Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli (original) (raw)
Abstract
Peptidoglycan is a cell-wall glycopeptide polymer that protects bacteria from osmotic lysis. Whereas in Gram-positive bacteria it also serves as scaffold for many virulence factors, in Gram-negative bacteria, peptidoglycan is an anchor for the outer membrane. For years, we have known the enzymes required for the biosynthesis of peptidoglycan; what was missing was the flippase that translocates the lipid-anchored precursors across the cytoplasmic membrane before their polymerization into mature peptidoglycan. Using a reductionist bioinformatics approach, I have identified the essential inner-membrane protein MviN (renamed MurJ) as a likely candidate for the peptidoglycan flippase in Escherichia coli. Here, I present genetic and biochemical data that confirm the requirement of MurJ for peptidoglycan biosynthesis and that are in agreement with a role of MurJ as a flippase. Because of its essential nature, MurJ could serve as a target in the continuing search for antimicrobial compounds.
Keywords: endosymbiont, genome, lysis, murein, reductionist
Almost all bacteria possess an extracytoplasmic glycopeptide polymer known as peptidoglycan (or murein) that is composed of glycan chains connected by peptide bridges (1–3). This mesh-like rigid structure protects bacteria from lysis caused by osmotic pressure and determines their shape. In addition, peptidoglycan serves as an anchor for virulence factors and cellular structures (4). For these reasons, and the fact that peptidoglycan is absent in humans but present in most bacteria, its biogenesis has been one of the preferred targets for antibiotics (5–7).
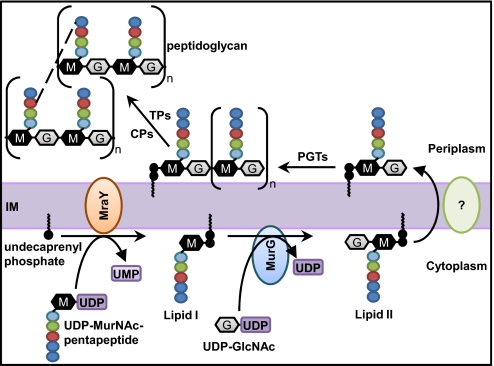
Biogenesis of peptidoglycan begins in the cytoplasm and culminates in the extracytoplasmic environment, where assembly and cross-linking of glycan chains occurs. Therefore, this process requires the transport of intermediates across the inner (or cytoplasmic) membrane (IM). In Escherichia coli, peptidoglycan biogenesis starts with the synthesis of the nucleotide precursors in the cytoplasm (Fig. 1) (8). The GlmSMU enzymes synthesize UDP-N-acetylglucosamine (UDP-GlcNAc), which is then transformed in step-wise fashion to UDP-N-acetylmuramic acid-pentapeptide (UDP-MurNAc-pentapeptide) by MurABCDEF. At the IM, MraY transfers the MurNAc-pentapeptide to the C55-lipid carrier undecaprenyl phosphate to form the undecaprenyl pyrophosphoryl-MurNAc-pentapeptide known as lipid I (1, 2). GlcNAc is next added to lipid I by the IM-associated MurG enzyme, resulting in lipid II (1, 2). Lipid II is then flipped across the IM by an unknown mechanism that involves an unidentified protein(s) (2, 9). Once at the periplasmic side of the IM, the penicillin-binding proteins (PBPs) use lipid II to catalyze the maturation of peptidoglycan into a mesh-like three-dimensional structure (3, 10).
Fig. 1.
Biogenesis of peptidoglycan in E. coli. After being synthesized by MurABCDEF in the cytoplasm, UDP-MurNAc-pentapeptide is transferred to undecaprenyl phosphate by MraY, generating lipid I. Next, MurG synthesizes lipid II by adding GlcNAc (from UDP-GlcNAc) to lipid I. Lipid II is translocated across the IM by an unknown flippase. At the periplasmic side of the IM, glycosyltransferases (PGTs) assemble and elongate glycan chains, and transpeptidases (TPs) cross-link their peptide chains (dashed lines). DD-carboxypeptidases (CPs) remove terminal D-Ala from the stem pentapeptide in mature peptidoglycan.
Maturation of peptidoglycan involves the assembly and elongation of the glycan chains by glycosyltransferases, cross-linking of their peptide chains by transpeptidases, and removal of the terminal D-Ala from the stem pentapeptide by DD-carboxypeptidases (Fig. 1) (3, 10). The peptidoglycan glycosyltransferases use lipid II to assemble and elongate the glycan chains. During this processive chain-elongation step, undecaprenyl pyrophosphate is released from the growing glycan chain as new lipid II precursors are added (11); the lipid carrier can then be recycled in a process that is not well understood but that is known to involve dephosphorylation by multiple phosphatases and transport across the IM (1, 12). In E. coli, recycled and newly synthesized undecaprenyl phosphate can be used in new rounds of peptidoglycan biosynthesis as well as in the transport across the IM of other cell envelope polysaccharides, such as lipopolysaccharides (LPS) and enterobacterial common antigen (ECA) (13, 14). Thus, although we do not understand how undecaprenyl pyrophosphate is flipped back across the IM for recycling, the only unknown factor required for an essential step unique to peptidoglycan biogenesis is the flippase that transports lipid II across the IM.
For decades, a large body of research has elucidated the steps of peptidoglycan biogenesis and the enzymes that catalyze them, except for how lipid II is translocated across the IM. Lipid II flippase is, therefore, the last essential player in peptidoglycan biogenesis that remains to be identified (2, 9). Because the search for the lipid II flippase using standard genetic and biochemical approaches has proven technically difficult and unsuccessful (2), I performed a reductionist bioinformatics search that led me to propose that MviN (renamed MurJ) is the putative lipid II flippase in E. coli.
Results
Bioinformatics Search for the E. coli Lipid II Flippase.
Recently, together with the T. J. Silhavy and D. Kahne laboratories, I searched for essential outer membrane (OM) biogenesis factors in E. coli using a reductionist bioinformatics approach that reduced the number of possible candidates from ≈2,000 proteins of unknown function to 3 (15). All three candidates have been shown to be the missing components of the machinery that transports LPS from the IM to the OM (15, 16). A key step of this approach was based on the realization that a factor that is essential for OM biogenesis in E. coli would be conserved in Gram-negative endosymbiotic bacteria despite these organisms having very small genomes. Even though evolution of endosymbionts is marked by extensive gene loss, their genomes should still encode all of the machinery required for building an OM (15).
Here, I applied a similar reductionist approach to identify the lipid II flippase of E. coli. If E. coli has a protein that is required for flipping peptidoglycan lipid II across the IM, this factor would be: (i) an IM protein, (ii) conserved among Gram-negative endosymbiotic bacteria that possess peptidoglycan, (iii) absent in bacteria that lack peptidoglycan, and (iv) essential in E. coli, unless there are two or more proteins that can perform this function. To conduct this search, I obtained the list of 963 predicted IM proteins (integral, bitopic, and lipoproteins) from EchoLOCATION (17). Next, I determined which of these 963 proteins are present in the proteomes of the two endosymbiotic bacteria Blochmannia floridanus and Buchnera aphidicola str. APS. I chose these endosymbionts because both are closely related to E. coli but their proteomes are ≈86% smaller than E. coli's; nevertheless, proteomes of both endosymbionts contain enzymes required for peptidoglycan biosynthesis, such as MraY and MurG. However, the cell envelopes of these endosymbionts are not identical, because the proteome of Bl. floridanus contains the proteins required for LPS synthesis and transport, whereas that of B. aphidicola str. APS does not (15, 18).
Using GeneVenn (19), I found that 854 of the 963 predicted IM proteins of E. coli are absent in both endosymbionts. Reflective of the intracellular lifestyle of these endosymbionts, this large subset of IM proteins include many known nutrient transporters, proteins involved in sensing the environment, such as histidine kinases, and factors involved in the biogenesis of surface polysaccharides, such as the _O_-antigen of LPS and ECA (data not shown).
This analysis also revealed that only 39 E. coli IM proteins are present in both Bl. floridanus and B. aphidicola str. APS. As expected, these 39 proteins include factors required for essential processes such as the insertion of IM proteins into the IM, protein transport across the IM, processing of signal sequences, transport of lipoproteins from the IM to the OM, peptidoglycan biosynthesis, and cell division [supporting information (SI) Table S1]. The essential IM protease FtsH and its regulators HflCK are also conserved as well as proteins involved in generating energy, phosphate transport, and cardiolipin biosynthesis (Table S1). More importantly, there are six proteins of unknown function (MviN, YajR, YhgN, YibN, YoaE, and YtfN) among the 39 conserved proteins that could be candidates for the peptidoglycan lipid II flippase (Table S1). I must clarify that the function of MviN is unknown; mviN (or mviS) was misnamed as a gene encoding a factor required for _m_ouse _vi_rulence, but it was later found that the mutation responsible for the virulence phenotype maps to nearby genes responsible for flagellum biogenesis (20). Therefore, because the Mvi nomenclature is a misnomer and for reasons described below, I propose to rename this factor MurJ.
Because a lipid II flippase should be absent in bacteria that lack peptidoglycan, I searched for conservation of these six proteins of unknown function among bacteria that do not produce peptidoglycan. I conducted Genomic BLAST (21) searches of MurJ (MviN), YajR, YhgN, YibN, YoaE, and YtfN in the proteomes of Mollicutes, Dehalococcoides, Candidatus Carsonella ruddii PV, and B. aphidicola str. Cc, bacteria known to lack peptidoglycan (22–24). Notably, B. aphidicola str. Cc belongs to the same species as the peptidoglycan producer B. aphidicola str. APS, and both are members of the Enterobacteriaceae family to which E. coli belongs. As controls for conservation among peptidoglycan producers, I included in the Genomic BLAST searches proteomes of two endosymbionts that belong to the γ_-Proteobacteria_ class, B. aphidicola str. APS and Candidatus Vesicomyosocius okutanii HA.
These Genomic BLAST analyses revealed that only MurJ was conserved among the two peptidoglycan-producing endosymbionts but absent in those aforementioned bacteria that lack peptidoglycan (Table 1). Interestingly, the cell division protein FtsW, a RodA homolog, has been postulated as a lipid II flippase candidate (25). However, Genomic BLAST searches revealed that the peptidoglycan producer V. okutanii lacks FtsW (but has RodA), whereas the peptidoglycan-less Mollicute Eubacterium dolichum DSM 3991 has two FtsW homologs, one of which is likely RodA (data not shown). Together, these data suggest that MurJ is the likely candidate for lipid II flippase in E. coli.
Table 1.
Conservation of proteins of unknown function in proteomes of bacteria that either lack or produce peptidoglycan (PG)
| Protein | Lack PG | Produce PG | ||||
|---|---|---|---|---|---|---|
| Mollicutes* | D. ethenogenes | B. aphidicola str. Cc | C. ruddii PV | B. aphidicola str. APS | V. okutanii HA | |
| MurJ | –† | – | – | – | 5.00e−126 | 4.00e−94 |
| YajR | – | 3.00e−05 | – | – | 1.00e−79 | 2.00e−73 |
| YhgN | – | 4.00e−16 | – | – | 9.00e−61 | – |
| YibN | 3.00e−04 | 1.00e−05 | 1.00e−03 | – | 1.00e−19 | 3.00e−07 |
| YoaE | 6.00e−14 | 5.00e−17 | 1.00e−73 | 1.00e−73 | 1.00e−173 | 3.00e−17 |
| YtfN | – | – | – | – | 1.00e−39 | – |
Functional Predictions Support MurJ (MviN) as the E. coli Lipid II Flippase Candidate.
The bioinformatics search described above identified MurJ as the only putative lipid II flippase in E. coli. Additional findings support this hypothesis. First, searches for predicted functional partners of MurJ, YajR, YhgN, YibN, YoaE, and YtfN using the STRING database (26) revealed that MurJ is the only of these six proteins predicted to be a functional partner of several proteins involved in peptidoglycan biosynthesis (Table S2). Second, if MurJ was the only lipid II flippase candidate in E. coli, it should be essential; indeed, attempts to delete murJ in E. coli, Burkholderia pseudomallei, and Sinorhizobium meliloti have failed (27–29). Third, by using ParAlign (30), MurJ homologs can be found in the archaeal Methanobacteriales and Methanosarcinales, which produce peptidoglycan-like cell-wall polymers whose biogenesis includes undecaprenyl-linked lipid intermediates (31). Likewise, MurJ homologs are also present in the plastid of the amoeba Paulinella chromatophora and in the moss Physcomitrella patens, both of which produce peptidoglycan (32, 33). Finally, MurJ belongs to the multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily (34). MurJ is the single member of the mouse virulence family (MVF) family, one of the four families that compose the MOP exporter superfamily (34). The MVF family has the greatest sequence similarity to members of the polysaccharide transporter (PST) family, which include WzxE, the flippase of the undecaprenyl-phosphate-linked saccharide precursor of ECA and WzxB (or RfbX), the LPS _O_-antigen flippase (34–36). In addition, the MOP exporter superfamily also encompasses the oligosaccharidyl-lipid flippase (OLF) family, which includes Rtf1p, a protein that was proposed to flip dolichyl diphosphate-GlcNAc2Man5 across the endoplasmic reticulum (37). Consequently, it had been proposed that members of the MVF family might export complex carbohydrates across the IM (34). Independently, Pfam classifies MurJ in the MviN_MATE clan (CL0222), which consists of four protein families that correspond to those members of the MOP exporter superfamily (38).
MurJ Is Essential in E. coli.
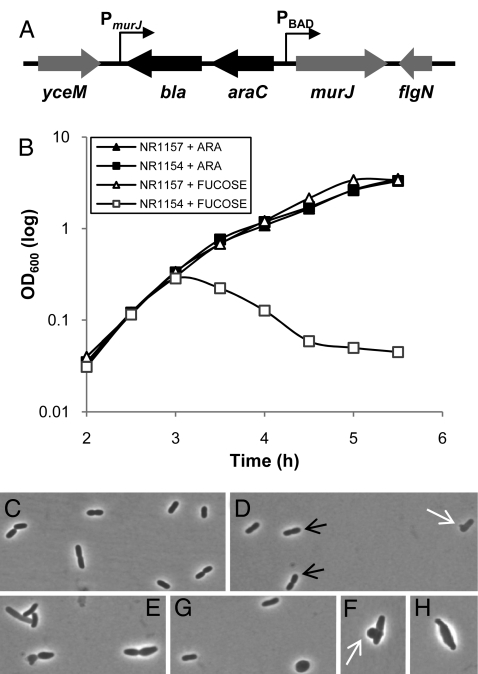
If MurJ is the peptidoglycan lipid II flippase in E. coli, as the compelling bioinformatic evidence provided above indicates, then two predictions can be made. MurJ should be an essential protein, and abolishing MurJ function should inhibit peptidoglycan biogenesis. Because attempts to delete murJ had failed (27), I built a MurJ-depletion strain where murJ expression is under the control of an inducible promoter. Specifically, I inserted the arabinose-inducible PBAD promoter 14 bp upstream of the murJ start codon, decoupling murJ from its own promoter (Fig. 2A). The growth of such a MurJ-depletion strain is dependent on the presence of the inducer arabinose in the growth medium (Fig. 2B), demonstrating that MurJ is essential in E. coli.
Fig. 2.
MurJ is essential. (A) Chromosomal organization of the MurJ-depletion strain. The PBAD promoter, araC and bla were inserted 14 bp upstream of the murJ start codon so that murJ expression is under the control of PBAD and decoupled from its native PmurJ promoter. (B) Growth (measured by OD600) of depletion strain NR1154 (MC4100 ara+ Δ_lysA_::kan murJ Ω(−14::bla araC PBAD) depends on the presence of arabinose in the medium. Addition of d-fucose, a nonmetabolizable analog of l-arabinose, causes cell lysis of NR1154 but not the MurJ+ parent strain NR1157 (MC4100 ara+ Δ_lysA_::kan). (C–H) Cell morphology (under magnification ×100 phase objective) of wild-type (NR1157, C) and MurJ-depletion (NR1154, D–H) strains grown in the presence of fucose. During MurJ depletion, whereas some cells appear wild type (black arrows), many are larger, irregularly shaped, and form large blebs (white arrows).
MurJ Is Required for Peptidoglycan Biogenesis in E. coli.
Peptidoglycan protects bacteria from lysis caused by osmotic pressure and determines their shape (1–3). Thus, if MurJ was required for peptidoglycan biogenesis, MurJ depletion would cause cell-shape defects and eventually cell lysis. Indeed, in the absence of arabinose, cell density of the MurJ-depletion strain decreases (Fig. 2B), indicative of cell lysis. In addition, during MurJ depletion, cell morphology becomes heterogeneous (Fig. 2 C–H). Cells can be irregularly shaped and larger, and they form blebs large enough to be observed by light microscopy (Fig. 2 D–H). Eventually, cells lyse, as evidenced by the appearance of irregularly shaped bacterial ghost envelopes (data not shown). However, because E. coli cells cannot be synchronized, cultures of this strain contain cells at various stages of depletion at any given time, so the severity of these phenotypes varies from cell to cell (Fig. 2 D–H).
When sucrose is present in the growth medium as an osmotic stabilizer to prevent cell lysis during depletion, MurJ-depleted cells become round, and they can reach higher cell densities than in the absence of sucrose (data not shown). If sucrose is removed from such cultures, the round MurJ-depleted cells immediately lyse (data not shown). Moreover, although sucrose can prevent lysis, the MurJ-depletion strain cannot form colonies on plates lacking arabinose even if sucrose is provided. Thus, as MurJ is gradually depleted, cell lysis can be prevented by adding sucrose to the growing media, and growth can be maintained temporarily. However, there is a threshold of MurJ level below which growth cannot be maintained even in the presence of sucrose. This is consistent with the recent finding that L-form-like E. coli induced by antibiotics requires low levels of peptidoglycan synthesis for growth (39).
To further demonstrate that MurJ depletion inhibits peptidoglycan biogenesis, I compared the amount of incorporation of 3H- diaminopimelic acid (DAP) into mature peptidoglycan (i.e., sacculus) in equivalent amount of cells from wild-type and MurJ-depleted cultures grown in the absence of arabinose. After four generations of growth in the absence of arabinose, the levels of 3H-DAP incorporation into mature peptidoglycan in the MurJ-depleted cultures were similar (112.0 ± 7.6%) to those in the wild-type strain. However, two to three generations later, even before cell density declined, the MurJ-depletion strain incorporated 70.3 ± 3.9% less 3H-DAP into mature peptidoglycan than wild type, demonstrating that MurJ is required for peptidoglycan biosynthesis.
As indicated above, MurJ-depleted cultures synthesize ≈30% of the amount of mature peptidoglycan that wild-type cultures do. Next, I analyzed the distribution of 3H-DAP incorporated among mature peptidoglycan and its nucleotide and lipid precursors in wild-type and MurJ-depleted cultures. These analyses revealed that depletion of MurJ causes not only the decrease in incorporation into mature peptidoglycan but also the accumulation of both nucleotide and lipid precursors (Table 2). Inhibition of the lipid II synthase MurG (Fig. 1) leads to both the accumulation of peptidoglycan nucleotide precursors and only a limited accumulation of the lipid precursor lipid I, likely because of the fact that MraY is believed to be reversible and because of the low availability of the undecaprenyl-phosphate lipid carrier (40). Because the lipid carrier is thought to be limiting in the cell (40), it is possible that inhibition of lipid II translocation across the IM could also result in the accumulation of nucleotide precursors, as reported here. Together, the data presented here show that MurJ catalyzes a step in peptidoglycan biosynthesis before polymerization but downstream, at least, of lipid I synthesis. Because the only missing step between lipid I synthesis and peptidoglycan polymerization is the translocation of lipid II across the IM, these data are consistent with a role for MurJ as the lipid II flippase in E. coli.
Table 2.
Distribution of 3H-DAP in wild-type and MurJ -depletion strains
| Species | Wild type* | MurJ depletion |
|---|---|---|
| PG | 85.2 ± 3.8 (12,054 ± 905) | 60.5 ± 5.5 (3,589 ± 582) |
| Nucleotides | 12.3 ± 3.9 (1,745 ± 451) | 31.3 ± 1.6 (1,838 ± 132) |
| Lipids | 2.4 ± 0.2 (351 ± 36) | 8.1 ± 3.9 (460 ± 132) |
Discussion
The reactions necessary for the biogenesis of peptidoglycan have been known for decades. The enzymes required for these reactions have all been identified. What has been missing is the factor that translocates the lipid II intermediate across the IM. Van Dam et al. (9) determined that this translocation step requires a protein(s) whose identity has long remained elusive. Using a reductionist bioinformatics approach, I accumulated compelling bioinformatics evidence suggesting that MurJ is the missing lipid II flippase. Genetic and biochemical data demonstrate that MurJ is indeed an essential E. coli protein required for peptidoglycan biosynthesis. Although additional biochemical studies are needed to demonstrate its lipid II flippase activity, the data presented here are in agreement with MurJ's performing such a function.
The reductionist bioinformatics approach of using a comparative analysis between the proteomes of E. coli and closely related endosymbionts has successfully identified four proteins that are essential for the biogenesis of the cell envelope of E. coli (15). The great diversity among bacteria and the increasing number of sequenced genomes offer an opportunity to conduct similar comparative analyses to identify missing components of known pathways. However, although bioinformatics has been instrumental in the identification of MurJ as the potential lipid II flippase, it does not provide insight into how MurJ might act as a flippase. For example, it does not provide information as to what energy source might drive MurJ activity or whether export of lipid II might be coupled to the import of any molecule across the IM. The fact that depletion of MurJ only results in a low level of accumulation of lipid intermediates could be the result of MurJ's also being involved in the recycling of undecaprenyl phosphate.
To understand MurJ activity, biochemical assays are needed. Because those available for flippase activity are difficult and controversial (37, 41), the identification of MurJ will hopefully aid in the development of better methods to measure flippase activity. This is an especially important goal for MurJ, because it represents an undeveloped target for broad-spectrum antibacterial agents.
Materials and Methods
Bacterial Strains.
Strains were derived from NR754, an araD+ revertant of MC4100 (15, 42). Alleles were transferred by using P1 transduction (43). A MurJ-depletion strain was constructed by recombineering as follows. Primers 5mviNPbad (5′-_CATAATCCAGGTAGAC TATTCGCCTCTTTCAGCGCCTGCCTTGCAGGCGTTTTGCCCGTGGGTCT_GTATATATGAGTAAACTTGGTCTG) and 3mviNPbad (5′-_CACGCGAAAACATGGTCATCGAGCTGA CGGCGGCCAGCGATTTTAATAAATTCATCGGTGTTCTAATCC_TCCTTAGAGCTCGAATTCCC) were used to amplify the _bla-araC_-PBAD region of pKD46 (44). The ends of the resulting PCR product are identical (italicized portion of primers) to the chromosomal region upstream of murJ. This PCR product was used for recombineering to insert bla-araC_-PBAD in the chromosome of DY378 (45) 14 bp upstream of the murJ start codon. The resulting strain (NR1151) was selected at 30°C on media containing ampicillin and arabinose and confirmed by PCR analysis. The murJ Ω(−14::bla araC PBAD) insertion of NR1151 was introduced into NR574 to construct NR1152 by using P1 transduction (43) by selecting ampicillin-resistant transductants in the presence of arabinose. To label with 3H-DAP, NR1157 and NR1154 were constructed by introducing the Δ_lysA::kan allele from the Keio collection (27) into NR754 and NR1152, respectively. The presence or absence of lysA did not affect any of the MurJ-dependent phenotypes.
Growth Conditions.
LB broth and agar were prepared as described (43). When appropriate, ampicillin (25 μg/ml), kanamycin (25 μg/ml), l-arabinose (0.2% wt/vol), d-fucose (0.05% wt/vol), and sucrose (0.23 M) were added. For labeling experiments, 100 μg/ml methionine, lysine, and threonine were added to growth media to reduce the internal pool of DAP (46). For data shown in Fig. 2B, 1 ml of overnight cultures grown with arabinose was pelleted, washed once with LB broth, and resuspended in 1 ml of LB broth. Fresh LB broth with either arabinose or fucose was inoculated with a 1:5,000 dilution of washed cultures and grown under aeration at 37°C. Growth was monitored by optical density at OD600.
Microscopy.
Cells grown in the presence of either arabinose or fucose were placed on a pad of 1% agarose (wt/vol) in M63 minimal medium (43) on a microscope slide and covered with a coverslip. Samples were examined under a Nikon 90i fluorescence microscope (magnification ×100 objective), and images were captured with a Rolera XR Fast 1394 camera (Qimaging).
Labeling with 3H-DAP.
Cells (NR1157 and NR1154) grown overnight in LB broth containing arabinose were washed as indicated above. Fresh LB broth containing fucose, methionine, lysine, and threonine was inoculated with a 1:100 dilution of washed cultures and grown under aeration at 37°C for 2 h to OD600 ≈0.6 (approximately four generations). Then, 1 ml of each culture was labeled as indicated below. At the same time, cells were diluted to OD600 ≈0.1 into a fresh culture containing fucose, methionine, lysine, and threonine and grown until OD600 ≈0.6. At that time, 1 ml of each culture was labeled for 15 min at 37°C with 5 μCi of 3H-DAP (60 Ci/mmol; American Radiolabeled Chemicals). At the end of the labeling period, cells were placed on ice and pelleted at 4°C for 5 min at 16,100 × g. Pellets were resuspended in 7 μl of ice-cold water and immediately frozen in dry ice until they were subjected to paper chromatography.
Cellular Distribution of 3H-DAP.
Labeled cells prepared as above were spotted onto Whatman 3MM paper, and labeled peptidoglycan precursors were separated by ascending chromatography by development with isobutyric acid:1 M NH4OH (5:3; vol/vol) for ≈16 h (47). Paper was dried and cut into 1-cm squares that were counted in vials containing scintillation mixture (Ready Safe; Beckman Coulter). Mature peptidoglycan remained at the origin, whereas nucleotide and lipid precursors ran at Rf values of 0.15–0.25 and 0.8–0.9, respectively (48). For each strain, counts corresponding to those Rf values were added, and their sum represented 100% of incorporated label. The percentage corresponding to each species (mature peptidoglycan, nucleotides, and lipid precursors) was calculated accordingly for each experiment, and data from three independent experiments were used to calculate the average and standard deviation. To compare the mature peptidoglycan synthesis in wild-type and MurJ-depleted cultures, cpm values at the origin were used. Comparison of incorporation between strains was done by calculating the percentage of incorporation in the MurJ-depletion strain with respect to wild type. Data from three independent experiments were used to calculate percentage average and standard deviation.
Proteome-Based Analysis.
For Genomic BLAST searches (21), genome sequences were obtained from GenBank for the following organisms: Blochmannia floridanus (BX248583); Buchnera aphidicola str. APS (BA000003); Aster yellows witches'-broom phytoplasma AYWB (CP000061); Eubacterium dolichum DSM 3991 (ABAW00000000); Mesoplasma florum L 1 (AE017263); Mycoplasma agalactiae PG2 (CU179680); Mycoplasma capricolum subsp. capricolum ATCC 27343 (CP000123); Mycoplasma gallisepticum R (AE015450); Mycoplasma genitalium G37 (L43967); Mycoplasma hyopneumoniae 232 (AE017332); Mycoplasma hyopneumoniae 7448 (AE017244); Mycoplasma hyopneumoniae J (AE017243); Mycoplasma mobile 163K (AE017308); Mycoplasma mycoides subsp_. mycoides_ SC str. PG1 (BX293980); Mycoplasma penetrans HF-2 (BA000026); Mycoplasma pneumoniae M129 (U00089); Mycoplasma pulmonis UAB CTIP (AL445566); Mycoplasma synoviae 53 (AE017245); Onion yellows phytoplasma OY-M (AP006628); Ureaplasma parvum serovar 3 str. ATCC 700970 (AF222894); Dehalococcoides ethenogenes 195 (CP000027); Buchnera aphidicola str. Cc (Cinara cedri) (CP000263); Candidatus Carsonella ruddii PV (AP009180); Candidatus Vesicomyosocius okutanii HA (AP009247). Genomic BLAST searches were conducted by using the BLASTP 2.2.18 program (49). In all searches, protein sequences for MurJ (MviN), YajR, YhgN, YibN, YoaE, and YtfN were obtained from EcoCyc (50).
Supplementary Material
Supporting Information
Acknowledgments.
I thank Thomas Silhavy for his help and support, Daniel Kahne and Juliana Malinverni for their helpful discussions, and Zemer Gitai for the use of his microscope. This work was supported by National Institute of General Medical Sciences Grant GM34821.
Note Added in Proof.
Inoue et al. have recently reported that MviN (MurJ) is involved in peptidoglycan synthesis (51).
Footnotes
The author declares no conflict of interest.
References
- 1.Bouhss A, Trunkfield AE, Bugg TD, Mengin-Lecreulx D. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol Rev. 2008;32:208–233. doi: 10.1111/j.1574-6976.2007.00089.x. [DOI] [PubMed] [Google Scholar]
- 2.van Heijenoort J. Lipid intermediates in the biosynthesis of bacterial peptidoglycan. Microbiol Mol Biol Rev. 2007;71:620–635. doi: 10.1128/MMBR.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 4.Dramsi S, Magnet S, Davison S, Arthur M. Covalent attachment of proteins to peptidoglycan. FEMS Microbiol Rev. 2008;32:307–320. doi: 10.1111/j.1574-6976.2008.00102.x. [DOI] [PubMed] [Google Scholar]
- 5.Koch AL. Bacterial wall as target for attack: Past, present, and future research. Clin Microbiol Rev. 2003;16:673–687. doi: 10.1128/CMR.16.4.673-687.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ostash B, Walker S. Bacterial transglycosylase inhibitors. Curr Opin Chem Biol. 2005;9:459–466. doi: 10.1016/j.cbpa.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 7.Walsh CT. Actions, Origins, Resistance. Washington, DC: Am Soc Microbiol Press; 2003. [Google Scholar]
- 8.Barreteau H, et al. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:168–207. doi: 10.1111/j.1574-6976.2008.00104.x. [DOI] [PubMed] [Google Scholar]
- 9.van Dam V, et al. Transmembrane transport of peptidoglycan precursors across model and bacterial membranes. Mol Microbiol. 2007;64:1105–1114. doi: 10.1111/j.1365-2958.2007.05722.x. [DOI] [PubMed] [Google Scholar]
- 10.Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol Rev. 2006;30:673–691. doi: 10.1111/j.1574-6976.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 11.Perlstein DL, Zhang Y, Wang TS, Kahne DE, Walker S. The direction of glycan chain elongation by peptidoglycan glycosyltransferases. J Am Chem Soc. 2007;129:12674–12675. doi: 10.1021/ja075965y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Ghachi M, Derbise A, Bouhss A, Mengin-Lecreulx D. Identification of multiple genes encoding membrane proteins with undecaprenyl pyrophosphate phosphatase (UppP) activity in Escherichia coli. J Biol Chem. 2005;280:18689–18695. doi: 10.1074/jbc.M412277200. [DOI] [PubMed] [Google Scholar]
- 13.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rick PD, et al. Characterization of the lipid-carrier involved in the synthesis of enterobacterial common antigen (ECA) and identification of a novel phosphoglyceride in a mutant of Salmonella typhimurium defective in ECA synthesis. Glycobiology. 1998;8:557–567. doi: 10.1093/glycob/8.6.557. [DOI] [PubMed] [Google Scholar]
- 15.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci USA. 2008;105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sperandeo P, et al. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J Bacteriol. 2008;190:4460–4469. doi: 10.1128/JB.00270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Misra RV, Horler RS, Reindl W, Goryanin II, Thomas GH. EchoBASE: An integrated post-genomic database for Escherichia coli. Nucleic Acids Res. 2005;33:D329–D333. doi: 10.1093/nar/gki028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 19.Pirooznia M, Nagarajan V, Deng Y. GeneVenn—A web application for comparing gene lists using Venn diagrams. Bioinformation. 2007;1:420–422. doi: 10.6026/97320630001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitt CK, Darnell SC, Tesh VL, Stocker BA, O'Brien AD. Mutation of flgM attenuates virulence of Salmonella typhimurium, and mutation of fliA represses the attenuated phenotype. J Bacteriol. 1994;176:368–377. doi: 10.1128/jb.176.2.368-377.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cummings L, et al. Genomic BLAST: Custom-defined virtual databases for complete and unfinished genomes. FEMS Microbiol Lett. 2002;216:133–138. doi: 10.1111/j.1574-6968.2002.tb11426.x. [DOI] [PubMed] [Google Scholar]
- 22.Galperin MY. The fuzzy border between a cell and an organelle. Environ Microbiol. 2006;8:2062–2067. doi: 10.1111/j.1462-2920.2006.01178.x. [DOI] [PubMed] [Google Scholar]
- 23.Maymo-Gatell X, Chien Y, Gossett JM, Zinder SH. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science. 1997;276:1568–1571. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- 24.Trachtenberg S. Mollicutes. Curr Biol. 2005;15:R483–R484. doi: 10.1016/j.cub.2005.06.049. [DOI] [PubMed] [Google Scholar]
- 25.Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Mering C, et al. STRING 7–recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006. doi: 10.1038/msb4100050. 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ling JM, Moore RA, Surette MG, Woods DE. The mviN homolog in Burkholderia pseudomallei is essential for viability and virulence. Can J Microbiol. 2006;52:831–842. doi: 10.1139/w06-042. [DOI] [PubMed] [Google Scholar]
- 29.Rudnick PA, Arcondeguy T, Kennedy CK, Kahn D. glnD and mviN are genes of an essential operon in Sinorhizobium meliloti. J Bacteriol. 2001;183:2682–2685. doi: 10.1128/JB.183.8.2682-2685.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saebo PE, Andersen SM, Myrseth J, Laerdahl JK, Rognes T. PARALIGN: Rapid and sensitive sequence similarity searches powered by parallel computing technology. Nucleic Acids Res. 2005;33:W535–W539. doi: 10.1093/nar/gki423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kandler O, Konig H. Cell wall polymers in Archaea (Archaebacteria) Cell Mol Life Sci. 1998;54:305–308. doi: 10.1007/s000180050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Machida M, et al. Genes for the peptidoglycan synthesis pathway are essential for chloroplast division in moss. Proc Natl Acad Sci USA. 2006;103:6753–6758. doi: 10.1073/pnas.0510693103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nowack EC, Melkonian M, Glockner G. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr Biol. 2008;18:410–418. doi: 10.1016/j.cub.2008.02.051. [DOI] [PubMed] [Google Scholar]
- 34.Hvorup RN, et al. The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur J Biochem FEBS. 2003;270:799–813. doi: 10.1046/j.1432-1033.2003.03418.x. [DOI] [PubMed] [Google Scholar]
- 35.Liu D, Cole RA, Reeves PR. An O-antigen processing function for Wzx (RfbX): A promising candidate for O-unit flippase. J Bacteriol. 1996;178:2102–2107. doi: 10.1128/jb.178.7.2102-2107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rick PD, et al. Evidence that the wzxE gene of Escherichia coli K-12 encodes a protein involved in the transbilayer movement of a trisaccharide-lipid intermediate in the assembly of enterobacterial common antigen. J Biol Chem. 2003;278:16534–16542. doi: 10.1074/jbc.M301750200. [DOI] [PubMed] [Google Scholar]
- 37.Helenius J, et al. Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 protein. Nature. 2002;415:447–450. doi: 10.1038/415447a. [DOI] [PubMed] [Google Scholar]
- 38.Finn RD, et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joseleau-Petit D, Liebart JC, Ayala JA, D'Ari R. Unstable Escherichia coli L forms revisited: Growth requires peptidoglycan synthesis. J Bacteriol. 2007;189:6512–6520. doi: 10.1128/JB.00273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mengin-Lecreulx D, Texier L, Rousseau M, van Heijenoort J. The murG gene of Escherichia coli codes for the UDP-N-acetylglucosamine: N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase involved in the membrane steps of peptidoglycan synthesis. J Bacteriol. 1991;173:4625–4636. doi: 10.1128/jb.173.15.4625-4636.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Helenius J, et al. Helenius et al. reply. Nature. 2008;454:E4–E5. [Google Scholar]
- 42.Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 43.Silhavy TJ, Berman ML, Enquist LW. Experiments with Gene Fusions. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1984. [Google Scholar]
- 44.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu D, et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wientjes FB, Pas E, Taschner PE, Woldringh CL. Kinetics of uptake and incorporation of meso-diaminopimelic acid in different Escherichia coli strains. J Bacteriol. 1985;164:331–337. doi: 10.1128/jb.164.1.331-337.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lugtenberg EJ, de Haan PG. A simple method for following the fate of alanine-containing components in murein synthesis in Escherichia coli. Antonie van Leeuwenhoek. 1971;37:537–552. doi: 10.1007/BF02218524. [DOI] [PubMed] [Google Scholar]
- 48.Ramey WD, Ishiguro EE. Site of inhibition of peptidoglycan biosynthesis during the stringent response in Escherichia coli. J Bacteriol. 1978;135:71–77. doi: 10.1128/jb.135.1.71-77.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karp PD, Riley M, Paley SM, Pellegrini-Toole A, Krummenacker M. Eco Cyc: Encyclopedia of Escherichia coli genes and metabolism. Nucleic Acids Res. 1999;27:55–58. doi: 10.1093/nar/27.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Inoue, et al. Involvement of an essential gene, mviN, in murein synthesis in Escherichia coli. J Bacteriol. 2008 doi: 10.1128/JB.00551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information