleus, which is important for initiation of actin polymerization (Pollard, 2007).
Our studies showed that mutations in genes encoding subunits of the Arp2/3 complex interfered with Atg9 function in growing conditions. This is consistent with the previous finding that an intact cytoskeletal network is essential for the Cvt pathway, but not for bulk autophagy (Reggiori et al., 2005a). In addition, we found that in arp2-1 mutant cells Atg9 movement was severely compromised; Atg9 was unable to localize to prApe1, suggesting a role for Arp2 in directing or regulating Atg9 movement to the cargo. Here, we propose that actin nucleation by the Arp2/3 complex promotes Atg9 movement, and the remodeling of actin structures serves as a scaffold that is necessary for the function of autophagic membranes under vegetative growth conditions.
MATERIALS AND METHODS
Strains and Media
The yeast strains used in this study are listed in Table 1. The arp2-1 mutant was described previously (Moreau et al., 1997). The arc15-10 mutant was identified through analysis of a yeast temperature-sensitive mutant collection provided by Dr. Charles Boone (University of Toronto). Yeast cells were grown in rich medium (YPD: 1% yeast extract, 2% peptone, and 2% glucose) or synthetic minimal medium (SMD: 0.67% yeast nitrogen base, 2% glucose, amino acids, and vitamins). Starvation experiments were performed in synthetic minimal medium lacking nitrogen (SD-N: 0.17% yeast nitrogen base without amino acids, and 2% glucose).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| ARP2-TAP | BY4742 ARP2-TAP::HIS3 | Euroscarf |
| BGY809 | MATa _his3, ura3, LYS2, arc35_Δ::KAN arc35-6::LEU | D'Agostino and Goode (2005) |
| BGY883 | MATa _ura3-52 leu2-3,112, ade2-101, his3_-Δ_200 arc40_Δ::HIS3, arc40-118::LEU2 | Bruce L. Goode |
| CCH014 | SEY6210 ATG9 H192L -3GFP::URA3 ARP2-3DsRed::_LEU2 atg9_Δ::HIS3 | This study |
| DDY1797 | MATα ura3-52 leu2-1 his3_-Δ_200 tub2-201 lys2-801 ade2-101 trp1_-Δ_63 arp2-1 | This study |
| DDY1960 | MATa las17-11::LEU2 ade2-101 his3_-Δ_200 lys2-801 leu2-3,112 ura3-52 | Duncan et al. (2001) |
| DDY2885 | MATa las17_Δ_WCA::HIS3 pan1_Δ_855-1480::_URA3 his3_-Δ_200 lys2-801 leu2-3,112 ura3-52 bar1_Δ::LEU2 | Toshima et al. (2005) |
| DDY3045 | MATa las17_Δ_WCA::_HIS3 his3_-Δ_200 leu2-3,112 ura3-52 bar1_Δ::LEU2 | Toshima et al., (2005) |
| FRY171 | SEY6210 ATG9PA::TRP1 | He et al. (2006) |
| IRA020 | ARP2-TAP::_HIS3 atg9_Δ::LEU2 | This study |
| IRA023 | ARP2-TAP::_HIS3 atg1_Δ::URA3 | This study |
| IRA028 | ARP2-TAP::_HIS3 atg11_Δ::KAN | This study |
| IRA029 | SEY6210 ATG9-3GFP::URA3 | This study |
| IRA030 | DDY1797 ATG9-3GFP::URA3 | This study |
| IRA031 | DDY1797 ATG9-3GFP::_URA3 atg1_Δ::HIS3 | This study |
| IRA033 | IRA029 promTPI1-ARP2-3DsRed::LEU2 | This study |
| IRA036 | DDY1797 PEX14-GFP::HIS3 S.k. | This study |
| IRA037 | IRA033 _atg1_Δ::HIS3 | This study |
| IRA038 | DDY1797 arp2-1-TAP::TRP1 | This study |
| IRA039 | SEY6210 promTPI1-ARP2-3DsRed::URA3 VRG4-GFP::HIS3 S.k. | This study |
| JGY086 | IRA030 promTPI1-arp2-1-3DsRed::LEU2 | This study |
| JKY007 | SEY6210 _atg9_Δ::HIS3 | He_, et al._ (2006) |
| PJ69-4A | MATα _leu2-3,112 trp1_-Δ_901 ura3-52 his3_-Δ_200 gal4_Δ _gal80_Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al. (1996) |
| RLY193 | MATα _ura3-52 leu2-3,112 his3_-Δ_200 arp3_Δ::HIS3 pW25-arp3-2-CEN | Winter_, et al._ (1997) |
| SEY6210 | MATα ura3-52 leu2-3,112 his3_-Δ_200 trp1_-Δ_901 lys2-801 suc2_-Δ_9 mel GAL | Robinson et al. (1988) |
| UNY23 | IRA029 _atg1_Δ::HIS3 | This study |
| UNY102 | SEY6210 TAP-ATG1::URA3 | Yorimitsu et al. (2006) |
| Y4963 | MATa arc15-10::KAN his3_Δ_1 leu2_Δ_0 ura3_Δ_0 met15_Δ_0 | This study |
| YAS1115 | MATα _pan1_Δ::HIS3 ade2-101 leu2-3,112 ura3-52 his3_-Δ_200 trp1-1 ppan1-3-URA3-CEN | Wendland et al. (1996) |
Polymerase chain reaction (PCR)-based integration of a DNA fragment encoding green fluorescent protein (GFP) at the 3′ end of PEX14 generated an arp2-1 mutant strain (IRA036) expressing chromosomally tagged Pex14-GFP for the pexophagy assay. The template for the integration cassette was pFA6a-GFP-HIS3 (Longtine et al., 1998). PCR verification and Western blot were used to confirm fusion protein expression.
Plasmids
To generate a strain expressing fluorescently tagged Atg9, the ATG9 gene was amplified by PCR from genomic DNA digested with XhoI and BamHI and cloned into the XhoI and BamHI sites of the integration vector pPG5-3xGFP (Boyd et al., 2004). The resulting plasmid, pATG9-3GFP(306), was linearized by digestion with BglII, and it was integrated at the ATG9 genomic locus. To tag Arp2 with DsRed, the ARP2 open reading frame was amplified from genomic DNA and first cloned into pSNA3416 (Reggiori and Pelham, 2001) by using HindIII and BamHI sites. This generated ARP2 under the control of the TPI1 promotor. Next, the TPI-ARP2 fragment was excised by digestion with XhoI and BamHI and introduced into the pRS305-3DsRed vector, which was created by cloning a BamHI–NotI fragment containing triple DsRed from pDsRed.M1x3 (a generous gift from Dr. Benjamin Glick, University of Chicago) into pRS305. The resulting integrative plasmid pTPIARP2-3DsRed(305) was linearized by digestion with EcoRI, and then it was integrated into the LEU2 gene locus.
To delete the ATG11, ATG1, and ATG9 genes, the entire coding regions were replaced by the Esherichia coli kan r or Schizosaccharomyces pombe HIS3 or LEU2 genes, respectively, by using PCR primers containing 60 bases of identity to the regions flanking the open reading frames. For the ATG1 gene disruption, the BamHI–ClaI fragment was cut from pΔatg1-URA (Abeliovich et al., 2003) and integrated into the ATG1 locus.
To generate two-hybrid selection plasmids, the ARP2 gene was cloned into pGBDU-C1 and pGAD-C1 vectors by using BamHI and SalI sites to generate the plasmids pGBDU-ARP2 and pGAD-ARP2, respectively. The two-hybrid plasmids pGBDU-ATG9 and pGAD-ATG9 have been described previously (Reggiori et al., 2005b).
The plasmids pAPG9(416), pATG1ts(414), pGFPAUT7(414), pGFPAUT7(416), and pRFPApe1(305) were described previously (Guan et al., 2001; Suzuki et al., 2001; Shintani et al., 2002; He et al., 2006).
Fluorescence Microscopy
Cells expressing fusion proteins with fluorescent tags were grown in SMD medium to mid-log phase. Fluorescence signals were visualized on a wide-field fluorescence inverted microscope (IX-70; Olympus America, Mellville, NY) equipped with a 100× oil numerical aperture (NA) 1.4 objective lens, and red fluorescent protein (RFP) and fluorescein isothiocyanate (FITC) filters. The images were captured by a Photometrix CoolSnap HQ camera (Photometrics, Tucson, AZ), and they were deconvolved using DeltaVision software (Applied Precision, Issaquah, WA).
For live-cell imaging, an ultrasensitive epifluorescence microscope (Nikon TE2000U-based dual camera system; 60× water immersion NA 1.2 objective lens) configured for high-speed acquisition of time-lapsed three-dimensional data were used. The dual charge-coupled device cameras (Cascade 2 cooled; Photometrics) and the filters were controlled by MetaMorph software (Molecular Devices, Sunnyvale, CA). The mid-log grown cells were immobilized on 1 mg/ml concanavalin A (Con A; Sigma-Aldrich, St. Louis, MO)-coated, acid-washed no. 1.5 coverglasses (Fisher Scientific, Pittsburgh, PA) mounted in a Leiden chamber (Harvard Apparatus, Cambridge, MA) as follows. An aliquot of the mid-log grown culture was added on the coverglass coated with Con A. After 2–3 min, the majority of the cells were adhered to the surface of Con A. The rest of the cells that did not attach were gently washed away with distilled water. The immobilized cells on the coverglass were placed into the Leiden chamber and covered with 1 ml of SMD medium. For the four-dimensional (4D) imaging, a Z-stack of seven optical sections spanning the entire cell was acquired either every 2 s or 10–20 s. The optical sections for each Z-stack were then collapsed to generate a two-dimensional-sum projection. The data were processed using ImageJ software (http://rsb.info.nih.gov/ij/).
Protein A Affinity Isolation
Cells were grown to OD600 = 0.8 in SMD. Cells (the equivalent of 30 ml at OD600 = 1.0) were harvested and resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM KCl, 5 mM MgCl2, 0.5% Nonidet-P40, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail). The detergent extracts were incubated with immunoglobulin (Ig)G-Sepharose beads for 2 h at 4°C. The beads were washed with lysis buffer six times and eluted in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer by incubating at 55°C for 15 min. The eluates were resolved by SDS-PAGE and immunoblotted with Atg9 antiserum or anti-protein A antibody.
Additional Assays
The pulse-chase analysis, GFP-Atg8 and Pex14-GFP processing assays, actin cytoskeleton staining with Texas Red-X phalloidin (TR phalloidin), and two-hybrid selection were carried out as described previously (Scott et al., 1997; Kim et al., 2001; Shintani and Klionsky, 2004b; Reggiori et al., 2005a). The western blots were quantified using ImageJ software.
Reagents
Antiserum to Ape1 was described previously (Klionsky et al., 1992). The anti-GFP monoclonal antibodies are from Covance Research Products (Princeton, NJ). SuperSignal West Pico Chemiluminescent Substrate was from Pierce Chemical (Rockford, IL). All other reagents were from Sigma-Aldrich or Fisher Scientific unless specified otherwise.
RESULTS
An arp2 Mutant Is Defective in Selective Autophagy
Recently, we reported that disruption of actin function results in a defect in the Cvt pathway (Reggiori et al., 2005a). To extend our investigation of the involvement of the actin cytoskeleton in selective autophagy, we performed a screen of mutants carrying temperature-sensitive (ts) alleles of essential genes encoding actin-binding proteins, regulating cytosolic actin rearrangements. Cells were monitored for vacuolar import and maturation of prApe1 at permissive and nonpermissive temperatures, which measures the selective Cvt pathway, and for processing of a GFP-tagged Atg8 chimera, which is an assay for nonspecific autophagy (Shintani and Klionsky, 2004b; Cheong et al., 2005).
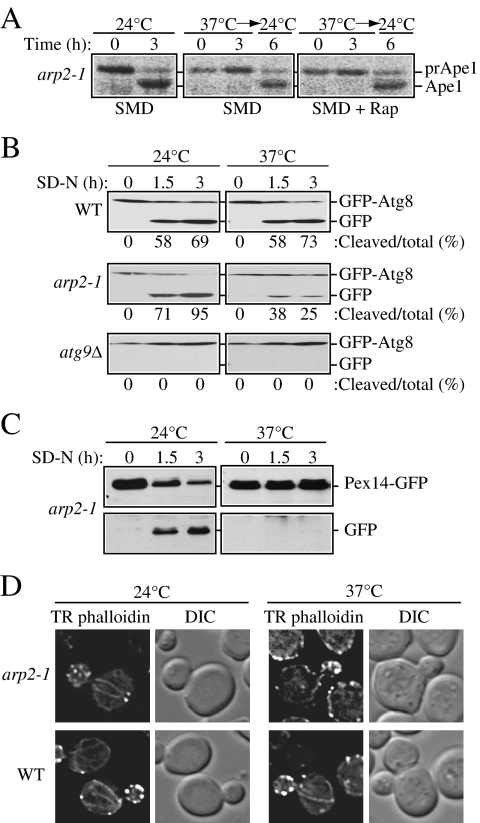
Ape1 maturation in the ts mutants was assessed by pulse-chase radiolabeling. The cells were grown to early log phase at a permissive 24°C temperature and shifted for 30 min to a nonpermissive temperature of 37°C before the addition of a radioactive label. After a 10-min pulse, a portion of the cell culture was subjected to a nonradioactive chase for 3 h at 37°C, and a final part of the culture was then incubated for 3 more h at 24°C (Figure 1A). When cells were incubated at the permissive temperature, essentially all of prApe1 was converted to the mature form, Ape1, by the end of a 3-h chase. In contrast, at the nonpermissive temperature, the conditional arp2 H330L mutant (hereafter referred to as arp2-1) displayed a strong defect in prApe1 maturation in growing conditions, with essentially all of the protein present as the precursor form (3-h time point in Figure 1A). We also examined cells treated with rapamycin to induce autophagy and observed a similar result; although autophagy is generally considered to be nonselective, import of prApe1 after rapamycin treatment or during starvation still occurs through a selective mechanism (Yorimitsu and Klionsky, 2005). Precursor Ape1 maturation was restored in arp2-1 cells after shifting back to 24°C, suggesting that the defect seen at the nonpermissive temperature was not due to loss of cell viability. These results suggest that the arp2-1 mutant is defective for the specific Cvt pathway.
Figure 1.
The arp2-1 mutant is defective in selective autophagy. (A) Precursor Ape1 processing is blocked in arp2-1 cells. The arp2-1 mutant cells were pulse-labeled for 10 min and subjected to a nonradioactive chase for 3 h at either 24°C (permissive temperature) or 37°C (nonpermissive temperature) in SMD medium, followed by an additional 3-h chase at 24°C as indicated. Rapamycin (Rap; 0.2 μg/ml) was added after pulse labeling, as indicated. Ape1 was immunoprecipitated and resolved by SDS-PAGE. The positions of precursor and mature Ape1 are marked. (B) Autophagy is delayed in the arp2-1 mutant. The wild-type, arp2-1, and _atg9_Δ strains carrying a plasmid expressing GFP-Atg8 [pGFPAUT7(414)] were grown in SMD at 24°C to early log phase, and then they were preincubated for 30 min either at 24 or 37°C and finally shifted to nitrogen-depleted (SD-N) medium to induce autophagy. Samples for the Western blot were taken before (0 h) and after 1.5 and 3 h of starvation. The numbers below each lane correspond to the percentage of cleaved (free) GFP. (C) Pexophagy is impaired in arp2-1 cells. The arp2-1 Pex14-GFP strain (IRA036) was grown at 24°C in oleate medium to induce massive peroxisome proliferation, then one portion of the culture was incubated for 3 h at 24°C and another portion at 37°C. Cells were then shifted to nitrogen starvation conditions to induce selective peroxisome degradation (pexophagy). Protein extracts were prepared at the time points indicated and resolved by SDS-PAGE, and after Western blot, membranes were probed with anti-GFP monoclonal antibody. Full-length chimeric Pex14-GFP (66 kDa) and free GFP (∼25 kDa) were detected as indicated. (D) The actin cytoskeleton morphology is altered in arp2-1 mutants. Fluorescence images of actin structure in wild-type and arp2-1 cells were collected after 3 h of incubation at 24 and 37°C. Cells were fixed, permeabilized, and subsequently incubated for 2 h at room temperature with TR phalloidin to stain the actin cytoskeleton.
Monitoring prApe1 maturation is not sufficient for measuring bulk, starvation-induced autophagy; prApe1 is a selective cargo that utilizes a receptor, and in some cases prApe1 import still can occur in conditions where bulk autophagy is blocked (Cheong et al., 2005). Accordingly, we also monitored GFP-Atg8 processing during starvation. Atg8 present on the inner side of the sequestering membrane remains associated with the completed autophagosome, and it is delivered to the vacuole inside the autophagic body; it is degraded in the vacuole lumen. The GFP moiety of chimeric GFP-Atg8 is relatively stable, so the appearance of free GFP reflects the rate of bulk autophagy. In wild-type cells there was a clear increase in free GFP after the induction of autophagy by shifting cells to medium lacking nitrogen, at both 24 and 37°C (Figure 1B). In contrast, the arp2-1 mutant showed a clear decrease in GFP-Atg8 processing at the nonpermissive temperature relative to the wild-type control. Nevertheless, arp2-1 cells were able to process GFP-Atg8 relative to the _atg9_Δ mutant, which displayed a complete block in autophagy (Figure 1B). Thus, we concluded that bulk autophagy is only partially blocked in the arp2-1 mutant. This finding is in agreement with our previous studies that indicated a requirement for actin function primarily in selective, but not bulk, autophagy (Reggiori et al., 2005a).
Because the defect in GFP-Atg8 processing in the conditions that induce bulk autophagy was less evident, we decided to focus our study on selective types of autophagy. Therefore, in addition to the Cvt pathway, we examined whether any other types of selective autophagy were affected by the conditional arp2-1 allele. Selective peroxisome degradation (pexophagy) is induced when cells are grown in conditions that induce massive peroxisome proliferation, and then they are shifted to conditions where peroxisomes are no longer needed for growth. The peroxisomal membrane protein Pex14 fused to GFP can be used as a marker to monitor peroxisome degradation, similar to GFP-Atg8 (Reggiori et al., 2005a). Free GFP was generated from arp2-1 mutant cells at the permissive temperature (Figure 1C). In contrast, Pex14-GFP processing in arp2-1 cells was completely blocked at a nonpermissive temperature, indicating a defect in pexophagy.
To verify the mutant phenotype of arp2-1, in particular the alteration of actin cytoskeleton, the latter was examined by the TR phalloidin actin-staining assay (Figure 1D). TR phalloidin is a fluorescent dye that associates with actin filaments and stains all filamentous actin structures in the cell. After staining, both actin cables and patches can be visualized by fluorescence microscopy. At 24°C, no difference was observed in actin organization between mutant and wild-type cells. In contrast, at 37°C, the mutant cells showed an abnormal actin distribution compared with wild-type cells; the cables were much less visible and the patches were more randomly distributed at the plasma membrane and they did not display as highly a polarized bud localization as seen in the wild type, where cortical actin patches are more concentrated at the site of growth. The mutant phenotype of arp2-1 that we observed at the nonpermissive temperature was consistent with previous reports. Finally, we sequenced the arp2-1 genomic locus after amplification by PCR, and we verified the presence of the mutation that alters the codon for histidine at position 330 to leucine (data not shown).
Localization of Atg9 Is Altered in the arp2-1 Conditional Mutant
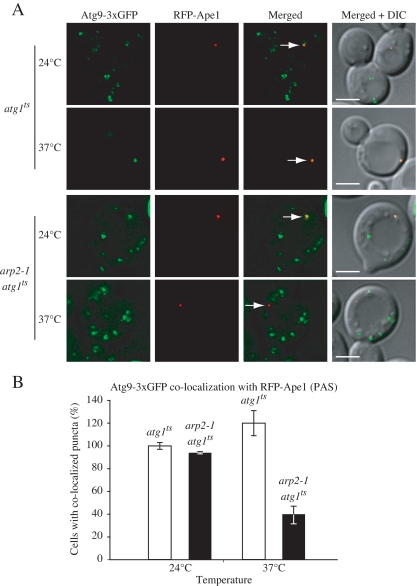
In contrast to other Atg proteins that are restricted to the perivacuolar vesicle nucleation site (the PAS), Atg9 localizes to multiple punctate structures. In wild-type cells, one of the Atg9 puncta corresponds to the PAS, whereas the other sites are peripheral to this structure; Atg9 cycles between the peripheral sites and the PAS (Reggiori et al., 2005b), possibly providing lipids to the forming autophagosomes or Cvt vesicles (Reggiori and Klionsky, 2005). The retrograde movement (from the PAS) of Atg9 requires the Atg1–Atg13 complex, Atg2, Atg18, and the phosphatidylinositol 3-kinase complex. In contrast, the actin cytoskeleton is required for the anterograde transport (to the PAS) of Atg9 (Reggiori et al., 2005a). Because Arp2 is an actin-related protein that is involved in intracellular trafficking we decided to test whether there was any defect in Atg9 cycling in the arp2-1 mutant. For this reason, we used the _T_ransport of _A_tg9 after _K_nocking out _A_TG1 (TAKA) assay (Cheong et al., 2005). This is an epistasis assay that examines the effect of a second mutation relative to _atg1_Δ with regard to Atg9 movement to the PAS; in _atg1_Δ cells, Atg9 is confined to the PAS.
We examined _atg1_Δ cells that harbored a plasmid encoding a temperature-sensitive allele of ATG1. This strain was chromosomally tagged with Atg9-3xGFP and RFP-Ape1; the tagged prApe1 was used to mark the location of the PAS. At the permissive temperature Atg9-3xGFP was seen in multiple dots, and one of them coincided with the RFP-Ape1 PAS marker (Figure 2A). When the atg1 ts cells were shifted to nonpermissive temperature, Atg9-3xGFP fluorescence collapsed into a single punctum, which completely colocalized with RFP-Ape1, similar to the result seen in _atg1_Δ cells (Reggiori et al., 2005b). In double mutant arp2-1 atg1 ts cells, at the permissive temperature, the Atg9-GFP fluorescence pattern looked the same as in atg1 ts cells. In contrast, at the nonpermissive temperature Atg9-3xGFP was excluded from the PAS, a phenotype that was clearly distinct from cells that harbored only the _atg1_Δ deletion (Figure 2B). Hence, Atg9 did not localize to the PAS in the arp2-1 conditional mutant when Atg1 was inactive, and we conclude that the trafficking of Atg9 to the PAS depends on Arp2 function; that is, Arp2 function was epistatic to that of Atg1.
Figure 2.
Anterograde transport of Atg9 is impaired in the arp2-1 mutant. (A) The _atg1_Δ and _arp2-1 atg1_Δ strains chromosomally-tagged with Atg9-3xGFP and RFP-Ape1, and carrying a centromeric plasmid (pATG1ts(414)) encoding a temperature-sensitive allele of ATG1 were grown to mid-log phase at 24 and 37°C and visualized by fluorescence microscopy. Arrows mark the site of the PAS. DIC, differential interference contrast. Scale bar, 2 μm. (B) Quantification of the Atg9-3xGFP and RFP-Ape1 colocalization from (A); colocalization was defined as one Atg9-3xGFP dot overlapping with the RFP-Ape1 signal. Approximately 50 cells expressing RFP-Ape1 were examined. The number of cells (40) where Atg9-3xGFP and RFP-Ape1 colocalized in the atg1 ts strain at 24°C was set to 100%. The values represent mean and standard deviation for 3 independent experiments.
Atg9 Movement Is Dependent upon Arp2
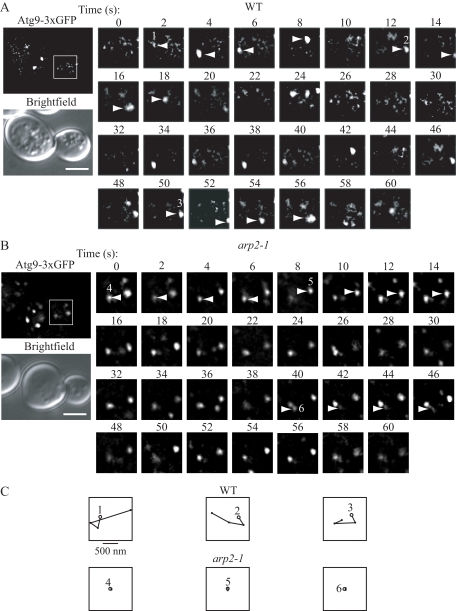
Previously, we showed that Atg9 localization was dynamic; Atg9 displays rapid movement at the peripheral sites in addition to its cycling to and from the PAS (Reggiori et al., 2005b). We next decided to extend our analysis by following Atg9 movement over time. To this end, we performed a time-lapse experiment in the strains that expressed chromosomally 3xGFP-tagged Atg9 in both the wild-type and the conditional arp2-1 mutant cells. Cells were grown at the permissive temperature, and then they were shifted for 30 min to nonpermissive conditions, and Atg9 movement was tracked by fluorescence wide-field microscopy. In the wild-type strain, small GFP-marked fluorescent dots appeared, moved, and fused with other, bigger dots. In addition, larger dots could be observed dividing and generating smaller dots. Overall, the Atg9-3xGFP dots were constantly moving, fusing, and dividing (Figure 3A). In contrast to the rapid movement of Atg9 fluorescent dots seen in the wild-type cells, there was very little movement observed at the nonpermissive temperature in arp2-1 cells (Figure 3, B and C). For each dot, the average velocity was calculated, and the values were compared between groups. The mean velocity for the arp2-1 strain was 40 ± 1.2 nm/s compared with 570 ± 32 nm/s for the wild type, with a two-sample t test indicating a significant difference (p ≤ 0.05). Although some of the Atg9-3xGFP dots in the arp2-1 strain appeared and/or disappeared over the time course of the analysis, we think these corresponded to puncta that were moving into and out of the focal plane due to Brownian motion; however, we cannot rule out the possibility that some of the puncta in the mutant strain displayed movement. Overall, it was clear that the majority of the dots in the arp2-1 strain were not changing location or that their velocities were significantly reduced compared with the wild-type strain. Therefore, we conclude that the movement of Atg9 at the peripheral sites was compromised in the arp2-1 conditional mutant.
Figure 3.
Movement of Atg9 is Arp2-dependent. Wild-type and arp2-1 cells were grown to mid-log phase and shifted to nonpermissive temperature for 30 min before visualization. Movement of Atg9-3xGFP patches was tracked by time-lapse fluorescence imaging as described in Materials and Methods. The presented images are still frames of the indicated boxed regions shown at 2× enlargement and taken with a time lapse of 2 s for 60-s duration. (A) In wild-type cells, the Atg9-3xGFP puncta rapidly changed their location accompanied by fusion or division. (B) In the arp2-1 conditional mutant, there was no displacement of the majority of Atg9-3xGFP puncta during 60 s. Bar, 2 μm. (C) The single Atg9-3xGFP puncta indicated in A and B were tracked in the wild-type Atg9-3xGFP puncta indicated in A and B were tracked in the wild-type versus arp2-1 strains. Spatial positions of the centers of puncta were determined in each frame (with 2-s intervals) of a movie using ImageJ software, and consecutive positions were connected by lines. Open, larger circles denote the first position of each punctum. Bar, 500 nm.
Arp2 Colocalizes with Atg9
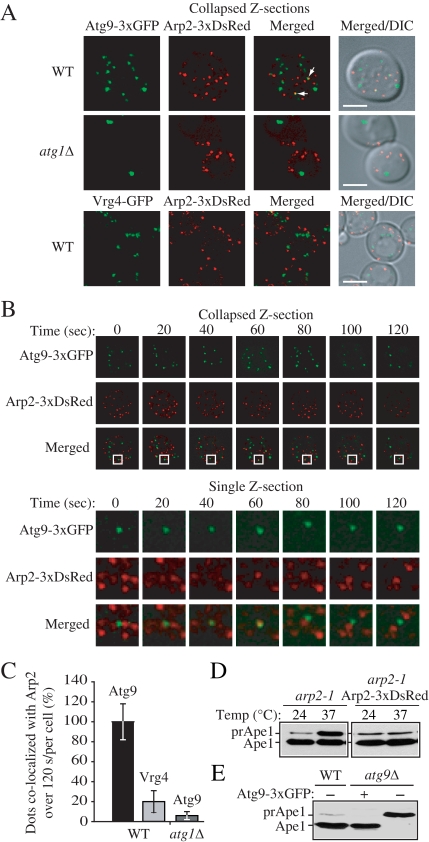
Atg9 movement at the peripheral sites, and its movement between these sites and the PAS, depends on Arp2 function, suggesting that these two proteins might interact. Accordingly, we decided to examine Arp2 localization relative to Atg9 by using a fluorescently tagged version of the protein. Using wide-field real-time microscopy with a short time lapse, we detected individual Atg9-3xGFP puncta that momentarily coincided with an Arp2-3xDsRed dot (Figure 4A). The projected image is a sum projection of Z-sections. To verify that this apparent colocalization was not a result of overlapping, but different, focal planes, we examined whether the Atg9 and Arp2 signals coincided in a single optical plane. We observed a brief colocalization that was imaged in a single Z-section (Figure 4B).
Figure 4.
Atg9 colocalizes with Arp2. (A) Atg9 and Arp2 were tracked in a single live cell in wild-type (IRA033) and _atg1_Δ (IRA037) cells expressing chromosomally tagged Atg9-3xGFP and TPI promoter-driven Arp2-3xDsRed, and Arp2 and chromosomally tagged Vrg4-GFP were monitored in wild-type cells (IRA039). Mid-log phase cells were analyzed by simultaneous two-color imaging combined in 4D (x,y,z,t). The RFP and GFP images were collected simultaneously with a time interval between Z-stacks acquisitions of 20 s for 120-s duration. A collapsed series of Z-sections is shown at the 60 s time point. Scale bar, 2 μm. (B) Still frames from a collapsed series of Z-sections and individual Z-sections at a single focal plane from a different cell demonstrate Atg9-3xGFP and Arp2-3xDsRed colocalization. The single Z-section images show the magnified view of the boxed area. (C) Quantification of the number of Atg9-3xGFP puncta colocalized with Arp2-3xDsRed in the wild-type and _atg1_Δ strains, and the number of Vrg4-GFP dots colocalized with Arp2-3xDsRed (n = 100). The number of Atg9 dots that colocalized with Arp2 in the wild-type strain per cell was set to 100%. (D) Arp2-3xDsRed is functional. arp2-1 cells expressing Arp2-DsRed or empty vector were grown to mid-log phase at 24°C, and then they were incubated for 3 h either at 24 or 37°C. Precursor Ape1 maturation was monitored by Western blot. The positions of precursor and mature Ape1 are indicated. (E) Atg9-3xGFP is functional. Wild-type cells or _atg9_Δ cells expressing Atg9-3xGFP or empty vector were grown to mid-log phase, and then they were examined by Western blot as described in C.
To determine whether this colocalization of Arp2 and Atg9 is a random event, using the same approach we examined Arp2-3xDsRed and Atg9-3xGFP colocalization in the _atg1_Δ strain background, as well as Arp2-3xDsRed and Vrg4-GFP colocalization in the wild-type background, and we carried out a statistical analysis. In the _atg1_Δ strain, Atg9 was mostly restricted to a single perivacuolar dot, which did not show colocalization with any of the multiple Arp2 puncta (Figure 4A). We chose to analyze Vrg4 because it is an integral membrane protein, as is Atg9, which localizes to the Golgi complex; there is no indication that Arp2 specifically localizes to the Golgi, and Vrg4 therefore serves as a nonspecific marker protein. Vrg4 chromosomally tagged with GFP was also present in multiple dots, the number of puncta being similar to Atg9-GFP; however, we found that it did not colocalize with Arp2 in wild-type cells (Figure 4A). Our statistical analysis revealed that the number of Arp2 puncta that colocalized with Atg9 in the wild-type strain (∼14%) was significantly higher than the number of dots of Arp2 that colocalized either with Vrg4 in the wild-type strain or with Atg9 in the _atg1_Δ strain (Figure 4C). Finally, we verified that the chimeric Arp2 and Atg9 proteins, with C-terminal fusions to 3xDsRed or 3xGFP, respectively were functional; Arp2 fused to 3xDsRed revealed normal cellular distribution as it was colocalized with GFP-tagged Abp1, a marker for actin patches (our unpublished data). In addition, the cell morphology and actin cytoskeleton organization were normal and the cells grew at rates similar to wild-type cells at 24°C as well as at 37°C (our unpublished data). Finally, both chimeras complemented arp2-1 or _atg9_Δ mutant strains, respectively, for maturation of prApe1 (Figure 4, D and E).
Arp2 Interacts with Atg9
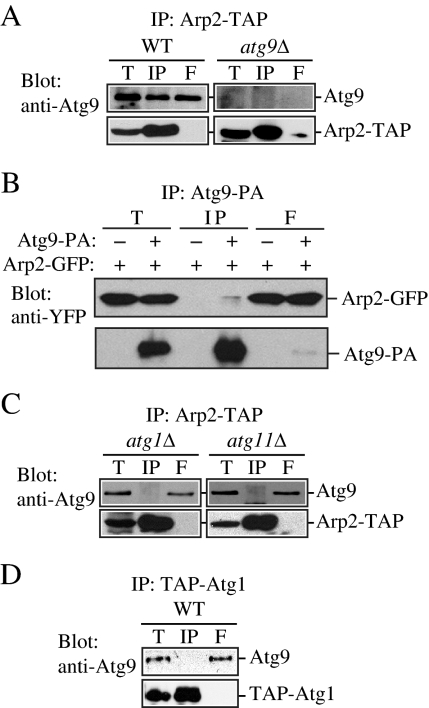
The fluorescence microscopy data suggest that Atg9 and Arp2 transiently colocalize. Accordingly, we decided to examine whether these proteins interact. To this end, we first performed a yeast two hybrid-based analysis. We found that yeast two-hybrid cells harboring plasmids encoding Arp2 and Atg9 showed growth on plates lacking histidine but not on plates lacking adenine, indicating that Arp2 could interact weakly with Atg9 under the conditions of this assay (our unpublished data). To further clarify the physiological significance of the Atg9–Arp2 interaction, we decided to test the interaction using a biochemical approach. The functionality of the chromosomally tagged Arp2 was verified by a prApe1 maturation assay (our unpublished data). Wild-type, _atg1_Δ, and _atg11_Δ cells expressing chromosomally TAP-tagged Arp2 and endogenous Atg9 were lysed, and the TAP-tagged protein was isolated with IgG-Sepharose beads. The presence of Arp2-TAP and Atg9 was detected by immunoblotting with anti-Atg9 antibody.
Approximately 25 ± 7% (n = 3) of the Atg9 pool was coimmunoprecipitated with Arp2-TAP, which suggests that the two proteins interact at a moderate level (Figure 5A). Nonetheless, this level of interaction was substantially higher than we expected based on our fluorescence microscopy data, which revealed that only a very small proportion of Atg9 puncta showed any colocalization with Arp2 (Figure 4). To investigate the nature of this apparent discrepancy, we repeated the affinity isolation with a strain harboring a centromeric plasmid allowing expression of ATG9 from an endogenous promoter but at a higher level, and we found a substantial increase in the amount of Atg9 protein bound to Arp2 (our unpublished data). Increasing the incubation time for the coimmunoprecipitation of the cell lysate gave a similar result (our unpublished data). Thus, the amount of Atg9 bound to Arp2 seems to be affected by the expression level and experimental conditions; Arp2 may bind a higher level of Atg9 during the in vitro affinity isolation, perhaps due to a loss of regulation that normally occurs in vivo. To verify the interaction between these two proteins, we repeated the affinity isolation in the reverse direction, using Atg9 tagged with protein A to coprecipitate Arp2-GFP. In this case, we detected a very weak, but reproducible, interaction between the two proteins (Figure 5B).
Figure 5.
Atg9 binds to Arp2. (A) Atg9 is coimmunoprecipitated with Arp2. Either wild-type cells containing chromosomally TAP-tagged Arp2 (Arp2-TAP, WT) or _atg9_Δ cells with chromosomally tagged Arp2-TAP (IRA020) were used for affinity isolation as described in Materials and Methods. Eluted polypeptides were separated by SDS-PAGE and detected with anti-Atg9 antiserum. The same amounts of the total lysate (T), immunoprecipitate (IP), and flow through (F) were loaded per gel lane. (B) Arp2 is coimmunoprecipitated with Atg9. A strain expressing chromosomally tagged Atg9-protein A (FRY171) and plasmid-based Arp2-GFP or a control strain (SEY6210) only expressing plasmid-based Arp2-GFP and protein A driven by the CUP1 promoter were used for affinity isolation as described in A. Protein bands were detected with anti-YFP antibody. (C) Endogenous Atg9 is not coimmunoprecipitated with Arp2 in the absence of Atg1 or Atg11. The indicated _atg1_Δ (IRA023) or _atg11_Δ (IRA028) Arp2-tagged strains were used for affinity isolation as in A and probed with anti-Atg9 antiserum. (D) Atg9 is not coimmunoprecipitated with Atg1. A strain with chromosomally TAP-tagged Atg1 (UNY102) served as a control for Atg9 affinity isolation. Eluted polypeptides were separated by SDS-PAGE and visualized by immunoblotting as described in A.
In contrast to wild-type cells, Atg9 was not coimmunoprecipitated with Arp2-TAP in the _atg1_Δ strain (Figure 5C). This result suggests that Arp2 is not localized at the PAS or that the two proteins do not interact at this site. Similarly, we did not detect an interaction between these two proteins in an _atg11_Δ strain (Figure 5C). The latter result was interesting because Atg11 is required for transit of Atg9 from the peripheral sites to the PAS (He et al., 2006). Thus, the absence of either Atg1 or Atg11 eliminated the interaction between Arp2 and Atg9, suggesting that this interaction is either directly or indirectly dependent on those proteins. Chromosomally TAP-tagged Atg1 did not coimmunoprecipitate with Atg9 (Figure 5D), suggesting that Atg1 may not directly be involved in the Arp2 interaction, and this analysis also served as an additional negative control. We conclude that Arp2 interacts with Atg9, and this interaction does not occur when Atg9 is restricted to the PAS in _atg1_Δ cells or when Atg9 cycling is blocked in _atg11_Δ cells. In addition, these results are consistent with our fluorescence microscopy data, which showed that the two proteins do not colocalize when Atg9 is restricted to a single PAS dot in _atg1_Δ cells.
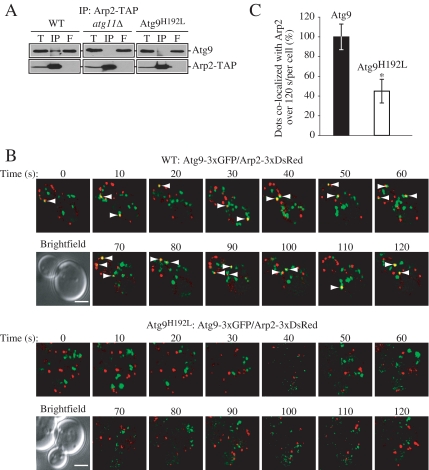
For _atg1_Δ cells, Atg9 is localized in a distinct site separate from Arp2; however, this is not the case in an _atg11_Δ strain. Accordingly, because of the lack of interaction in the _atg11_Δ cells, we hypothesized that the interaction of Atg9 with Arp2 might be dependent on Atg11. To test this, we examined the interaction in the Atg9H192L mutant, in which the codon for histidine at position 192 was altered to leucine; this mutation causes loss of interaction between Atg9 and Atg11 (He et al., 2006), and we asked the question of whether the loss of Atg9–Atg11 interaction affected the interaction between Atg9 and Arp2. First, we tested the Atg9–Arp2 interaction by coimmunoprecipitation, and we found that Atg9H192L was essentially not coimmunoprecipitated with Arp2-TAP (Figure 6A). Next, we performed a time-lapse experiment and examined Atg9 and Arp2 colocalization in strains expressing either wild-type Atg9 or Atg9H192L tagged with 3xGFP (Figure 6B). We used wide-field real-time microscopy with a short time lapse to learn more about the nature of the Atg9–Arp2 interaction. Each projected image shown is a sum projection of Z-sections. We saw that in the wild-type strain an individual Atg9-3xGFP punctum momentarily coincided with an Arp2-3xDsRed dot usually for <10 s, although we occasionally detected colocalization for more than 10 s (Figure 6B). In contrast, in cells expressing the mutant Atg9H192L-3xGFP, dots rarely coincided with Arp2. Quantification of the results revealed that colocalization was significantly reduced with the Atg9H192L mutant, which is consistent with our coimmunoprecipitation results (Figure 6C).
Figure 6.
A mutant Atg9 that loses the ability to interact with Atg11 does not bind Arp2. (A) Endogenous Atg9 is coimmunoprecipitated with Arp2 in a wild-type strain but not in the absence of Atg11 or in a mutant of Atg9 that loses the ability to interact with Atg11. The indicated wild-type (ARP2-TAP, WT), _atg11_Δ (IRA028), or Atg9H192L (IRA020 harboring pATG9H192L) strains were used for affinity isolation as in Figure 5. (B) Mid-log phase cells were analyzed by simultaneous two-color imaging combined in 4D (x,y,z,t). The RFP and GFP images were collected simultaneously with a time interval between Z-stacks acquisitions of 10- for 120-s duration. Still frames from collapsed series Z-sections examine Atg9-3xGFP and Arp2-3xDsRed colocalization (indicated by arrows) in the wild-type Atg9-3xGFP or mutant Atg9H192L-3xGFP strain. Bar, 2 μm. (C) Quantification of the number of Atg9-3xGFP dots colocalized with Arp2-3xDsRed per cell (n = 79) in the wild-type versus Atg9H192L mutant strain. Fifteen of a total of 105 Atg9-3xGFP dots displayed in 13 frames taken during a 120-s time course colocalized with Arp2 in the wild-type strain; this number was set to 100%. *p < 0.05 value indicates that a statistically significantly higher number of Atg9-3xGFP dots colocalized with Arp2-3xDsRed compared with the mutant Atg9H192L-3xGFP.
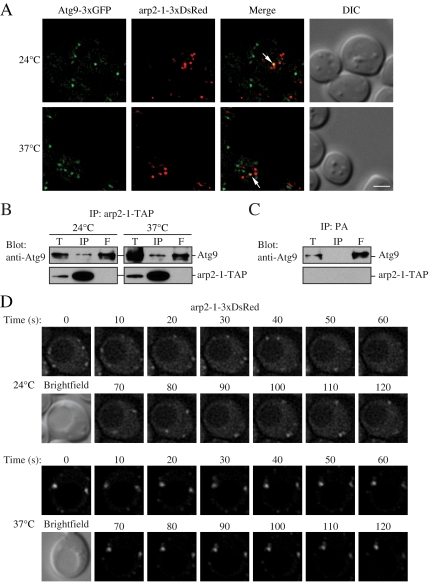
Because we found that Atg9 localization at the PAS and movement were compromised in the arp2-1 mutant (Figures 2 and 3), we extended our analysis of Atg9–Arp2 interaction by examining the ability of Atg9 to interact with the mutant arp2-1 protein. When chromosomally tagged Atg9-3xGFP and arp2-1-3xDsRed were expressed at either 24 or 37°C, we detected limited colocalization of the two proteins (Figure 7A), similar to the level detected with Arp2-3xDsRed (Figure 4). The two proteins also coprecipitated; TAP-tagged arp2-1 was able to coprecipitate Atg9, whereas Atg9 was not affinity isolated by protein A alone (Figure 7, B and C). We then examined movement of arp2-1-3xDsRed at the permissive and nonpermissive temperatures, and we found that the mutant protein displayed a dramatic reduction in movement at the nonpermissive temperature (Figure 7D), essentially the same as that seen with Atg9-3xGFP in the arp2-1 mutant at 37°C (Figure 3B). Thus, the defect in Atg9 movement in the arp2-1 mutant does not seem to be the result of an inability of the two proteins to interact, but rather from the inherent defect in the dynamics of the arp2-1 mutant protein at the nonpermissive temperature.
Figure 7.
Atg9 interacts with arp2-1, which is defective in movement. (A) Atg9-3xGFP and arp2-1-3xDsRed colocalize. arp2-1 cells expressing chromosomally tagged Atg9-3xGFP and arp2-1-3xDsRed (JGY086) were grown to mid-log phase at 24°C, and then they were shifted to 37°C for 30 min. Samples were collected before and after the temperature shift, and then they were analyzed by microscopy as described in Materials and Methods. Colocalization of Atg9-3xGFP and arp2-1-3xDsRed in a single Z-section is indicated by arrows. (B) The arp2-1 protein interacts with Atg9. Cells (IRA038) expressing the chromosomally TAP-tagged arp2-1 protein were grown in SMD medium at 24°C, and then one aliquot of the culture was shifted to 37°C for 3 h. Cells were collected, and affinity isolation was performed as in Materials and Methods. The same amounts of total lysate (T), immunoprecipitate (IP), and flow through (F) were separated by SDS-PAGE and detected by anti-Atg9 antiserum. Bar, 2 μm. (C) Atg9 is not coimmunoprecipitated with protein A (PA) alone. Wild-type cells expressing CUP1 promoter-driven PA were used as a negative control for Atg9 interaction. Cells were cultured at 30°C and analyzed by affinity isolation as described in B. (D) arp2-1-DsRed is defective in movement at the nonpermissive temperature. Mid-log phase cells expressing arp2-1-3xDsRed (JGY086) were grown and imaged as in described in A with a time interval between Z-stack acquisitions of 10 s for 120-s duration. Still frames from collapsed series Z-sections examine arp2-1-3xDsRed movement at 24 and 37°C. Bar, 2 μm.
The Arp2/3 Complex Is Required for Selective Autophagy
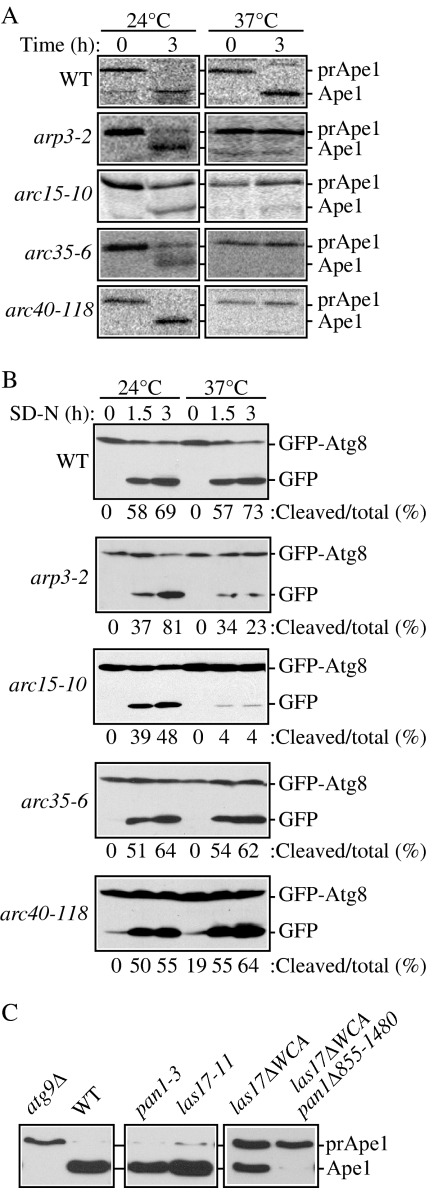
Arp2 is one component of the Arp2/3 complex, which is composed of seven subunits: Arp2, Arp3, Arc15/p15, Arc18/p18/p21, Arc19/p19, Arc35/p35, and Arc40/p40. All of the subunits of this complex, except for Arc18, are encoded by essential genes. The Arp2/3 complex is required for proper actin cytoskeleton organization, and its subunits colocalize with cortical actin structures. Therefore, we hypothesized that the Arp2/3 complex, not just Arp2, might be required for selective autophagy. To test this hypothesis, we examined prApe1 maturation by pulse-chase radiolabeling in strains with mutations in genes encoding Arp2/3 subunits. At the permissive temperature, the arp3-2, arc15-10, arc35-6, and arc40-118 mutants were able to mature some or all of the prApe1 (Figure 8A). At the nonpermissive temperature, there was a severe block in prApe1 maturation in these mutant strains, reflecting a defect in the Cvt pathway.
Figure 8.
The Arp2/3 complex is necessary for selective autophagy. (A) Precursor Ape1 maturation is blocked in the arp3-2 (RLY193), arc15-10 (Y4963), arc35-6 (BGY0809), and arc40-118 (BGY0883) strains. The cells were grown to mid-log phase at 24°C, and then they were incubated for 3 h either at 24 or 37°C. Precursor Ape1 maturation was monitored with pulse-chase radiolabeling followed by immunoprecipitation with antiserum to Ape1. The positions of precursor and mature Ape1 are indicated. (B) GFP-Atg8 processing is delayed in some strains with mutations in Arp2/3 complex subunits. The wild-type, arp3-2, arc15-10, arc35-6, and arc40-118 strains transformed with a GFP-Atg8 plasmid [pGFPAUT7(414) or pGFPAUT7(416)] were grown in SMD to mid-log phase at permissive temperature and preincubated for 30 min either at 24 or 37°C before being shifted to starvation conditions (SD-N). At the indicated time points, aliquots were taken and protein extracts were analyzed by Western blot by using anti-GFP antibody. The positions of GFP-Atg8 and free GFP are indicated along with the percentage of free GFP. (C) Mutations in Arp2/3 complex regulatory proteins block selective autophagy. The wild-type (SEY6210) and _atg9_Δ (JKY007) strains were grown to mid-log phase at 30°C, and protein extracts were prepared for controls. The pan1-3 (YAS1115), las17-11 (DDY1960), las17_Δ_WCA (DDY3045) and las17_Δ_WCA pan1_Δ_855-1480 (DDY2885) strains were grown to mid-log phase at 24°C, shifted to 37°C for 60 min, and examined by Western blot as described in A.
We next examined bulk autophagy by using the GFP-Atg8 processing assay. In contrast to the Cvt defective phenotype, the block in nonspecific autophagy was less severe than that seen in an atg mutant; all of the mutants showed at least partial generation of free GFP after the induction of autophagy, although the arp3-2 and arc15-10 defects were relatively severe similar to that seen with arp2-1 (Figures 1A and 8B). Arp2 and Arp3 are the two main subunits of the Arp2/3 complex, and they play a central role by forming an initial nucleus for actin polymerization. Differences in severity among the other components of the complex may reflect the particular alleles available for analysis.
Finally, we examined mutant forms of two proteins that regulate the activity of the Arp2/3 complex, Las17 and Pan1. Both the pan1-3 and las17-11 mutants displayed normal processing of prApe1 (Figure 8C); however, these two proteins are partially redundant in function (Toshima et al., 2005). Therefore, we next examined versions of these proteins that are defective for activating and/or binding the Arp2/3 complex (Toshima et al., 2005). The _las17_ΔWCA mutant displayed an ∼50% block in prApe1 maturation, whereas the double _las17_ΔWCA _pan1_Δ855-1480 mutant was completely defective for the Cvt pathway based on prApe1 processing (Figure 8C). Overall, these results suggest that the function of the Arp2/3 complex is required particularly for selective autophagy.
DISCUSSION
The Arp2/3 Complex Is Required for Selective Autophagy
Previously, we showed that the actin cytoskeleton is required for selective autophagy. The cycling of Atg9 in conditional actin mutants, or in cells treated with the actin-depolymerizing drug latrunculin A, is defective. In particular, the anterograde transport of Atg9 from peripheral sites in the cell to the site of vesicle formation requires a functional actin cytoskeleton. Accordingly, we screened yeast strains with mutations in genes encoding actin binding proteins to understand how actin, and associated factors, regulates the anterograde movement of Atg9. In the current study, we found that mutations in subunits of the Arp2/3 complex caused a severe defect in transport of the specific autophagy marker precursor Ape1 in growing conditions (Figures 1A and 8A). Similarly, we found that pexophagy, another type of selective autophagy, was defective in arp2-1 mutant cells (Figure 1C). In contrast, this mutant processed GFP-Atg8, indicating that bulk or nonspecific autophagy was relatively unaffected (Figure 1B). This finding is consistent with our previous results where an intact cytoskeletal network was essential only for selective autophagy (Reggiori et al., 2005a). The Arp2/3 complex consists of seven subunits: Arp2, Arp3, Arc15/p15, Arc18/p18/p21, Arc19/p19, Arc35/p35, and Arc40/p40. This complex functions to nucleate the synthesis of actin filaments. We found that mutations in any of the Arp2/3 complex components that we tested displayed a similar phenotype; all of these mutants were defective for prApe1 import, but they displayed relatively normal kinetics for nonspecific autophagy (Figure 8) although there were subunit-specific differences that may reflect the severity of the defect in particular alleles of the various mutants.
Atg9 Cycling Is Regulated by Arp2 during Selective Autophagy
Unlike other intracellular trafficking mechanisms, autophagy uses double-membrane vesicles that are formed de novo. One of the most important questions concerning autophagy is the mechanism used to compose the autophagosomes, and the source of the lipids that supply the site of autophagosome assembly, the PAS. Atg9, is a transmembrane protein that cycles between peripheral structures and the PAS, and it is therefore proposed to play a key role in membrane delivery during the assembly process (Reggiori and Klionsky, 2005). Transfer of Atg9 to the cargo site involves Atg11, Atg23, Atg27, and actin (He et al., 2006, Legakis et al., 2007, Yen et al., 2007); however, the mechanism by which actin mediates Atg9 anterograde movement is not known. We found that in arp2-1 mutant cells, Atg9 movement was severely compromised; Atg9 was unable to colocalize with the PAS marker prApe1, suggesting a role for Arp2 in directing or regulating Atg9 anterograde movement to the PAS (Figure 2).
Real-time analysis of Atg9 dynamics in arp2-1 cells revealed that patches of Atg9 located at the peripheral sites are immobile, in contrast to their rapid movement in wild-type cells, implying that Arp2 is required for Atg9 movement (Figure 3). These data suggest that Arp2, presumably in conjunction with other subunits of the Arp2/3 complex, may function in some aspect of vesicle movement involving Atg9, by analogy with the role of Arp2 in other processes involving membrane movement such as endocytosis (Liu et al., 2006, Takenawa and Suetsugu, 2007). The components of the Arp2/3 complex display transient interaction with forming endocytic vesicles. Accordingly, we examined potential interactions between Atg9 and Arp2.
Using real-time three-dimensional (4D)-microscopy, we found that Atg9 and Arp2 display transient colocalization (Figure 4). We used affinity isolation to verify that the colocalization corresponded to an actual interaction (Figure 5). These events most likely take place at the peripheral patches where Atg9 colocalizes with Arp2 but not at the PAS, which is the ultimate destination, because we did not detect colocalization or direct interaction between these two proteins in an _atg1_Δ mutant where Atg9 is confined to the PAS (Figures 4 and 5). There was also a lack of interaction between Atg9 and Arp2 in an _atg11_Δ strain or in an Atg11 mutant that is defective in binding Atg9 (Figure 6), even though Atg9 is restricted to the peripheral sites in this mutant. Therefore, this result suggests that Atg11 may directly or indirectly mediate the interaction between Atg9 and Arp2. In contrast to the results with the _atg11_Δ strain, Atg9 still interacts with the arp2-1 mutant protein, even at the nonpermissive temperature (Figure 7). Therefore, lack of movement in the arp2-1 mutant does not reflect an inability of Atg9 to interact with arp2-1, but rather it is due to the defect in the function of the arp2-1 protein. These data support a model where actin nucleation events direct the movement of Atg9 from peripheral sites to allow the delivery of membranes to the cargo, and expansion of the forming autophagosome under vegetative growth conditions.
How Does Arp2/3 Function in Selective Autophagy?
Arp2/3 and its activators such as Las17 (the yeast homologue of the Wiskott–Aldrich syndrome protein, N-WASP), possibly in complex with the type 1 myosins Myo3 and Myo5, Abp1, or Pan1 initiate the assembly of actin filaments (Pollard, 2007, Winder and Ayscough, 2005). This proceeds in such a way that actin monomers (Act1) are added to the barbed ends of the Arp2 and Arp3 subunits of the Arp2/3 complex. Arp2 and Arp3 have a tertiary structure similar to that of actin itself. Thus, when the Arp2/3 complex binds the first actin monomer, it allows creation of the nucleus, which is a growing end during actin filament elongation. This growing end of the filament is directed toward a membrane surface, the specific place where a vesicle will bud off. For example, during endocytosis the elongation of growing actin filaments pushes against the plasma membrane, providing the driving force that results in membrane invagination and subsequent vesicle propulsion, eventually leading to vesicle scission and movement away from the donor membrane (Ayscough, 2005, Takenawa and Suetsugu, 2007).
We propose that during autophagy the Arp2/3 complex functions in a similar manner, providing the force that is required for anterograde Atg9 movement. The Arp2/3 proteins may allow Atg9 and associated membrane to “bud” off the membrane source at the peripheral sites. As a result, Atg9 is directed in an Atg11-, Atg23- and Atg27-dependent mechanism, presumably along actin cables, to the prApe1 cargo at the PAS. The specific autophagy factors such as Atg19 and Atg11, and perhaps other molecular components, serve as adaptors between the Cvt cargo and the actin cytoskeleton. We are just beginning to understand the mechanism of Atg9 trafficking. Our knowledge primarily derives from studies in yeast; however, it is worth noting that the Arp2/3 complex, its activators and Atg9 are highly conserved through all eukaryotes, including humans. Not much is known about the relationship between the actin cytoskeleton and autophagy in higher eukaryotic systems. Further analysis of Atg9 cycling and its interactions with actin-binding proteins, will unravel the essential mechanisms of cytoskeleton function in specific types of autophagy.
ACKNOWLEDGMENTS
We thank Dr. Benjamin Glick (University of Chicago) for providing DsRed and for helpful advice about fluorescence microscopy and Drs. Charles Boone, David Drubin (University of California, Berkeley), Susan Ferro-Novick (Yale University), Bruce Goode (Brandeis University), Rong Li (The Stowers Institute), and Howard Riezman (University of Geneva) for providing strains and/or plasmids. We are grateful to the members of the Klionsky lab for helpful discussions, and we thank Dr. Usha Nair for technical assistance. We also thank Dr. Andrei Kindzelski for help with live-cell imaging. The 3D and 4D imaging were performed in the Center for Live Cell Imaging (University of Michigan Medical School). This work was supported by National Institutes of Health Public Health Service grant GM-53396 to (D.J.K.).
Abbreviations used:
Atg
autophagy-related
Cvt
cytoplasm to vacuole targeting
GFP
green fluorescent protein
PAS
phagophore assembly site
prApe1
precursor aminopeptidase I
TR phalloidin
Texas Red-X phalloidin
ts
temperature sensitive.
Footnotes
REFERENCES
- Abeliovich H., Zhang C., Dunn W. A., Jr, Shokat K. M., Klionsky D. J. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol. Biol. Cell. 2003;14:477–490. doi: 10.1091/mbc.E02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aplin A., Jasionowski T., Tuttle D. L., Lenk S. E., Dunn W. A., Jr Cytoskeletal elements are required for the formation and maturation of autophagic vacuoles. J. Cell. Physiol. 1992;152:458–466. doi: 10.1002/jcp.1041520304. [DOI] [PubMed] [Google Scholar]
- Ayscough K. R. Coupling actin dynamics to the endocytic process in Saccharomyces cerevisiae. Protoplasma. 2005;226:81–88. doi: 10.1007/s00709-005-0107-5. [DOI] [PubMed] [Google Scholar]
- Baba M., Osumi M., Scott S. V., Klionsky D. J., Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham C. L., Brumell J. H. Autophagy recognizes intracellular Salmonella enterica serovar Typhimurium in damaged vacuoles. Autophagy. 2006;2:156–158. doi: 10.4161/auto.2825. [DOI] [PubMed] [Google Scholar]
- Boldogh I. R., Nowakowski D. W., Yang H. C., Chung H., Karmon S., Royes P., Pon L. A. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol. Biol. Cell. 2003;14:4618–4627. doi: 10.1091/mbc.E03-04-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh I. R., Yang H. C., Nowakowski W. D., Karmon S. L., Hays L. G., Yates J. R., III, Pon L. A. Arp2/3 complex and actin dynamics are required for actin-based mitochondrial motility in yeast. Proc. Natl. Acad. Sci. USA. 2001;98:3162–3167. doi: 10.1073/pnas.051494698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd C., Hughes T., Pypaert M., Novick P. Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J. Cell Biol. 2004;167:889–901. doi: 10.1083/jcb.200408124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Yorimitsu T., Reggiori F., Legakis J. E., Wang C.-W., Klionsky D. J. Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M. I., Gutierrez M. G., Romano P. S. The two faces of autophagy: Coxiella and Mycobacterium. Autophagy. 2006;2:162–164. doi: 10.4161/auto.2827. [DOI] [PubMed] [Google Scholar]
- D'Agostino J. L., Goode B. L. Dissection of Arp2/3 complex actin nucleation mechanism and distinct roles for its nucleation-promoting factors in Saccharomyces cerevisiae. Genetics. 2005;171:35–47. doi: 10.1534/genetics.105.040634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan M. C., Cope M. J., Goode B. L., Wendland B., Drubin D. G. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat. Cell Biol. 2001;3:687–690. doi: 10.1038/35083087. [DOI] [PubMed] [Google Scholar]
- Dunn W. A., Jr, Cregg J. M., Kiel J. A. K. W., van der Klei I. J., Oku M., Sakai Y., Sibirny A. A., Stasyk O. V., Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- Fehrenbacher K. L., Yang H. C., Gay A. C., Huckaba T. M., Pon L. A. Live cell imaging of mitochondrial movement along actin cables in budding yeast. Curr. Biol. 2004;14:1996–2004. doi: 10.1016/j.cub.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Guan J., Stromhaug P. E., George M. D., Habibzadegah-Tari P., Bevan A., Dunn W. A., Jr, Klionsky D. J. Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol. Biol. Cell. 2001;12:3821–3838. doi: 10.1091/mbc.12.12.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M., Noda T., Baba M., Ohsumi Y. Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic. 2005;6:56–65. doi: 10.1111/j.1600-0854.2004.00245.x. [DOI] [PubMed] [Google Scholar]
- He C., Song H., Yorimitsu T., Monastyrska I., Yen W.-L., Legakis J. E., Klionsky D. J. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J. Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Huang W.-P., Klionsky D. J. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissova I., Salin B., Schaeffer J., Bhatia S., Manon S., Camougrand N. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy. 2007;3:329–336. doi: 10.4161/auto.4034. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J. Autophagy. Curr. Biol. 2005;15:R282–R283. doi: 10.1016/j.cub.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., et al. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., Cueva R., Yaver D. S. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legakis J. E., Yen W.-L., Klionsky D. J. A cycling protein complex required for selective autophagy. Autophagy. 2007;3:422–432. doi: 10.4161/auto.4129. [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D. J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Liu J., Kaksonen M., Drubin D. G., Oster G. Endocytic vesicle scission by lipid phase boundary forces. Proc. Natl. Acad. Sci. USA. 2006;103:10277–10282. doi: 10.1073/pnas.0601045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Klionsky D. J. Protein turnover via autophagy: implications for metabolism. Annu. Rev. Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- Moreau V., Galan J. M., Devilliers G., Haguenauer-Tsapis R., Winsor B. The yeast actin-related protein Arp2p is required for the internalization step of endocytosis. Mol. Biol. Cell. 1997;8:1361–1375. doi: 10.1091/mbc.8.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyol R., Haeckel A., Ritter A., Qualmann B., Kessels M. M. Regulation of N-wasp and the Arp2/3 complex by abp1 controls neuronal morphology. PLoS ONE. 2007;2:e400. doi: 10.1371/journal.pone.0000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T. D. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Klionsky D. J. Autophagosomes: biogenesis from scratch? Curr. Opin. Cell Biol. 2005;17:415–422. doi: 10.1016/j.ceb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska L., Shintani T., Klionsky D. J. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2005a;16:5843–5856. doi: 10.1091/mbc.E05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Pelham H.R.B. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 2001;20:5176–5186. doi: 10.1093/emboj/20.18.5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Shintani T., Nair U., Klionsky D. J. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005b;1:101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein D. C., Gestwicki J. E., Murphy L. O., Klionsky D. J. Potential therapeutic applications of autophagy. Nat. Rev. Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- Scott S. V., Baba M., Ohsumi Y., Klionsky D. J. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J. Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen P. O., Berg T. O., Blankson H., Fengsrud M., Holen I., Stromhaug P. E. Structural aspects of autophagy. Adv. Exp. Med. Biol. 1996;389:103–111. doi: 10.1007/978-1-4613-0335-0_12. [DOI] [PubMed] [Google Scholar]
- Shintani T., Huang W.-P., Stromhaug P. E., Klionsky D. J. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D. J. Autophagy in health and disease: a double-edged sword. Science. 2004a;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D. J. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 2004b;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa T., Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- Toshima J., Toshima J. Y., Martin A. C., Drubin D. G. Phosphoregulation of Arp2/3-dependent actin assembly during receptor-mediated endocytosis. Nat. Cell Biol. 2005;7:246–254. doi: 10.1038/ncb1229. [DOI] [PubMed] [Google Scholar]
- Welch M. D., Rosenblatt J., Skoble J., Portnoy D. A., Mitchison T. J. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- Wendland B., McCaffery J. M., Xiao Q., Emr S. D. A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J. Cell Biol. 1996;135:1485–1500. doi: 10.1083/jcb.135.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder S. J., Ayscough K. R. Actin-binding proteins. J. Cell Sci. 2005;118:651–654. doi: 10.1242/jcs.01670. [DOI] [PubMed] [Google Scholar]
- Winter D., Podtelejnikov A. V., Mann M., Li R. The complex containing actin-related proteins Arp2 and Arp3 is required for the motility and integrity of yeast actin patches. Curr. Biol. 1997;7:519–529. doi: 10.1016/s0960-9822(06)00223-5. [DOI] [PubMed] [Google Scholar]
- Yang Z., Huang J., Geng J., Nair U., Klionsky D. J. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol. Biol. Cell. 2006;17:5094–5104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen W.-L., Legakis J. E., Nair U., Klionsky D. J. Atg27 is required for autophagy-dependent cycling of Atg9. Mol. Biol. Cell. 2007;18:581–593. doi: 10.1091/mbc.E06-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D. J. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Nair U., Yang Z., Klionsky D. J. Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Qi H., Taylor R., Xu W., Liu L. F., Jin S. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy. 2007;3:337–346. doi: 10.4161/auto.4127. [DOI] [PubMed] [Google Scholar]