Dysfunction in ankyrin-B-dependent ion channel and transporter targeting causes human sinus node disease (original) (raw)
Abstract
The identification of nearly a dozen ion channel genes involved in the genesis of human atrial and ventricular arrhythmias has been critical for the diagnosis and treatment of fatal cardiovascular diseases. In contrast, very little is known about the genetic and molecular mechanisms underlying human sinus node dysfunction (SND). Here, we report a genetic and molecular mechanism for human SND. We mapped two families with highly penetrant and severe SND to the human ANK2 (ankyrin-B/AnkB) locus. Mice heterozygous for AnkB phenocopy human SND displayed severe bradycardia and rate variability. AnkB is essential for normal membrane organization of sinoatrial node cell channels and transporters, and AnkB is required for physiological cardiac pacing. Finally, dysfunction in AnkB-based trafficking pathways causes abnormal sinoatrial node (SAN) electrical activity and SND. Together, our findings associate abnormal channel targeting with human SND and highlight the critical role of local membrane organization for sinoatrial node excitability.
Keywords: calcium, trafficking, arrhythmia, cytoskeleton
Cardiac pacing is orchestrated by a small group of specialized excitable cells termed the sinoatrial node (SAN). The importance of SAN activity for vertebrate physiology is clearly illustrated by dysfunction in SAN activity in human disease. Sinus node dysfunction (SND) causes “sick sinus syndrome”, which includes sinus bradycardia, sinus arrest or exit block, combinations of sinoatrial and atrioventricular nodal defects, and atrial tachyarrhythmias (1–3). SND may occur at all ages, but is most prevalent in the elderly (1:600 cardiac patients >65 years) (4). In fact, SND is the reason for over half of permanent pacemakers (>1 million worldwide) at an annual cost of nearly two billion dollars (in the United States alone) (5). Moreover, SND is an independent predictor of serious cardiovascular disease and death (6). Although common in adults with acquired heart disease, following surgical correction for congenital heart disease or during antiarrhythmic therapy, SND is also present in patients without identifiable cardiac abnormities or associated conditions (7, 8). These findings and observations from twin studies support the role of genetic factors in SND.
Genetic variants in a number of genes, mostly encoding ion channels, predispose a fraction of the population to atrial and ventricular arrhythmias (9). The identification of these variants has enabled early diagnosis and treatment of potentially fatal disease. In contrast, the genetic and molecular mechanisms underlying human SND are essentially unknown. Moreover, because of the experimental difficulty of working with primary SAN cells (low number, unique molecular, structural, and electrical properties compared with atrial/ventricular myocytes), our understanding of SAN biology is limited.
Ankyrins are adapter proteins required for targeting channels and transporters in diverse cells (10). Dysfunction in ankyrin-based pathways has been linked with human disease, including hemolytic anemia (11) and ventricular arrhythmias (12). Here, we associate a genetic and molecular mechanism for human SND with dysfunction in the AnkB pathway. We mapped two large families with highly penetrant and severe SND to the human ANK2 locus (encodes AnkB), and demonstrate that the identified genetic variant represents a loss-of-function mutation in SAN cells. We demonstrate that AnkB is highly expressed in the SAN, and AnkB activity is essential for the posttranslational organization of SAN channels and transporters. Mice lacking AnkB expression phenocopy human ANK2 SND displayed sinus bradycardia and heart rate variability. Dysfunction in AnkB-based channel/transporter trafficking at the level of the single SAN cell leads to loss of normal cell Ca2+ handling and automaticity. These results implicate ANK2 as an unexpectedly common SAN disease gene, associate a unique class of “channelopathy” with abnormal ion channel-targeting in specialized SAN cells, and highlight the critical role of local membrane organization for SAN excitability.
Results
ANK2 Gene Variants in Human SND.
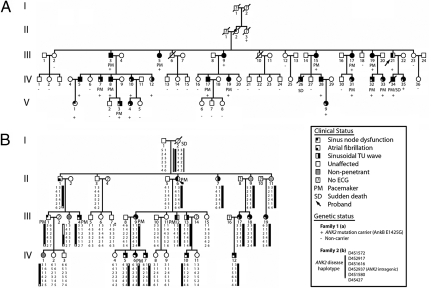
In Family 1 [Fig. 1A, supporting information (SI) Table S1], the index patient (IndIII-21) was identified because of SND and atrial fibrillation (AF). His son (IndIV-34), also affected by SND and AF, died suddenly at age 18 while being awakened. The first episode of sudden death in Family 1 occurred in a 12-year-old boy (IndIV-26) after exercise. These events were the starting point for familial screening that allowed identification of 74 members (see Fig. 1A). Among these members, 25 were affected by SND. In these patients, the rhythm originated from the SAN in 7, the coronary sinus in 7, and junctional escape rhythm was recorded in 12 patients. Thirteen family members were affected by AF (5 paroxysmal, 8 permanent; mean onset, 40 ± 18 years). SND led to pacemaker implantation in 14 patients (mean age for implantation, 34 ± 17 years). A typical ECG trace of SND in a child and an in utero echocardiogram of the same patient (IndV-3) are shown in Fig. S1 a and b. Twenty-three individuals were also affected by abnormal ventricular repolarization characterized by a prominent sinusoidal TU wave leading to a prolonged QTU interval. Sequencing all available patients, 24 of 26 presenting with SND or prolonged QTU interval plus 23 unaffected and 1 undetermined (II-3), identified 25 mutation carriers (AnkB-E1425G, ref. 12), whereas all unaffected family members were noncarriers. The AnkB-E1425G mutation is located in the AnkB spectrin-binding domain and affects ankyrin-binding activity for membrane partners including NCX1, Na/K ATPase, and IP3R (12, 13). The heart rate was lower in ANK2 mutation carriers than in noncarriers (56 ± 15 bpm versus 85 ± 24 bpm; P < 0.001). In 22 carriers, repolarization was characterized by a sinusoidal TU wave (mean QTU for 22 carriers, 619 ± 114 ms) consistent with the known role of AnkB mutations in ventricular long QT sydrome (12).
Fig. 1.
SND in human kindreds with ANK2 allele variants. (A) Family 1: Affected patients carry an AnkB-E1425G mutation depicted by a plus, whereas noncarriers are depicted by a minus. Other individuals did not undergo genetic testing. Note that at least 23 of 25 variant carriers (92%) display SND. (B) Family 2: Affected patients carry a common haplotype depicted by a black bar at the ANK2 locus. Markers D4S1572 and D4S427 delimitate the disease haplotype to a 16.5 cM interval (recombinations for patients III-9 and II-1, respectively). Squares represent males and circles represent females.
In Family 2 (Fig. 1B, Table S2), the index patient (IndII-6) was identified because of supraventricular and ventricular arrhythmias associated with SND leading to a pacemaker implantation (age 51). Familial screening allowed identification of 44 members (see Fig. 1B, Table S2). Thirteen were affected by SND. In these patients, the rhythm was from the SAN in 10 and from the coronary sinus in 3. Three family members were affected by AF (two paroxysmal, one permanent; mean age for onset, 48 ± 12 years). SND led to pacemaker implantation in six patients (mean age for implantation, 30 ± 18 years). Examples of abnormal ECG patterns are shown in Fig. S1_c_ (SND) and Fig. S1_d_ (AF). Twelve individuals were also affected by an abnormal sinusoidal repolarization with a prominent U wave and prolongation of the QTU interval, similar to our findings in Family 1. Based on the presence of SND, AF, or abnormal repolarization, we classified 16 family members as affected. Echocardiography examination identified five cases of atrial septal defect. Given the phenotypic similarities between the two families, we genotyped six microsatellite markers at the ANK2 locus. Among the 36 members of Family 2 included in the study, 20 were carriers of a common haplotype at the ANK2 locus, whereas 16 were noncarriers (see Fig. 1B). The maximum LOD score was obtained for marker D4S1616 and showed evidence for a strong linkage (Zmax = 5.9, θ = 0). All patients considered as affected were carriers of the ANK2 disease haplotype. One patient (IndI-2), who was an obligate carrier of this disease haplotype, experienced sudden death while sleeping at age 43. No ECG is available and no autopsy was performed. Only four patients were nonpenetrant. The similarities of the phenotype with the original kindred, with a marked SND and a LQT4-like morphology of the T wave and an overwhelming linkage with ANK2 locus, are strongly in favor of an AnkB defect. Although no ANK2 mutation has yet been identified in any ANK2 exons or splicing donor/acceptor site, immunoblot of a muscle biopsy obtained following pacemaker implantation of patient III-1 revealed a striking decrease in AnkB expression compared with samples from two unaffected individuals (Fig. S2). These data suggest that the Family 2 ANK2 variant resides in a promoter/enhancer sequence in uncharacterized 300 kb of the ANK2 gene sequence upstream of ANK2 exon 1, and significantly reduces AnkB expression, similar to AnkB+/− mice. Among the carriers of the ANK2 disease haplotype, the heart rate was lower than in noncarriers (57 ± 14 bpm versus 77 ± 15 bpm; P < 0.001). In 12 carriers, ventricular repolarization was characterized by a sinusoidal TU wave (mean QTU for these 12 carriers, 552 ± 54 ms). Finally, heart rates were similar (56 ± 15 bpm versus 57 ± 14 bpm) between the carriers of families 1 and 2. Taken together, these data demonstrate that mutant human ANK2 alleles associated with reduced AnkB expression or AnkB loss-of-function are strongly associated with severe human SND.
AnkB Is Required for SAN Function.
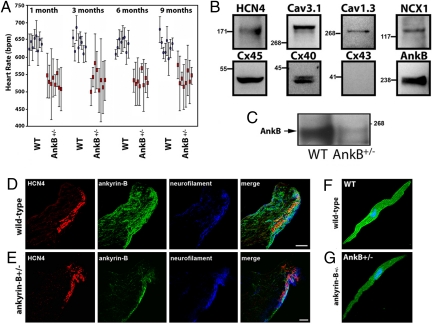
Radiotelemetry was used to assess SAN function in conscious unrestrained mice heterozygous for AnkB (AnkB+/− mice). AnkB+/− mice backcrossed 18 generations displayed pronounced bradycardia compared with WT littermates at all ages (Fig. 2A; 1, 3, 6, and 9 months), consistent with observations in human ANK2 gene variant carriers. In addition to bradycardia, we observed striking variability in resting heart rate of AnkB+/− mice compared with WT littermates (see Fig. 2A). Therefore, loss of an ANK2 allele causes SND in mice, consistent with data from humans heterozygous for a mutant AnkB allele (see Fig. 1 and Fig. S1).
Fig. 2.
Ankyrin-B is expressed in human and mouse SAN and AnkB+/− mice display severe SND. (A) Adult AnkB− mice exhibit significant bradycardia and heart rate variability. Data represents mean ± SD for eight mice/genotype. (B) Immunoblots of human SAN tissue for SAN resident proteins HCN4, Cav3.1, Cav1.3, NCX1, and Cx45. Note that Cx43 is not a SAN-resident protein. We observed decreased expression of AnkB in all SAN preparations. (C) AnkB is expressed in SAN of the WT mouse heart and is significantly reduced in AnkB+/− adult mice. Equal protein loading was assessed by blotting for unrelated protein (data not shown, NHERF1). (D and E) AnkB is expressed in the mouse SAN. WT and AnkB+/− mouse sections were immunolabeled for AnkB and SAN markers HCN4 and neurofilament, and imaged by using identical protocols. E indicates loss of AnkB expression in AnkB+/− mice. (Scale bars, 10 μm.) (F and G) Expression of AnkB in isolated WT and AnkB+/− SAN cells. (Scale bars, 10 μm.)
AnkB Is Enriched in SAN.
Immunoblots of isolated human SAN lysates revealed expression of AnkB (Fig. 2B). SAN proteins HCN4, Cav1.3, Cav3.1, NCX1, and connexins 45 and 40 were observed in parallel SAN blots (see Fig. 2B). Moreover, connexin 43 expression was not observed in human SAN blots (found in atria, but not central SAN; refs. 14–17), demonstrating accurate dissection of the SAN region from surrounding right atria. We observed AnkB expression in adult mouse SAN (Fig. 2C). AnkB+/− mice displayed significant reduction in SAN AnkB expression (see Fig. 2C) (>50% reduction, n = 3; P < 0.01). Confocal imaging revealed that AnkB is expressed in the SAN as denoted by SAN marker proteins HCN4 and neurofilaments (Fig. 2D). Parallel staining experiments of AnkB+/− mice revealed significant reduction of SAN AnkB (Fig. 2E). No changes in expression of HCN4 or neurofilament were observed in AnkB+/− SAN (see Fig. 2 D and E). Moreover, no gross abnormalities in SAN morphology were detected in AnkB+/− mice.
AnkB is localized on the cell membrane of isolated SAN cells (Fig. 2F). We also observed AnkB expression in a striated pattern corresponding to the SAN cell M-line (SAN cells do not have T-tubules; ref 18). AnkB expression is reduced and heterogeneous in AnkB+/− SAN cells (Fig. 2G) compared to WT controls. Therefore, AnkB is enriched in SAN, is present at the SAN membrane surface, and is reduced in AnkB+/− SAN cells.
AnkB Is Required for Targeting SAN Channels and Transporters.
Immunoblots of WT and AnkB+/− SAN cells (Fig. S3_a_) revealed reduced expression of Na/Ca exchanger (NCX1) and Na/K ATPase (NKA, both reduced 30–40%; P < 0.05; n = 3, each n = 3 per genotype). Additionally, IP3 receptor (IP3R) expression was reduced ≈40% in AnkB+/− SAN (see Fig. S3_a_). In contrast, Cav1.2, Cav1.3, RyR2, HCN4, Cav3.1, Cx45, NHERF1, and NKAβ1–2 expression levels were unchanged in AnkB+/− SAN (see Fig. S3_a_). Connexin 43 expression was not observed in SAN cell lysates (see Fig. S3_a_).
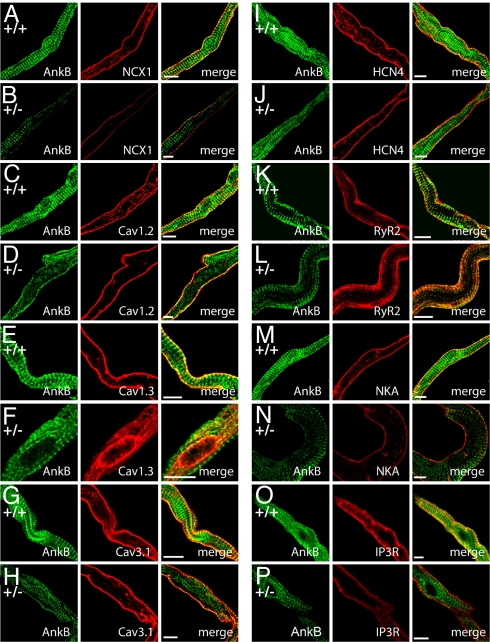
Loss of NCX1, NKA, and IP3R in AnkB+/− SAN cells is paralleled by abnormal localization of NCX1 (Fig. 3 A and B), NKA (Fig. 3 m and n), and IP3R (Fig. 3 O and P) in isolated AnkB+/− SAN. Unexpectedly, we also observed striking differences in distribution of Cav1.3 in AnkB+/− SAN. Cav1.3 expression in AnkB+/− SAN cells was limited to an internal perinuclear distribution, in contrast to the homogenous membrane distribution in WT cells (Fig. 3 E and F). We observed no difference in the localization of other SAN proteins including RyR2 (Fig. 3 K and L), Cav1.2 (Fig. 3 C and D), Cav3.1 (Fig. 3 G and H), HCN4 (Fig. 3 I and J), and connexin 40 (data not shown). Abnormal expression and targeting of Cav1.3, NCX1, IP3R, and NKA in AnkB+/− SAN cells is likely the result of a posttranslational event, as no significant difference in the mRNA levels of these transcripts or other key SAN transcripts was identified by TaqMan Low Density Arrays (Tables S3 and S4). Together, these data support a role for AnkB in the posttranslational targeting and protein stability of Cav1.3, NCX1, IP3R, and NKA in SAN. In support of this role, ankyrins have previously been demonstrated to be essential for the posttranslational stability of membrane proteins in other cell types (19).
Fig. 3.
NCX1, IP3R, Na/K ATPase, and Cav1.3 membrane expression is affected in AnkB+/− SAN cells. (A–P) Confocal imaging of SAN cells from WT and AnkB+/− mice. SAN cells were immunolabeled and imaged by using identical protocols. Note that NCX1 (A and B), Na/K ATPase (NKA) (M and N), and IP3R (O and P) immunolabeling is generally reduced across the cell, whereas Cav1.3 immunostaining is concentrated near the perinuclear region of AnkB+/− SAN cells (E and F). WT and AnkB+/− SAN cells displayed no difference in the expression or localization of Cav1.2 (C and D), Cav3.1 (G and H), HCN4 (I and J), RyR2 (K and L), or connexin 40 (data not shown). (Scale bars, 10 μm.)
Human ANK2 SND Variant E1425G Is a Loss-of-Function Mutation in SAN.
Abnormal SAN ion channel/transporter phenotypes are caused by monogenic loss of AnkB, as exogenous viral expression of AnkB into AnkB+/− SAN cells normalized the distribution of AnkB-associated proteins (NCX1 in Fig. S3 b–d). Moreover, replacement with AnkB harboring the human E1425G mutation was unable to rescue abnormal NCX1 targeting in AnkB+/− SAN, even though the mutant AnkB (E1425G) was properly expressed and localized in AnkB+/− SAN cells (Fig. S3_e_). These data indicate that a full compliment of AnkB is necessary for the membrane expression of Cav1.3, NCX1, NKA, and IP3R in SAN cells, and suggest that the E1425G mutation, causing clinical SAN dysfunction in Family 1 patients, abolishes this activity.
AnkB Is Required for INCX and ICa,L.
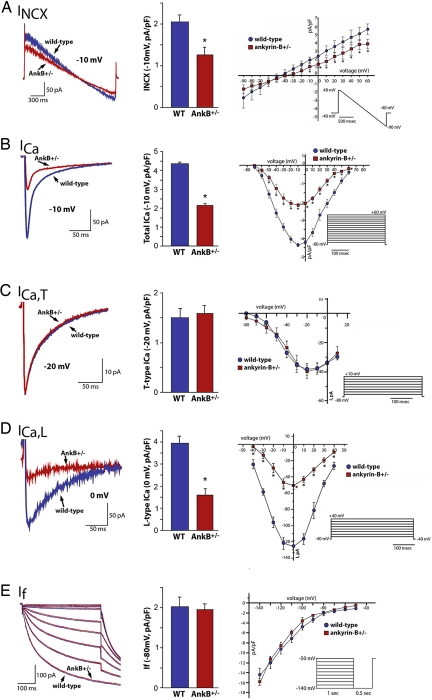
Based on the loss of membrane localization of NCX1 and CaV1.3 we observed in AnkB+/− SAN cells, we predicted that NCX1 (INCX) and CaV1.3 (ICa,L) currents would be diminished. INCX was significantly reduced in AnkB+/− SAN cells (Fig. 4A) (reduced ≈50% at nearly all positive voltages, n = 10; P < 0.05), consistent with reduced expression and abnormal localization of NCX1 in AnkB+/− SAN (see Fig. 3 and Fig. S3_a_). Total Ca2+ current (ICa) density was reduced ≈50% in AnkB+/− SAN cells at nearly all voltages (Fig. 4_B_) (_n_ = 8, _P_ < 0.05), similar to INCX. In SAN, ICa is comprised of T-type, low voltage-activated (ICa,T), and L-type, high voltage-activated (ICa,L) currents (20). In contrast to total ICa, we observed no significant difference in ICa,T density (Cav3.1, 3.2) between WT and AnkB+/− SAN (Fig. 4_C_). In contrast, ICa,L in AnkB+/− SAN was significantly reduced (>60% reduced, n = 10; P < 0.05) compared with WT SAN cells (Fig. 4D). Finally, If current was similar in WT and AnkB+/− cells (Fig. 4E) (n = 8). Therefore, our functional data strongly support a role for AnkB in the targeting and activity of specific SAN channels and transporters.
Fig. 4.
Reduced INCX and ICa,L in AnkB+/− SAN cells. Reduction of AnkB leads to reduced NCX1 and L-type Ca2+ currents. (A) INCX density is significantly lower in isolated AnkB+/− SAN cells compared to WT at voltages greater than 0 mV (n = 12, P < 0.05). Raw trace and bar graph represent current at −10 mV. (B) ICa density is reduced significantly in isolated AnkB+/− SAN cells compared to WT cells at all voltages tested (n = 10, P < 0.05). Raw trace and bar graph represent current at −10 mV. (C and D) T-type Ca2+ current is unchanged between WT and AnkB+/− SAN cells (n = 10, NS), whereas L-type Ca2+ current is dramatically reduced in AnkB+/− SAN cells (n = 10, P < 0.05). Raw traces and bar graphs represent current at −20 mV (ICa,T) and 0 mV (_I_Ca,L). (E) WT and AnkB+/− SAN cells display similar _I_f current (n = 10, NS). Bar graph represents current at −80 mV.
AnkB Is Required for SAN Ca2+ Homeostasis.
Cytosolic Ca2+ handling is critical for normal SAN function, and cellular Ca2+ entry by ICa,L is necessary for generation of normal cardiac pacemaker activity in the SAN (21). Inactivation of Cav1.3 in mice leads to significant reduction in SAN ICa,L (≈70%), reduced SAN rate, and spontaneous SAN arrhythmias (22). Recent findings also demonstrate the importance of NCX1 activity for SAN function (23). Knockout of NCX1 in mice is lethal at 11 days postcoitum, at least in part because of lack of a beating heart (24). Therefore, abnormal NCX1 and Cav1.3 expression and function in AnkB+/− SAN cells (see Fig. 3 and Fig. S3_a_) are likely to contribute to the mechanism of SND in AnkB+/− mice and in patients. Because NCX is a major pathway for cellular Ca2+ removal, we predicted that loss of NCX1 should lead to loss of normal SAN Ca2+ homeostasis in AnkB+/− mice.
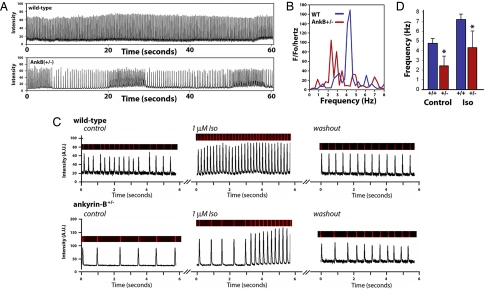
We examined potential defects from Ca2+ handling in AnkB+/− SAN cells by performing confocal imaging on spontaneous Ca2+ transients. Recordings were obtained from freshly dissected SAN tissue explants and isolated cells. Isolated WT SAN cells exhibited synchronous Ca2+ transients (Fig. 5A). Single AnkB+/− SAN cells and AnkB SAN explants displayed pronounced reduction in rate compared with WT cells (+/+: 4.4 ± 0.3 Hz; AnkB+/−: 2.6 ± 0.8 Hz; n = 10 per genotype from five animals per genotype; P < 0.05) (see Fig. 5A). Moreover, AnkB+/− SAN cells displayed striking heterogeneity in rate (Fig. 5 A and B), in agreement with human and mouse data. Fourier transform of rate data revealed a single rate frequency for WT SAN cells (4.3 Hz) (see Fig. 5B). In contrast, AnkB+/− cells displayed enhanced rate variability, seen as two prominent frequencies, both lower than WT rates (≈2.2 Hz/≈2.9 Hz). Finally, AnkB+/− SAN cells reduced frequency of Ca2+ release remained delayed and irregular even in the presence of β-adrenergic stimulation (n = 8, P < 0.05) (Fig. 5 C and D). Isoproterenol application to AnkB+/− SAN cells increased SAN rate, but not to levels observed in isoproterenol-treated WT cells (see Fig. 5 C and D). In fact, AnkB+/− SAN cells treated with isoproterenol displayed increased rate variability following treatment (see Fig. 5 C and D). Therefore, a full complement of AnkB is required for SAN Ca2+ homeostasis. Moreover, abnormal NCX1 and Cav1.3 membrane expression likely underlie abnormal Ca2+ handling phenotypes.
Fig. 5.
AnkB is required for SAN Ca2+ homeostasis. (A) Rate and frequency of Ca2+ transients from isolated SAN cells of WT and AnkB+/− mice were measured by using Fluo-3 AM and confocal imaging. Note reduced rate and extreme rate variability in AnkB+/− SAN cells. (B) Fourier transformation of pooled data from eight independent experiments from rate and frequency measurements in A. Note that AnkB+/− SAN explants display increased power density at ≥2 dominant frequencies. (C) Ca2+ transients of WT and AnkB+/− mouse SAN. AnkB+/− cells display increased cycle length and inconsistent response to isoproterenol. (D) Mean data from isoproterenol experiments. (n = 8 per genotype, P < 0.05.)
AnkB Is Required for SAN Electrical Activity.
Observed dysfunction in INCX and ICa,L (see Fig. 4), as well as likely aberrant Na/K ATPase and IP3R membrane function (see Fig. 3), predict that AnkB+/− SAN cells display abnormal electrical activity. We measured action potentials in single isolated WT and AnkB+/− SAN cells by using the perforated-patch technique to preserve SAN cell homeostasis and signaling pathways. Spontaneous cell membrane potential oscillations (after-polarizations) were observed in a significant percentage of single AnkB+/− SAN cells (≈18% of AnkB+/− cells) (Fig. S4). Spontaneous after-depolarizations were only rarely observed in WT cells (≈3% cells). Together, our data strongly link AnkB function with normal SAN electrical activity.
Discussion
The genetics of human SND are poorly defined. Loss-of-function variants in the human ANK2 are the cause of the congenital type 4 long QT syndrome (LQTS) (12, 25, 26). Variant carriers display risk of tachycardia, syncope, and sudden death (12, 26). Our findings associate ANK2 with a second cardiovascular disease, human SND. SND is nearly completely penetrant in individuals with ANK2 linkage and is observed for all ages, including in utero. AnkB is the only non-ion channel protein implicated in human SND and the first example of SND disease based on dysfunction in intracellular Ca2+ regulation. Therefore, our recent results associate SAN disease with abnormal ion channel and transporter targeting and highlight the critical role for local membrane organization for SAN excitability.
ANK2 dysfunction may play a role in the genesis of SND and sudden death in the general human population. Recent findings linked a major locus for resting heart rate in the general population to human chromosome 4q (27). The locus site was mapped near D4S2394 ([LOD] score = 3.9) (27), a site that overlaps the location of ANK2. It is noteworthy that a heart-rate locus in rat has been mapped to a homologous gene region that contains the rat Ank2 gene (chromosome 2, D2Rat62-247, LOD = 2.9) (28, 29). Our findings suggest that both loci represent heart-rate inconsistency because of variability in ANK2.
To date, ANK2 gene variants have been linked with catecholaminergic polymorphic ventricular tachycardia (CPVT), AF, defects in conduction, and severe SND (9, 12, 25, 26). A key unanswered question is how AnkB mutations may result in such a diverse collection of clinical phenotypes. Our findings demonstrate that AnkB-based pathways are critical for calcium regulation in multiple, electrically excitable, cardiac-cell types (ventricular, SAN). Therefore, we predict that analogous defects in AnkB-based pathways in different cardiac-cell types likely account for the breadth and diversity of clinical phenotypes observed in ANK2 variant carriers (LQTS, CPVT, SND). However, similar to other disease phenotypes, we recognize that genetic modifiers, unidentified compounding mutations, and a host of environmental factors likely play central roles in defining the ultimate cardiac phenotype of ANK2 variant carriers (e.g., LQTS versus SND). Data from additional probands (and family members) will be critical for resolving genotype/phenotype relationships for ANK2 variant carriers.
In summary, our findings identify a unique genetic basis for human SND and reinforce the importance of ankyrin-based targeting pathways for regulating the physiology of excitable cells. As SND is an independent risk factor for mortality, these findings identify an unexpected cardiovascular disease susceptibility gene in ANK2. Moreover, these findings suggest the exciting potential of ankyrin pathways as targets for future therapies for diseases of excitable cells.
Materials and Methods
Clinical Investigation.
See SI Methods.
Genetic Analysis.
Family 1 genetic analysis has been previously published (12). Analysis for Family 2 is described in SI Methods.
Mouse ECG Recordings.
Heart rates of ambulatory animals were determined by averaging resting heart rates (n = 6 for each mouse) of WT and AnkB+/− conscious mice over 2 h, taken at a similar time each day (12). Heart rates for eight mice from each genotype were monitored for each age (1, 3, 6, and 9 months).
SAN Preparation and Electrophysiological Recordings.
See SI Methods.
SAN Preparation for Ca2+ Imaging and Biochemistry.
See SI Methods.
Statistical Analysis.
Data were analyzed by using either paired two-tailed t tests or two-way analysis of variance, and P values <0.05 were considered significant. Data are expressed as means ± standard deviation.
Supplementary Material
Supporting Information
Acknowledgments.
This work was supported by the National Institutes of Health Grants HL084583 and HL083422 (to P.J.M.), HL079031, HL62494, and HL70250 (to M.E.A.), and HL090905 (to L.-S.S.); American Heart Association Grant 0635056N (to L.-S.S.); the Pew Scholars Trust (P.J.M.); a Fondation Leducq TransAtlantic Network of Excellence Grant (05 CVD 01, Preventing Sudden Death); and a grant from Association Française contre les Myopathies (to J.J.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Mangrum JM, DiMarco JP. The evaluation and management of bradycardia. N Engl J Med. 2000;342:703–709. doi: 10.1056/NEJM200003093421006. [DOI] [PubMed] [Google Scholar]
- 2.Kusumoto FM, Goldschlager N. Cardiac pacing. N Engl J Med. 1996;334:89–97. doi: 10.1056/NEJM199601113340206. [DOI] [PubMed] [Google Scholar]
- 3.DiFrancesco D. Pacemaker mechanisms in cardiac tissue. Annu Rev Physiol. 1993;55:455–472. doi: 10.1146/annurev.ph.55.030193.002323. [DOI] [PubMed] [Google Scholar]
- 4.Dobrzynski H, Boyett MR, Anderson RH. New insights into pacemaker activity: Promoting understanding of sick sinus syndrome. Circulation. 2007;115:1921–1932. doi: 10.1161/CIRCULATIONAHA.106.616011. [DOI] [PubMed] [Google Scholar]
- 5.Lamas GA, et al. The mode selection trial (MOST) in sinus node dysfunction: Design, rationale, and baseline characteristics of the first 1,000 patients. Am Heart J. 2000;140:541–551. doi: 10.1067/mhj.2000.109652. [DOI] [PubMed] [Google Scholar]
- 6.Palatini P, Casiglia E, Julius S, Pessina AC. High heart rate: A risk factor for cardiovascular death in elderly men. Arch Intern Med. 1999;159:585–592. doi: 10.1001/archinte.159.6.585. [DOI] [PubMed] [Google Scholar]
- 7.Schulze-Bahr E, et al. Pacemaker channel dysfunction in a patient with sinus node disease. J Clin Invest. 2003;111:1537–1545. doi: 10.1172/JCI16387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milanesi R, Baruscotti M, Gnecchi-Ruscone T, DiFrancesco D. Familial sinus bradycardia associated with a mutation in the cardiac pacemaker channel. N Engl J Med. 2006;354:151–157. doi: 10.1056/NEJMoa052475. [DOI] [PubMed] [Google Scholar]
- 9.Lehnart SE, et al. Inherited Arrhythmias: A National Heart, Lung, and Blood Institute and Office of Rare Diseases Workshop Consensus Report about the diagnosis, phenotyping, molecular mechanisms, and therapeutic approaches for primary cardiomyopathies of gene mutations affecting ion channel function. Circulation. 2007;116:2325–2345. doi: 10.1161/CIRCULATIONAHA.107.711689. [DOI] [PubMed] [Google Scholar]
- 10.Bennett V, Baines AJ. Spectrin and ankyrin-based pathways: Metazoan inventions for integrating cells into tissues. Physiol Rev. 2001;81:1353–1392. doi: 10.1152/physrev.2001.81.3.1353. [DOI] [PubMed] [Google Scholar]
- 11.Lux SE, et al. Hereditary spherocytosis associated with deletion of human erythrocyte ankyrin gene on chromosome 8. Nature. 1990;345:736–739. doi: 10.1038/345736a0. [DOI] [PubMed] [Google Scholar]
- 12.Mohler PJ, et al. Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature. 2003;421:634–639. doi: 10.1038/nature01335. [DOI] [PubMed] [Google Scholar]
- 13.Mohler PJ, Davis JQ, Bennett V. Ankyrin-B coordinates the Na/K ATPase, Na/Ca exchanger, and InsP(3) receptor in a cardiac T-tubule/SR microdomain. PLoS Biol. 2005;3:e423. doi: 10.1371/journal.pbio.0030423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coppen SR, et al. Connexin45, a major connexin of the rabbit sinoatrial node, is co-expressed with connexin43 in a restricted zone at the nodal-crista terminalis border. J Histochem Cytochem. 1999;47:907–918. doi: 10.1177/002215549904700708. [DOI] [PubMed] [Google Scholar]
- 15.Musa H, et al. Heterogeneous expression of Ca2+ handling proteins in rabbit sinoatrial node. J Histochem Cytochem. 2002;50:311–324. doi: 10.1177/002215540205000303. [DOI] [PubMed] [Google Scholar]
- 16.Davis LM, Rodefeld ME, Green K, Beyer EC, Saffitz JE. Gap junction protein phenotypes of the human heart and conduction system. J Cardiovasc Electrophysiol. 1995;6:813–822. doi: 10.1111/j.1540-8167.1995.tb00357.x. [DOI] [PubMed] [Google Scholar]
- 17.Boyett MR, et al. Connexins in the sinoatrial and atrioventricular nodes. Advances in Cardiology. 2006;42:175–197. doi: 10.1159/000092569. [DOI] [PubMed] [Google Scholar]
- 18.Bogdanov KY, Vinogradova TM, Lakatta EG. Sinoatrial nodal cell ryanodine receptor and Na+-Ca2+ exchanger: Molecular partners in pacemaker regulation. Circ Res. 2001;88:1254–1258. doi: 10.1161/hh1201.092095. [DOI] [PubMed] [Google Scholar]
- 19.Mohler PJ, et al. Inositol 1,4,5-trisphosphate receptor localization and stability in neonatal cardiomyocytes requires interaction with ankyrin-B. J Biol Chem. 2004;279:12980–12987. doi: 10.1074/jbc.M313979200. [DOI] [PubMed] [Google Scholar]
- 20.Maltsev VA, Vinogradova TM, Lakatta EG. The emergence of a general theory of the initiation and strength of the heartbeat. J Pharmacol Sci. 2006;100:338–369. doi: 10.1254/jphs.cr0060018. [DOI] [PubMed] [Google Scholar]
- 21.Kodama I, et al. Regional differences in the role of the Ca2+ and Na+ currents in pacemaker activity in the sinoatrial node. Am J Physiol. 1997;272:H2793–H2806. doi: 10.1152/ajpheart.1997.272.6.H2793. [DOI] [PubMed] [Google Scholar]
- 22.Mangoni ME, et al. Functional role of L-type Cav1.3 Ca2+ channels in cardiac pacemaker activity. Proc Natl Acad Sci USA. 2003;100:5543–5548. doi: 10.1073/pnas.0935295100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bogdanov KY, et al. Membrane potential fluctuations resulting from submembrane Ca2+ releases in rabbit sinoatrial nodal cells impart an exponential phase to the late diastolic depolarization that controls their chronotropic state. Circ Res. 2006;99:979–987. doi: 10.1161/01.RES.0000247933.66532.0b. [DOI] [PubMed] [Google Scholar]
- 24.Wakimoto K, et al. Targeted disruption of Na+/Ca2+ exchanger gene leads to cardiomyocyte apoptosis and defects in heartbeat. J Biol Chem. 2000;275:36991–36998. doi: 10.1074/jbc.M004035200. [DOI] [PubMed] [Google Scholar]
- 25.Mohler PJ, et al. Defining the cellular phenotype of “ankyrin-B syndrome” variants: Human ANK2 variants associated with clinical phenotypes display a spectrum of activities in cardiomyocytes. Circulation. 2007;115:432–441. doi: 10.1161/CIRCULATIONAHA.106.656512. [DOI] [PubMed] [Google Scholar]
- 26.Mohler PJ, et al. A cardiac arrhythmia syndrome caused by loss of ankyrin-B function. Proc Natl Acad Sci USA. 2004;101:9137–9142. doi: 10.1073/pnas.0402546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin LJ, et al. Major quantitative trait locus for resting heart rate maps to a region on chromosome 4. Hypertension. 2004;43:1146–1151. doi: 10.1161/01.HYP.0000122873.42047.17. [DOI] [PubMed] [Google Scholar]
- 28.Jaworski RL, et al. Heart rate and blood pressure quantitative trait loci for the airpuff startle reaction. Hypertension. 2002;39:348–352. doi: 10.1161/hy0202.103419. [DOI] [PubMed] [Google Scholar]
- 29.Alemayehu A, Breen L, Krenova D, Printz MP. Reciprocal rat chromosome 2 congenic strains reveal contrasting blood pressure and heart rate QTL. Physiol Genomics. 2002;10:199–210. doi: 10.1152/physiolgenomics.00065.2002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information