High-Throughput T7 LIC Vector for Introducing C-Terminus Poly-Histidine Tags with Variable Lengths without Extra Sequences (original) (raw)
. Author manuscript; available in PMC: 2010 Jan 1.
Published in final edited form as: Protein Expr Purif. 2008 Sep 14;63(1):58–61. doi: 10.1016/j.pep.2008.09.005
Abstract
Immobilized metal ion affinity chromatography (IMAC) has become one of the most popular protein purification methods for recombinant proteins with a hexa-histidine tag (His-tag) placed at the C- or N- terminus of proteins. Nevertheless, there are always difficult proteins that show weak binding to the metal chelating resin and thus low purity. These difficulties are often overcome by increasing the His-tag to 8 or 10 histidines. Despite their success, there are only few expression vectors available to easily clone and test different His-tag lengths. Therefore, we have modified Escherichia coli T7 expression vector pET21a to accommodate ligation-independent cloning (LIC) that will allow easy and efficient parallel cloning of target genes with different His-tag lengths using a single insert. Unlike most LIC vectors available commercially, our vectors will not translate unwanted extra sequences by engineering the N-terminal linker to anneal before the open reading frame, and the C-terminal linker to anneal as a His-tag.
Keywords: Histidine tag of variable lengths, Ligation independent cloning, Immobilized metal ion affinity chromatography, T7 bacterial expression, High-throughput protein purification
Introduction
Immobilized metal ion affinity chromatography (IMAC) has become one of the most popular recombinant protein purification methods [1–5]. By adding six histidines at the N- or C- terminus of a protein, this allows the metal affinity resin charged with nickel or some other transition metal ion to selectively capture the target protein by metal-ligand covalent bonding. By increasing the concentration of imidazole in the purification buffer or stripping the metals off the resins with strong metal chelators such as EDTA, easy controlled release of captured proteins is achieved. Furthermore, the interaction between the resin and protein can also occur in buffer conditions containing urea thus allowing on-column refolding [6–10].
Despite all the benefits of IMAC, there can be problems like weak or no interaction between the resin and target. This is often caused by concealed His-tags, and could be alleviated by switching the position of the His-tag to the other terminus of the protein. For some cases this does not help either, presumably because the “free” portion of the His-tag is still not long enough for strong binding. We encountered this situation with an E. coli chemotaxis receptor. This can be resolved by increasing the binding affinity of the target protein thus leading to increased purity of the target. Many researchers have increased the length of His-tags on their targets with good results [11–15]. Despite these successes, there are no expression vectors that allow easy and efficient cloning of proteins with different His-tag lengths. Therefore, we modified EMD Biosciences’ pET21a, an E. coli T7 protein expression vectors to allow introduction of variable length His-tags using ligation-independent cloning (LIC).
Materials and Methods
Materials
The vectors were derived from E. coli expression vector pET21a and expression was performed using BL21(DE3) Star E. coli cells (Novagen, La Jolla, CA). All synthesized oligos and PCR primers were from Integrated DNA Technologies (Coralville, IA) and dNTP, dGTP and dCTP were from Promega (Madison, WI). Deep Vent DNA Polymerase, T4 DNA Polymerase, and restriction enzymes StuI came from New England Biolabs (Beverly, MA). Mutations were introduced using QuikChange II Site-Directed Mutagenesis (Stratagene, La Jolla, CA). MachT1 DH5α E. coli competent cells were used for cloning (Invitrogen, Carlsbad, CA). Carbenecillin (United States Biological, Swampscott, MA) was used at a concentration of 100 µg/mL. n-Dodecyl-beta-D-maltopyranoside was purchased from Anatrace (Maumee, OH). The HisTrap 5mL column was purchased from GE Healthcare.
Construction of His-tag Length Variable LIC Vector pJL
The vectors pJL-H6, pJL-H8, and pJL-H10 were derived from pL, a pET21a derived LIC vector for C-term His-tag cloning (Not published). Briefly, pL vector was constructed by modifying the multiple cloning site of pET21a to allow LIC cloning of the gene with a C-terminal six histidine tag. All three vectors were made by QuikChange site directed mutagenesis of pL using primers, pJL-H6 : 5’-GAAGGAGATATAAGGCCTCACCATCACCATCATCACTGAGATCCGG-3’ and 5’-CCGGATCTCAGTGATGATGGTGATGGTGAGGCCTTATATCTCCTTC-3’, pJL-H8 : 5’-GAAGGAGATATAAGGCCTCACCATCACCATCATCACCACCACTGAG-3’ and 5’-CTCAGTGGTGGTGATGATGGTGATGGTGAGGCCTTATATCTCCTTC-3’, and pJL-H10 : 5’-GAAGGAGATATAAGGCCTCACCATCACCATCATCACCACCACCACCACTGAG-3’ and 5’-CTCAGTGGTGGTGGTGGTGATGATGGTGATGGTGAGGCCTTATATCTCCTTC-3’. All constructs were confirmed by DNA sequencing.
Parallel Cloning and Expression of Tsr Receptor in pJL with Different His-tag Lengths
LIC ready vector stocks were made by digesting each pJL vector with StuI and then purified by agarose gel elecrophoresis. The purified excised vectors were made LIC ready by treating with T4 DNA polymerase with dCTP. The LIC ready vectors were stored at −20°C until use. E. coli serine chemotaxis receptor (Tsr) was amplified from DH5α genomic DNA using Deep Vent DNA polymerase and primers 5’- GAAGGAGATATAAGGATGTTAAAACGTATCAAAATTGTGACC-3’ and 5’-ATGATGGTGATGGTGAAATGTTTCCCAGTTCTCCTCG-3’. In these primers, 5’-GAAGGAGATATAAGG-3’ is the N-terminal annealing sequence and 5’-ATGATGGTGATGGTG-3’ is the C-terminal annealing sequence for the LIC reaction. The C-terminal annealing sequence is compatible with all three pJL-H6, pJL-H8, and pJL-H10 vectors to form hexa-, octa-, and deca- histidine tags, respectively. For cloning hepta-, nona-, undeca- histidine tags fused Tsr, Tsr was amplified using 5’-ATGGTGATGGTGGTGAAATGTTTCCCAGTTCTCCTCG-3’ as the C-terminal primer where 5’-ATGGTGATGGTGGTG-3’ is the insert linker sequence. Purified PCR products were treated with T4 DNA polymerase and dGTP, and mixed with LIC ready vector at a 1 to 2 ratio (vol/vol) of vector to insert (estimated 20 ng to 40 ng, respectively) before transformation into MachT1 DH5α cells. A single colony was picked, plasmid purified by miniprep, and sequence confirmed. Expression level was tested using Studier’s auto-inducing media [16]. Briefly, pJL clones and the native pJL vector (negative control) were transformed into BL21(DE3) Star cells, plated onto a Medium phosphate, aspartate (D) containing 17 Amino acid supplemented, Glucose containing (MDAG) plate with Carbenicillin. A single colony was inoculated into 2 mL ZYM-5052 autoinduction media. The culture was incubated at 37 °C overnight with shaking at 200 RPM. Ten µL of the overnight culture was mixed with 3X Laemmli sample buffer, boiled for 5 minutes, then run on 4–20% Tris-HCl SDS-PAGE gel. The gel was visualized using 0.12% Coomassie Blue R-250 stain. Expression levels per liter were estimated by determining the concentration of the 60 kDa presumptive Tsr expression band by comparison to the 75 kDa molecular marker band (which is estimated to be 1 ug). Expression levels per gram of wet weight cells were estimated by converting the expression level per liter using the conversion factor of 8 grams per liter of wet cells. This factor is empirically determined by harvesting about a dozen liters of ZYM-5052 expressed pET21(a) Tsr.
Immobilized Nickel Ion Affinity Chromatography of pJL Tsr with Different His-tag Lengths
Each of the pJL Tsr with different His-tag lengths were grown in 2 L ZYM-5052, harvested by centrifugation and stored in −80 °C until use. Frozen cell pastes were resuspended in an ice cold buffer (20 mM Tris pH 8, 0.1 M NaCl, 1 mM PMSF), and cells were disrupted by a microfluidizer (Microfluidics, Newton, MA). The detergent n-dodecyl-beta-D-maltopyranoside (DDM) was directly added to the cell lysate (1% w/v final concentration), and stirred for a half hour to extract membrane proteins, including Tsr. The detergent extracted cell lysate was cleared by centrifugation at 100,000 g. The imidazole concentration of cleared lysate was raised to 40 mM before loading into a Histrap 5 mL column equilibrated with an equilibration buffer (20 mM Tris pH 8, 0.1 M NaCl, 40 mM imidazole and 0.02% w/v DDM). The column was washed with 70 mL of an equilibration buffer before elution. Tsr was eluted in 40 to 300 mM imidazole gradient in 50 mL elution volume while collecting 3 mL fractions. Protein concentrations of eluates from UV 280 nm absorbance peak fractions were determined by Bradford protein assay, and sample fraction purities were visualized by running peak fractions in 12% Tris-HCl SDS-PAGE gel.
Results and Discussion
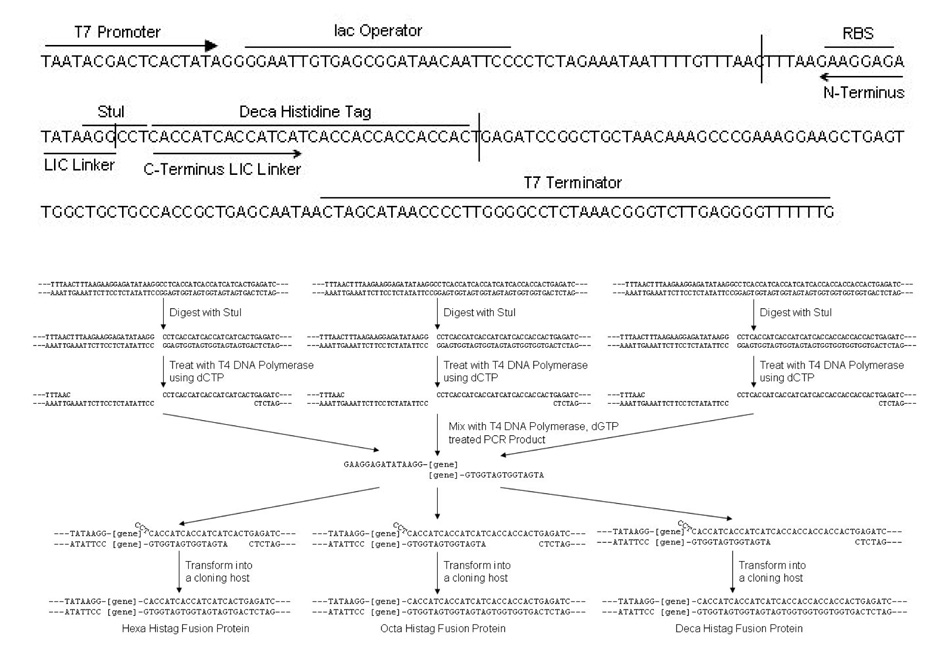
pL, the parent of pJL, was intended to facilitate the high-throughput cloning, expression, and purification of transmembrane proteins with a C-terminal His-tag without any unwanted cloning sequence in the open-reading frame (ORF) in the cloned gene. We realized that pL could be further improved by allowing one insert to generate different length variations of His-tag fusions (Figure 1) for optimizing purification of difficult protein targets that do not or weakly bind to the IMAC resins as demonstrated by many researchers [11–15]. For pJL, the N-terminal annealing sequence for the insert is 5’-GAAGGAGATATAAGG-3’ and the cloner must include the start codon for the gene to be translated. The C-terminal annealing sequence for the insert (5’-ATGATGGTGATGGTG-3’) generates a hexa-(pJL-H6), octa (pJL-H8), and deca (pJL-H10) histidine tag fusion protein. LIC ready pJL vectors can be prepared by excising with StuI, and treating with T4 DNA polymerase with dCTP. LIC ready inserts can be prepared by treating with T4 DNA polymerase with dGTP.
Figure 1. Cloning Site of pJL and the LIC Reaction Scheme.
pJL was derived from pL by changing the MCS by making three different variations to accommodate three different His-tag lengths. These vector variations are made to allow simultaneous cloning of proteins with three different His-tag lengths. The length of the His-tag can be further modified by inserting or removing histidine codons in the PCR primers of the C-terminus LIC linker.
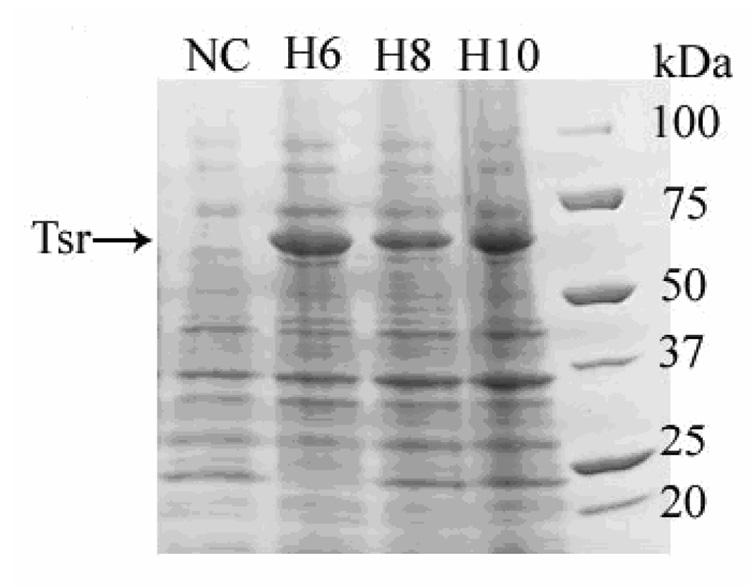
Cloning E. coli serine chemotaxis receptor (Tsr) yielded all three constructs with the correct length of His-tags (Data not shown). This demonstrates successful cloning into pJL. Furthermore, by inserting a single extra histidine before the linker sequence allows cloning of hepta-, nona-, and undeca- histidine tags fused proteins. On the other hand, by inserting the stop codon before the linker sequence prevents translation of the His-tag demonstrating complete His-tag length control. pJL also allows protein translation rate control by changing the length of the spacer between RBS and the start codon [18] by shortening or lengthening the 3’ end of the insert’s N-terminal LIC linker. Testing expression in BL21(DE3) Star in ZYM autoinduction media showed clear protein expression at the correct molecular weight for E. coli serine chemotaxis receptor (Figure 2). Expression level reached up to 200 mg/L or 25 mg/g of wet cells.
Figure 2. SDS-PAGE Gel of Expression Testing of pJL Clones in BL21(DE3) Star using ZYM autoinducing media.
Expression of E. coli serine chemotaxis receptor (Tsr) in pJL vectors. 4–20% SDS-PAGE stained in Coomassie R-250 stain: Lane NC: Negative control, empty pJL-H6 vector; Lane H6: pJL-H6.Tsr; Lane H8: pJL-H8.Tsr; Lane H10: pJL-H10.Tsr. Tsr is a 59 kDa protein. All samples were grown in 2 mL ZYM-5052 media at 37°C, 200 RPM with overnight shaking (~16 hours). Estimated level of expressions are Tsr H6 – 200 mg/L or 25 mg/g of wet cells, Tsr H8 – 100 mg/L or 13 mg/g of wet cells, and Tsr H10 – 200 mg/L or 25 mg/g of wet cells.
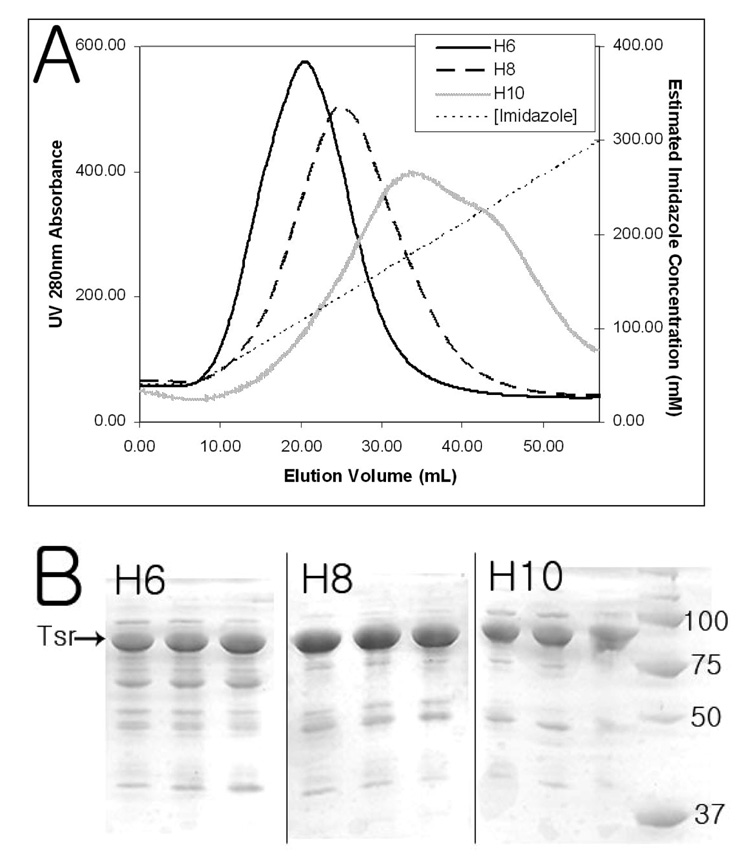
Immobilized nickel ion affinity chromatography of Tsr with six, eight, and ten histidine tag demonstrates an increase in the binding affinity as the length of the His-tag increases (Figure 3A). The elution peak shifts toward a higher imidazole concentration as the His-tag length increases. The elution peak also broadens as it shifts to longer His-tag lengths. The purity of the UV 280 nm maximum fractions, determined by a SDS-PAGE gel, indicates a small increase in the purity as the length of the His-tag increases. This indicates a possibility that impurities interact with Tsr. These interactions may be disrupted at very high salt concentration because an increase in the NaCl concentration to 2 M after lysis and during IMAC raises the purity of Tsr significantly (Data not yet published). However, the clear correlation between the binding affinity and His-tag length demonstrates the utility of increasing the His-tag length for protein targets that elute during the column loading and washing.
Figure 3. IMAC of pJL. Tsr with Different His-tag Lengths.
A) UV 280 nm absorbance profile of different His-tag length Tsr elution. H6 – six histidine tag; H8 – eight histidine tag; H10 – ten histidine tag; [Imidazole] – estimated imidazole concentration in mM (corresponds to the right Y-axis scale). The UV absorbance of H10 was scaled up by 1.7X to match integrated peak areas of H6 and H8 for easier comparison of peak height and broadness. As the length of a His-tag increases, the eluting imidazole concentration and broadness of elution peak increases. UV absorbance maxima corresponds to 110 mM (H6), 133 mM (H8), and 180 mM (H10) imidazole.
B) 12% SDS-PAGE gel of peak fractions. Each lane corresponds to 3 mL fractions collected at UV absorbance peaks which corresponds to the elution volume, H6 – Lane 1 :12 to 15 mL, Lane 2 : 15 to 18 mL, Lane 3 : 18 to 21 mL; H8 – Lane 1 : 21 to 24 mL, Lane 2 : 24 to 27 mL, Lane 3 : 27 to 30 mL; H10 – Lane 1 : 30 to 33 mL, Lane 2 : 33 to 36 mL, Lane 3 : 36 to 39 mL. Eight micrograms of a total protein, determined by Bradford assay, were loaded on each lane except the last lane (marker lane). The overall purity increases slightly as the length of a His-tag increases indicating a possibility that impurities interact with Tsr.
Conclusion
We have successfully cloned and tested target expression with variable His-tag lengths in a bacterial T7 expression system. The vector pJL is derived from pET21a to accommodate LIC ligation and cloning to increase the throughput while allowing researchers to generate expression clones without any unwanted extra sequences and to control the length of the C-terminal His-tag using a single LIC ready insert. Expression of various clones in BL21(DE3) Star in ZYM autoinduction media confirmed the expression of correct MW proteins with identical expression levels as when cloned into pET21a. The immobilized nickel ion affinity chromatography of a membrane protein serine chemotaxis receptor demonstrates the clear correlation of the His-tag length and binding affinity with a slight increase in the purity. pJL will be a good vector for optimizing the purification of difficult protein targets that shows a weak binding affinity to IMAC columns.
Acknowledgments
We thank the UC Berkeley DNA Sequencing Facility for sequencing analysis and Drs. In-Geol Choi, Jose Henrique Pereira, and Rosalind Kim for their advice. This work was supported by National Institute of Health GM 62412.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Porath J, Carlsson J, Olsson I, Belfrage G. Metal chelate affinity chromatography, a new approach to protein fractionation. Nature. 1975;258:598–599. doi: 10.1038/258598a0. [DOI] [PubMed] [Google Scholar]
- 2.Porath J. Immobilized metal ion affinity chromatography. Protein Expr. Purif. 1992;3:263–281. doi: 10.1016/1046-5928(92)90001-d. [DOI] [PubMed] [Google Scholar]
- 3.Gutierrez R, del Valle E, Galan M. Immobilized metal-ion affinity chromatography: status and trends. Separation. Purif. Rev. 2007;36:71–111. [Google Scholar]
- 4.Gaberc-Porekar V, Menart V. Potential for using histidine tags in purification of proteins at large scale. Chem. Eng. Tech. 2005;28:1306–1314. [Google Scholar]
- 5.Changa G. Twenty-five years of immobilized metal ion affinity chromatograph: past, present and future. Jour. Biochem. Biophys. Met. 2001;49:313–334. doi: 10.1016/s0165-022x(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 6.Jungbauer A, Kaar W, Schlegl R. Folding and refolding of proteins in chromatographic beds. Curr. Opin. Biotech. 2004;15:487–494. doi: 10.1016/j.copbio.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Schauer S, Luer C, Moser J. Large scale production of biologically active Escherichia coli glutamyl-tRNA reductase from inclusion bodies. Protein Expr. Purif. 2003;31:271–275. doi: 10.1016/s1046-5928(03)00184-0. [DOI] [PubMed] [Google Scholar]
- 8.Glynou K, Ioannou PC, Christopoulos TK. One-step purification and refolding of recombinant photoprotein aequorin by immobilized metal-ion affinity chromatography. Protein Expr. Purif. 2003;27:384–390. doi: 10.1016/s1046-5928(02)00614-9. [DOI] [PubMed] [Google Scholar]
- 9.Lemercier G, Bakalara N, Santarelli X. On-column refolding of an insoluble histidine tag recombinant exopolyphosphatase from Trypanosoma brucei overexpressed in Escherichia coli. J Chrom. B Anal. Tech. Bio. Life Sci. 2003;786:305–309. doi: 10.1016/s1570-0232(02)00745-6. [DOI] [PubMed] [Google Scholar]
- 10.Oganesyan N, Kim S-H, Kim R. On-column protein refolding for crystallization. J. Struct. Funct. Genomics. 2005;6:177–182. doi: 10.1007/s10969-005-2827-3. [DOI] [PubMed] [Google Scholar]
- 11.Grisshammer R, White J, Trinh L, Shiloach J. Large-scale Expression and Purification of a G-protein-coupled Receptor for Structure Determination – An Overview. J. Struct. Func. Genomics. 2005;6:159–153. doi: 10.1007/s10969-005-1917-6. [DOI] [PubMed] [Google Scholar]
- 12.Yeliseev A, Wong K, Soubias O, Gawrisch K. Expression of human peripheral cannabinoid receptor for structural studies. Protein Sci. 2005;14:2638–2653. doi: 10.1110/ps.051550305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohnaty A, Wiener M. Membrane protein expression and production: effects of polyhistidine tag length and position. Protein Expr. Purif. 2004;33:311–325. doi: 10.1016/j.pep.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Rattenholl A, Pappano W, Koch M, Keene D, Kadler K, Sasaki T, Timpl R, Burgeson R, Greenspan D, Bruckner-Tuderman L. Proteinases of the bond morphogenetic protein-1 family convert procollagen VII to mature anchoring fibril collagen. J. Biol. Chem. 2002;277:26372–26378. doi: 10.1074/jbc.M203247200. [DOI] [PubMed] [Google Scholar]
- 15.Kim S, Sharma S, Hoskins J, Wickner S. Interaction of the DnaK and DnaJ chaperone system with a native substrate P1 RepA. J. Biol. Chem. 2002;277:44778–44783. doi: 10.1074/jbc.M206176200. [DOI] [PubMed] [Google Scholar]
- 16.Studier F. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41(1):207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 17.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 18.Ringquist S, Shinedling S, Barrick D, Green L, Binkley J, Stormo G, Gold L. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol. Microbio. 1992;6:1219–1229. doi: 10.1111/j.1365-2958.1992.tb01561.x. [DOI] [PubMed] [Google Scholar]