Endosomal Targeting of MEK2 Requires RAF, MEK Kinase Activity and Clathrin-Dependent Endocytosis (original) (raw)
. Author manuscript; available in PMC: 2009 Sep 24.
Abstract
To study spatiotemporal regulation of the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK1/2) signaling cascade in living cells, a HeLa cell line in which MAPK kinase of ERK kinase (MEK) 2 (MAPK kinase) was knocked down by RNA interference and replaced with the green fluorescent protein (GFP)-tagged MEK2 was generated. In these cells, MEK2–GFP was stably expressed at a level similar to that of the endogenous MEK2 in the parental cells. Upon activation of the EGF receptor (EGFR), a pool of MEK2–GFP was found initially translocated to the plasma membrane and then accumulated in a subset of early and late endosomes. However, activated MEK was detected only at the plasma membrane and not in endosomes. Surprisingly, MEK2–GFP endosomes did not contain active EGFR, suggesting that endosomal MEK2–GFP was separated from the upstream signaling complexes. Knockdown of clathrin by small interfering RNA (siRNA) abolished MEK2 recruitment to endosomes but resulted in increased activation of ERK without affecting the activity of MEK2–GFP. The accumulation of MEK2–GFP in endosomes was also blocked by siRNA depletion of RAF kinases and by the MEK1/2 inhibitor, UO126. We propose that the recruitment of MEK2 to endosomes can be a part of the negative feedback regulation of the EGFR–MAPK signaling pathway by endocytosis.
Keywords: EGF receptor, endocytosis, endosome, ERK, MEK
Binding of EGF to the EGF receptor (EGFR) at the cell surface induces receptor dimerization and tyrosine phosphorylation of its carboxyl-terminal tail (reviewed in 1). Recruitment of proteins bearing phosphotyrosine recognition domains to the receptor tail results in the activation of several signal transduction cascades. One of the major signaling pathways triggered by EGFR leads to the activation of mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK1/2). This pathway is initiated by binding of the Grb2 adaptor protein to the receptor. Grb2 is constitutively associated with the Ras GTP exchange factor (GEF), Son of Sevenless (SOS). Interaction of the Grb2–SOS complex with the receptor results in the activation of the membrane-anchored Ras guanine triphosphatase (2). Subsequently, the GTP-loaded Ras recruits to the membrane and activates RAF kinases (MAPK kinase kinases), which in turn phosphorylates and activates MAPK kinase of ERK kinase (MEK)1/2 (MAPK kinase) (reviewed in 3). The function of MEK is to phosphorylate ERK. Both MEK and ERK exist as two isoforms (MEK1 and 2, ERK1 and 2) that are considered to be functionally redundant with only minor differences reported. ERK1/2 phosphorylates a large number of substrates that are components of various cellular machineries present in the cytosol and nucleus (4,5).
EGF binding to the receptor also induces rapid internalization of ligand–receptor complexes using clathrin-dependent and clathrin-independent mechanisms (reviewed in 6). Internalized EGFR is then efficiently sorted to lysosomes for degradation, which results in downregulation of the EGFR protein. This downregulation is considered to be the major negative feedback regulatory loop of EGFR signaling (reviewed in 7). However, accumulating evidence suggests that endocytosis is more than just a mechanism for signal termination. Specifically, it has been proposed that endocytosis is necessary for the full and prolonged activation of ERK1/2 and that endosomal membranes may serve as the platforms for the formation of MAPK signaling complexes (8,9). A number of studies demonstrated that EGFR remains associated with Grb2 and Shc adaptors in endosomes (10–12). Other components of the MAPK signaling cascade, such as SOS and RAS, have also been detected in their active forms in endosomes (11–14). It has been reported that activation of RAF-1 triggers movement of cytosolic MEK to endosomes (15). Although the bulk of MEK was cytosolic, activated MEK was detected in endosomes by cell fractionation and immunofluorescence (16,17). It has also been suggested that the endosomal localization of MEK1 is important for prolonged ERK activation (18).
Inhibition of endocytosis by expression of dominant-negative mutants of dynamin (K44A) abolished activation of MEK1/2 and ERK1/2 by EGF (9,19,20). However, this effect of the dynamin mutant on EGF-induced ERK activity was not observed in other studies (21). Given that it was demonstrated that overexpression of the dynamin K44A mutant may have effects unrelated to endocytosis, the role of endocytosis in activation of MEK and ERK remains unclear [discussed in Sorkin and Von Zastrow (22)]. Moreover, while the evidence for the endosomal localization of the receptor complexes containing upstream components of the MAPK pathway includes live cell microscopy and is, in general, convincing, the endosomal localization of MEK and ERK has not been demonstrated in living cells. The data demonstrating the localization of MEK and ERK in endosomes are limited and obtained using indirect immunofluorescence and cell fractionation.
Hence, we analyzed the localization of MEK2 in living cells stimulated with EGF. The majority of the previous studies using live cell microscopy utilized proteins tagged with various green fluorescent protein (GFP) variants overexpressed in cells. However, overexpression of the proteins involved in the MAPK pathway often yields outcomes that are opposite to their proposed function (23,24). Apparently, such overexpression leads to the sequestration of the scaffold proteins and formation of non-specific complexes, therefore affecting signal regulation. To prevent these potential problems, we used a knockdown and reconstitution (KDAR) strategy to generate a cell line that constitutively expresses MEK2–GFP at physiological levels and lacks endogenous MEK2. The live cell microscopic analysis revealed that upon EGFR activation, a pool of MEK2–GFP accumulates in a subset of early and late endosomal compartments that lack EGFR. Analysis of the mechanisms underlying the targeting of MEK2 suggested that the endosomal localization of MEK2–GFP requires clathrin-dependent endocytosis, the presence of an upstream kinase, RAF, and the catalytic activity of MEK and that endosomal accumulation of MEK2 may serve as a part of negative regulation of the signaling process by endocytosis.
Results
Replacement of MEK2 by MEK2–GFP in HeLa cells
To study the localization of MEK2 in living cells, GFP fusion proteins of MEK2 were prepared by attaching GFP to either the N-terminus or the C-terminus of MEK2 and transiently expressed in HeLa cells. The functionality of the fusion proteins was tested by measuring the extent of MEK2 phosphorylation induced by the EGF treatment of serum-starved cells. Whereas GFP–MEK2 had low activity (data not shown), the phosphorylation of MEK2 tagged with GFP at the C-terminus (MEK2–GFP) was similar to that of the endogenous MEK1/2 (Figure 1A). Therefore, MEK2–GFP was used in all subsequent experiments. Similar fluorescent-protein-tagged fusion proteins of MEK1 have been previously shown to be functional (25).
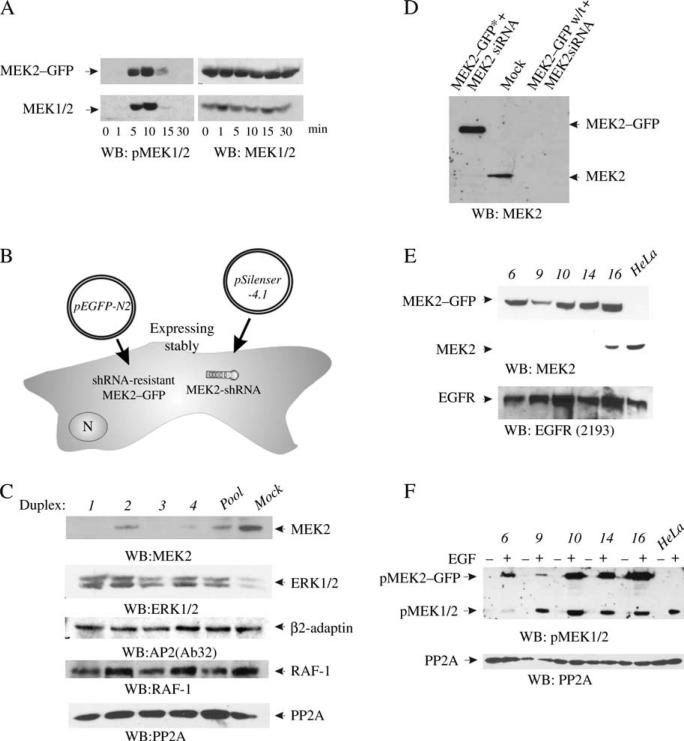
Figure 1. Replacement of MEK2 with MEK2–GFP in HeLa cells.
A) HeLa cells transiently expressing MEK2–GFP were serum starved and treated with 50 ng/mL EGF for indicated times at 37°C. The lysates were probed for activated MEK1/2 (pMEK1/2) and total MEK1/2. B) Schematic representation of the KDAR method for generation of cell lines stably expressing MEK2–GFP (see Materials and Methods). N, nucleus. C) HeLa cells were transiently transfected with four individual siRNA duplexes or SMARTpool of these duplexes, lysed and MEK2 was detected in lysates by western blotting. Lysates were also probed for ERK1/2, β2-adaptin, RAF-1 and PP2A (loading control). D) HeLa cells were transiently transfected with MEK2–GFP resistant to siRNA duplex 3 (MEK2–GFP*) or wild-type MEK2–GFP (MEK2–GFP w/t) together with MEK2 siRNA duplex 3. Mock, transfection with nontargeting siRNA. MEK2 was detected in cell lysates using MEK2 antibodies. E) MEK2–GFP and endogenous MEK2 were detected in HeLa/MEK2–GFP clones and parental HeLa cells (HeLa) using MEK2 antibodies. EGFR was detected using antibodies 2193. F) Individual clones of HeLa/MEK2–GFP cells and parental HeLa cells (HeLa) were starved and then incubated without (–) or with 10 ng/mL EGF (+) for 5 min at 37°C. The lysates were probed for activated MEK1/2 (pMEK1/2) and PP2A (loading control).
MEK2–GFP transiently expressed in HeLa cells displayed a cytosolic distribution in serum-starved cells. In cells treated with EGF, MEK2–GFP was also predominantly localized in the cytosol, and no translocation of MEK2–GFP to the plasma or intracellular membranes was observed (data not shown). We interpreted the absence of the expected membrane localization as the result of a high total expression level of MEKs, given that MEK2–GFP was expressed in the presence of endogenous MEK1/2. Therefore, to prevent mistargeting and non-specific interactions of MEK2–GFP because of overexpression, HeLa cells in which endogenous MEK2 was knocked down by stable vector-mediated small hairpin RNA (shRNA) expression with simultaneous stable expression of MEK2–GFP were generated. This approach was termed KDAR (see Figure 1B for schematic explanation). First, the most efficient small interfering RNA (siRNA) sequence (3) targeting MEK2 among four siRNA duplexes was identified (Figure 1C). Duplex 3 did not affect expression of several signaling and endocytosis proteins (Figure 1C) or influence the morphology and growth rates of cells (data not shown). Therefore, this duplex was chosen for further experiments.
MEK2–GFP mutant resistant to duplex 3 (MEK2–GFP*) was prepared in which six ‘silent’ mutations were introduced. These mutations rendered MEK2–GFP* resistant to shRNA without changing the amino acid sequence (Figure 1D). To generate a stable cell line expressing MEK2–GFP at the expense of endogenous MEK2, MEK2 shRNA was coexpressed with MEK2–GFP* (see Materials and Methods). Efficient selection of clones expressing both plasmids was possible because the pSilencer™ 4.1-MEK2 and the MEK2–GFP vectors contain different selective markers.
Figure 1E shows western blot detection of MEK2–GFP and endogenous MEK2 in HeLa cells stably expressing MEK2 shRNA and MEK2–GFP constructs. Several clones depleted of MEK2 and stably expressing MEK2–GFP at a level comparable to or exceeding the expression level of endogenous MEK2 in parental HeLa cells were examined (referred to as HeLa/MEK2–GFP cells) (Figure 1E). EGF treatment resulted in the activation of MEK2–GFP in all these clones (Figure 1F). Surprisingly, clone 6 displayed low activity corresponding to endogenous MEK1/2, suggesting that in addition to knockdown of MEK2, clone 6 expressed partially reduced level of MEK1. We took advantage of the observation that the total activity of endogenous MEK1/2 in these cells was very low and the activity of MEK2–GFP was comparable to the total activity of MEK1/2 in the parental HeLa cells (Figure 1F). Thus, clone 6 was chosen as the main cell line for subsequent experiments.
Targeting of MEK2–GFP to the plasma membrane and cytosolic vesicles in EGF-stimulated cells
Live cell fluorescence microscopy of serum-starved HeLa/MEK2–GFP cells revealed that MEK2–GFP was diffusely distributed in the cytosol (Figure 2A). In contrast, chemical fixation resulted in apparent localization of MEK2–GFP at the cell edges (plasma membrane) in these cells (data not shown), which emphasizes the importance of imaging analysis in living cells.
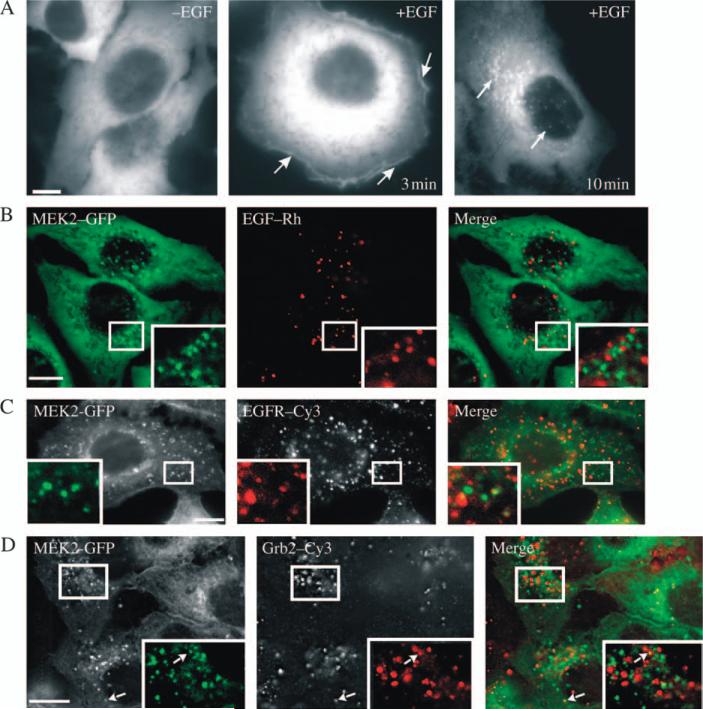
Figure 2. Localization of MEK2–GFP relative to EGF and EGFR – I.
A) Serum-starved HeLa/MEK2–GFP (clone 6) cells were treated with EGF (10 ng/mL) for 3 and 10 min at 37°C. The living cells were imaged before and after EGF treatment. The arrows show examples of MEK2–GFP localization at the plasma membrane and in intracellular vesicles. B) Serum-starved HeLa/MEK2–GFP cells were treated with 10 ng/mL EGF–Rh for 10 min at 37°C and imaged using fluorescein isothiocyanate (for GFP) and Cy3 (for Rh) channels. C) Serum-starved HeLa/MEK2–GFP cells were treated with 10 ng/mL EGF, fixed, permeabilized and stained with the EGFR antibody 528. D) Serum-starved HeLa/MEK2–GFP cells were treated with 10 ng/mL EGF at 37°C, fixed, permeabilized and stained with the Grb2 antibody. Insets in (B–D) show high-magnification images of the regions of the cell indicated by white rectangles. Scale bars, 10 μm.
Upon activation of EGFR at 37°C, MEK2–GFP was rapidly translocated to the plasma membrane (Figure 2A). Plasma membrane accumulation of MEK2–GFP was typically observed during the first 5–10 min of cell stimulation with EGF. After 7–10 min of EGF treatment, MEK2–GFP was seen to accumulate in the intracellular vesicular structures (Figure 2A). Such an accumulation of MEK2–GFP in vesicles reached a maximum level within 10 min following EGF stimulation and was maintained for additional 20–30 min. Careful examination of starved cells revealed that a small population of cells also contained a few MEK2–GFP vesicles; however, EGF treatment resulted in a dramatic increase in the number of cells with GFP-decorated vesicles as well as the number of vesicles per individual cell. MEK2–GFP-containing vesicles were located mostly in the perinuclear region. The majority of these vesicles appeared as fluorescent dots of approximately similar intensity, although ‘donut’-like profiles of MEK2–GFP-decorating structures with visible limiting membrane could be occasionally observed. Some vesicles were relatively static, while most of the vesicles showed rapid lateral and directed movement over long distances. Often, several MEK2–GFP-containing vesicles were sequentially moving along a single track within a short time, characteristic of microtubule-dependent motility of endosomes (QuickTime Movie S1).
MEK2–GFP vesicles do not accumulate active EGFR
To examine whether MEK2–GFP accumulated in endosomes containing internalized EGFR, the cells were stimulated with EGF–rhodamine (Rh). Surprisingly, MEK2–GFP vesicles were not colocalized with endosomal EGF–Rh in living cells during 5–60 min of continuous endocytosis at 37°C (Figure 2B). The absence of EGFR in MEK2–GFP vesicles was confirmed in fixed cells stained with EGFR antibodies (Figure 2C). Furthermore, immunofluorescence staining of Grb2 adaptor, known to be associated with the EGF-activated EGFR in endosomes (11), was used to compare the localization of EGFR signaling complexes with MEK2–GFP. EGF stimulation resulted in rapid recruitment of Grb2 to endosomes (Figure 2D). Whereas single vesicles containing both MEK2–GFP and Grb2 could be rarely detected, the majority of MEK2–GFP vesicles and Grb2-containing endosomes did not colocalize. Altogether, the data presented in Figure 2 suggest that MEK2–GFP vesicles do not contain EGF:EGFR signaling complexes.
The absence of EGFR in MEK2–GFP vesicles could be because of limitation in the detection of EGFR expressed at a relatively low level in HeLa cells. To further explore the role of EGFR levels, EGFR–monomeric red fluorescent protein (mRFP) was transiently expressed in HeLa/MEK2–GFP cells and the cells were stimulated with EGF–Alexa647. As expected, EGF–Alexa647 and EGFR–mRFP were highly colocalized in endosomes (Figure 3A,B). Interestingly, EGFR–mRFP was detected in MEK2–GFP-containing vesicles, indicating that these vesicles are likely to be of an endosomal nature and that EGFR can be accumulated in this endosomal compartment only under conditions of receptor overexpression. However, analysis of numerous images revealed virtually no overlap of EGF–Alexa647 with MEK2–GFP in endosomes (Figure 3A,B), although colocalization of EGF–Alexa647 and MEK2–GFP was observed in the plasma membrane ruffles (Figure 3C). These data suggest that EGFR–mRFP reached MEK2–GFP endosomes at the stage of post-endocytic trafficking following the dissociation of EGF–Alexa647 from the receptor and inactivation of the receptor.
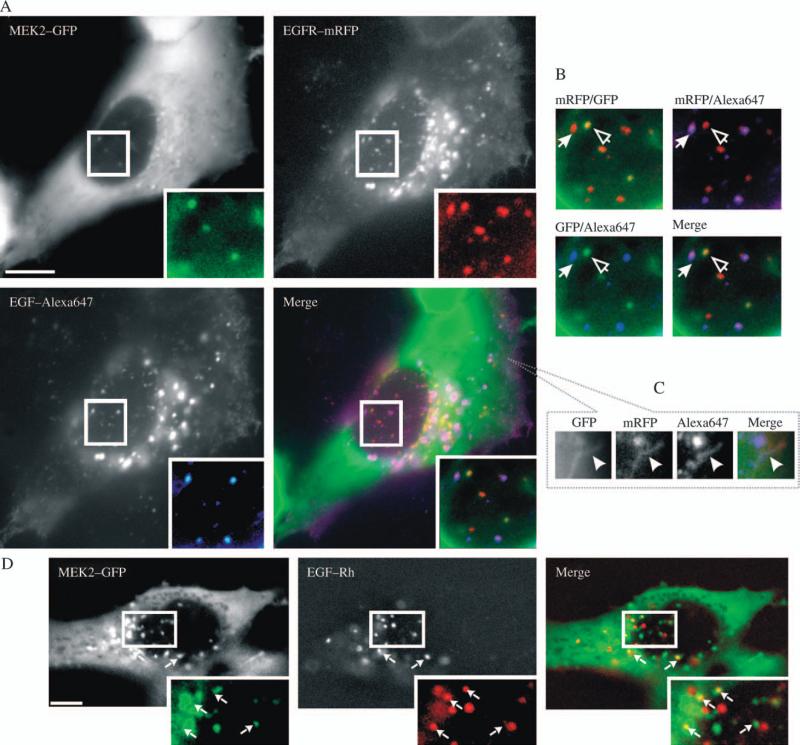
Figure 3. Localization of MEK2–GFP relative to EGF and EGFR – II.
A) HeLa/MEK2–GFP cells were transfected with EGFR–mRFP and after 2 days, serum starved and treated with 10 ng/mL EGF–Alexa647 for 10 min at 37°C. Fluorescein isothiocyanate (FITC) (for GFP), Cy3 (for mRFP) and Cy5 (for Alexa647) filter channels were used for imaging of living cells. Insets show high-magnification images of the regions of the cell indicated by white rectangles. Scale bar, 10 μm. B) High-magnification images of the regions of the cell presented in (A) shown to highlight colocalization of MEK2–GFP with EGFR–mRFP (open arrowheads), EGFR–mRFP with EGF–Alexa647 (solid arrowheads) and absence of colocalization of EGF–Alexa647 and MEK2–GFP. C) Blowout of the peripheral region of the cell in (A) at the plane of the plasma membrane. Arrow points on the ruffle-like structure containing MEK2–GFP, EGFR–mRFP and EGF–Alexa647. D) Serum-starved HeLa/MEK2–GFP cells were incubated with 10 ng/mL EGF–Rh for 90 min at 20°C. Living cells were imaged using FITC (for GFP) and Cy3 (for Rh) channels. Insets show high-magnification images of the regions of the cell indicated by white rectangles. Arrows point on endosomes containing MEK2–GFP and EGF–Rh. Scale bar, 10 μm.
The experiments with overexpressed EGFR suggested that the colocalization of the endogenous receptors with MEK2–GFP was difficult to detect because internalized endogenous receptors could transiently pass through the compartments containing MEK2–GFP. To examine this possibility, HeLa/MEK2–GFP cells were stimulated with EGF–Rh at 20°C, conditions allowing slow internalization and recycling but preventing receptor sorting to late endosomes and lysosomes. Figure 3D shows that partial colocalization of MEK2–GFP and EGF–Rh in endosomes was readily detectable at low temperature, thus supporting the possibility that EGFR rapidly traffics through the MEK2–GFP endosomes at 37°C on the way to late compartments.
MEK2–GFP is not phosphorylated in endosomes
MEK2–GFP activity was examined by indirect immunofluorescence using phospho-MEK1/2 (pMEK1/2) antibodies. Figure 4 shows that active MEK staining colocalized with MEK2–GFP was detected at the plasma membrane (cell edges) of the EGF-treated cells. In contrast, we have not been able to demonstrate the presence of phosphorylated MEK in endosomes despite trying several pMEK1/2 antibodies and different protocols of immunofluorescence staining with these antibodies (Figure 4). We noticed that the fixation procedure used to stain cells with pMEK1/2 antibodies resulted in a substantial loss of the MEK2–GFP signal associated with endosomes. Thus, absence of detectable active MEK2–GFP in these endosomes could be in part attributed to the reduced amount of MEK2–GFP in endosomes of fixed cells. However, relative concentrations of MEK2–GFP (mean fluorescence intensity per pixel) at the plasma membrane and endosomes in fixed cells were comparable, indicating that the difference in the pMEK signal reflects the difference in the activities of these pools of MEK2–GFP. The inability to detect MEK activity in endosomes was not because of the deficiencies of our experimental protocol because EGF-induced activation of MEK2 chimeric protein targeted to early endosomes by the attachment of the FYVE domain could be easily detected (Figure S1).
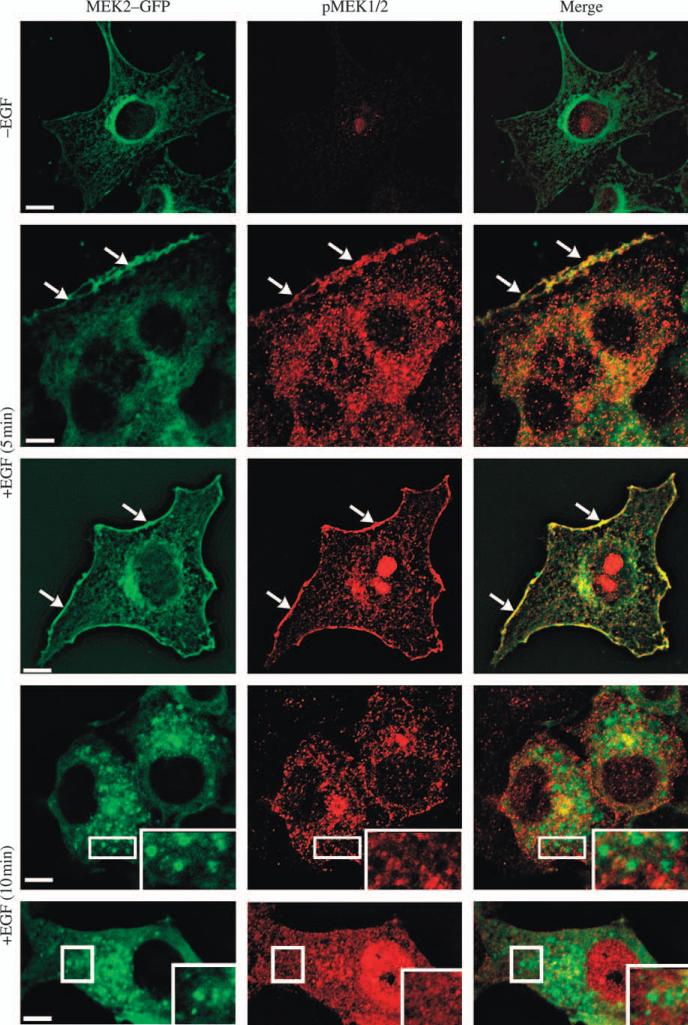
Figure 4. Localization of phosphorylated MEK in the plasma membrane but not in endosomes.
Serum-starved HeLa/MEK2–GFP cells were untreated (–EGF) or treated with 10 ng/mL EGF for 5 and 10 min at 37°C and fixed. The cells were then processed for immunofluorescence staining with pMEK1/2 antibodies and secondary Cy3 donkey anti-rabbit antibodies. GFP and Cy3 were detected using fluorescein isothiocyanate and Cy3 filter channels. The examples of phosphorylated MEK2–GFP located at cell edges are shown by arrows. Insets represent high-magnification images of the regions of the cell indicated by white rectangles. Staining with pMEK1/2 antibodies often results in non-specific nuclear signal seen in untreated and EGF-treated cells. Therefore, two examples of cells treated with EGF with and without nuclear staining are shown. Scale bars, 10 μm.
MEK2–GFP endosomes contain markers of early and late endosomal compartments
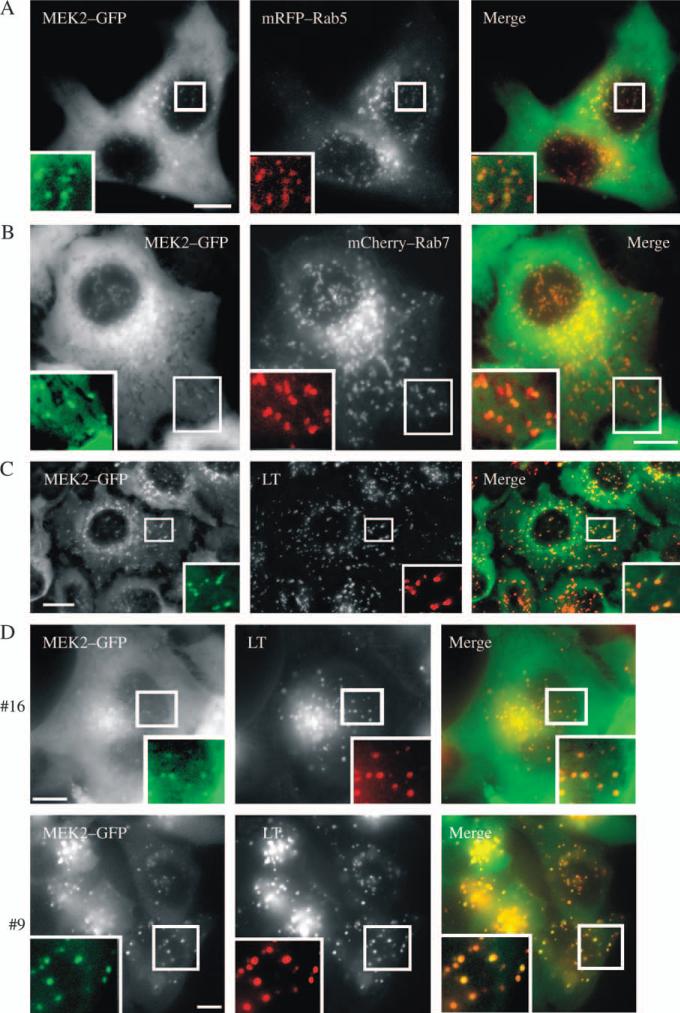
To better define MEK2–GFP-containing endosomes, we examined whether MEK2–GFP-containing endosomes colocalized with markers of early endosomes (Rab5/EEA.1), late endosomes and lysosomes (Rab7 and LysoTracker™) in living cells. HeLa/MEK2–GFP cells were transiently transfected with mRFP–Rab5 or mCherry–Rab7 or preloaded with LysoTracker. MEK2–GFP compartments were found to colocalize with a small subset of early and late endosomes labeled with mRFP–Rab5 and mCherry–Rab7, respectively (Figure 5A,B). MEK2–GFP vesicles were also partially colocalized with the expressed EEA.1–mRFP fusion protein, another marker of early endosomes (26), but not well colocalized with internalized transferrin–Texas Red (Tfr–TR), a marker of early and recycling endosomes (data not shown). Furthermore, the presence of MEK2–GFP in late endosomal compartments was confirmed by the demonstration of the substantial colocalization of GFP with LysoTracker (Figure 5C). Essentially, similar results were obtained in experiments with other clones of HeLa/MEK2–GFP cells (9 and 16) (Figure 5D and Figure S2). The ratio of endosomal and cytosolic pools of MEK2–GFP was the highest in clone 9, expressing the lowest total level of MEK2–GFP. Altogether, the data presented in Figures 2–5 demonstrated that upon EGFR activation, MEK2 is activated at the plasma membrane and also recruited to a subset of early and late endosomal compartments that do not accumulate internalized EGFR.
Figure 5. Characterization of endosomes containing MEK2–GFP.
HeLa/MEK2–GFP cells were transfected with mRFP–Rab5 (A) or mCherry–Rab7 (B). Serum-starved cells were treated with EGF (10 ng/mL) for 10 min at 37°C. In (C), untransfected HeLa/MEK2–GFP cells were preloaded with 75 nm LysoTracker (LT) for 30 min and treated with EGF as in (A). In (D), clones 9 and 16 of HeLa/MEK2–GFP cells were preloaded with 75 nm LysoTracker for 30 min and treated with EGF as in (A). Living cells were imaged through fluorescein isothiocyanate and Cy3 filter channels. Insets show high-magnification images of the regions of the cell indicated by white rectangles. Scale bars, 10 μm.
Depletion of p14 does not affect endosomal recruitment of MEK2–GFP
The presence of ERK1/2 in late endosomes has been previously demonstrated (18). The latter study proposed that MEK1 and ERK1/2 are recruited to late endosomes by the MEK1-specific scaffold protein MEK1 partner 1 (MP1), which is in turn recruited to late endosomes through interaction with p14, a late-endosome-resident protein (18,27). Disruption of the MP1–p14 complex or depletion of either protein by siRNA affected ERK recruitment to late endosomes and reduced the duration of ERK activation (18).
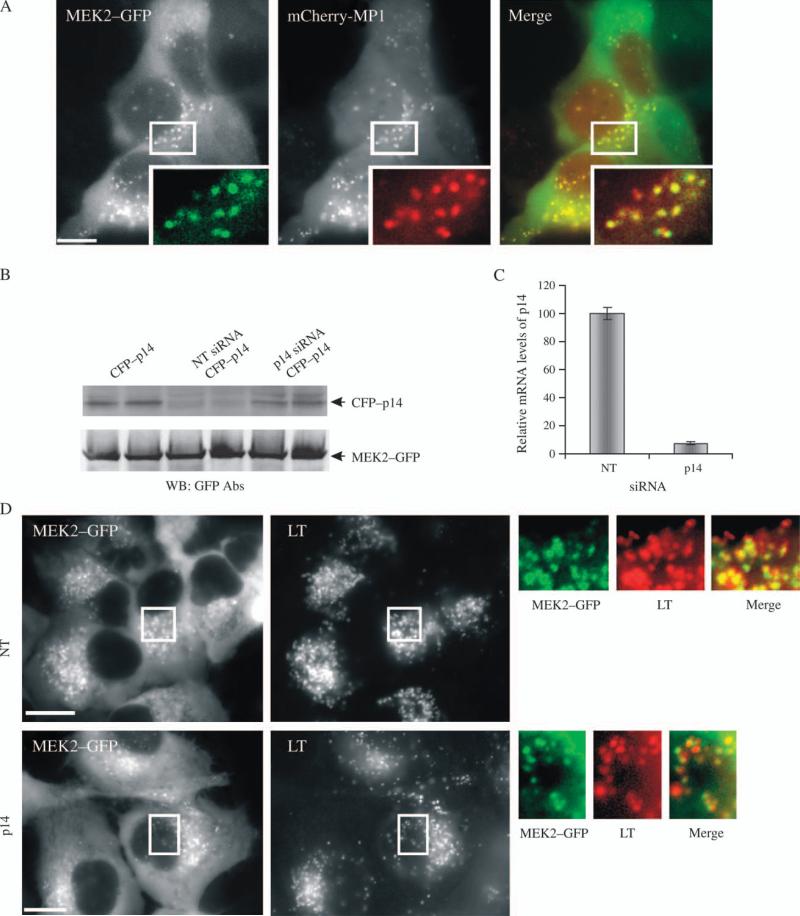
To test whether the MP1–p14 complex also participates in the MEK2 targeting to endosomes, HeLa/MEK2–GFP cells were transiently transfected with mCherry–MP1. As shown in Figure 6A, MEK2–GFP and mCherry–MP1 were well colocalized. A proper targeting of mCherry–MP1 was confirmed by its colocalization with yellow fluorescent protein (YFP)–p14 in endosomes of living cells where the interaction of these two proteins was detected by fluorescence resonance energy transfer (Figure S3). These experiments suggested that MEK2–GFP and MP1–p14 complex are located in the same endosomes.
Figure 6. Effect of p14 siRNA on MEK2–GFP recruitment to vesicles.
A) HeLa/MEK2–GFP cells were transfected with mCherry–MP1, serum starved and treated with 10 ng/mL EGF for 10 min at 37°C. Cells were imaged through fluorescein isothiocyanate and Cy3 filter channels. Insets show high-magnification images of the regions of the cell indicated by white rectangles. Scale bar, 10 μm. B) HeLa/MEK2–GFP cells were cotransfected with p14–CFP plasmid and either nontargeting (NT) siRNA or duplex targeting p14. Total lysates were probed for p14–CFP and MEK2–GFP (loading control) using GFP antibodies. C) Total RNA was extracted from HeLa/MEK2–GFP cells transfected with either NT siRNA or siRNA duplex targeting p14 (p14), and quantitative RT-PCR was performed using specific p14 primers. The data are presented as per cent of p14 mRNA levels in cells transfected with NT siRNA (mean ± SD, n = 2). D) HeLa/MEK2–GFP cells were transfected with p14 or NT siRNA. Starved were pretreated 75 nm LysoTracker (LT) for 30 min and then with 10 ng/mL EGF for 10 min at 37°C. Images were acquired from living cells. Insets show high-magnification images of the regions of the cell indicated by white rectangles. Scale bar, 10 μm.
To further explore the role of MP1–p14 complex in endosomal localization of MEK2–GFP, the effect of siRNA knockdown of p14 on MEK2–GFP endosomal localization was analyzed. Because of the unavailability of the commercial antibodies to p14, the efficiency of p14 knockdown was confirmed using cyan fluorescent protein (CFP)–p14 transiently expressed in parental HeLa cells (Figure 6B) and quantitative real-time polymerase chain reaction (RT-PCR) (Figure 6C). Live cell microscopy analysis of HeLa/MEK2–GFP cells, which were preloaded with LysoTracker (to label the total population of late endosomes/lysosomes) and transfected with p14 siRNA, revealed no obvious decrease in the extent of the recruitment of MEK2–GFP to endosomes compared with cells transfected with the control nontargeting siRNA (Figure 6D). At the same time, p14 depletion did not affect the general pattern of LysoTracker labeling of endosomes, although it resulted in a smaller average size (or lower brightness) and an increased number of MEK2–GFP endosomes per cell, consistent with the putative role of p14 in late endosome biogenesis (28). Knockdown of p14 partially reduced the extent of EGF-induced ERK1/2 activation as revealed by western blotting and immunofluorescence staining with the phospho-ERK1/2 (pERK1/2) antibody (Figure S4). Therefore, the data presented in Figure 6 suggest that the MP1–p14 complex does not appear to be necessary for the endosomal targeting of MEK2–GFP, which is in agreement with the notion that MP1 is a MEK1-specific scaffold (29).
Clathrin-dependent endocytosis is required for MEK2 recruitment to endocytic vesicles
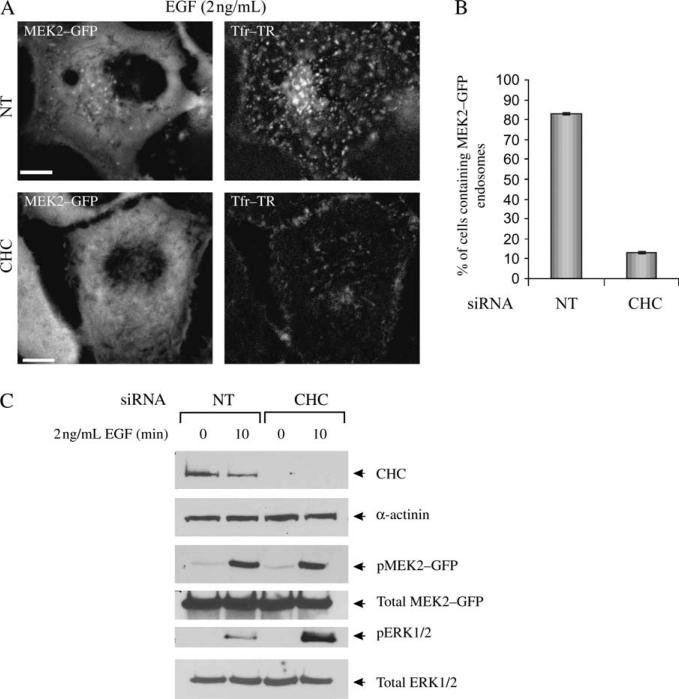
To investigate the role of clathrin-dependent endocytosis in MEK2–GFP recruitment to endosomes, clathrin heavy chain (CHC) siRNA was depleted by siRNA. Knockdown of CHC was previously demonstrated to efficiently block the endocytosis of EGF (by 65–70%) and transferrin (by 70–80%) (30). In our experiments, the blockade of endocytosis of Tfr–TR, known to internalize through coated pits, was used as an evidence of the efficiency of CHC depletion in the individual cells. The cells were stimulated with the low concentration of EGF (2 ng/mL), conditions favoring internalization of EGF–receptor complexes predominantly through clathrin-coated pits. Live cell microscopy analysis showed that CHC siRNA dramatically abolished targeting of MEK2–GFP to endosomes (Figure 7A,B). These data demonstrated that clathrin-dependent processes are required for MEK2–GFP recruitment to endosomes. Surprisingly, the amount of phosphorylated MEK2–GFP was not altered by CHC depletion (Figure 7C). Moreover, ERK1/2 activity was elevated in cells where clathrin-dependent endocytosis was inhibited (Figure 7C). Therefore, clathrin-dependent endocytosis is not required for MEK and ERK activation in HeLa/MEK2–GFP cells.
Figure 7. Effect of CHC siRNA on MEK2–GFP recruitment to endosomes.
A) HeLa/MEK2–GFP cells were transfected with CHC or non-targeting (NT) siRNAs and serum starved. In (A), the cells were incubated with 2 ng/mL EGF and 5 μg/mL Tfr–TR for 10 min at 37°C and imaged through fluorescein isothiocyanate and Cy3 filter channels. Scale bar, 10 μm. B) Multiple images from the experiments exemplified in (A) were visually analyzed, and the percentage of cells containing MEK2–GFP endosomes was calculated (±SD). The data are representative of three independent experiments. C) The cells were treated with EGF as in (A) and lysed. The lysates were probed for CHC, α-actinin (loading control), pMEK1/2, total MEK1/2, pERK1/2 and total ERK1/2.
RAF kinases and MEK activity are necessary for MEK2 localization in endosomes
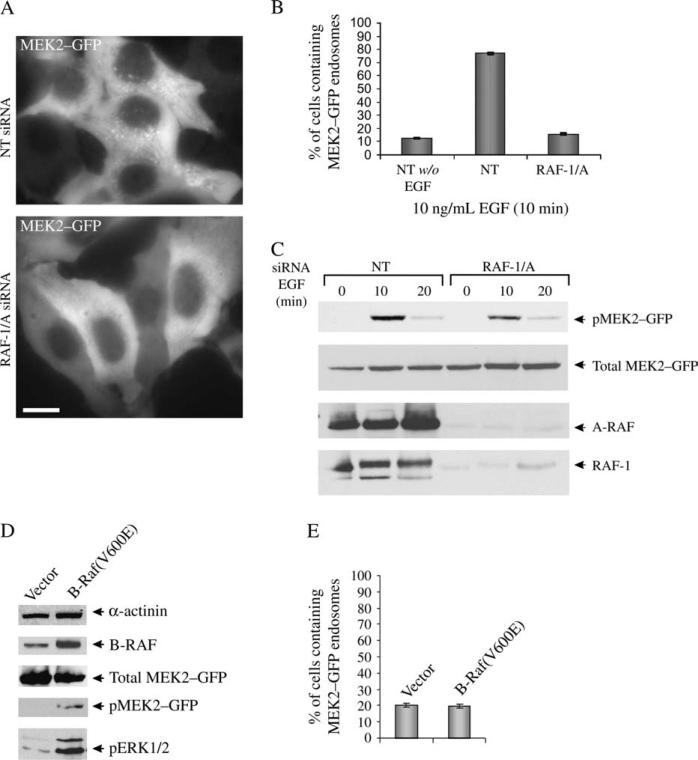
To further analyze the mechanisms of MEK2 targeting to endosomes, we tested whether an upstream MEK kinase, RAF, is necessary for endosomal localization of MEK2–GFP. In HeLa cells, RAF-1 has been reported to activate both MEK1 and MEK2 isoforms, whereas A-RAF is thought to selectively activate MEK1 in response to EGF (31,32). However, depletion of RAF-1 by siRNA resulted in an elevated MEK2–GFP activity and did not affect MEK2–GFP endosomal targeting (data not shown). Therefore, RAF-1 and A-RAF were simultaneously depleted by specific siRNAs. Figure 8A shows that the knockdown of two RAF isoforms significantly reduced the number of cells containing MEK2–GFP endosomes and dramatically reduced a number of detectable endosomes with MEK2–GFP per individual cell (Figure 8A,B). Interestingly, only a partial (about 40%) inhibition of EGF-dependent MEK2–GFP phosphorylation was observed in cells depleted of RAF-1 and A-RAF, presumably because of the presence of B-RAF (Figure 8C). If this assumption is correct, B-RAF must not be necessary for the MEK2–GFP targeting to endosomes but may be sufficient to mediate the activation of total cellular MEK2–GFP. To test this hypothesis, HeLa/MEK2–GFP cells were transiently transfected with the constitutively active B-RAF V600E mutant (33). Although elevated MEK2–GFP and ERK1/2 activities were detected in cells overexpressing B-RAF/V600E (Figure 8D), expression of B-RAF mutant did not result in increased targeting of MEK2–GFP to endosomes (Figure 8E). Therefore, RAF-1 and A-RAF are required for the recruitment of a pool of MEK2–GFP to endosomes, while these isoforms are dispensable for the activation of MEK2 by EGFR.
Figure 8. Effect of A-RAF/RAF-1 siRNA on MEK2–GFP recruitment to endosomes.
A) HeLa/MEK2–GFP cells were transfected with siRNAs, A-RAF and RAF-1 (RAF-1/A) or nontargeting (NT) siRNA. Starved cells were treated with 10 ng/mL of EGF for 10 min at 37°C. Images were acquired from living cells. Scale bar, 10 μm. B) Multiple images from the experiments exemplified in (A) were visually analyzed, and the percentage of cells containing MEK2–GFP endosomes was calculated (±SD). The data are representative of six independent experiments. C) The cells were treated as in (A), lysed and the lysates were probed by western blotting with antibodies to pMEK1/2, MEK1/2, A-RAF and RAF-1. D) HeLa/MEK2–GFP cells were transfected with pBabe-puro empty vector (vector) or pBabe-puro encoding B-RAF mutant V600E [B-RAF(V600E)]. The cells were serum starved and lysed. The lysates were probed by western blotting with antibodies to pMEK1/2, MEK1/2, pERK1/2, B-RAF and α-actinin (loading control). E) Multiple images from the experiments described in (D) were analyzed, and the percentage of cells containing MEK2–GFP endosomes was calculated (±SD). The data are representative of two independent experiments.
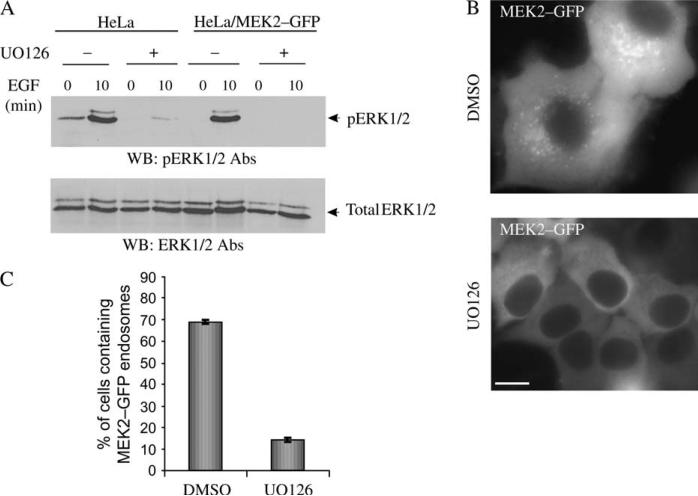
We next addressed whether the activity of MEK is necessary for its recruitment to endosomes using a MEK1/2 inhibitor UO126. This inhibitor blocks the transfer of phosphate by MEK1/2 to ERK1/2, thus resulting in inhibition of the MEK kinase activity toward ERK (34). In control experiments, reduced phosphorylation of ERK1/2 was observed in EGF-stimulated HeLa/MEK2–GFP cells treated with UO126 (Figure 9A). Figure 9B shows that in cells treated with UO126, both the number of cells containing MEK2–GFP endosomes and the number of endosomes per cell were dramatically reduced. Targeting of MEK2–GFP to the endosomes in cells treated with UO126 was significantly reduced compared with that in control cells (Figure 9C). These data indicate that the kinase activity is required for MEK2–GFP recruitment to endosomes.
Figure 9. Effect of UO126 on MEK2–GFP recruitment to endosomes.
A) Serum-starved HeLa and HeLa/MEK2–GFP cells were treated with UO126 or dimethyl sulfoxide (DMSO) (vehicle) for 2 h at 37°C. The cells were then incubated with 10 ng/mL of EGF at 37°C, lysed and the activated ERK1/2 proteins were detected by western blot using pERK1/2 antibodies. B) Serum-starved HeLa/MEK2–GFP cells were treated with UO126 or DMSO and then with EGF as in (A). Images were acquired from living cells. Scale bar, 10 μm. C) Multiple images from the experiments exemplified in (B) were visually analyzed, and the percentage of cells containing MEK2–GFP endosomes was calculated (±SD). The data are representative of four independent experiments.
Discussion
Development of the methodologies to follow localization of signaling proteins tagged with various GFP variants (XFPs) by live cell microscopy led to the discovery of many new insights into the mechanisms of spatial and temporal regulation of signal transduction processes (35). For instance, XFP-fused EGFR and components of the MAPK activation cascade have been utilized to study the regulation of this signaling pathway (11,36,37). However, most of these studies analyzed localization of overexpressed XFP fusion proteins in the presence of an endogenous counterpart. Such overexpression often leads to aberrant localization and activities of the signaling proteins (11,38,39). The KDAR method, involving the replacement of an endogenous protein by its XFP-fused version, allows a more realistic analysis of protein localization during signal transduction.
In this study, the KDAR method was used to generate HeLa/MEK2–GFP cells that express physiological levels of MEK2–GFP in the absence of endogenous MEK2. Microscopic analysis of these cells revealed rapid, EGF-induced translocation of MEK2–GFP to the plasma membrane and endosomes. Activated MEK was previously detected at the plasma membrane and endocytic vesicles by cell fractionation and immunofluorescence methods (16,17). However, the amounts of MEK found to be associated with these membrane compartments were very small. Given that it is technically difficult to purify endosomal fractions free of the plasma membrane from cultured cells, the detection of MEK2–GFP in endosomes in our live cell studies allowed better characterization of these endosomal compartments. Interestingly, MEK2–GFP was localized to a subset of endosomes that contained markers of early (Rab5) and late (Rab7 and LysoTracker) compartments. It is possible that vesicles containing MEK2–GFP represented an ‘intermediate’ compartment that contained both Rab5 and Rab7. It has been predicted that there is a progressive conversion of Rab5 endosomes into Rab7 endosomes involving the Rab7 GEF–class C VPS/HOPS complex, which also interacts with Rab5 (40,41). Alternatively, MEK2–GFP could be recruited to separate subsets of early and late endosomes.
Surprisingly, EGF and active EGFR were not detected in MEK2–GFP endosomes in cells stimulated with EGF (Figures 2 and 3). This result seems to somewhat disagree with the demonstration of colocalization of p14, phosphorylated ERK and transiently expressed EGFR–GFP fusion in late endosomes of HeLa cells by Teis et al. (18). However, when overexpressed, the EGFR–mRFP fusion protein was colocalized with MEK2–GFP in our experiments as well, although the receptors present in MEK2–GFP endosomes were unoccupied by the ligand (Figure 3). Continuous incubation of cells with EGF at low temperature was the only condition that allowed detection of a considerable colocalization of EGF and MEK2–GFP in endosomes (Figure 3). At this temperature, EGFR is slowly internalized and recycled, but the sorting of internalized receptors to late endosomes is severely inhibited. Therefore, in agreement with the presence of both Rab5 and Rab7 in MEK2–GFP endosomes, these endosomes may represent an ‘intermediate’ compartment that participates in rapid transition of a limited pool of EGFR from early to late endosomes. We suggest that because of the low expression level, fast endocytosis and endosomal sorting of endogenous EGFR in the variant of HeLa cells used in our laboratory, activated EGFR was not accumulated in MEK2–GFP endosomes under physiological conditions (37°C).
The finding of MEK2–GFP in late endosomes in our study is reminiscent of the findings of the MAPK scaffold complex, involving the MP1–p14 complex, in late endosomes by Huber and coworkers (18,27). Although more likely to be an interacting protein than a ‘true’ scaffold because of its small size, MP1 has been shown to interact specifically with MEK1 but not with MEK2 (29). In our experiments, MEK2–GFP was highly colocalized with the heterologously expressed MP1 and p14 complex, suggesting that three proteins are localized in the same endosomal compartments. However, siRNA depletion of p14 did not inhibit endosomal recruitment of MEK2–GFP (Figure 6), suggesting that it is unlikely that endosomal MEK2 is a part of the MP1–p14 scaffold complex. Therefore, we suggest in this study that a mechanism other than an interaction with the MP1–p14 scaffold regulates MEK2 recruitment to endosomes. Clathrin-mediated endocytosis can be a part of this mechanism because siRNA knockdown of CHC blocked targeting of MEK2–GFP to endosomes (Figure 7). Another component that appears to be required for the MEK2 recruitment to endosomes is RAF kinases. Simultaneous depletion of both RAF-1 and A-RAF had a major effect on MEK2–GFP endosomal localization (Figure 8A–C). At the same time, only a partial decrease of the MEK2–GFP phosphorylation level was observed in cells depleted of RAFs, which can be explained by the presence of B-RAF (Figure 8D,E). Thus, it is possible that a stoichiometric amount of RAF kinases is necessary for MEK2 recruitment to endosomes, whereas a much smaller amount of RAF is sufficient to activate MEK1/2.
Finally, in addition to the requirement of endocytosis and RAF, localization of MEK2 in endosomes required the catalytic activity of MEK (Figure 9). Phosphorylation of ERK by MEK is known to be necessary and sufficient for the dissociation of the MEK–ERK complex, and the dissociation of the complex is required for the activity of ERK (42). Thus, MEK2 localization in endosomes may require dissociation of ERK from MEK2 upon ERK phosphorylation. Another possibility is that the endosomal targeting of MEK2 requires its deactivating phosphorylation by active ERK (43,44) and therefore could be expected to be disrupted if ERK is not activated. In this scenario, phosphorylation and activation of ERK lead to the negative feedback regulation of MEK through its phosphorylation by ERK (43,44).
What is the role of endosomal MEK2–GFP? Because no active MEK was detected in MEK2–GFP endosomes in our experiments (Figure 4), it is logical to assume that this pool of MEK2 does not participate in signaling. While the inability to detect active MEK2 in endosomes could be because of technical limitations, the most likely interpretation of the data is that endosomal compartmentalization of MEK2 may serve to negatively regulate EGFR–MAPK signaling cascade. This hypothesis is supported by the observation that MEK2–GFP-containing endosomes represent a pool of endosomes distinct from the classical ‘signaling’ endosomes that contain active EGFR, adaptor-like Grb2 and active Ras. Thus, endosomal recruitment of MEK2 may result in the spatial separation of MEK2 from EGFR signaling complexes. The possibility that endosomal MEK2 does not participate in EGFR signaling in our model cell system is consistent with our data demonstrating that clathrin-mediated endocytosis is not necessary for MEK2 activation but is required for its endosomal accumulation (Figure 7). Moreover, the observation that clathrin knockdown results in increased ERK1/2 activation without changing the activity of MEK (Figure 7) suggests that in the presence of clathrin, a pool of ERK1/2 can be inaccessible for phosphorylation by active MEK2 at the plasma membrane because of the association of this pool of ERK1/2 with inactive MEK2 in endosomes.
In summary, live cell microscopy revealed localization of a pool of MEK2 in a distinct subset of endosomes in EGF-stimulated cells, which may serve as negative feedback regulation of EGFR–MAPK signaling cascade. While a more precise characterization of endosomal MEK2 complexes will be performed in future studies, our data demonstrate that targeting of MEK2 to endosomes requires the interaction of MEK2 with both the upstream and the downstream components of the ERK activation cascade and is regulated by a clathrin-dependent process.
Materials and Methods
Chemicals and antibodies
EGF was obtained from Collaborative Research. EGF conjugated to Rhodamine (EGF–Rh), EGF conjugated to Alexa fluor 647 streptavidin (EGF–Alexa647), human transferrin conjugated to Texas Red (Tfr–TR) and LysoTracker were purchased from Molecular Probes. Antibodies to MEK1/2, MEK2, ERK1/2, pERK1/2, pMEK1/2 and A-RAF were purchased from Cell Signaling Technology. RAF-1 antibodies were purchased from Santa Cruz Biosciences, and B-RAF antibodies were kindly provided by Dr R Nemenoff (University of Colorado Denver Health Sciences Center, UCDHSC). EGFR antibodies (2913) were kindly provided by Dr L Beguinot (DIBIT Rafaele, Milan, Italy). Rabbit polyclonal antibody Ab32 specific to β-adaptins was described previously (45), α-actinin antibodies were purchased from Chemicon, Inc. and CHC antibodies (TD1) were purchased from American Type Culture Collection. Monoclonal Grb2 antibodies were purchased from NeoMarkers (LabVision). Protein phosphatase 2A (PP2A) antibodies were from BD Biosciences. GFP antibodies were purchased from Zymed, Inc. Pfu polymerase was purchased from Stratagene. Various siRNAs were purchased from Dharmacon.
Expression plasmids
The full-length human MEK2 in pcDNA3.1 was kindly provided by Dr L Heasley (UCDHSC). pBabe-puro plasmid encoding B-RAF mutant V600E was kindly provided by Dr C. J. Der (University of North Carolina, Chapel Hill, NC, USA). To generate the GFP-tagged version of MEK2, a forward primer containing a _Xho_I site and a reverse primer containing an _Eco_RI site after the stop codon were used to amplify the human MEK2 sequence by PCR. The mCherry complementary DNA (cDNA) was kindly provided by Dr R. Tsien (University of California San Diego, La Jolla, CA, USA). To generate the pmCherry-C1 expression vector, mCherry full-length cDNA was amplified by PCR and ligated into pEYFP-C1 (Clontech) using _Bam_HI and _Bsr_GI restriction sites. To generate EEA.1 mRFP fusion proteins, a fragment corresponding to amino acid residues 1098–1411 of EEA.1 (further referred as EEA.1–mRFP) was amplified by PCR and ligated into pmRFP-C3 by using _Hin_dIII and _Bam_HI restriction sites. GFP-tagged canine Rab7 plasmid was kindly provided by Dr A. Wandinger-Ness (University of New Mexico, Albuquerque, NM, USA). To generate the mCherry fusion protein, a full-length Rab7 sequence was moved from GFP–Rab7 into pmCherry-C1 using _Kpn_I and _Xho_I restriction sites. YFP–p14 and CFP–MP1 plasmids were kindly provided by Dr L. A. Huber (University of Innsbruck, Innsbruck, Austria). To generate the mCherry–MP1 fusion protein, the CFP sequence in CFP–MP1 was substituted for mCherry plasmid using _Bcl_I and _Hin_dIII restriction sites. The mRFP–Rab5 and EGFR–mRFP plasmids were described previously (46).
‘SMARTpool’ and four individual duplexes to human MEK2 were obtained from Dharmacon and used for transient transfection. To silence the expression of other proteins, SMARTpools (Dharmacon) of siRNAs were used. To generate a vector, stably expressing shRNA pSilencer-4.1-CMVhygro plasmid (Ambion, Inc.) was used. A set of oligonucleotides corresponding to the MEK2-specific sequence of siRNA duplex 3 (5′-UCCAGGAGUUUGUCAATAA-3′) containing _Hin_dIII and _Bam_HI restriction sites was synthesized. The following primers were used: 5′-GATCCGGAAGCTGATCCACCTTGATTCAAGAGATCAAGGTGGATCAGCTTCCTTA-3′ and 5′-AGCTTAAGGAAGCTGATCCACCTTGATCTCTTGAATCAAGGTGGATCAGCTTCCG 3-′. Oligonucleotides were annealed according to the manufacturer's recommendations and ligated into pSilencer-4.1-CMVhygro using _Bam_HI and _Hin_dIII restriction sites. The pSilencer-4.1-MEK2 construct was verified by dideoxynucleotide sequencing. Point mutations in MEK2–GFP construct were introduced using a QuickChange site-directed mutagenesis kit according to the manufacturer's directions (Stratagene). Silent mutations changed the DNA sequence but not the amino acid sequence, making the constructs resistant to shRNA knockdown. The following primers were used: 5′-GTTCACCCCCGACTTTCAAGAATTCGTGAACAAATGCCTCATCAAG-3′ and 5′-CTTGATGAGGCATTTGTTCTCGAATTCTTGAAAGTCGGGGGTGAAC-3′. The construct was verified by dideoxynucleotide sequencing.
To silence the expression of p14, a previously published siRNA duplex (5′-CCAAGGAGAGACCGUGGGCUUUU-3′) was used (18).
Cell culture and DNA transfections
Human cervical carcinoma HeLa cells were grown in DMEM containing 10% FBS. The transfections of DNA constructs were performed using Effectene (Qiagen). Expression of GFP-fused proteins was confirmed by western blotting as described below. The stably expressing cell lines were selected by growing the transfected cells in the presence of G418 (0.4 mg/mL) and hygromycin (0.2 mg/mL).
siRNA transfections
To silence protein expression by RNA interference, HeLa cells were seeded in 12-well plates (50–60% confluent, 1 mL of DMEM/FBS per well) at least 20 h before transfection. siRNA transfections were performed at 24- to 36-h intervals according to the manufacturer's recommendations using Dharmafect reagent 1 (Dharmacon). Three to four days after transfection, the confluent cells were trypsinized, and half of the cells were seeded on glass-bottom dishes (MatTek) for immunofluorescence/microscopy analysis, while the other half was used for western blot analysis. The cells were then incubated in serum-free and phenol-red-free medium containing 0.2% BSA for 20 h prior to the microscopy experiments.
For siRNA–DNA coexpression experiments, the cells were cotransfected with 3 μL of 20 μm siRNA and 1.5 μg of a DNA plasmid using an Optifect transfection kit (Invitrogen). The efficiency of the siRNA knockdown was validated by western blotting.
Western blot analysis
The cells were solubilized in Triton-X-100/glycerol lysis buffer and subjected to electrophoresis and western blotting as described previously (47). To detect phosphorylated MEK and ERK by immunoblotting, the cells were directly lysed in the sample loading buffer. Several X-ray films were analyzed to determine the linear range of the chemiluminescence signals, and the quantifications were performed using densitometry analysis mode of AlphaEaseFC software (Alpha Innotech Co.).
Immunofluorescence staining
Cells grown on glass-bottom dishes were either treated or not treated with 10 ng/mL of EGF and washed with Ca2+-, Mg2+-free PBS. The cells were then fixed with freshly prepared 4% paraformaldehyde (Electron Microscopy Sciences) for 10 min at room temperature and permeabilized using 0.2% Triton-X-100 or methanol for 5 min at room temperature. Immunostaining was performed according to manufacturer's recommendations for the pMEK1/2 and pERK1/2 antibodies. The coverslips were mounted into Mowiol (Molecular Probes), and a Z-stack of x–y images was obtained and deconvoluted using epifluorescence Mariannas™ workstation (Intelligent Imaging Innovation) as described previously (48).
Fluorescence imaging of living cells
The cells were replated 24 h before the experiment onto 35-mm glass-bottom dishes and kept in serum-free and phenol-red-free medium containing 0.2% BSA for 16–20 h before experiments. The cells were imaged at room temperature using Mariannas workstation. The detection of GFP fluorescence was performed using fluorescein isothiocyanate filter channel, mRFP and mCherry; rhodamine fluorescence using mRFP or Cy3 channel and Alexa647 fluorescence using CY5 channel and an 86004BS dichroic mirror (Chroma) (46). Images were acquired using 2 × 2 binning mode.
To quantify the endosomal localization of MEK2–GFP, several three-dimensional images were acquired under each experimental condition. The cells were categorized based on visual inspection into two groups: (i) cells containing at least one endosome decorated by GFP and (ii) cells containing no GFP-decorated endosomes. The number of cells containing endosomal MEK2–GFP was expressed as per cent of total cells analyzed.
Real-time quantitative polymerase chain reaction
Seventy-two hours after the transfection of p14 siRNA, total RNA was isolated from HeLa/MEK2–GFP cells using an RNeasy kit (Qiagen) according to the manufacturer's instructions. Aliquots containing equal amounts of messenger RNA (mRNA) were subjected to RT-PCR analysis. Quantitative RT-PCR was performed using the Applied Biosystems (ABI) Prism 7700 HT sequence detection system. Sequence-specific TaqMan probes and primer set for the p14 gene were obtained from ABI (TaqMan® Gene Expression Assays), and RT-PCR was performed according to the manufacturer's protocol. Finally, 18_S_ rRNA was used as an internal control (TaqMan Gene Expression Assays). Samples were normalized against the internal control, and the results were presented as percentage of p14 mRNA recovered from cells transfected with nontargeting siRNA. The data represent the mean ± standard deviation from two independent experiments.
Figure S1: A) Schematic representation of the MEK2–GFP-FYVE chimeric protein. B) Serum-starved HeLa cells stably coexpressing pSilencer4.1-MEK2 siRNA and MEK2–GFP-FYVE (Methods) were treated with 10 ng/mL EGF for 0 (–EGF) and 10 min at 37°C (+EGF) and fixed. The cells were then processed for immunofluorescence with pMEK1/2 antibodies and secondary Cy3 donkey anti-rabbit antibodies. GFP and Cy3 were detected using fluorescein isothiocyanate and Cy3 filter channels. Scale bar, 10 μm.
Figure S2: HeLa/MEK2–GFP cell clones (9 and 16) were transfected with mCherry–Rab7. Serum-starved cells were treated with EGF (10 ng/mL) for 10 min at 37°C. Living cells were imaged through fluorescein isothiocyanate and Cy3 filter channels. Scale bars, 10 μm.
Figure S3: A) Schematic representation of mCherry–MP1 and p14–YFP fusion proteins. B) p14–YFP was coexpressed with mCherry–MP1 in HeLa cells. Fluorescence resonance energy transfer (FRET) images were obtained from living cells as described in ‘Supplementary Methods’. FRETC (FCYR) was calculated as described in ‘Supplementary Methods’ and presented as a pseudocolor image. A.l.u.f.i., arbitrary linear units of fluorescence intensity. Insets represent high-magnification images of the regions of the cell indicated by white rectangle. Scale bar, 10 μm.
Figure S4: A) HeLa/MEK2–GFP cells were transfected with p14 or control (NT) siRNA. Serum-starved HeLa/MEK2–GFP cells were untreated (–EGF) or treated with 10 ng/mL EGF for 10 min at 37°C (+EGF) and fixed. The cells were then processed for immunofluorescence staining with pERK1/2 antibodies and secondary Cy3 donkey anti-rabbit antibodies. GFP and Cy3 were detected using fluorescein isothiocyanate and Cy3 filter channels. Scale bars, 10 μm. B) HeLa/MEK2–GFP cells were transfected with p14 or control (NT) siRNA. The cells were serum starved and treated with 10 ng/mL EGF for indicated times at 37°C. The lysates were probed for activated ERK1/2 (pERK1/2) and total ERK1/2 (ERK1/2).
Movie S1: HeLa/MEK2–GFP cells were treated with 10 ng/mL EGF for 10 min at 37°C as described in Figure 2A. Time-lapse images were acquired every 3 seconds (total time – 2 min) at room temperature.
Supplementary Material
SF
SM
1
Acknowledgments
This study was supported by American Cancer Society grant RSG-00-247-04-CSM and National Institutes of Health grants CA089151-08 and GM059570. We thank Melissa Adams for help in the manuscript preparation, Dr P. David for critical reading of the manuscript, Drs L. Beguinot, L. Heasley, R. Tsien, A. Wandinger-Ness, R. Nemenoff, C. J. Der, G. L Johnson and L. A. Huber for the kind gifts of the reagents and Uma Pugazhenthi (UCCC Gene Expression Shared Core Service) for technical help.
Footnotes
Additional Supporting Information may be found in the online version of this article:
- Endosomal targeting of Mek2 requires Raf, Mek kinase activity and clathrin-dependent endocytosis
- Supplementary Methods
- Plasmids
- Immunofluorescence staining
- FRET microscopy
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 2.Rozakis-Adcock M, Fernley R, Wade J, Pawson T, Bowtell D. The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1. Nature. 1993;363:83–85. doi: 10.1038/363083a0. [DOI] [PubMed] [Google Scholar]
- 3.Cobb MH. MAP kinase pathways. Prog Biophys Mol Biol. 1999;71:479–500. doi: 10.1016/s0079-6107(98)00056-x. [DOI] [PubMed] [Google Scholar]
- 4.Kolch W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem J. 2000;2:289–305. [PMC free article] [PubMed] [Google Scholar]
- 5.Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter G. The EGF receptor: a nexus for trafficking and signaling. Bioessays. 2000;22:697–707. doi: 10.1002/1521-1878(200008)22:8<697::AID-BIES3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 7.von Zastrow M, Sorkin A. Signaling on the endocytic pathway. Curr Opin Cell Biol. 2007;19:436–445. doi: 10.1016/j.ceb.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miaczynska M, Pelkmans L, Zerial M. Not just a sink: endosomes in control of signal transduction. Curr Opin Cell Biol. 2004;16:400–406. doi: 10.1016/j.ceb.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Vieira AV, Lamaze C, Schmid SL. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 10.Sorkin A. The endocytosis machinery. J Cell Sci. 2000;24:4375–4376. doi: 10.1242/jcs.113.24.4375. [DOI] [PubMed] [Google Scholar]
- 11.Jiang X, Sorkin A. Coordinated traffic of Grb2 and Ras during epidermal growth factor receptor endocytosis visualized in living cells. Mol Biol Cell. 2002;13:1522–1535. doi: 10.1091/mbc.01-11-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Guglielmo GM, Baass PC, Ou WJ, Posner BI, Bergeron JJ. Compartmentalization of SHC, GRB2 and mSOS, and hyperphosphorylation of Raf-1 by EGF but not insulin in liver parenchyma. EMBO J. 1994;13:4269–4277. doi: 10.1002/j.1460-2075.1994.tb06747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy S, Wyse B, Hancock JF. H-Ras signaling and K-Ras signaling are differentially dependent on endocytosis. Mol Cell Biol. 2002;22:5128–5140. doi: 10.1128/MCB.22.14.5128-5140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prior IA, Hancock JF. Compartmentalization of Ras proteins. J Cell Sci. 2001;114:1603–1608. doi: 10.1242/jcs.114.9.1603. [DOI] [PubMed] [Google Scholar]
- 15.Rizzo MA, Shome K, Watkins SC, Romero G. The recruitment of Raf-1 to membranes is mediated by direct interaction with phosphatidic acid and is independent of association with Ras. J Biol Chem. 2000;275:23911–23918. doi: 10.1074/jbc.M001553200. [DOI] [PubMed] [Google Scholar]
- 16.Pol A, Calvo M, Enrich C. Isolated endosomes from quiescent rat liver contain the signal transduction machinery. Differential distribution of activated Raf-1 and Mek in the endocytic compartment. FEBS Lett. 1998;441:34–38. doi: 10.1016/s0014-5793(98)01517-8. [DOI] [PubMed] [Google Scholar]
- 17.Rizzo MA, Kraft CA, Watkins SC, Levitan ES, Romero G. Agonist-dependent traffic of raft-associated Ras and Raf-1 is required for activation of the mitogen-activated protein kinase cascade. J Biol Chem. 2001;276:34928–34933. doi: 10.1074/jbc.M105918200. [DOI] [PubMed] [Google Scholar]
- 18.Teis D, Wunderlich W, Huber LA. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev Cell. 2002;3:803–814. doi: 10.1016/s1534-5807(02)00364-7. [DOI] [PubMed] [Google Scholar]
- 19.Kranenburg O, Verlaan I, Moolenaar WH. Dynamin is required for the activation of mitogen-activated protein (MAP) kinase by MAP kinase kinase. J Biol Chem. 1999;274:35301–35304. doi: 10.1074/jbc.274.50.35301. [DOI] [PubMed] [Google Scholar]
- 20.DeGraff JL, Gagnon AW, Benovic JL, Orsini MJ. Role of arrestins in endocytosis and signaling of alpha2-adrenergic receptor subtypes. J Biol Chem. 1999;274:11253–11259. doi: 10.1074/jbc.274.16.11253. [DOI] [PubMed] [Google Scholar]
- 21.Johannessen LE, Ringerike T, Molnes J, Madshus IH. Epidermal growth factor receptor efficiently activates mitogen-activated protein kinase in HeLa cells and Hep2 cells conditionally defective in clathrin-dependent endocytosis. Exp Cell Res. 2000;260:136–145. doi: 10.1006/excr.2000.5004. [DOI] [PubMed] [Google Scholar]
- 22.Sorkin A, Von Zastrow M. Signal transduction and endocytosis: close encounters of many kinds. Nat Rev Mol Cell Biol. 2002;3:600–614. doi: 10.1038/nrm883. [DOI] [PubMed] [Google Scholar]
- 23.Morrison DK. KSR: a MAPK scaffold of the Ras pathway? J Cell Sci. 2001;114:1609–1612. doi: 10.1242/jcs.114.9.1609. [DOI] [PubMed] [Google Scholar]
- 24.Cacace AM, Michaud NR, Therrien M, Mathes K, Copeland T, Rubin GM, Morrison DK. Identification of constitutive and ras-inducible phosphorylation sites of KSR: implications for 14-3-3 binding, mitogen-activated protein kinase binding, and KSR overexpression. Mol Cell Biol. 1999;19:229–240. doi: 10.1128/mcb.19.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furuno T, Hirashima N, Onizawa S, Sagiya N, Nakanishi M. Nuclear shuttling of mitogen-activated protein (MAP) kinase (extracellular signal-regulated kinase (ERK) 2) was dynamically controlled by MAP/ERK kinase after antigen stimulation in RBL-2H3 cells. J Immunol. 2001;166:4416–4421. doi: 10.4049/jimmunol.166.7.4416. [DOI] [PubMed] [Google Scholar]
- 26.Galperin E, Sorkin A. Visualization of Rab5 activity in living cells by FRET microscopy and influence of plasma-membrane-targeted Rab5 on clathrin-dependent endocytosis. J Cell Sci. 2003;116:4799–4810. doi: 10.1242/jcs.00801. [DOI] [PubMed] [Google Scholar]
- 27.Wunderlich W, Fialka I, Teis D, Alpi A, Pfeifer A, Parton RG, Lottspeich F, Huber LA. A novel 14-kilodalton protein interacts with the mitogen-activated protein kinase scaffold mp1 on a late endosomal/lysosomal compartment. J Cell Biol. 2001;152:765–776. doi: 10.1083/jcb.152.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teis D, Taub N, Kurzbauer R, Hilber D, de Araujo ME, Erlacher M, Offterdinger M, Villunger A, Geley S, Bohn G, Klein C, Hess MW, Huber LA. P14-MP1-MEK1 signaling regulates endosomal traffic and cellular proliferation during tissue homeostasis. J Cell Biol. 2006;175:861–868. doi: 10.1083/jcb.200607025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaeffer HJ, Catling AD, Eblen ST, Collier LS, Krauss A, Weber MJ. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science. 1998;281:1668–1671. doi: 10.1126/science.281.5383.1668. [DOI] [PubMed] [Google Scholar]
- 30.Huang F, Khvorova A, Marshall W, Sorkin A. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J Biol Chem. 2004;279:16657–16661. doi: 10.1074/jbc.C400046200. [DOI] [PubMed] [Google Scholar]
- 31.Wu X, Noh SJ, Zhou G, Dixon JE, Guan KL. Selective activation of MEK1 but not MEK2 by A-Raf from epidermal growth factor-stimulated Hela cells. J Biol Chem. 1996;271:3265–3271. doi: 10.1074/jbc.271.6.3265. [DOI] [PubMed] [Google Scholar]
- 32.'O'Neill E, Kolch W. Conferring specificity on the ubiquitous Raf/MEK ignaling pathway. Br J Cancer. 2004;90:283–288. doi: 10.1038/sj.bjc.6601488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao H, Muniz-Medina VM, Mehta H, Thomas NE, Khazak V, Der CJ, Shields JM. Context-dependent roles of mutant B-Raf signaling in melanoma and colorectal carcinoma cell growth. Mol Cancer Ther. 2007;6:2220–2229. doi: 10.1158/1535-7163.MCT-06-0728. [DOI] [PubMed] [Google Scholar]
- 34.English JM, Cobb MH. Pharmacological inhibitors of MAPK pathways. Trends Pharmacol Sci. 2002;23:40–45. doi: 10.1016/s0165-6147(00)01865-4. [DOI] [PubMed] [Google Scholar]
- 35.Tsien RY, Miyawaki A. Seeing the machinery of live cells. Science. 1998;280:1954–1955. doi: 10.1126/science.280.5371.1954. [DOI] [PubMed] [Google Scholar]
- 36.Sorkin A, McClure M, Huang F, Carter R. Interaction of EGF receptor and grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Curr Biol. 2000;10:1395–1398. doi: 10.1016/s0960-9822(00)00785-5. [DOI] [PubMed] [Google Scholar]
- 37.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL II, Cox AD, Philips MR. Ras signaling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 38.Morrison DK, Davis RJ. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu Rev Cell Dev Biol. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 39.Janssen RA, Kim PN, Mier JW, Morrison DK. Overexpression of kinase suppressor of Ras upregulates the high-molecular-weight tropomyosin isoforms in ras-transformed NIH 3T3 fibroblasts. Mol Cell Biol. 2003;23:1786–1797. doi: 10.1128/MCB.23.5.1786-1797.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 41.Seals DF, Eitzen G, Margolis N, Wickner WT, Price A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homo-typic vacuole fusion. Proc Natl Acad Sci U S A. 2000;97:9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pouyssegur J, Volmat V, Lenormand P. Fidelity and spatio-temporal control in MAP kinase (ERKs) signaling. Biochem Pharmacol. 2002;64:755–763. doi: 10.1016/s0006-2952(02)01135-8. [DOI] [PubMed] [Google Scholar]
- 43.Brunet A, Pages G, Pouyssegur J. Growth factor-stimulated MAP kinase induces rapid retrophosphorylation and inhibition of MAP kinase kinase (MEK1). FEBS Lett. 1994;346:299–303. doi: 10.1016/0014-5793(94)00475-7. [DOI] [PubMed] [Google Scholar]
- 44.Eblen ST, Slack-Davis JK, Tarcsafalvi A, Parsons JT, Weber MJ, Catling AD. Mitogen-activated protein kinase feedback phosphorylation regulates MEK1 complex formation and activation during cellular adhesion. Mol Cell Biol. 2004;24:2308–2317. doi: 10.1128/MCB.24.6.2308-2317.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sorkin A, McKinsey T, Shih W, Kirchhausen T, Carpenter G. Stoichiometric interaction of the epidermal growth factor receptor with the clathrin-associated protein complex AP-2. J Biol Chem. 1995;270:619–625. doi: 10.1074/jbc.270.2.619. [DOI] [PubMed] [Google Scholar]
- 46.Galperin E, Verkhusha VV, Sorkin A. Three-chromophore FRET microscopy to analyze multiprotein interactions in living cells. Nat Methods. 2004;1:209–217. doi: 10.1038/nmeth720. [DOI] [PubMed] [Google Scholar]
- 47.Jiang X, Huang F, Marusyk A, Sorkin A. Grb2 regulates internalization of EGF receptors through clathrin-coated pits. Mol Biol Cell. 2003;14:858–870. doi: 10.1091/mbc.E02-08-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galperin E, Sorkin A. Visualization of Rab5 activity in living cells using FRET microscopy. Methods Enzymol. 2005;403:119–134. doi: 10.1016/S0076-6879(05)03011-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SF
SM
1