The bHLH/Per-Arnt-Sim transcription factor SIM2 regulates muscle transcript myomesin2 via a novel, non-canonical E-box sequence (original) (raw)
Abstract
Despite a growing number of descriptive studies that show Single-minded 2 (Sim2) is not only essential for murine survival, but also upregulated in colon, prostate and pancreatic tumours, there is a lack of direct target genes identified for this basic helix–loop–helix/PAS transcription factor. We have performed a set of microarray experiments aimed at identifying genes that are differentially regulated by SIM2, and successfully verified that the Myomesin2 (Myom2) gene is SIM2-responsive. Although SIM2 has been reported to be a transcription repressor, we find that SIM2 induces transcription of Myom2 and activates the Myom2 promoter sequence when co-expressed with the heterodimeric partner protein, ARNT1, in human embryonic kidney cells. Truncation and mutation of the Myom2 promoter sequence, combined with chromatin immunoprecipitation studies in cells, has lead to the delineation of a non-canonical E-box sequence 5′-AACGTG-3′ that is bound by SIM2/ARNT1 heterodimers. Interestingly, in immortalized human myoblasts knock down of Sim2 results in increased levels of Myom2 RNA, suggesting that SIM2 is acting as a repressor in these cells and so its activity is likely to be highly context dependent. This is the first report of a direct SIM2/ARNT1 target gene with accompanying analysis of a functional response element.
INTRODUCTION
The Single-minded 2 (SIM2) protein is a member of the basic helix–loop–helix/Per-Arnt-Sim homology (bHLH/PAS) group of transcriptional regulators that are characteristically involved in mediating a variety of developmental events such as angiogenesis, neurogenesis and tracheal formation, and cellular responses to environmental stimuli including hypoxia, toxic pollutants and the light/dark cycle (1–3). Similar to most bHLH/PAS family members, both of the murine Single-minded genes (Sim1 and Sim2) are essential for post-natal survival in mice. (4–6). Their Drosophila homologue, dSim, encodes a positively acting, master regulator of central nervous system (CNS) midline development (7–10). As with dSIM, SIM2 appears to have key neurological functions. Sim2 is expressed in brain regions where it is required to produce a full complement of anterior hypothalamic cells expressing thyrotropin-releasing hormone and somatostatin (11,12) and for the correct development of mammillary body neurons (13). The Sim2 homozygous mutant phenotype, however, is complex and not immediately informative. Sim2−/− mice die soon after birth due to a breathing defect involving abnormal rib protrusions that attach aberrantly to intercostal muscle, a hypoplastic diaphragm and eventual tears in the pleural mesothelium (5). The majority of mutant mice also develop congenital scoliosis associated with asymmetric rib growth and a second group has reported additional secondary palatal closure and craniofacial defects in Sim2 mutant mice on a slightly different genetic background (5,6). Extensive in situ hybridization experiments have identified a tissue-specific pattern of Sim2 transcript expression in the diencephalon, kidney, craniofacial structures, limbs, ribs and skeletal muscle of the developing mouse that is maintained in the adult, with highest expression in the kidneys, skeletal muscle and brain (5,14–18). Interestingly, the location of the human Sim2 gene (hSim2) in the Down syndrome critical region of the human genome, coupled with learning defects found in mice overexpressing Sim2 (19,20), suggest it may play some role in the complex aetiology of Down syndrome. Despite this impressive array of descriptive data there remains a deficit of mechanistic, functional analysis of the SIM2 protein.
Group I members of the bHLH/PAS family (including SIM2) are usually signal or spatiotemporally induced and require heterodimerization with an ARNT partner protein, in order to recognize their cognate DNA element and affect transcription of target genes. Unlike the majority of bHLH/PAS factors that contain strong transactivation domains (including dSIM), the murine Sim2 gene product is a potent repressor of transcription in mammalian one-hybrid experiments (21). Whilst the native SIM2/ARNT DNA response element has yet to be elucidated, the dSIM/dARNT complex recognizes CNS midline enhancer elements [CME, 5′-(G/A)(T/A)ACGTG-3′] often present in multiple copies in target genes (22). Expression of a short isoform of mammalian SIM2 or truncation mutants, in which the C-terminal repression regions of SIM2 have either been totally removed or replaced with a constitutive transactivation domain, results in activation of CME-driven reporter genes in transient transfection assays in mammalian cells (23–25). The SIM2/ARNT heterodimer has also been shown to bind hypoxic response element (HRE, 5′-TACGTG-3′) sequences in a reporter gene context in cells (26) and so a reasonable prediction is that direct target genes of SIM2 will contain hexameric sequences that are identical or very similar to the core 5′-ACGTG-3′ sequence found in CME and HRE sequences.
Interestingly, the short isoform of hSIM2 (hSIM2s) has been shown to be expressed selectively in colon, prostate and pancreatic carcinomas but not in corresponding normal tissues (27,28). This hSim2s splice variant encodes a 570 amino acid protein (29) that contains only one of the two SIM2 repression regions and a unique C-terminal sequence of 44 residues. More excitingly, antisense inhibition of hSim2s results in inhibition of cancer cell growth and induction of apoptosis, leading to speculation that it may be a useful tumour marker and possible therapeutic target (28,30). While most attention has focused on the upregulation of hSim2s, transcripts encoding the long isoform of hSim2 (hSim2) have also been detected selectively in pancreatic tumours and cell lines but not in normal tissue (28). Conversely, hSim2s expression has been reported to be higher in normal breast epithelium and cell lines than breast tumour tissue and derived cell lines, indicating that this misregulation may be tissue-type specific (31). Despite the fundamental biological roles of the SIM proteins and apparent misregulation in specific tumour types, extremely little is known of their mechanisms of action. In an effort to elucidate the function of SIM2, we show that the transcription factor is nuclear localized in adult mouse skeletal muscle and we have used a microarray approach to identify SIM2-responsive transcripts. From this list of potential SIM2 targets, we analyse the muscle and kidney expressed Myomesin2 (Myom2) in greater detail, uncovering a non-canonical E-box sequence, 5′-AACGTG-3′, as a SIM2/ARNT binding site in the Myom2 promoter. Interestingly, although this non-canonical E-box sequence contains the core HRE sequence, it is rarely found in hypoxia inducible genes, and consistent with this observation the Myom2 promoter is not induced by HIF-1α/ARNT1. Surprisingly, given the repressive behaviour of SIM2 in several transcriptional assays (14,21), the long and short isoforms of SIM2 activate transcription of the Myom2 promoter with ARNT1 in human embryonic kidney cells. However, knockdown of Sim2 in immortalized human myoblasts leads to an increase in Myom2 levels. This suggests that SIM2 can regulate transcription of target genes, either positively or negatively, in a context-specific manner. This is the first report of an endogenous SIM2/ARNT response element in a bona fide direct target gene identified for SIM2.
MATERIALS & METHODS
Construction of expression and reporter vectors
The entire hSim2s cDNA sequence was amplified in three segments from cDNA derived from HEK293 cells using primers SIM2-1 (5′CTA AGC TAG CAT GAA GGA GAA GTC C) with SIM2-184 (5′GTG GAT GAC CTT GTA TCC) for fragment 1, SIM2-149F (5′CTG CTC CAA GAG TAC GAG) with SIM2-480R (5′CTC AGG AAA AAG CGT GCC) for fragment 2 and SIM2-456F (5′AAA AGC CAA TGT TGC CGG) with hSIM2sEcoRV-R (5′GAC TGA TAT CCT TAG AAG CAG AAA GAG G) for fragment 3. Fragment 1 digested with NheI/BstBI and fragment 2 digested with BstBI/BamHI were inserted into NheI/BamHI digested pEF/mSIM2(Myc)2/IRESpuro (26). The NheI/BamHI fragment from this vector and fragment 3 digested with BamHI/EcoRV were ligated into NheI/EcoRV digested pEF/mSIM2(Myc)2/IRESpuro to produce a mammalian expression vector for Myc-epitope tagged short form of hSim2, pEF/hSIM2s(Myc)2/IRESpuro. pEF/hARNT1/bos and pEF/hARNT1/IRESneo were generated by insertion of the untagged human ARNT1 cDNA BamHI fragment from pGEM7Arnt into BamHI digested pEF-bos-cs (32), or linearised pEF/IRESneo (M.Kleman). A bacterial expression vector for thioredoxin/6xHis (trx/His) tagged murine SIM2 (residues 1–231) was generated by PCR amplification of Sim2 cds to include an in frame 5′ EcoRI site and 3′ stop codon with XhoI site and insertion of this fragment into EcoRI/XhoI digested pET-32a (Novagen, Gibbstown, NJ, USA). The p_Myom2__1.3 kb-LUC vector was generated by PCR amplification of a 1.3 kb fragment of human genomic DNA sequence corresponding to the human Myom2 promoter region (−1178/+ 149) and insertion into SmaI/XhoI digested pGL3-Basic (Promega, Madison, WI, USA). Directed mutagenesis of this reporter at Site1 (5′AACGTG′3 to 5′AAAAAG′3) generated p_Myom2__1.3 kb_Site1mut-LUC, Site 2 (5′AACGTG′3 to 5′AAAAAG′3) p_Myom2__1.3 kb_Site2mut-LUC and both sites p_Myom2__1.3 kb_Site1&2mut-LUC. Sequential truncation of the Myom2 promoter produced Δ1 (−695/+149), Δ2 (−245/+149), Δ3 (−151/+149) and Δ4 (−91/+149) by PCR amplification of the relevant sections of the Myom2 promoter and subsequent subcloning into KpnI/XhoI digested pGL3-Basic (Promega). The pHGTD-PBasic-LUC plasmid containing 733 bp of the HGTD-P promoter was generated as described for HGTD-P-luc-2 (33), except that the promoter fragment was cloned into the pGL3-Basic vector (Promega). All other expression or reporter vectors have been previously described (26,34).
Microarray
Generation and propagation of the 293TControl, 293TSIM2 and 293TSIM2/AD cell lines used has been previously described (26). Labelled cDNA samples derived from RNA samples from each of the 293T stable cell lines were hybridized in a pairwise fashion to slides printed with the 8000 clone human Research Genetics cDNA library. Each comparison was replicated and each duplicate comparison performed with a dye swap to allow correction for dye labelling biases, resulting in four separate comparisons (i.e. four slides). Each replicate was separately reverse transcribed and labelled, following the microarray experimental procedure outlined at http://www.microarray.adelaide.edu.au/. Good consistency was observed between replicate spots containing the same clone on the same slide, between slides replicated with the same dye pairing and between slides hybridized with dye swapped samples. Statistical analysis of normalized microarray data involved the use of Spot (http://experimental.act.cmis.csiro.au/Spot/index.php) and a statistics and graphical package R (http://www.r-project.org/) in combination with a data analysis package, LIMMA (http://bioinf.wehi.edu.au/limmaGUI/index.html#Rpkgs).
Cell culture and production of double stable cell lines expressing SIM2 or hSIM2s and ARNT1
Human embryonic kidney 293a or 293T cells were routinely maintained at 37°C, 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) (JRH Biosciences, Lenexa, KS, USA). To produce stably transfected polyclonal lines, subconfluent 293a cells were transfected with blank pEF/IRESpuro, or pEF/SIM2(Myc)2/IRESpuro or pEF/hSIM2s(Myc)2/IRESpuro with pEF/IRESneo or pEF/hARNT1/IRESneo, respectively and transfected cells were selected and expanded with up to 10 µg/ml puromycin (Sigma, St. Louis, MO, USA) and 1mg/ml G418 (Sigma). Immortalized human myoblast cell line, LHCN-M2, was cultured to confluency in growth medium as described (35). At confluency (Day 1), medium was changed to growth media without hepatocyte growth factor and with 20 nM dexamethasone, 0.5% FBS and 10 mg/l insulin and 550 mg/l transferrin to induce fusion of the cells. Cells were transfected with 10 nM control siRNA or siRNAs targeting Sim2 or Myom2 (Ambion, Austin, TX, USA) using HiPerfect on days 1 and 4 as per the manufacturer's instructions (Qiagen, Valencia, CA, USA). By Day 7, multinucleate cells were present in the culture at low frequency (∼10% of cells) and cells were not yet fully elongated or fully differentiated into myotubes.
Immunofluorescence
Fresh-frozen adult mouse hind limb muscle tissue samples were sectioned using a cryostat at −20°C, 8 μm sections collected on SuperFrost Plus slides (Menzel-Glaser, Portsmouth, NH, USA) and immediately fixed in freshly prepared 4% paraformaldehyde for 6 min at room temperature. Sections were washed three times in 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.1% Triton X-100 (TBST) for 5 min and then blocked by incubation with 10% FBS in TBST for 1–3 h at room temperature. Following this, sections were incubated overnight at 4°C in a humid chamber with goat anti-hSIM2 (sc-8716, lot C1505, Santa Cruz Biotechnology, Santa Cruz, CA, USA, 1:50), rabbit anti-mouse MYOM1-EH (R2-α-moEH, 1:1000), mouse anti-mouse MYOM2 (AA259, 1:2) or anti-hSIM2 and anti-mouse MYOM2 or anti-myogenin (Chemicon, Billerica, MA, USA, 1:100) antibodies together, all diluted in 10% FBS in TBST. After washing three times in TBST for 5 min, sections were incubated with the following secondary antibodies diluted 1:1000 in 10% FBS/TBST with bisBenzimide H stain for 1 h at room temperature in a dark container; donkey anti-goat Alexa Fluor 594 (Invitrogen), goat anti-mouse IgA-FITC (Sigma) or donkey anti-rabbit Alexa Fluor 488 (Invitrogen). Secondary incubations for sections co-stained for SIM2 and MYOM2 or SIM2 and myogenin were performed sequentially, with the donkey anti-goat Alexa Fluor 594 first and the goat anti-mouse IgA-FITC second with three, 5 min TBST washes in between. Sections were subsequently washed three times in TBST, mounted in Vectashield media (Vector, Burlingame, CA, USA) or SlowFade mountant (Invitrogen) and examined using a Nikon Eclipse E800 microscope.
RNA expression analysis
Total RNA from adult mouse tissues or lines was prepared using RNAwiz reagent (Ambion) or RNABee (Teltest, Friendswood, TX, USA) according to the manufacturer's instructions with an additional DNAse treatment step to prevent genomic DNA contamination of samples. Each RNA sample (1 μg) was reverse transcribed using Superscript and the resulting cDNA samples PCR amplified using primers or primer/probe sets (Applied Biosystems, Foster City, CA, USA) specific for Sim2, Myom2 and the control GAPDH.
Immunoblotting
Nuclear extracts were prepared as previously described (36), whilst whole-cell lysates were generated using passive lysis buffer in accordance with the manufacturer's instructions for luciferase reporter studies (Promega). Lysates were subjected to SDS–PAGE (7.5% gel) and then transferred to nitrocellulose using a semidry blotter (Biorad, Hercules, CA, USA). Proteins were detected with the anti-Myc 9E10 or 71D10 monoclonal antibodies (Cell Signalling) or an anti-ARNT1 rabbit polyclonal serum raised against residues 1–142 of human ARNT1 (#51).
Luciferase reporter assays
The Myom2 and HGTD-P luciferase reporter plasmids described above, were used in conjunction with the control pGL3-Basic-LUC (Promega) vector and the internal control plasmid pRL-SV40 (Promega). Subconfluent 293a or 293T cells were transfected 12–24 h after plating using FuGENE6 (Boehringer Mannheim) as per the manufacturer's instructions. Each transfection contained 200 ng/well of Firefly Luciferase reporter and 20 ng internal control Renilla pRL-SV40, together with 5–250 ng/well of each expression plasmid or blank expression plasmid necessary to normalize the amount of DNA transfected. Cells were lysed in Passive Lysis Buffer (Promega) 48 h post-transfection and lysates were analysed for luciferase activity using the Dual Luciferase Reporter assay (Promega) according to the manufacturer's directions.
Electrophoretic mobility shift assays
Bacterial expression of thioredoxin/6xHis (trx/His) tagged ARNT1 (residues 1–362) and partial purification using nickel affinity chromatography has been previously described (34). Co-expression and isolation of the trx/His SIM2 (residues 1–231) with trx/His ARNT1 (1–362) was performed in a similar manner except that cells were lysed in 20 mM sodium phosphate (pH 7.2), 150 mM NaCl, 10 mM imidazole and the protein eluting with 250 mM imidazole from the nickel affinity column was immediately desalted using a PD10 column into 20 mM Tris–HCl (pH7.5), 0.1 mM EDTA, 10% glycerol, 50 mM NaCl, 0.2 mM dithiothreitol. Double stranded, fluorescein-labelled DNA probes were composed of the previously published Ebox sequence (34), or a portion of the human Myom2 promoter containing the sequence 5′-AATCACGATTCAAAACTGGAACGTGTCCTTTCTGAGTCCC-3′ or the mutant Myom2 probe with sequence 5′-AATCACGATTCAAAACTGGAAAAAGTCCTTTCTGAGTCCC-3′ (GeneWorks, Adelaide, SA, Australia). Binding reaction conditions were identical to those previously specified for the HRE probe (34) containing 1 mM PMSF and 8 nM FAM-labelled DNA probe and with pre-immune or a polyclonal antibody against the N-terminus of ARNT1 (Bambi) added to the reaction as indicated. Reactions were quickly prepared in siliconized microfuge tubes and incubated in the dark for 20 min at room temperature. Protein/DNA complexes were resolved on a 7% polyacrylamide, 5% glycerol, 25 mM Tris–HCl (pH 8.3), 25 mM boric acid and 0.5 mM EDTA gel that had been precooled overnight at 4°C and DNA was visualized with an FX molecular imager (BioRad).
Chromatin immunoprecipitation assay
Chromatin extracts were prepared from 293 hSIM2s/ARNT stable cells (∼9 × 106 cells/immunoprecipitation) as described in the EZ ChIP protocol (Upstate). To solubilize chromatin and shear DNA, six 10 s sonication pulses interspersed with 20 s rest periods were performed on ice using a Sonifier Cell Disruptor 200 (Branson). Chromatin immunoprecipitation experiments were performed using the EZ ChIP kit (Upstate) as per the manufacturer's instructions with 5 µg of goat anti-hSIM2 antibody (C15, Santa Cruz Biotech.) or mouse anti-Myc epitope (4A6, Upstate) for the specific immunoprecipitations and 5 µg goat IgG (Santa Cruz Biotech.) or mouse IgG (Upstate) for the non-specific controls. A total of 3 µl of eluted DNA was added to each PCR using primers designed to amplify Myom2 promoter sequences (Site 1F 5′-AAGGATTAGAACTCATGGAGGAG A-3′, Site 1R 5′-AGGAAAAGCTTCTTGTTCAA ACTG-3′, Site 3F 5′-GCGTCTTTCTCGTATCTG ATCTTC-3′, Site 3R 5′-CTTAAGAAACTCCAC GTTCTCACA-3′) or the control GAPDH primers included with the EZ ChIP kit (Upstate).
RESULTS
Identification of transcripts differentially regulated by SIM2 in human embryonic kidney cells using microarray technology
In an effort to understand the role of SIM2 in the kidney and at other sites of expression, we undertook a microarray-based experiment to identify transcripts that are differentially regulated by SIM2. As SIM2 is one of the few bHLH/PAS family members to act as a repressor in cell culture experiments (14,21), we made use of the fusion protein, SIM2/AD, in which the SIM2 repression regions have been replaced by the constitutive transactivation domain of the DR, to expedite identification of genes that may be transcriptionally regulated by SIM2. The transcript profiles from 293T polyclonal stable cell lines engineered to be control cells or to express SIM2 or SIM2/AD (26), were examined for differentially expressed transcripts by three two-way comparisons between the different cell lines. Many transcripts of interest were identified in this manner, particularly those that are expressed in tissues that also express Sim2, such as the skeletal muscle transcript Myom2 and those with roles in tumourigenesis. A list of the top 100 most differentially expressed transcripts is supplied (Supplementary material). This article focuses on our examination of the relationship between SIM2 and one of the differentially regulated transcripts, M_yom2_.
SIM2AD fusion protein activates transcription of the Myom2 promoter
The protein product of Myom2 (MYOM2, also known as the M-protein) is a 165 kDa structural component of sarcomeric myofibrils (37,38). It belongs to the intracellular group of the immunoglobulin superfamily of proteins and interacts with the giant protein, TITIN, found in striated muscles and myosin (39,40). The role of MYOM2 as a structural component of the M-band in striated muscle has been investigated in detail using electron microscopy studies, but the expression and function of MYOM2 in other non-muscle tissues has not been examined. Interestingly, both Sim2 and Myom2 RNAs are detected in the adult mouse kidney and also in the human embryonic kidney cell line, 293a (Figures 2B and 5B).
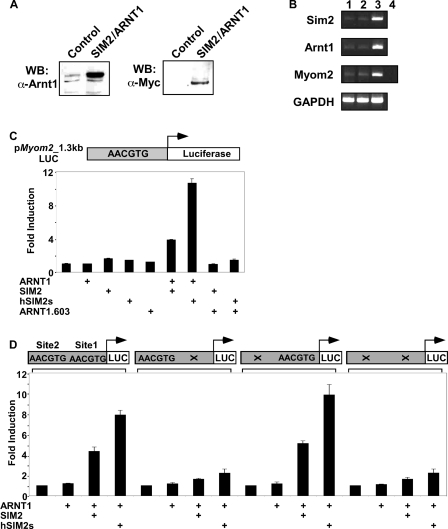
Figure 2.
Expression of the SIM2/ARNT1 heterodimer induces Myom2 transcription in human embryonic kidney cells through a non-canonical E-box. (A) Nuclear extracts prepared from the 293Control or 293SIM2/ARNT1 cell lines were subjected to SDS–PAGE and western blot using either anti-ARNT1 polyclonal or anti-Myc monoclonal antibodies. (B) total RNA samples prepared from untransfected 293a cells (lane 1), 293Control (lane 2) or 293SIM2/ARNT1 (lane 3) double stable cells, were reverse transcribed and the resultant cDNA samples were PCR-amplified in the linear range using primers specific for Sim2, Arnt1, Myom2 and the internal control, GAPDH. Control Myom2 reactions containing RNA samples as template were also performed (lane 4). C and D, 293 cells were co-transfected for 48h with the _Myom2_-promoter containing reporter plasmid and expression vectors as indicated and then assayed for luciferase activity. Luciferase activity was normalized against a Renilla luciferase internal control and results are depicted as fold induction over the backbone pGL3-Basic-LUC reporter for each transfection. Results shown are either from a single experiment performed in triplicate with standard deviation, which is representative of four repeats of this set of transfections (C) or from three independent experiments performed in triplicate with standard deviation (D).
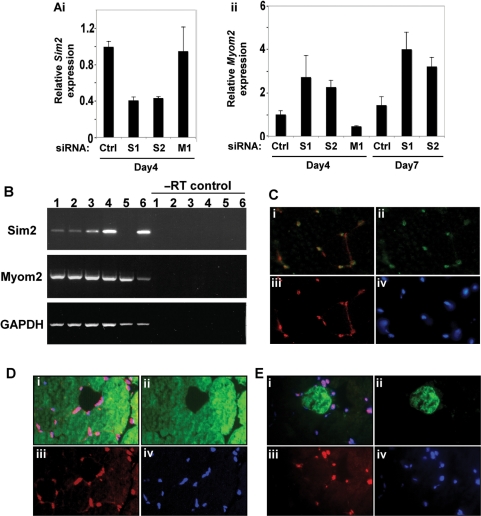
Figure 5.
SIM2 represses Myom2 expression in human myoblasts but is coexpressed with MYOM2 in adult mouse muscle. (A) Knockdown of Sim2 in LHCN-M2 myoblasts results in increased Myom2 expression. Level of Sim2 (i) and Myom2 (ii) RNA in LHCN-M2 cells treated with control (ctrl), Myom2 siRNA (M1) or either of two independent Sim2 siRNAs (S1 and S2) determined by real-time PCR. Experiment is representative of a set of three repeats, Sim2 and Myom2 expression is relative to GAPDH expression and error bars depict standard deviation. (B) Total RNA samples prepared from adult mouse muscle tissues (lane 1 abdominal muscle, lane 2 diaphragm, lane 3 forelimb muscle, lane 4 hindlimb muscle, lane 5 heart) and kidney (lane 6) were reverse transcribed and the resultant cDNA samples were PCR-amplified in the linear range using primers specific for Sim2, Myom2 and the internal control, GAPDH. (C–E) Multiple muscle fibres are presented in the figure in cross section and each fibre is surrounded by multiple nuclei. (C) SIM2 is expressed in myonuclei of adult mouse muscle. Composite image (i) of cryostat sections of adult mouse hind limb muscle immunostained for myogenin (ii) and SIM2 proteins (iii), showing nuclei visualised with bis-Benzimide H (iv). (D) SIM2 is expressed in both MYOM2 positive and negative muscle fibres. Composite image (i) of cryostat sections of adult mouse hind limb muscle immunostained for MYOM2 (ii, fast fibres) and SIM2 proteins (iii), showing nuclei visualized with bis-Benzimide H (iv). (E) composite image (i) of cryostat sections of adult mouse hind limb muscle immunostained for EH-MYOM1 (ii, one positive fibre in view) and SIM2 proteins (iii), showing nuclei visualised with bis-Benzimide H (iv).
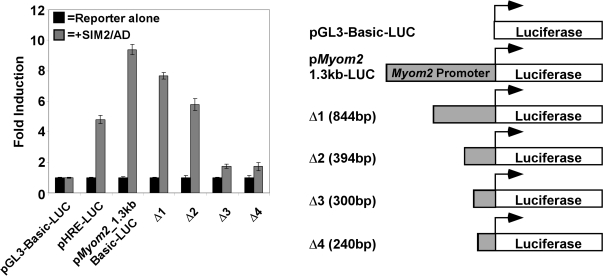
We have previously shown that SIM2/ARNT1 heterodimers can recognize HRE sequences from the erythropoietin (epo) enhancer in a reporter gene context, but unlike HIF-1α/ARNT1 complexes, do not activate transcription of the reporter in 293T cells (26). As mentioned earlier, the SIM2/AD fusion protein functions as a transactivator and has been used as a tool to examine possible SIM2 target genes. Transfection of the SIM2/AD chimera clearly activates transcription of the positive control HRE-reporter in 293T cells (Figure 1). To examine whether the Myom2 gene was responsive to SIM2 at a transcriptional level, a fragment of the human Myom2 promoter (1.3 kb upstream from the transcription start) was cloned into a luciferase reporter plasmid. Expression of SIM2/AD activated transcription of this Myom2_-reporter to a higher level than the HRE-reporter (Figure 1). Potential SIM2/ARNT-binding sites in the Myom2 promoter region were delineated by sequential deletion of the promoter fragment in the luciferase reporter (Figure 1), with reporter activation by SIM2/AD decreasing gradually as the promoter fragment was shortened (compare Δ1 and Δ2 with p_Myom2_1.3 kb-LUC, Figure 1). Significantly, there was a marked loss of activity upon truncation of a 95 bp section of promoter (Figure 1, compare Δ2 and Δ3), suggesting that it may contain SIM2 responsive sites. Sequence analysis of this fragment identified a hexameric sequence that is similar, but not identical to the HRE and CME core consensus sequence of 5′-TACGTG-3′. We have termed this Site 1 (5′-AACGTG-3′).
Figure 1.
Myom2 promoter is induced by SIM2/AD expression. Delineation of required promoter regions and a potential SIM2 responsive site. Subconfluent 293T cells were co-transfected with luciferase reporter plasmids and a blank or SIM2/AD expression plasmid as indicated. After 48 h luciferase activity was assayed and relative luciferase activities normalized to the value for each reporter alone and presented as a fold induction. Results depicted are from an experiment performed in triplicate with standard deviation, which is representative of four repeats of this set of transfections.
SIM2/ARNT expression upregulates the endogenous Myom2 transcript in HEK293a cells
As the Myom2 promoter was induced by expression of the SIM2/AD chimera, we wished to ascertain the effect of expression of the wild-type SIM2/ARNT1 heterodimer on Myom2 transcript levels. As human embryonic kidney 293a cells endogenously express the Myom2 transcript they were used to generate double stable cell lines expressing elevated levels of both ARNT1 and SIM2. Expression of Myc-epitope tagged SIM2 and ARNT1 was confirmed by western blot analyses of lysates from the stable cell lines (Figure 2A). Surprisingly, given the reported role of the SIM2 protein as a transcriptional repressor, the co-expression of both ARNT1 and SIM2 dramatically increases the level of the endogenous Myom2 transcript, compared to control stable cell lines containing blank expression vectors or untransfected 293a cells (Figure 2B).
SIM2/ARNT heterodimers activate transcription from the human Myom2 promoter through a non-canonical E-box sequence
Consistent with the change in Myom2 transcript levels in our 293SIM2/ARNT1 stable cell lines, the _Myom2_-reporter is induced with transient co-expression of the long or short isoform of SIM2 and ARNT1 in 293 cells, but not with ARNT1 alone (Figure 2C). ARNT1 levels appear to be limiting in 293 cells as SIM2 alone also does not activate the Myom2 reporter (Figure 2C). Co-expression of SIM2 with a truncation mutant of ARNT1 lacking the C-terminal activation domain, ARNT1.603, results in abrogation of reporter induction, showing that the transactivation domain of ARNT1 is required for this activation of transcription (Figure 2C). This leads to the proposal that SIM2/ARNT1 heterodimers may operate as activators of Myom2 transcription by utilizing the ARNT1 transactivation domain.
To test whether the Myom2 promoter Site1 is a functional SIM2/ARNT1 binding site, we mutated this site in the Myom2 reporter construct. An identical 5′-AACGTG-3′ hexameric sequence was found 860 bp upstream of Site1 in the Myom2 promoter, termed Site2, and this was also independently mutated. Intriguingly, mutation of Site 1 but not Site 2 resulted in a dramatic decrease in the induction of this reporter gene by SIM2/ARNT1 (Figure 2D), suggesting that Site1 is necessary for SIM2/ARNT1 activation of Myom2 transcription.
SIM2/ARNT1 complexes recognize Myom2 promoter sequences in vitro and in human kidney cells
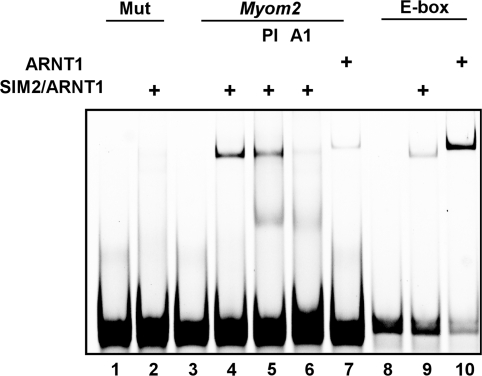
To investigate whether the SIM2/ARNT1 heterodimer could directly recognise DNA sequences found in the Myom2 promoter, in vitro studies using the EMSA technique with a 40 bp probe sequence from the Myom2 promoter encompassing the Site 1 sequence were performed. Bacterially co-expressed and partially purified thioredoxin/6xHis (trx/His) tagged SIM2 (residues 1–231) and ARNT1 (residues 1–362) formed a complex with the Myom2 probe that could be specifically blocked with an anti-ARNT1 antibody but not the pre-immune serum (Figure 3, lanes 4–6). This gel-retarded complex did not form when the partially purified proteins were mixed with a probe in which Site1 had been mutated (Figure 3, lane 2, 5′-AACGTG-3′ mutated to 5′-AAAAAG-3′). The trx/His-tagged ARNT1(1–362) protein expressed alone and partially purified also forms a shifted complex with the Myom2 probe; however, this homodimer clearly runs slightly higher on the non-denaturing gel than the heterodimer and is of much lower intensity (Figure 3, compare lanes 4 and 7). By comparison, the same amount of partially purified trx/His-ARNT1(1–362) produces a much stronger intensity shifted band with its cognate DNA binding element, the E-box, than the co-expressed and purified trx/His-SIM2 (1–231) and ARNT1 (1–362, Figure 3, compare lanes 9 and 10). These in vitro DNA-binding assays are consistent with SIM2/ARNT1 heterodimers recognising the Site1 5′-AACGTG-3′ sequence in order to regulate the transcription of the Myom2 gene.
Figure 3.
SIM2/ARNT1 dimers recognise the 40 bp Myom2 promoter sequence in vitro. Bacterially co-expressed and partially purified, thioredoxin/6xHis tagged N-terminal portions of SIM2 and ARNT1 or ARNT1 alone were bound to fluorescently labelled 40bp Myom2, Myom2mut or E-box probes and subjected to non-denaturing PAGE. The presence of ARNT1 in the gel shifted complex was visualized by incubation with anti-ARNT1 serum (A1) compared to control pre-immune serum (PI).
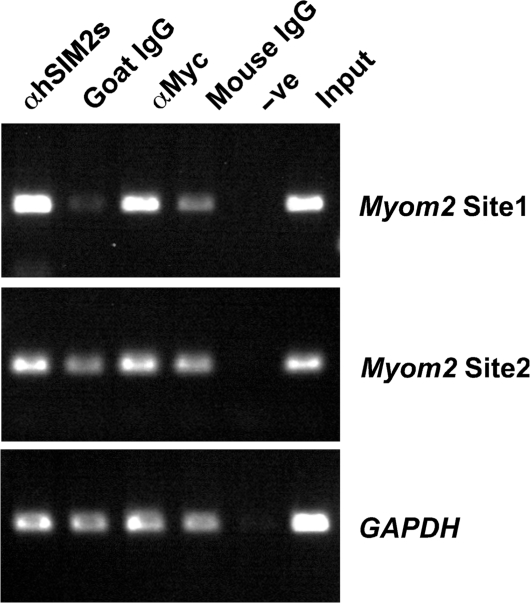
To further support these observations, chromatin immunoprecipitation experiments were performed using 293a cells engineered to express both ARNT1 and Myc-epitope tagged hSIM2s. Endogenous Myom2 promoter DNA containing the Site 1 sequence was highly enriched in the specific immunoprecipitation for hSIM2s using an anti-hSIM2s antibody or an antibody that recognises the Myc-epitope tag of hSIM2s, compared to the non-specific immunoglobulin controls (Figure 4, Site 1 panel, compare lane 1 with lane 2; lane 3 with lane 4). There was no enrichment of the control DNA sequences from the GAPDH gene in the chromatin immunoprecipitation samples (Figure 4). Likewise, PCR-amplification of Site2 in the Myom2 promoter containing an identical core sequence of 5′-AACGTG-3′ that is 860 bp upstream of Site 1, did not indicate any enrichment in the hSIM2s specific immunoprecipitations at this site (Figure 4). This confirms that hSIM2s can recognise the Myom2 promoter in human cells and specifically binds to DNA at Site 1.
Figure 4.
The endogenous Myom2 promoter is bound by SIM2 in human cells. Chromatin extracts from 293 cells stably expressing ARNT1 and Myc-epitope tagged hSIM2s were immunoprecipitated with antibodies recognising hSIM2s (αhSIM2s) or the Myc tag (αMyc) or an equal amount of the non-specific immunoglobulin controls (Goat IgG and Mouse IgG). DNA eluted from the chromatin immunoprecipitations was examined by PCR amplification using primers designed to amplify the Site 1 or Site 3 (containing an identical hexamer site to Site 1 but 850 bp upstream) sequences from the Myom2 promoter or the control GAPDH sequence. Input contains DNA isolated from the chromatin extract and –ve indicates a control eluate in which no chromatin extract was added to the immunoprecipitation.
Depletion of Sim2 in cultured human myoblasts affects endogenous Myom2 transcript levels
The immortalized human myoblast cell line LHCN-M2 was generated by introduction of hTERT and cyclin-dependent kinase 4 into human satellite cells (35). These cells express both Sim2 and Myom2 and so we utilized this cell system to investigate the effect of decreasing Sim2 levels on Myom2 expression in the context of a muscle cell. LHCN-M2 myoblast cells were transfected with a control siRNA, either of two independent siRNAs designed to target Sim2 or an siRNA against Myom2. Knock down of Sim2 RNA levels with each Sim2 siRNA was observed by real-time PCR assay 3 days post-siRNA transfection, compared to treatment with control or Myom2 targeted siRNAs (Figure 5Ai). This decrease in Sim2 levels was accompanied by an increase in Myom2 expression after both a single (Day4) or double (Day7) siRNA treatment (Figure 5Aii). Thus, Myom2 transcript expression is becoming derepressed by decreasing Sim2 levels, suggesting Sim2 represses Myom2 expression in LHCN-M2 human myoblasts.
The muscle structural protein Myomesin2 is co-expressed with SIM2
The expression of Myom2 is regulated through mammalian muscle development. It is transiently expressed in the very early stages of sarcomerogenesis in cardiomyocytes and later in skeletal muscle cells, is perinatally downregulated and then reappears in adult striated cardiac and fast fibers only in rodents (41–43). The tight temporal and spatial expression of the MYOM2 protein appears to mimic the expression of the transcript (41). Myom2 has recently been identified as a transcriptional target of Myocyte enhancer factor 2 (MEF2) family members (44), although control of Myom2 fibre-type specific expression remains to be fully elucidated. Contradictory to our data from cultured human myoblasts, in which SIM2 appears to be repressing Myom2, at a gross tissue level the Sim2 and Myom2 transcripts appear to be co-expressed in various adult mouse muscles (with the exception of the heart, Figure 5B lane 5) by reverse-transcription–PCR. We wished to address this discrepancy by determining whether they are co-expressed at a cellular level in normal tissue. The cellular localization of the SIM2 protein appears to be constitutively nuclear when the protein is overexpressed in transformed human cell lines (26,45), however, the localisation and expression of the endogenous protein has not been examined in normal tissue. Clear, nuclear localized expression of SIM2 was obvious throughout sections from adult mouse hindlimb muscle, but absent in control sections incubated with secondary antibody alone or with a non-specific primary (Figure 5C, D and E iii, data not shown). SIM2 colocalises with a member of the muscle regulatory factor family, myogenin, illustrating that SIM2 is indeed found in myonuclei (Figure 5Ci). Interestingly, given our data that SIM2 can act both positively and negatively on Myom2 expression in different cell types (46), SIM2 positive nuclei are found surrounding fibres that express MYOM2 (Figure 5Di) and also nuclei surrounding fibres that are not expressing MYOM2. MYOM2 protein expression predominantly marks fast fibres in the adult mouse (46), and so SIM2 expression from this colocalisation data is not fibre type specific. The Myomesin1 (Myom1) gene is a closely related homologue of Myom2 and encodes a particular isoform, EH-MYOM1, that is expressed in a complementary fashion to MYOM2 in mice (46). SIM2 appears to be expressed in nuclei surrounding EH-MYOM1 positive and negative fibres (Figure 5E), consistent with the notion that SIM2 expression does not appear to be fibre-type regulated in adult mouse muscle.
Myom2 promoter is specifically responsive to SIM2/ARNT1 despite binding site similarities to HRE sequences
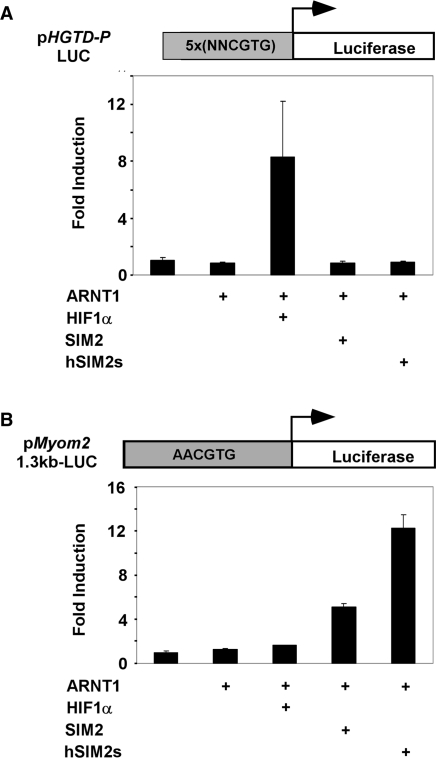
Previous experiments have demonstrated that SIM2/ARNT1 heterodimers can recognise the same HRE sequences in a reporter construct that are bound by hypoxically induced HIF-1α/ARNT1 complexes (26). These experiments utilise reporter constructs containing 4 tandem copies of the 18-nucleotide HRE sequence from the erythropoietin gene, with the core sequence of 5′-TACGTG-3′. Given this similarity in possible DNA recognition motifs between HIF-1α and SIM2 complexes, the Myom2 promoter was tested for responsiveness to HIF-1α. While co-expression of HIF-1α and ARNT1 in 293 cells significantly induces the hypoxically responsive p_HGTD-P_-LUC reporter gene, neither long or short isoforms of SIM2 with ARNT1 induced transcription from this reporter (Figure 6A). In contrast, both SIM2 isoforms can induce the _Myom2_-reporter in partnership with ARNT1, whilst co-expression of HIF-1α and ARNT1 or treatment with the hypoxia mimetic, 2,2′-dipyridyl, do not activate the Myom2 promoter sequence (Figure 6B, data not shown). Use of full-length promoter constructs rather than truncated, multimerized binding sites has allowed the elucidation of promoter choice specificity between HIF-1α and SIM2 complexes.
Figure 6.
The Myom2 promoter is not activated by HIF-1α/ARNT1. 293 cells were cotransfected for 40 h with the p_HGTD_-P_-LUC (A) or p_Myom2_1.3kbBasic-LUC (B) reporter plasmids and expression vectors as indicated and lysates were assayed for luciferase activity. Luciferase activity was normalised against a Renilla luciferase internal control and results are depicted as fold induction over the reporter alone for each transfection. Results depicted are from a single experiment performed in triplicate with standard deviation, which is representative of repeats of this set of transfections.
DISCUSSION
Despite the suggestion that misregulated SIM2 protein may play a role in the etiology of Down syndrome and particular solid tumours, very little is known about its normal function, mechanism of action and, as yet, there have been no target genes validated for this transcription factor with an accompanying description of the functional SIM2/ARNT1 response element. Surprisingly, whilst the tissue-specific expression pattern of Sim2 RNA in the mouse has been extensively mapped, with predominant levels in the developing and adult kidneys, brain and skeletal muscle (5,14–18) the endogenous SIM2 protein has not been visualised in normal tissue. We demonstrate here that native SIM2 protein appears to be constitutively nuclear localized in adult mouse hind limb muscle (Figure 5). This is consistent with the notion that unlike other bHLH/PAS family members such as the ubiquitously expressed DR and HIF-1α, SIM2 activity is controlled by spatiotemporal expression rather than signal regulated activation (26,47). Whilst there are defects noted in the formation of one particular muscle (the diaphragm) in Sim2−/− mice, the role of Sim2 in other muscles is not immediately apparent as no obvious skeletal muscle phenotype is reported for the Sim2 homozygous mutant mice in their limited post-natal life span (5,6).
In an effort to elucidate the function of the SIM2 protein, we have identified a host of transcripts that are differentially regulated with expression of SIM2 and/or the chimeric activator SIM2/AD in human embryonic kidney cells using a microarray screening approach. Given this large pool of potential SIM2 responsive genes, we have studied the interaction between SIM2 and transcription of the Myom2 gene in greatest detail. Excitingly, co-expression of SIM2 (long or short) with ARNT1 in 293 cells, led to the induction of a reporter gene controlled by 1.3 kb of Myom2 promoter sequence (Figure 2). This follows on from a recent report showing that the long and short isoforms of SIM2, in partnership with ARNT1, can activate transcription from a reporter driven by Drosophila CME sequences (25). In contrast, the _Myom2_-reporter is not induced by expression of HIF-1α/ARNT1 (Figure 6), and so despite the likelihood that SIM2 responsive sequences will resemble HRE-like sequences, this response appears to be SIM2 specific.
Truncation studies of the 1.3 kb Myom2 promoter led to the identification of a 95 bp fragment that is primarily, but not totally, responsible for the induction of this promoter with SIM2/AD (Figure 1). Informatic analysis of this 95bp sequence identified a 6bp sequence termed Site 1 (5′-AACGTG-3′) that is highly similar to the core HRE and CME consensus sequences [5′-ACGTG-3′]. Mutation of this sequence at both the ARNT1-binding half site and the partner protein half site (5′-AAAAAG-3′) resulted in a dramatically decreased induction of the 1.3 kb Myom2 reporter with either SIM2 isoform and ARNT1 (Figure 2). Mutation of a second possible DNA binding site (Site 2) had no effect on reporter gene induction by SIM2/ARNT1 or hSIM2s/ARNT1. In vitro gel shift assays indicate that the SIM2/ARNT1 dimer can recognise a 40 bp probe derived from the Myom2 promoter containing the Site 1 sequence, but not a mutant probe in which Site 1 is mutated (Figure 3). Likewise in cells, Site 1 sequences are enriched in hSIM2s-bound chromatin (Figure 4). Together these data suggest that Myom2 is a direct target of SIM2 and that SIM2/ARNT1 heterodimers recognize the Site 1 non-canonical E-box sequence. This is the first report of a bona fide target gene for SIM2 with an accompanying analysis of the endogeous binding site for SIM2. There are likely to be additional SIM2/ARNT1 binding sites in the 1.3 kb Myom2 fragment studied as truncation of approximately 450 bp of promoter sequence preceding the Site 1 sequence containing one 5′-AACGTG-3′, a 5′-TACGTG-3′ and three 5′-AGCGTG-3′ sequences, leads to a decrease in the induction of the _Myom2_-reporter with SIM2/AD and mutation of Site 1 leads to a significant, but not total, loss of induction with hSIM2s/ARNT1 (Figures 1 and 2).
Knockdown of SIM2 in immortalized human myoblasts leads to an upregulation of Myom2 transcript levels. This suggests that SIM2 is repressing transcription of the target Myom2 in cultured human myoblasts and is in concordance with previous reports of SIM2 acting as a potent transcriptional repressor (21). However, our experiments in human embryonic kidney cells and those of Metz and colleagues (25) illustrate the ability of the SIM2/ARNT heterodimer to also activate transcription. This activation is dependent on the presence of the ARNT1 C-terminal transactivation domain (Figure 2). This re-emphasises the common theme of transcription factors being capable of acting both positively and negatively depending on cellular context, promoter architecture and DNA binding site sequences. Indeed during the process of muscle differentiation, MEF2 factors initially interact with class II HDACs and repress target gene expression until activated by post-translational modification at later stages of muscle differentiation (48). MYOM2 and SIM2 are co-expressed in adult hind limb muscle, although unlike the majority of MYOM2 expression, SIM2 does not appear to be constrained to fast fibre types as it is also expressed in nuclei surrounding EH-MYOM1 positive fibres (Figure 5). Fibre type specification is dependent on the intricate coordination of a number of different pathways involving the hedgehog family members, calcineurin signaling and other transcription factors such as MEF2 family members and the serum response factor, Oct-1 (44,49–51). Thus whilst the Myom2 promoter is SIM2-responsive, SIM2 is likely to contribute to Myom2 transcription in partnership with other factors, particularly as consensus MEF2 binding sequences are present in the Myom2 promoter and MEF2 has been implicated in controlling fibre type transcriptional programs (41,52).
This article identifies the first SIM2/ARNT1 DNA response element (Site 1 5′-AACGTG-3′). The definition of a consensus SIM2/ARNT1 DNA response element awaits the verification of more target sequences; however, point mutation of the first base of the hexameric sequence in our reporter system revealed that, while other bases are also tolerated, an A at position 1 is required for full activation of the reporter gene (data not shown). It will be interesting to see whether SIM2, like other bHLH/PAS proteins, is capable of binding to a variety of similar but not identical core DNA sequences (41,44,49–51).
Whilst this article was in review, Laffin and his colleagues (53) published a description of hSIM2s as a transcriptional repressor of the newly identified target gene, SLUG. The authors localize hSIM2s binding to a 553 base pair fragment of the SLUG promoter, but do not definitively identify the actual site(s) that hSIM2s is recognizing. The sequence contains similar yet not identical sequences to Site1 (5′-AACGTG-3′) that we have identified here. A more detailed analysis of the actual hSIM2s binding site will be of great interest to enable the definition of a consensus SIM2 response element.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
[Supplementary Data]
ACKNOWLEDGEMENTS
We are indebted to Dr Anna Tsykin and Assoc. Prof. Patty Solomon (Applied Mathematics, University of Adelaide, Adelaide, Australia) for statistical analysis of microarray data, Christopher Esk (University of California San Francisco) for expertise with myoblast cell culture, Prof. L. Poellinger for the ARNT1 antibody Bambi and use of cell imaging equipment (Karolinska Institute, Stockholm, Sweden), Dr Irina Agarkova (Swiss Federal Institute of Technology, Zurich, Switzerland) and Prof. Dieter Furst (University of Potsdam, Potsdam, Germany) for MYOM2 and EH-MYOM1 antibodies. This work was supported by the ARC Special Centre for the Molecular Genetics of Development and Anti-Cancer SA. Funding to pay the Open Access publication charges for this article was provided by the ARC Special Centre for the Molecular Genetics of Development.
Conflict of interest statement. None declared.
REFERENCES
- 1.Crews ST, Fan CM. Remembrance of things PAS: regulation of development by bHLH-PAS proteins. Curr. Opin. Genet. Dev. 1999;9:580–587. doi: 10.1016/s0959-437x(99)00003-9. [DOI] [PubMed] [Google Scholar]
- 2.Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- 3.Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell. Biol. 2004;36:189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- 4.Michaud JL, Rosenquist T, May NR, Fan CM. Development of neuroendocrine lineages requires the bHLH-PAS transcription factor SIM1. Genes Dev. 1998;12:3264–3275. doi: 10.1101/gad.12.20.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goshu E, Jin H, Fasnacht R, Sepenski M, Michaud JL, Fan CM. Sim2 mutants have developmental defects not overlapping with those of Sim1 mutants. Mol. Cell. Biol. 2002;22:4147–4157. doi: 10.1128/MCB.22.12.4147-4157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shamblott MJ, Bugg EM, Lawler AM, Gearhart JD. Craniofacial abnormalities resulting from targeted disruption of the murine Sim2 gene. Dev. Dyn. 2002;224:373–380. doi: 10.1002/dvdy.10116. [DOI] [PubMed] [Google Scholar]
- 7.Crews ST, Thomas JB, Goodman CS. The Drosophila single-minded gene encodes a nuclear protein with sequence similarity to the per gene product. Cell. 1988;52:143–151. doi: 10.1016/0092-8674(88)90538-7. [DOI] [PubMed] [Google Scholar]
- 8.Nambu JR, Franks RG, Hu S, Crews ST. The single-minded gene of Drosophila is required for the expression of genes important for the development of CNS midline cells. Cell. 1990;63:63–75. doi: 10.1016/0092-8674(90)90288-p. [DOI] [PubMed] [Google Scholar]
- 9.Nambu JR, Lewis JO, Wharton K.A., Jr., Crews ST. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- 10.Franks RG, Crews ST. Transcriptional activation domains of the single-minded bHLH protein are required for CNS midline cell development. Mech Dev. 1994;45:269–277. doi: 10.1016/0925-4773(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 11.Michaud JL, Boucher F, Melnyk A, Gauthier F, Goshu E, Levy E, Mitchell GA, Himms-Hagen J, Fan CM. Sim1 haploinsufficiency causes hyperphagia, obesity and reduction of the paraventricular nucleus of the hypothalamus. Hum. Mol. Genet. 2001;10:1465–1473. doi: 10.1093/hmg/10.14.1465. [DOI] [PubMed] [Google Scholar]
- 12.Goshu E, Jin H, Lovejoy J, Marion JF, Michaud JL, Fan CM. Sim2 contributes to neuroendocrine hormone gene expression in the anterior hypothalamus. Mol. Endocrinol. 2004;18:1251–1262. doi: 10.1210/me.2003-0372. [DOI] [PubMed] [Google Scholar]
- 13.Marion JF, Yang C, Caqueret A, Boucher F, Michaud J. Sim1 and Sim2 are required for the correct targeting of mammillary body axons. Development. 2005;132:5527–5537. doi: 10.1242/dev.02142. [DOI] [PubMed] [Google Scholar]
- 14.Ema M, Morita M, Ikawa S, Tanaka M, Matsuda Y, Gotoh O, Saijoh Y, Fujii H, Hamada H, Kikuchi Y, et al. Two new members of the murine Sim gene family are transcriptional repressors and show different expression patterns during mouse embryogenesis. Mol. Cell. Biol. 1996b;16:5865–5875. doi: 10.1128/mcb.16.10.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan CM, Kuwana E, Bulfone A, Fletcher CF, Copeland NG, Jenkins NA, Crews S, Martinez S, Puelles L, Rubenstein JL, et al. Expression patterns of two murine homologs of Drosophila single-minded suggest possible roles in embryonic patterning and in the pathogenesis of Down syndrome. Mol. Cell. Neurosci. 1996;7:1–16. doi: 10.1006/mcne.1996.0001. [DOI] [PubMed] [Google Scholar]
- 16.Moffett P, Dayo M, Reece M, McCormick MK, Pelletier J. Characterization of msim, a murine homologue of the Drosophila sim transcription factor. Genomics. 1996;35:144–155. doi: 10.1006/geno.1996.0333. [DOI] [PubMed] [Google Scholar]
- 17.Yamaki A, Noda S, Kudoh J, Shindoh N, Maeda H, Minoshima S, Kawasaki K, Shimizu Y, Shimizu N. The mammalian single-minded (SIM) gene: mouse cDNA structure and diencephalic expression indicate a candidate gene for Down syndrome. Genomics. 1996;35:136–143. doi: 10.1006/geno.1996.0332. [DOI] [PubMed] [Google Scholar]
- 18.Ema M, Suzuki M, Morita M, Hirose K, Sogawa K, Matsuda Y, Gotoh O, Saijoh Y, Fujii H, Hamada H, et al. cDNA cloning of a murine homologue of Drosophila single-minded, its mRNA expression in mouse development, and chromosome localization. Biochem. Biophys. Res. Commun. 1996a;218:588–594. doi: 10.1006/bbrc.1996.0104. [DOI] [PubMed] [Google Scholar]
- 19.Chrast R, Scott HS, Madani R, Huber L, Wolfer DP, Prinz M, Aguzzi A, Lipp HP, Antonarakis SE. Mice trisomic for a bacterial artificial chromosome with the single-minded 2 gene (Sim2) show phenotypes similar to some of those present in the partial trisomy 16 mouse models of Down syndrome. Hum. Mol. Genet. 2000;9:1853–1864. doi: 10.1093/hmg/9.12.1853. [DOI] [PubMed] [Google Scholar]
- 20.Ema M, Ikegami S, Hosoya T, Mimura J, Ohtani H, Nakao K, Inokuchi K, Katsuki M, Fujii-Kuriyama Y. Mild impairment of learning and memory in mice overexpressing the mSim2 gene located on chromosome 16: an animal model of Down's syndrome. Hum. Mol. Genet. 1999;8:1409–1415. doi: 10.1093/hmg/8.8.1409. [DOI] [PubMed] [Google Scholar]
- 21.Moffett P, Reece M, Pelletier J. The murine Sim-2 gene product inhibits transcription by active repression and functional interference. Mol. Cell. Biol. 1997;17:4933–4947. doi: 10.1128/mcb.17.9.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wharton KA, Jr, Franks RG, Kasai Y, Crews ST. Control of CNS midline transcription by asymmetric E-box-like elements: similarity to xenobiotic responsive regulation. Development. 1994;120:3563–3569. doi: 10.1242/dev.120.12.3563. [DOI] [PubMed] [Google Scholar]
- 23.Ooe N, Saito K, Mikami N, Nakatuka I, Kaneko H. Identification of a novel basic helix-loop-helix-PAS factor, NXF, reveals a Sim2 competitive, positive regulatory role in dendritic-cytoskeleton modulator drebrin gene expression. Mol. Cell. Biol. 2004;24:608–616. doi: 10.1128/MCB.24.2.608-616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moffett P, Pelletier J. Different transcriptional properties of mSim-1 and mSim-2. FEBS Lett. 2000;466:80–86. doi: 10.1016/s0014-5793(99)01750-0. [DOI] [PubMed] [Google Scholar]
- 25.Metz RP, Kwak HI, Gustafson T, Laffin B, Porter WW. Differential transcriptional regulation by mouse single-minded 2s. J. Biol. Chem. 2006;281:10839–10848. doi: 10.1074/jbc.M508858200. [DOI] [PubMed] [Google Scholar]
- 26.Woods SL, Whitelaw ML. Differential activities of murine single minded 1 (SIM1) and SIM2 on a hypoxic response element. Cross-talk between basic helix-loop-helix/per-Arnt-Sim homology transcription factors. J. Biol. Chem. 2002;277:10236–10243. doi: 10.1074/jbc.M110752200. [DOI] [PubMed] [Google Scholar]
- 27.Deyoung MP, Scheurle D, Damania H, Zylberberg C, Narayanan R. Down's syndrome-associated single minded gene as a novel tumor marker. Anticancer Res. 2002;22:3149–3157. [PubMed] [Google Scholar]
- 28.DeYoung MP, Tress M, Narayanan R. Identification of Down's syndrome critical locus gene SIM2-s as a drug therapy target for solid tumors. Proc. Natl Acad. Sci. USA. 2003a;100:4760–4765. doi: 10.1073/pnas.0831000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chrast R, Scott HS, Chen H, Kudoh J, Rossier C, Minoshima S, Wang Y, Shimizu N, Antonarakis SE. Cloning of two human homologs of the Drosophila single-minded gene SIM1 on chromosome 6q and SIM2 on 21q within the Down syndrome chromosomal region. Genome Res. 1997;7:615–624. doi: 10.1101/gr.7.6.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeYoung MP, Tress M, Narayanan R. Down's syndrome-associated single minded 2 gene as a pancreatic cancer drug therapy target. Cancer Lett. 2003b;200:25–31. doi: 10.1016/s0304-3835(03)00409-9. [DOI] [PubMed] [Google Scholar]
- 31.Kwak HI, Gustafson T, Metz RP, Laffin B, Schedin P, Porter WW. Inhibition of breast cancer growth and invasion by single-minded 2s. Carcinogenesis. 2007;28:259–266. doi: 10.1093/carcin/bgl122. [DOI] [PubMed] [Google Scholar]
- 32.Goldman LA, Cutrone EC, Kotenko SV, Krause CD, Langer JA. Modifications of vectors pEF-BOS, pcDNA1 and pcDNA3 result in improved convenience and expression. Biotechniques. 1996;21:1013–1015. doi: 10.2144/96216bm10. [DOI] [PubMed] [Google Scholar]
- 33.Lee MJ, Kim JY, Suk K, Park JH. Identification of the hypoxia-inducible factor 1 alpha-responsive HGTD-P gene as a mediator in the mitochondrial apoptotic pathway. Mol. Cell. Biol. 2004;24:3918–3927. doi: 10.1128/MCB.24.9.3918-3927.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapman-Smith A, Lutwyche JK, Whitelaw ML. Contribution of the PAS domains to DNA binding by the basic helix-loop-helix PAS transcriptional regulators. J. Biol. Chem. 2003;279:5353–5362. doi: 10.1074/jbc.M310041200. [DOI] [PubMed] [Google Scholar]
- 35.Zhu CH, Mouly V, Cooper RN, Mamchaoui K, Bigot A, Shay JW, Di Santo JP, Butler-Browne GS, Wright WE. Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: consequences in aging muscle and therapeutic strategies for muscular dystrophies. Aging Cell. 2007;6:515–523. doi: 10.1111/j.1474-9726.2007.00306.x. [DOI] [PubMed] [Google Scholar]
- 36.Lees MJ, Whitelaw ML. Multiple roles of ligand in transforming the dioxin receptor to an active basic helix-loop-helix/PAS transcription factor complex with the nuclear protein Arnt. Mol. Cell. Biol. 1999;19:5811–5822. doi: 10.1128/mcb.19.8.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masaki T, Takaiti O. M-protein. J. Biochem. 1974;75:367–380. doi: 10.1093/oxfordjournals.jbchem.a130403. [DOI] [PubMed] [Google Scholar]
- 38.Obermann WMJ, Gautel M, Steiner F, Van der Ven PFM, Weber K, Furst DO. The structure of the sarcomeric M band:Localisation of defined domains of myomesin, M-protein and the 250kDa carboxyterminal region of titin by immunoelectron microscopy. J. Cell. Biol. 1996;134:1441–1453. doi: 10.1083/jcb.134.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obermann WMJ, van der ven PFM, Steiner F, Weber K, Furst DO. Mapping of a myosin-binding domain and a regulatory phosphorylation site in M-protein, a structural protein of the sarcomeric M band. Mol. Biol. Cell. 1998;9:829–840. doi: 10.1091/mbc.9.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Der Ven PF, Obermann WM, Weber K, Furst DO. Myomesin, M-protein and the structure of the sarcomeric M-band. Adv. Biophys. 1996;33:91–99. doi: 10.1016/0065-227x(96)81666-2. [DOI] [PubMed] [Google Scholar]
- 41.Steiner F, Weber K, Furst DO. Structure and expression of the gene encoding murine M-protein, a sarcomere-specific member of the immunoglobulin superfamily. Genomics. 1998;49:83–95. doi: 10.1006/geno.1998.5220. [DOI] [PubMed] [Google Scholar]
- 42.Carlsson E, Grove BK, Wallimann T, Eppenberger HM, Thornell LE. Myofibrillar M-band proteins in rat skeletal muscles during development. Histochemistry. 1990;95:27–35. doi: 10.1007/BF00737225. [DOI] [PubMed] [Google Scholar]
- 43.Grove BK, Cerny L, Perriard JC, Eppenberger HM. Myomesin and M-protein: expression of two M-band proteins in pectoral muscle and heart during development. J. Cell. Biol. 1985;101:1413–1421. doi: 10.1083/jcb.101.4.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Potthoff MJ, Arnold MA, McAnally J, Richardson JA, Bassel-Duby R, Olson EN. Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2c. Mol. Cell. Biol. 2007;27:8143–8151. doi: 10.1128/MCB.01187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaki A, Kudoh J, Shimizu N, Shimizu Y. A novel nuclear localization signal in the human single-minded proteins SIM1 and SIM2. Biochem. Biophys Res. Commun. 2004;313:482–488. doi: 10.1016/j.bbrc.2003.11.168. [DOI] [PubMed] [Google Scholar]
- 46.Agarkova I, Schoenauer R, Ehler E, Carlsson L, Carlsson E, Thornell LE, Perriard JC. The molecular composition of the sarcomeric M-band correlates with muscle fiber type. Eur. J. Cell. Biol. 2004;83:193–204. doi: 10.1078/0171-9335-00383. [DOI] [PubMed] [Google Scholar]
- 47.Ward MP, Mosher JT, Crews ST. Regulation of bHLH-PAS protein subcellular localization during Drosophila embryogenesis. Development. 1998;125:1599–1608. doi: 10.1242/dev.125.9.1599. [DOI] [PubMed] [Google Scholar]
- 48.Lu J, McKinsey TA, Zhang CL, Olson EN. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell. 2000;6:233–244. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 49.Allen DL, Weber JN, Sycuro LK, Leinwand LA. Myocyte enhancer factor-2 and serum response factor binding elements regulate fast Myosin heavy chain transcription in vivo. J. Biol. Chem. 2005;280:17126–17134. doi: 10.1074/jbc.M501207200. [DOI] [PubMed] [Google Scholar]
- 50.Te KG, Reggiani C. Skeletal muscle fibre type specification during embryonic development. J. Muscle Res. Cell. Motil. 2002;23:65–69. doi: 10.1023/a:1019940932275. [DOI] [PubMed] [Google Scholar]
- 51.Spangenburg EE, Booth FW. Molecular regulation of individual skeletal muscle fibre types. Acta Physiol. Scand. 2003;178:413–424. doi: 10.1046/j.1365-201X.2003.01158.x. [DOI] [PubMed] [Google Scholar]
- 52.Wu H, Naya FJ, McKinsey TA, Mercer B, Shelton JM, Chin ER, Simard AR, Michel RN, Bassel-Duby R, Olson EN, et al. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J. 2000;19:1963–1973. doi: 10.1093/emboj/19.9.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laffin B, Wellberg E, Kwak H, Burghardt R, Metz R, Gustafson T, Schedin P, Porter W. Mol. Cell Biol. 2008;28:1936–1946. doi: 10.1128/MCB.01701-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]