The SIRT2 Deacetylase Regulates Autoacetylation of p300 (original) (raw)
. Author manuscript; available in PMC: 2009 Nov 7.
SUMMARY
Autoacetylation of the p300 histone acetyltransferase controls the transition between VP16-mediated chromatin acetylation and preinitiation complex (PIC) assembly. Currently, it is unknown if and how autoacetylated p300 is deacetylated. We found that the NAD+-dependent histone deacetylase SIRT2 deacetylates p300 in vitro and in cells. SIRT2 deacetylates lysine residues in the catalytic domain of p300 and restores binding of p300 to the PIC. RNAi-mediated depletion or chemical inhibition of SIRT2 in cells results in accumulation of acetylated p300. The altered ac-p300/p300 ratio in SIRT2-depleted cells results in decreased p300 recruitment to an integrated VP16-responsive gene and inhibition of transcription. We conclude that p300 undergoes a dynamic cycle of autoacetylation and deacetylation.
Keywords: p300, SIRT2, Co-Activator, Mediator, Deacetylation, Acetylation, Autoacetylation
INTRODUCTION
Post-translational modification (PTM) of chromatin regulatory proteins through auto- or trans-modification has recently emerged as an important regulatory paradigm. Autoacetylation of the p300 (ac-p300) histone acetyltransferase (HAT) regulates its function by inducing a conformational change that reduces the affinity of p300 for preinitiation complexes formed by VP16 (Black et al., 2006). Automethylation of the histone methyltransferase G9a creates a docking site for the chromodomain of HP1 (Chin et al., 2007; Sampath et al., 2007). Auto-ADP-ribosylation of PARP-1 abrogates its interaction with chromatin, which releases it from active promoters (Kim et al., 2004). Finally, trans-acetylation of Rsc4 by SAGA leads to auto-inhibition of RSC by preventing its bromodomains from interacting with acetylated histones (VanDemark et al., 2007). Although it is clear that modification of chromatin regulatory factors can have a profound impact on their activities, it is unclear how such modifications are regulated.
The p300 coactivator is ubiquitously expressed and important in a wide range of biological processes including cell growth, differentiation, and survival (reviewed in (Giles et al., 1998; Goodman and Smolik, 2000)). In addition to acetylation of histone tails, p300 acetylates itself and non-histone proteins including p53, c-myc, HMG proteins, and nuclear receptors (Bergel et al., 2000; Faiola et al., 2005; Goodman and Smolik, 2000; Gu and Roeder, 1997). Therefore, understanding the regulation of p300 function and recruitment is crucial for understanding how histone and non-histone protein acetylation contribute to cellular processes.
We previously observed that autoacetylation of p300 induces a conformational change that facilitates the transition between chromatin modification and preinitiation complex assembly mediated by the activator VP16 (Black et al., 2006). Autoacetylation causes p300 to dissociate from complexes it forms with GAL4-VP16 and Mediator. Since autoacetylation occurs rapidly in vitro in the presence of acetyl-coA, we found it perplexing that p300 is largely unacetylated in mammalian cell extracts or in purified preparations isolated from baculovirus-infected SF9 cells, whereas the p300 HAT domain expressed in bacteria is heavily acetylated. These observations suggested the possibility that eukaryotic cells deacetylate p300 in vivo although the identity of the deacetylase is unknown.
We attempted to identify a p300 deacetylase in extracts from HeLa cells. Our study revealed the presence of an NAD+-dependent p300 deacetylase. Analysis of the seven mammalian NAD+-dependent deacetylases, the Sirtuin family, revealed that SIRT2 was uniquely capable of deacetylating autoacetylated p300 in vitro. Knockdown of SIRT2 increased cellular ac-p300 levels and affected activation of a model gene. Our results demonstrate that p300 undergoes a dynamic cycle of acetylation and deacetylation.
RESULTS
SIRT2 is a p300 Deacetylase
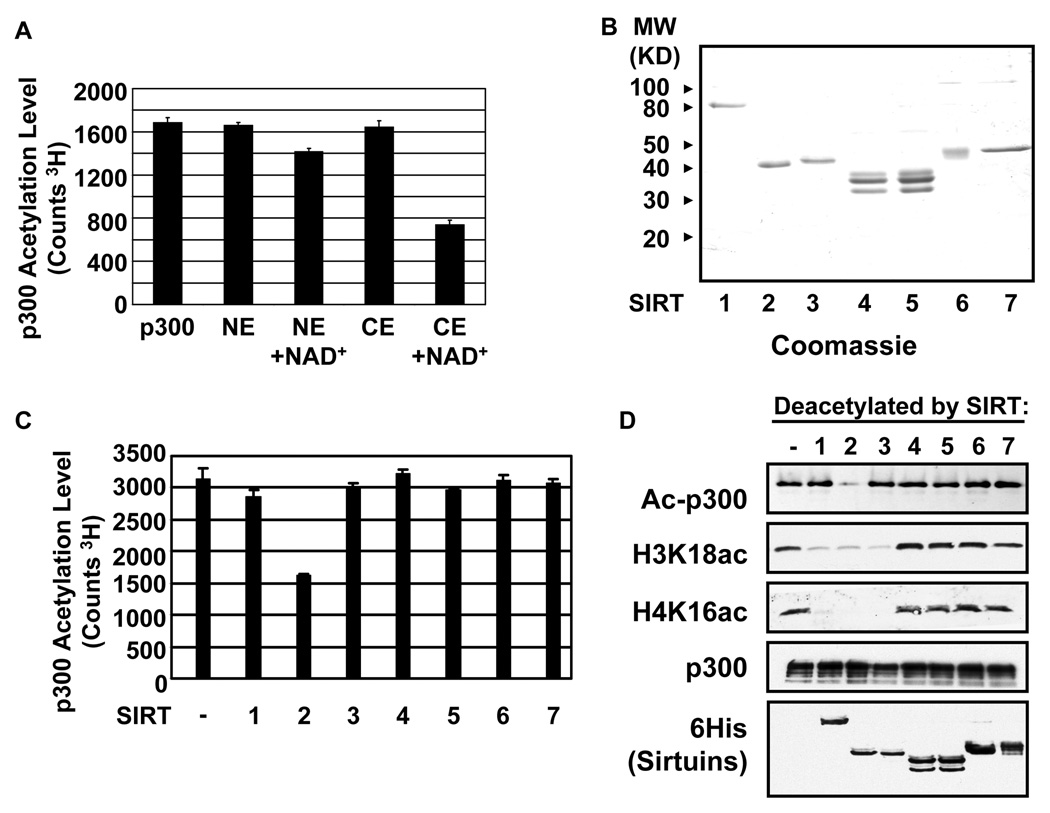
Our inability to detect ac-p300 in either our recombinant baculovirus-synthesized preparation or in HeLa cell extracts suggested that eukaryotic cells are actively deacetylating p300. This possibility was first tested by assaying HeLa cell cytoplasmic (CE) and nuclear (NE) extracts for p300 deacetylase activity. Deacetylation of 3H-ac-p300 was monitored by loss of radioactivity in a P81 filter binding assay. NE was unable to significantly deacetylate p300 (Figure 1A). In contrast, CE led to approximately 50% deacetylation of p300 when the reaction was supplemented with NAD+. The data indicate that HeLa cells contain an NAD+-dependent p300 deacetylase.
Figure 1.
SIRT2 Deacetylates p300. A.) Cellular extracts contain an NAD+ dependent p300 deacetylase activity. Dialyzed nuclear (NE) or cytoplasmic extract (CE) from equal number of cells were incubated with 3H ac-p300 in the presence of NAD+ as indicated. Error bars represent the standard deviation. B.) Coomassie-blue stained gel of purified Sirtuins (300ng). C.) SIRT2 deacetylates p300 in vitro. Filter binding assays of purified Sirtuins incubated with 3H ac-p300. Error bars represent the standard deviation. D.) SIRT1, 2 and 3 deacetylate histones. Sirtuins were incubated with NAD+, ac-p300 and HeLa core histones. Deacetylation was monitored by immunoblot.
The list of human NAD+-dependent deacetylases is limited to the seven members of the mammalian Sirtuin family, named SIRT1-7(Michan and Sinclair, 2007). SIRT1-7 were cloned and expressed from baculoviral expression vectors. Each family member was purified to near homogeneity (Figure 1B) and tested in our p300 deacetylation assay. Surprisingly, only SIRT2 was able to significantly deacetylate p300 (Figure 1C). In reactions containing acetylated histones and ac-p300, SIRT1, 2 and 3 deacetylated H4K16 and H3K18, but only SIRT2 deacetylated p300 as measured using an antibody specific to acetyl-lysine (Figure 1D). No deacetylation substrates have been reported for SIRT4, 5, and 7, and SIRT6 did not deacetylate H3K18 or H4K16 in our assay.
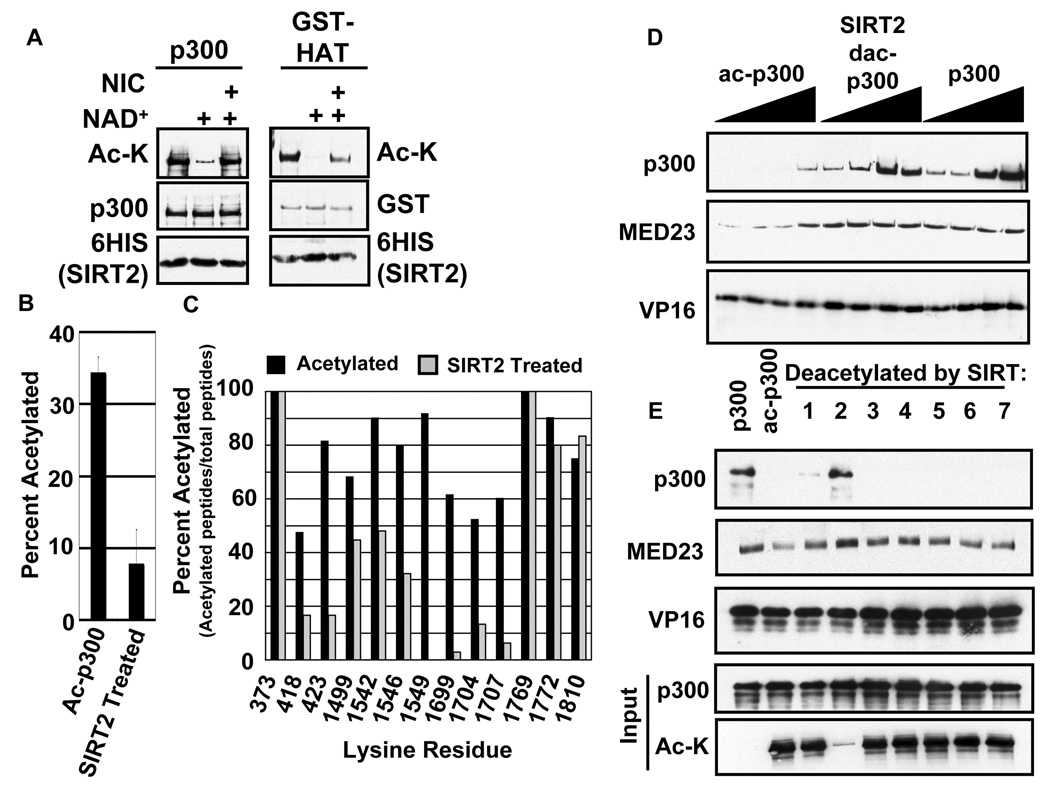
To further verify that SIRT2 is indeed acting as a p300 deacetylase, we analyzed its activity on ac-p300 and its response to the Sirtuin inhibitor nicotinamide. Treatment with SIRT2 and NAD+ led to deacetylation of ac-p300 (Figure 2A left panel). Nicotinamide, a competitive inhibitor byproduct of the Sirtuin deacetylation reaction (Sauve et al., 2006), was sufficient to prevent deacetylation of ac-p300 (Figure 2A). SIRT2 interacts with HDAC6 to deacetylate K40 on tubulin (North et al., 2003). However, HDAC6 did not deacetylate p300 nor modulate SIRT2 activity in our assays (Figure S1). Furthermore, treatment of U2OS cells with TSA resulted in only a weak increase in ac-p300 while treatment with Nicotinamide substantially increased ac-p300 (Figure S2). Additionally, TSA combined with Nicotinamide did not substantially increase ac-p300 levels above nicotinamide treatment alone.
Figure 2.
SIRT2 deacetylation of p300 restores p300 binding to GAL4-VP16-Mediator complexes. A.) SIRT2 deacetylates p300. Ac-p300 or ac-p300 HAT domain treated with SIRT2 in the presence of NAD+ or Nicotinamide (NIC) as indicated. Deacetylation was assayed by immunoblot with anti-acetyl-lysine antibody. B.) Amount of lysine acetylation detected in p300 MudPIT analyses. The percent lysine acetylation represents the number of spectral counts for acetylated lysine residues divided by the total spectral counts for lysine residues. The data is expressed as an average +/− standard deviation. A two-tailed student's t-test was performed and gave a p-value = 0.009691. C.) Representative lysines from MudPIT analysis. Data are shown as percent of peptides acetylated divided by the total number of peptides identified for each residue. D.) Immobilized template assay. Immobilized template was prebound by GAL4-VP16 followed by incubation with Mediator and p300, ac-p300, or SIRT2 treated ac-p300 as indicated. Binding was analyzed by immunoblot. Titrations represent 2-fold steps. E.) SIRT2 restores p300 binding to Mediator. Immobilized DNA was prebound by GAL4-VP16 and Mediator followed by incubation with p300, ac-p300, or SIRT1-7 treated ac-p300 as indicated. Input indicates levels of p300 and ac-p300 included in each immobilized template reaction.
SIRT2 Deacetylates the p300 HAT Domain
Our previous work and that of the Cole lab suggested that the HAT domain of p300 is a significant site of autoacetylation (Black et al., 2006; Karanam et al., 2006; Thompson et al., 2004). Furthermore, autoacetylation induced a conformational change in the HAT domain as measured by protease footprinting. This conformational change in p300 has since been verified by structural analysis (Arif et al., 2007). We thus attempted to determine if SIRT2 was able to deacetylate the p300 HAT domain. The recombinant GST-p300-HAT domain purified from bacteria is hyperacetylated (Thompson et al., 2004) (Figure 2A). SIRT2 was able to deacetylate the p300 HAT domain (residues 1066–1707) and was sensitive to nicotinamide inhibition (Figure 2A, right panel). Taken together these data indicate that SIRT2 interacts with and deacetylates the p300 HAT domain.
The HAT domain of p300 contains 55 lysine residues thereby precluding analysis of important deacetylation sites by systematic mutagenesis. We therefore addressed the specificity of SIRT2 for deacetylation of specific lysines by mass spectrometry analysis. We subjected ac-p300 and SIRT2 treated ac-p300 to Multidimensional Protein Identification Technology (MudPIT) to identify lysine residues in p300 that were acetylated and deacetylated. In addition, we analyzed an unmodified p300 sample (not incubated with Ac-CoA) and found very low levels of background acetylation in the purified protein (Supplementary Table 1). We were able to detect peptides for 103 out of 108 lysines in p300. Out of the 103 lysine residues detected, 67 were found to be acetylated at least once (Supplemental Table 1 and Supplemental Table 2). When comparing the autoacetylated p300 to the SIRT2-treated p300, we found that there was a 4-fold decrease in the total percentage of acetylated lysines with the addition of SIRT2 (Figure 2B, Supplementary Table 1). Surprisingly, SIRT2 exhibited preference for different lysines in p300 (Figure 2C, Supplemental Table 1). For example lysines 1542, 1546, 1549, 1699, 1704 and 1707 at the C-terminal end of the HAT domain were deacetylated greater than 40%, while K1499 was deacetylated by approximately 20%, and K1769, K1772, K1810 were not deacetylated (Figure 2C). SIRT2 also deacetylated lysines in the N terminus of p300 including K418 and K423 but not K373.
Previous work by the Cole lab using only the HAT domain of p300 has demonstrated that the loop region between amino acids 1550 and 1560 is heavily acetylated (Karanam et al., 2006). In the autoacetylated samples, we were able to detect near 100% acetylation of this region. In fact, the spectra obtained for residues 1550 through 1560 accounted for 41.25% of the acetyl-lysine spectra in the sample, indicating that it is one of the most highly acetylated regions in p300 (Supplemental Table 1). However, upon SIRT2 treatment, no peptides from this region were detected. The lack of these peptides in the SIRT2 sample is likely due to significant deacetylation of this region. Acetylation of lysine residues neutralizes their positive charge and also prevents their digestion by trypsin. Upon deacetylation, the highly charged nature of this region would prevent its identification by MudPIT using the parameters as described in the Experimental Procedures. Our data show that SIRT2 has strong preference for deacetylation of residues within the HAT domain. These results raise the question of how p300 deacetylation affects its recruitment to the PIC.
SIRT2 Deacetylation of p300 Restores Binding to GAL4-VP16-Mediator
We have previously shown that autoacetylated p300 has reduced affinity for GAL4-VP16-Mediator complexes (Black et al., 2006). We hypothesized that deacetylation of p300 by SIRT2 should restore affinity for GAL4-VP16-Mediator complexes. To test this hypothesis, we utilized an immobilized template assay. As previously described, p300 binds cooperatively to GAL4-VP16-Mediator complexes, while ac-p300 recruitment is reduced by 9 fold (Figure 2D). Treatment of the ac-p300 with SIRT2 and NAD+ restores p300 recruitment and stimulates Mediator binding as previously observed. This effect was mainly specific to SIRT2 as the only other Sirtuin to exhibit even weak restoration of ac-p300 binding was SIRT1 (Figure 2E). We were unable to detect recruitment of SIRT2 to the immobilized template (data not shown) suggesting that SIRT2 does not remain stably associated with p300 following deacetylation. Our data demonstrate that SIRT2 deacetylates specific residues in p300 that restore binding to GAL4-VP16-Mediator complexes in vitro.
Depletion of SIRT2 Increases Cellular ac-p300 Concentration
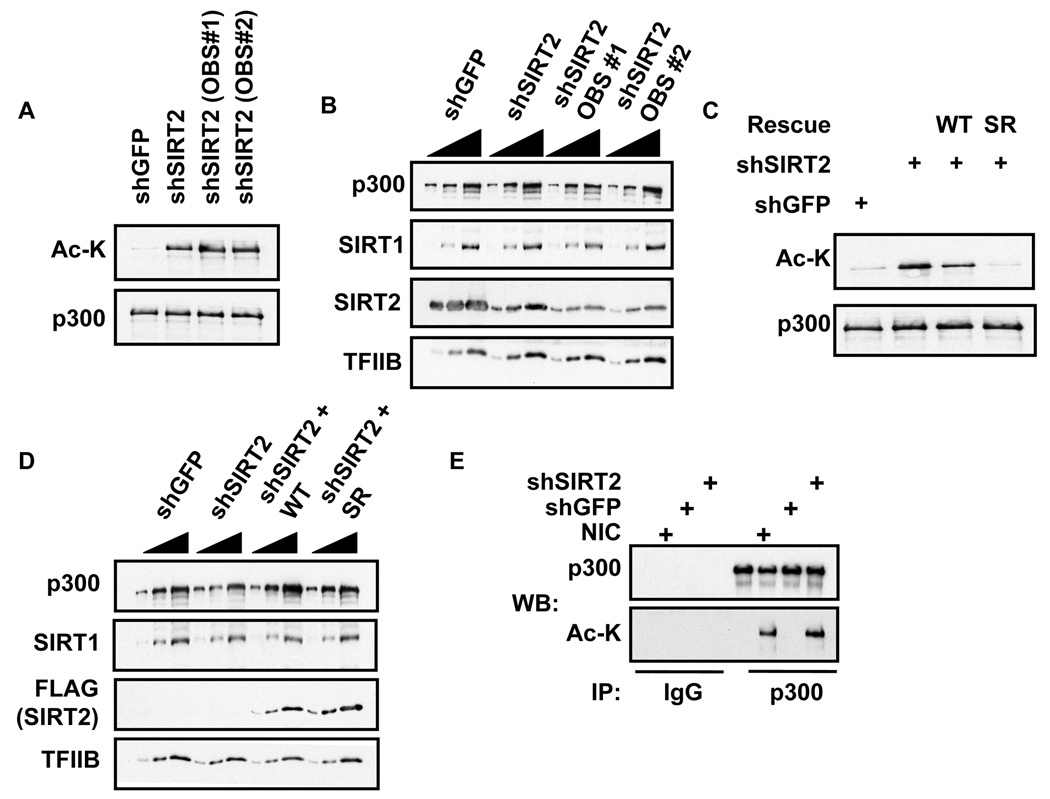
To determine whether SIRT2 deacetylates p300 in vivo we ablated SIRT2 expression by shRNA-mediated knockdown. We treated U2OS cells with shSIRT2 (Vaquero et al., 2006) and two shRNAs from Open Biosystems (OBS #1 and OBS #2) directed against two additional distinct sites in SIRT2. We immunoprecipitated cellular p300 from shRNA-treated cell extracts and assayed ac-p300 levels by immunoblotting with the acetyl-lysine antibody. Depletion of SIRT2 by the three different shRNAs resulted in significant increases in cellular ac-p300 (Figure 3A). Furthermore, the 2 to 4-fold knockdown of SIRT2 protein level by these constructs did not alter p300 or SIRT1 protein levels (Figure 3B).
Figure 3.
SIRT2 deacetylates p300 in vivo. A.) Depletion of SIRT2 by shRNA increases ac-p300. U2OS cells treated with shGFP, OBS #1, OBS #2 and shSIRT2. The p300 was immunoprecipitated and analyzed by acetyl-lysine immunoblot. The immunoblot signals correspond to mRNA levels measured by qPCR (data not shown) B.) Knockdown of SIRT2. Whole cell extracts from panel a immunoblotted for SIRT2 levels. Titrations are 3-fold steps. C.) Expression of shRNA resistant FLAG-SIRT2 in U2OS cells restores p300 deacetylation. Immunoprecipitated p300 analyzed for acetylation by immunoblot. WT= wildtype SIRT2, SR= shRNA resistant SIRT2. D.) Expression of FLAG-SIRT2 and SR FLAG-SIRT2. Whole cell extract immunoblots. Titrations are 3-fold steps. E.) Depletion or inhibition of SIRT2 increases cellular ac-p300. U2OS Cells treated with Nicotinamide (NIC), shGFP, or shSIRT2 were lysed and p300 immunoprecipitated. Immunoprecipitates were immunoblotted with anti acetyl-lysine antibody.
To preclude the possibility of off-target effects of the shSIRT2, we rescued the shSIRT2 knockdown with an shRNA-resistant SIRT2 clone. Overexpression of wildtype (WT) FLAG-SIRT2 led to a modest decrease in ac-p300 in shRNA-treated cells, while the shRNA-resistant (SR) FLAG-SIRT2 led to a significant decrease in ac-p300 (Figure 3C). This effect was not due to changes in p300 protein levels (Figure 3D). Finally, we compared the increase in ac-p300 resulting from treatment with shSIRT2 to treatment with nicotinamide. Even though nicotinamide inhibits the entire Sirtuin family, it resulted in a similar increase in ac-p300 levels to SIRT2 depletion (Figure 3E). We conclude that SIRT2 functions as the predominant p300 deacetylase in cells.
Depletion of SIRT2 Diminishes VP16-dependent Gene Expression
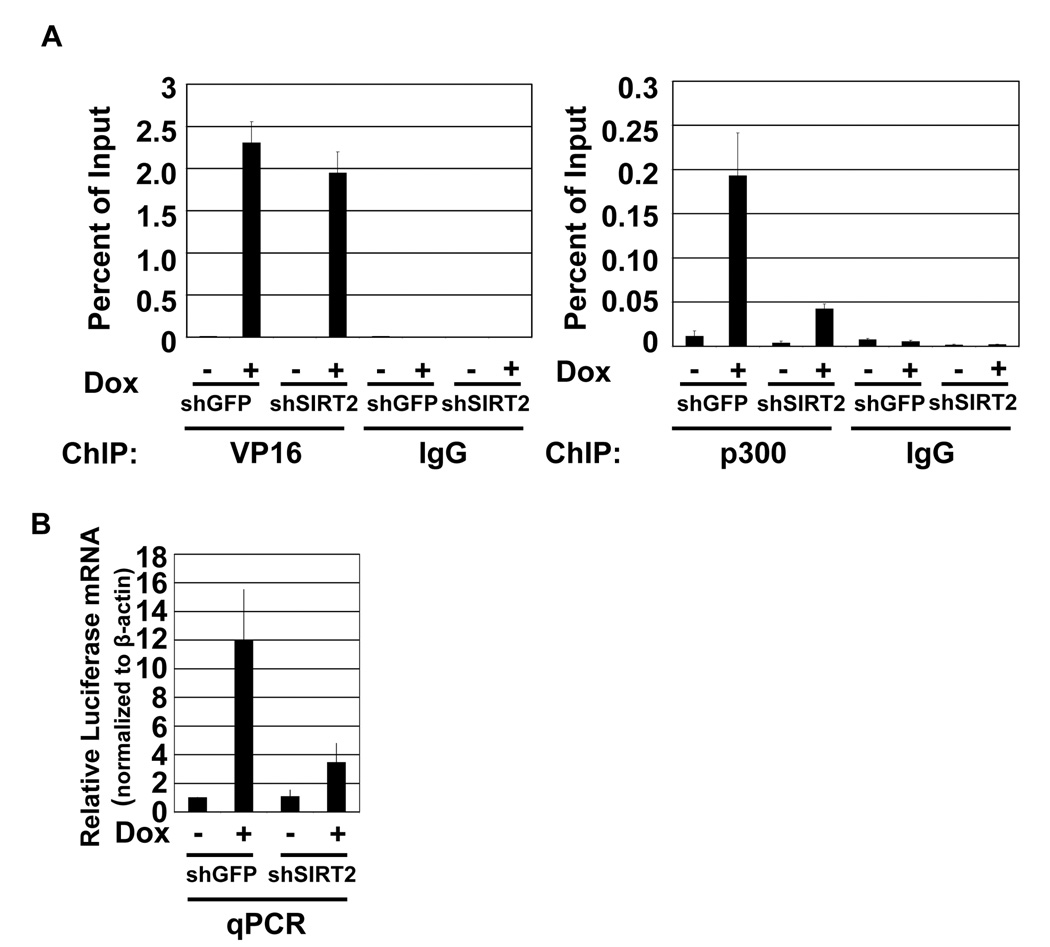
The increased cellular concentration of ac-p300 following shRNA depletion of SIRT2 or inhibition by nicotinamide demonstrates that SIRT2 is acting as a p300 deacetylase in cells. One prediction of this result is that increased levels of ac-p300 would inhibit VP16-mediated transcriptional activation. We have previously demonstrated that p300 is recruited to the promoter region of the integrated Luciferase gene in Tet-ON-VP16 U2OS cells (Black et al., 2006). Addition of doxycycline leads to binding of the Tet repressor-VP16 fusion protein allowing synchronous activation of the reporter gene. We previously determined that peak p300 binding occurs 180 minutes after adding doxycycline. Using this time point, we assayed p300 recruitment by chromatin immunoprecipitation (ChIP) in shRNA-treated cells. Doxycycline treatment of Tet-ON U2OS cells containing shGFP resulted in normal recruitment of the Tet-VP16 fusion protein and p300 (Figure 4A). However, treatment with shSIRT2 resulted in a four-fold decrease in p300 recruitment without significantly affecting Tet-VP16. The decreased recruitment of p300 correlated with a 3.4-fold reduction in Luciferase expression in the presence of shSIRT2 (p=0.031) (Figure 4B). The data demonstrate that SIRT2 is acting as a p300 deacetylase and that increased ac-p300/p300 ratios reduce VP16-mediated gene expression.
Figure 4.
Ablation of SIRT2 regulates VP16-dependent recruitment of p300 and gene expression. A.) Increased acetylated p300 levels result in decreased p300 recruitment. qPCR of ChIP analysis of p300 binding to Luciferase promoter in Tet-ON U2OS cells treated with shSIRT2 shRNAs. Error bars represent standard deviation of three biological replicates. B.) Loss of p300 recruitment inhibits Luciferase expression. qPCR analysis of Luciferase RNA was normalized to β-Actin (p=.031 using two-tailed student’s T test). Error bars represent standard deviation of three biological replicates.
DISCUSSION
Autoacetylation of p300 represents a transcriptional regulatory checkpoint in vitro. However, it was unclear if the acetylation status of p300 is regulated in cells, and whether altering the balance between p300 and ac-p300 regulates transcription in vivo. Our study demonstrates that this balance has important consequences on VP16-dependent transcription.
Although surprising, the observation that an NAD+-dependent deacetylase activity resides in the cytoplasmic fraction of HeLa cell extracts agrees with the predominantly cytoplasmic localization of SIRT2 (North and Verdin, 2007; Vaquero et al., 2006). Though the majority of SIRT2 is localized to the cytoplasm, SIRT2 undergoes constant shuttling into and out of the nucleus and becomes predominantly nuclear during mitosis (North and Verdin, 2007; Vaquero et al., 2006). We have also observed nuclear-cytoplasmic shuttling of SIRT2 in 293 and U2OS cells by immunofluorescence and cell fractionation (Figures S3, S4 and S5). SIRT2 becomes predominantly nuclear upon treatment with the nuclear export inhibitor Leptomycin B. Thus, SIRT2 and p300 can localize to the same cellular compartment. Furthermore, we observe modest, yet significant, colocalization between p300 and SIRT2 in LMB-treated cells (Figure S4). The nuclear-cytoplasmic shuttling of SIRT2 raises the question of where deacetylation of p300 is occurring. We have been unable to detect any significant redistribution of p300 in response to depletion or inhibition of SIRT2 (data not shown) leading us to speculate that deacetylation is occurring in the nucleus.
MudPIT analysis revealed a significant, but not complete, loss of acetylated lysine residues upon treatment with SIRT2. The fact that incomplete deacetylation of p300 is sufficient to restore binding raises the question of how many residues need to be acetylated/deacetylated to affect p300 affinity for the PIC. One possibility is there are a few critical residues that determine p300 affinity for GAL4-VP16-Mediator complexes. Alternatively, once a threshold acetylation level is achieved the affinity of p300 is altered. A cell could specify different transcriptional programs by controlling the dynamic ratio of ac-p300/p300 through SIRT2. Consistent with this idea, p300 isolated from cells treated with HDAC inhibitors interacts more tightly with p53 (Stiehl et al., 2007) and ATF-2 preferentially interacts with ac-p300 in vitro (Karanam et al., 2007). The dynamics of p300 autoacetylation and deacetylation by SIRT2 provide another level of precision in the fine-tuning of gene regulation programs.
We note that one group has also identified lysine residues 1020 and 1024 in p300 as targets of SIRT1 deacetylation in 293 cells (Bouras et al., 2005). It is unlikely that these are p300 autoacetylation sites in vitro as we failed to detect significant acetylation of these residues (Supplemental Table 1). Instead, they may be targets of other HATs such as PCAF. Furthermore, SIRT1 and SIRT6 knockout mouse embryonic fibroblasts still exhibit strong increases of ac-p300 in response to nicotinamide treatment suggesting SIRT1 and SIRT6 are not critical determinants of cellular ac-p300 levels (Figure S6). Lysines 1020 and 1024 also serve as sumoylation sites and are important for p300-mediated repression involving HDAC6. HDAC6 interacts with these sumoylated residues for recruitment to repressed genes (Girdwood et al., 2003).
In light of the discoveries that p300 is autoacetylated, G9a is automethylated, Rsc4 is acetylated, and PARP-1 is auto ADP-ribosylated it is becoming apparent that posttranslational modification of chromatin modifying complexes represents an additional level of transcriptional control. It is possible that these modifications will be recognized by additional effector proteins utilizing the same domains that recognize epigenetic marks on histone tails, as is the case where automethylated G9a is recognized by the HP1 chromodomain (Sampath et al., 2007). Thus, it is likely that SIRT2 is not the only histone de-modifying enzyme to play a role in the dynamics of coactivator post-translational modification.
EXPERIMENTAL PROCEDURES
Preparation of Extracts
Cytoplasmic and Nuclear Extracts were prepared following standard protocols (Dignam et al., 1983). Extracts were dialyzed against 0.1M Buffer D with two buffer changes prior to use in deacetylation assays.
Protein Purification
GAL4-VP16, p300, and Mediator were purified as described previously (Black et al., 2006). Sirtuin clones were obtained from RZPD and cloned into pFastBac with a 6XHIS tag (Invitrogen) and purified by metal ion affinity chromatography (Black et al., 2006). GST-p300-HAT was purchased from Upstate.
Filter Binding Deacetylation Assays
Autoacetylated p300 was generated by incubating p300 with 3H ac-CoA (Moravek) for two hours at 30°C; the excess ac-CoA was removed by dialysis. 3H ac-p300 (50 ng) was incubated with Sirtuins (100ng) or extracts (100µg) in Deacetylation Buffer (20mM HEPES pH 7.9, 67 mM KCl, 0.2mM EDTA, 5mM MgCl2) supplemented with 1mM NAD+ for two hours at 30°C. Reactions were spotted on P81 Whatman discs and placed in 0.85% phosphoric acid for five minutes. The discs were washed twice with 0.85% phosphoric acid and once in acetone. The discs were dried and counted.
Deacetylation Assays
Sirtuins were incubated with 2µg HeLa core histones and 100ng ac-p300 in deacetylation buffer for two hours at 30°C in the presence of NAD+. Following deacetylation, proteins were analyzed by immunoblot.
MudPIT Analysis
Purified p300 was treated with 100 µM Ac-CoA (Sigma) in 0.1M Buffer D with 5mM MgCl2 for three hours at 30°C. Ac-p300 was dialyzed away from the excess ac-CoA as above. The ac-p300 was divided into two separate reactions one treated with NAD+ (ac-p300) and the other SIRT2 and NAD+. The deacetylation reaction was performed for two hours at 30°C. Following deacetylation, the samples were treated with Benzonase (0.1U, Sigma) and precipitated in 20% TCA. Three separate preparations were digested with either LysC/Trypsin (Roche/Promega), Chymotrypsin (Roche), or GluC (Roche) and analyzed by MudPIT analysis. Detailed procedures for MudPIT analysis can be found in the Supplemental Material.
Immobilized Template Assay
Immobilized template assays were performed as described in (Black et al., 2006). The following antibodies were used: p300 (Santa Cruz sc-583), ac-lysine (Upstate), 6His (Sigma), TFIIB (Tantin et al., 1996), SIRT2 (Fisher), VP16 (Tantin et al., 1996) FLAG (Sigma), H3K18ac (Kurdistani Lab), H4K16ac (Grunnstein Lab), SIRT1 (Abcam), MED23 (BD Pharmingen), H3K9ac (Abcam), H3 (Abcam). 30ng of p300, ac-p300, or SIRT2 treated ac-p300 were incubated with immobilized templates (40 fmol) prebound with 40ng GAL4-VP16 and 300ng Mediator.
RNAi
A shRNA targeting SIRT2 using the previously identified sequence (Vaquero et al., 2006) was cloned into the RVGP plasmid (shSIRT2) (Ramirez-Carrozzi et al., 2006). RNAi control experiments were performed using a short hairpin targeting GFP. shRNAs were transfected into the Tet-ON U2OS cell line (Clontech) using Lipofectamine 2000. 12 hours post transfection cells were selected with puromycin (100µg/ml) for 48 hours before harvesting. The SIRT2 shRNA resistant clone was created by Quick Change (Stratagene) of three non consecutive point mutations in the coding sequence to render it resistant to the shRNA (sequence available upon request). The shRNA constructs OBS#1 and OBS#2 from Open Biosystems correspond to TRCN0000040221 and TRCN0000040222 respectively. For rescue experiments, U2OS Tet-ON cells were transfected with shRNA plasmid as above. 48 hours later, cells were re-transfected with additional shRNA plasmid and the indicated FLAG-SIRT2 construct. Cells were harvested 48 hours later.
Immunoprecipitation
5×106 cells were lysed in RIPA buffer (20mM Tris pH 7.5, 150mM NaCl, 1% NP-40) containing 0.5mM PMSF, 2mM Nicotinamide (Sigma), and 10µM Sodium Butyrate (Sigma). Lysate from 2×106 cells was diluted with IP buffer (20mM Tris pH 7.5, 150mM NaCl, 0.1% NP-40) and incubated overnight with 4µg of the indicated antibody. Antibody complexes were recovered on magnetic protein A beads (Invitrogen) and washed five times with IP buffer containing 0.25% NP-40. Immunoprecipitated material was analyzed by immunoblot.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed following the Upstate/Millipore protocol. Briefly 4×106 cells were resuspended in lysis buffer and sonicated to generate fragments ranging from 250–500 base pairs. Lysate from 5×105 cells was used per immunoprecipitation. The immunoprecipitated DNA was purified (Promega Gel Purification Kit) and analyzed by qPCR. Each ChIP was performed on a minimum of three biological replicates.
qPCR
Real time quantitative PCR (qPCR) was conducted using a Stratagene MX3000P thermal cycler and SYBR Green Universal Master Mix with ROX (Roche). Each sample was amplified in duplicate reactions and the threshold values were averaged. For RNA analysis all samples were normalized to Actin. For ChIP, each sample was calculated as a percentage of its corresponding input amplified at the same time. Primer sequences used for RNA and ChIP analysis are available upon request
Supplementary Material
01. SUPPLEMENTAL DATA.
Supplemental Material contains six figures, two charts, two tables and accompanying experimental procedures.
02
03
ACKNOWLEDGEMENTS
We thank Xuan Liu for the p300 baculovirus, Joan Conaway for FLAG-hIntersex HeLa cell line, Jerry Workman for p5SG5E4T, Karolin Luger for Xenopus histone expression vectors, Steve Smale for empty shRNA vectors and shGFP vector, Raul Mostoslavsky for SIRT1 and SIRT6 KO MEFs, and Bi-Ching Sang for HDAC6 clones. We thank Matt McBrian for technical assistance. This work was supported by grants from the National Institutes of Health to M.C. (GM074701). J.C.B. was supported by a Ruth L. Kirschstein National Research Award (GM07185). T.K. was supported by a fellowship from The Nakajima Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Arif M, Kumar GV, Narayana C, Kundu TK. Autoacetylation induced specific structural changes in histone acetyltransferase domain of p300: probed by surface enhanced Raman spectroscopy. J Phys Chem B. 2007;111:11877–11879. doi: 10.1021/jp0762931. [DOI] [PubMed] [Google Scholar]
- Bergel M, Herrera JE, Thatcher BJ, Prymakowska-Bosak M, Vassilev A, Nakatani Y, Martin B, Bustin M. Acetylation of novel sites in the nucleosomal binding domain of chromosomal protein HMG-14 by p300 alters its interaction with nucleosomes. J Biol Chem. 2000;275:11514–11520. doi: 10.1074/jbc.275.15.11514. [DOI] [PubMed] [Google Scholar]
- Black JC, Choi JE, Lombardo SR, Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- Bouras T, Fu M, Sauve AA, Wang F, Quong AA, Perkins ND, Hay RT, Gu W, Pestell RG. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J Biol Chem. 2005;280:10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- Chin HG, Esteve PO, Pradhan M, Benner J, Patnaik D, Carey MF, Pradhan S. Automethylation of G9a and its implication in wider substrate specificity and HP1 binding. Nucleic Acids Res. 2007;35:7313–7323. doi: 10.1093/nar/gkm726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faiola F, Liu X, Lo S, Pan S, Zhang K, Lymar E, Farina A, Martinez E. Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol Cell Biol. 2005;25:10220–10234. doi: 10.1128/MCB.25.23.10220-10234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles RH, Peters DJ, Breuning MH. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- Girdwood D, Bumpass D, Vaughan OA, Thain A, Anderson LA, Snowden AW, Garcia-Wilson E, Perkins ND, Hay RT. P300 transcriptional repression is mediated by SUMO modification. Mol Cell. 2003;11:1043–1054. doi: 10.1016/s1097-2765(03)00141-2. [DOI] [PubMed] [Google Scholar]
- Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Karanam B, Jiang L, Wang L, Kelleher NL, Cole PA. Kinetic and mass spectrometric analysis of p300 histone acetyltransferase domain autoacetylation. J Biol Chem. 2006;281:40292–40301. doi: 10.1074/jbc.M608813200. [DOI] [PubMed] [Google Scholar]
- Karanam B, Wang L, Wang D, Liu X, Marmorstein R, Cotter R, Cole PA. Multiple roles for acetylation in the interaction of p300 HAT with ATF-2. Biochemistry. 2007;46:8207–8216. doi: 10.1021/bi7000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- North BJ, Verdin E. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS ONE. 2007;2:e784. doi: 10.1371/journal.pone.0000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath SC, Marazzi I, Yap KL, Sampath SC, Krutchinsky AN, Mecklenbrauker I, Viale A, Rudensky E, Zhou MM, Chait BT, Tarakhovsky A. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell. 2007;27:596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Wolberger C, Schramm VL, Boeke JD. The Biochemistry of Sirtuins. Annu Rev Biochem. 2006 doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- Stiehl DP, Fath DM, Liang D, Jiang Y, Sang N. Histone deacetylase inhibitors synergize p300 autoacetylation that regulates its transactivation activity and complex formation. Cancer Res. 2007;67:2256–2264. doi: 10.1158/0008-5472.CAN-06-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantin D, Chi T, Hori R, Pyo S, Carey M. Biochemical mechanism of transcriptional activation by GAL4-VP16. Methods Enzymol. 1996;274:133–149. doi: 10.1016/s0076-6879(96)74013-2. [DOI] [PubMed] [Google Scholar]
- Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, et al. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol. 2004;11:308–315. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- VanDemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol Cell. 2007;27:817–828. doi: 10.1016/j.molcel.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01. SUPPLEMENTAL DATA.
Supplemental Material contains six figures, two charts, two tables and accompanying experimental procedures.
02
03