An evaluation of custom microarray applications: the oligonucleotide design challenge (original) (raw)
Abstract
The increase in feature resolution and the availability of multipack formats from microarray providers has opened the way to various custom genomic applications. However, oligonucleotide design and selection remains a bottleneck of the microarray workflow. Several tools are available to perform this work, and choosing the best one is not an easy task, nor are the choices obvious. Here we review the oligonucleotide design field to help users make their choice. We have first performed a comparative evaluation of the available solutions based on a set of criteria including: ease of installation, user-friendly access, the number of parameters and settings available. In a second step, we chose to submit two real cases to a selection of programs. Finally, we used a set of tests for the in silico benchmark of the oligo sets obtained from each type of software. We show that the design software must be selected according to the goal of the scientist, depending on factors such as the organism used, the number of probes required and their localization on the target sequence. The present work provides keys to the choice of the most relevant software, according to the various parameters we tested.
INTRODUCTION
The way we work with microarrays has changed with the increase in feature resolution, the availability of multipack formats and the flexibility of custom slide production from commercial providers. These new formats open up possibilities to reduce the cost of experiments and the amount of useless data, in comparison with classical methods that use arrays from catalogues. These applications typically focused on the study of a ‘limited’ number of probes using a custom design of microarray slides. Various biological questions can be addressed with such a microarray design. For example, it is possible to focus on the precise expression of a selected set of genes or transcription units. It also permits work on a specific sub-region of larger genomes by using a tiling design. It finally allows for study of a large number of experimental conditions, for example to screen mutant libraries, or to perform kinetic experiments useful to understanding how a biological system behaves.
The main problem that must be faced for such custom microarray applications is the oligonucleotide design step. A lot of parameters must be taken into account to ensure that the selected oligonucleotides offer the best specificity and sensitivity. Fortunately, numerous tools have been developed to perform this design step, and many are available to the academic community. They cover a wide range of applications, from screening for microbial communities by the design of a minimal primer set (1–3) to the design of PCR primers (4–6), short oligonucleotides (7–9), more specific oligonucleotides such as LNA (10) or overgo sequences (11) and tiling arrays (12–16). The most difficult task is to choose among all these solutions.
Here we present an overview of oligonucleotide design software focusing on the design of long oligonucleotides (more than 40-mer) for tiling or the analysis of a custom set of biological entities, for organisms whose complete genome sequence is available. We limited our selection to freely accessible tools for academics, and we focused on long oligonucleotides, because several studies (8,17,18) show that they seem to provide the best compromise between specificity and sensitivity. The first part of our study was to perform a comparative evaluation of the available solutions based on a set of criteria including: ease of installation, user-friendly access, number of parameters and settings available. All the information we provide was gathered from the original publication, associated website and software documentation when available. The second step was to then submit two real cases to a selection of software candidates. First, we designed a set of probes dedicated to the study of the developing mouse nervous system. Second, we created a tiling microarray against a fungal genome. Finally, we compared the oligo sets obtained from each software candidate in order to select the method best adapted to our needs.
MATERIALS AND METHODS
Custom Mus musculus oligo set design
We downloaded the transcript sequence file (release 21) from the RefSeq ftp site (ftp://ftp.ncbi.nih.gov/refseq/M_musculus/mRNA_Prot/) and retrieved the transcript sequence files for our 1421-gene collection. The 10 oligonucleotide programs were used to design one oligonucleotide for each of our 1421 genes with a preferential size of 50-mer. We used the parameters described in Supplementary Table S2 to run each program.
Tiling for Trichoderma reesei
We downloaded the unmasked FASTA file of T. reesei genome v.2.0 from the Department of Energy Joint Genome Institute website (JGI): http://genome.jgi-psf.org/Trire2/Trire2.home.html. We designed a T. reesei tiling array of 60-mer oligonucleotides (oligo length), each 150 bp (oligo distance), using OligoTiler from its web interface (http://tiling.gersteinlab.org/OligoTiler/oligotiler.cgi). We set up the advanced parameters as follows: IR region = 5, IR require = 3 and repeat region overlap = 4.
We obtained ArrayDesign from the author website (http://www.ebi.ac.uk/~graef/arraydesign/). We then created sequence windows of 150 bp every 91 bp along the 87 scaffolds, using tools from exonerate software (19). We computed minimal unique prefix by setting the MAX_PREFIX_LENGTH variable at 15. Finally, we launched the oligonucleotide selection using the following parameters: a minimal uniqueness score of 0, an offset to shift window over unit for a uniqueness score of 1, a _T_m value range between 60°C and 80°C, a G number cut-off set to 15, a percent palindromic filter of 40%, a maximum number of synthesis cycles allowed of 185 and no deviation of probe length allowed.
Measurement of designed oligonucleotide properties
Since the design programs do not use the same computation method, we computed the _T_m values with the Unafold (20) suite (melt.pl script) once the designs were done, so that all the _T_m values could be compared. As the melt.pl script requires a hybridization temperature, we fixed it to 65°C, which follows our lab protocol. We set the DNA concentration to 0.00001 M, the sodium concentration to 1 M and the magnesium concentration to 0 M. We also used the Unafold suite to compute the free energies of the most probable secondary structures for each software design. We used the hybrid.pl script with a hybridization temperature set to 65°C and the same DNA and salt concentrations as for the melt.pl program. Finally, we used the melt.pl program to compute the free energy of the duplex formed by the oligonucleotide and the best off-target hit. To find the best off-target hit of each probe, we used BLAST via the NCBI standalone blastn program with the following parameters: X = 30 and W = 11.
Since the target sequence is oriented from the 5′-end to the 3′-end, the distance of the probe 5′ to the target 3′ was computed by subtracting the relative position in the sequence of the probe 5′ from the total length of the sequence.
For the tiling design, the interval between oligonucleotides was calculated by measuring the distance between the first position (start) of two consecutive oligonucleotides. To calculate the number of designed oligonucleotides by transcripts, we first obtained the ‘Filtered Models’ transcript file from the JGI, which is the filtered set of models representing the best gene model for each locus. We selected all ‘exon’ features from this file, and we calculated the number of oligonucleotides fully included in each ‘exon’. For each transcript, we next merged this exon information and obtained the total number of oligonucleotides.
To estimate the first Kane's parameter, we launched WU-BLAST (21) on each oligonucleotide using the reference database (Mouse RefSeq or T. reesei genome) with the following parameters: expectation threshold for reporting database hits sets to 1.2 (E), seed word length for the ungapped BLAST algorithm sets to 11 (W), negative penalty score for mismatch nucleotides in the BLASTN search mode sets to –3 (N), penalty for a gap of length 1 to 3, per-residue penalty for extending a gap to 3 and not creating gapped alignments. From the output files, we counted the number of hits that presented more than 75% identity, for an alignment length that was equal to the probe size.
OLIGONUCLEOTIDE DESIGN FOR A CUSTOM SET OF SEQUENCES
The goal of the oligonucleotide design software is to find the best oligonucleotide(s) for each biological entity (gene, exon, etc.) among a user-defined set. Usual input parameters are as follows: on one side are the sequences whose expression we want to record using microarrays (called the target data set), and on the other side are the ones representing all the sequences that could potentially cross-hybridize (called the non-target data set). The aim is to obtain one (or more) specific probe(s) for each sequence of the target data set. This process requires dealing with structural parameters, including sequence complementarity of the oligonucleotides for their targets, and with more complex thermodynamic properties like the melting temperature of the duplex (_T_m) and the ability of the probe to form stable secondary structures that avoid formation of the duplex. Here we focus our overview on 20 available tools that allow the design of long oligonucleotides for target sequences providing a non-target sequences dataset (Table 1 and Supplementary Table 1). From this point on, we will use the word ‘probe’ to define the oligonucleotides spotted on the array and the word ‘target’ for the products found in samples hybridized to the array which can bind to the probe.
Table 1.
Description of the parameters involved in the oligonucleotide specificity and the way they are calculated
| Design program | Cross-hybridization assessment | Management of low complexity region | Design orientation | Number of probes designed by gene |
|---|---|---|---|---|
| ArrayOligoSelector (37) | Blast and thermodynamic calculations | Filter out probes using a lossless compression score | Probes ranking according to the 3′ end distance | Chosen by the user |
| CommOligo (38) | Thermodynamic calculations and Kane's specifications | Masking sequences with nucleotide repeats | Design starting from the 3′- or 5′-end | Chosen by the user |
| GoArrays (39) | Blast and Kane's specifications | Filter out probes with prohibited sequences | Read input sequences from the 3′ end | Chosen by the user |
| HPD (40) | Multiple alignment and hierarchical clustering (ClustaW) | No masking or filtering | No localization specified | All probes reaching selection criteria |
| Mprime (41) | Blast (wublast) | No masking or filtering | Probes weighted towards 3′-end (optional region specification) | Chosen by the user |
| OliD (42) | Blast | Filter out probes with trinucleotide repeats | Preference given to the 3′-end proximity | Chosen by the user |
| OligoArray (43) | Blast and thermodynamic calculations | Masking sequences with nucleotide repeats | Distance to the 3′-end specified by the user (max 1500) | Chosen by the user |
| Oligodb (44) | Blast | Masking sequences using DUST with BLAST | No localization specified | Chosen by the user |
| OligoFaktory (45) | Blast | Masking sequences using DUST with BLAST | Design starting from the 3′- or 5′-end | Chosen by the user (up to 3) |
| OligoPicker (46) | Blast and repetitive sequence stretches | Masking sequences using DUST with BLAST | Design choosen for the 3′- or the 5′-end | Chosen by the user (up to 5) |
| OligoWiz (47) | Blast and thermodynamic calculations | Filter out probes using a low complexity score calculation | Localization score based on centre, 5′- or 3′-distance | All probes reaching selection criteria |
| Oliz (48) | Blast and Kane's specifications | No masking or filtering | Probes designed in the 3′-UTR | All probes reaching selection criteria |
| Osprey (49) | Position Specific Scoring Matrix and Gribskov profiles | Masking sequences with nucleotide repeats | 5′- or 3′-bias as the last constraint | All probes reaching selection criteria |
| PICKY (50) | Suffix array approach using Kane's specifications and thermodynamic calculations | Masking sequence with a low complexity using suffix array structure | No localization specified | Chosen by the user (up to 5) |
| PROBEmer (51) | Suffix array approach | No masking or filtering | Optional range of positions | All probes reaching selection criteria |
| Probesel (52) | Suffix array approach and thermodynamic calculation | No masking or filtering | No localization specified | One probe by target gene |
| ProbeSelect (53) | Suffix array approach and thermodynamic calculation | Filter out probes with nucleotide repeats | No localization specified | Chosen by the user |
| ROSO (54) | Blast | Filter out probes with nucleotide repeats | Optional probe localization in a 3′ or 5′ range | Chosen by the user |
| SEPON (55) | Blast | No masking or filtering | Penalty given based on 3′ distance | Chosen by the user |
| YODA (56) | Custom sequence similarity search tool (SeqMatch) | Filter out probes with prohibited sequences | Selection of 5′-end, 3′-end, centre or all probes | All probes reaching selection criteria |
Specificity
Specificity is defined according to the ability of the probe to bind to non-target sequences in the hybridization sample. Cross-hybridization is usually one major source of non-specificity. Most of the programs (12 out of 20, see Table 1) use the BLAST algorithm or one of its derivatives to search for cross-hybridization. Some programs (ROSO, MPrime, ArrayOligoSelector and OligoWiz) use BLAST to set a homology threshold that determines whether the oligonucleotide is specific. Kane's recommendations for cross-hybridization have set a reference to discard probes that have a total percent identity >75–80% with a non-target sequence, or probes with contiguous stretches of identity >15 nt with a non-target sequence (17,22). Half of the programs that use the BLAST algorithm to evaluate cross-hybridization (ArrayOligoSelector, GoArrays, OligoArray, OligoPicker, OligoWiz and Oliz) apply these recommendations. Thermodynamic calculations are also used to evaluate cross-hybridization (ArrayOligoSelector, OligoArray and OligoWiz). Calculating the binding-free energy of the duplex formed by the probe and the non-target sequence gives an indication of this duplex's stability. Since the duplex is bound to the slide surface, and not free in solution, all authors agree that this calculation is approximative. Nevertheless, it can be considered as a good criterion to rank probe candidate according to their cross-hybridization ability, and thus evaluating their relative specificity. The most commonly used formula is the nearest neighbour (NN) model (23). To overcome BLAST similarity search limitation and improve the performances, PICKY, PROBEmer, Probesel and ProbeSelect apply a suffix array approach to the representation of the data set space (24). The suffix array data structure allows the exhaustive localization of common sub-strings instead of performing a first multi-alignment, since the main limitation is due to memory storage of the suffix structure. As for the BLAST algorithm, the cross-hybridization evaluation is supplemented using thermodynamic calculations (Probesel, ProbeSelect, PICKY) and Kane's specifications on sequence similarity (PICKY). Finally, four programs apply a custom methodology to evaluate specificity. CommOligo uses a global alignment and predicts cross-hybridization using three measurements between probe and non-targets: minimum binding-free energy, maximum sequence identity and continuous stretch. HPD relies on a multiple-alignment and a hierarchical clustering approach to generate specific probe candidates. Osprey and YODA each used their own method to evaluate sequence similarity.
Low-complexity sequences may also affect probe specificity. Regions that contain such sequences are often masked. Therefore, no probe candidates can be designed in those parts of the genome (Table 1). A lot of programs apply a filter or mask to nucleotide repeats (CommOligo, OliD, OligoArray, Osprey, ProbeSelect and ROSO). Others offer the possibility to discard regions defined by the user as prohibited (GoArrays and YODA). Low-complexity regions can also be defined by more complex calculations based either on a lossless compression algorithm for ArrayOligoSelector, or the properties of the suffix array structure for PICKY, or a custom complexity score for OligoWiz. Low-complexity regions can finally be masked using the DUST program (25), which is included in the tools that use the BLAST algorithm for cross-hybridization assessment. Oligodb, OligoFaktory and OligoPicker clearly state that this low-complexity filtering option is used with BLAST. This is not the case for the other ‘BLAST-based’ tools, even if the option can be chosen when the BLAST program is launched locally.
Oligonucleotide specificity can also be influenced by design orientation, which partly depends on the retro-transcription priming method used. For prokaryotic sequences, retro-transcription primers are usually random hexanucleotides, while poly(T) are used in the case of eukaryotic sequences. Thus 5′ ends and 3′ ends are over-represented in hybridization samples of prokaryotic or eukaryotic sequences, respectively. In addition, the 3′UTR of the mRNA is considered the less-conserved region in eukaryotic sequences (26). Consequently, choosing 3′UTRs for oligonucleotide design reduces the probability of cross-hybridization with close paralogues, but the potential alternative polyadenylation signals found in 3′UTR must be carefully taken into account. In contrast, probes localized in the 5′ region may cause a higher risk of potential cross-hybridization and alternative splicing. Table 1 displays the various design-orientation preferences for each software program. A lot of them are clearly oriented towards the 3′-end (ArrayOligoSelector, GoArrays, Mprime, OliD, OligoArray, Oliz and SEPON). Oliz software has even been specifically created for the design of probes located in the 3′-UTR region. Only a few tools allow for the customization of the oligonucleotide design orientation. To do so, OligoWiz applies a localization score based on distance to the centre, or to one of the ends (5′or 3′), of the sequence. OligoFaktory, OligoPicker, ROSO, PROBEmer and Osprey let the user localize the designed probes in a 3′-or 5′-range, while YODA displays either all of the non-overlapping oligonucleotides or only the ones located in a precise region (centre, 3′ or 5′) of the sequence.
The last way to influence specificity is to modify the number of probes per gene. The comparison of all probes that cover each gene can help to interpret the expression of various isoforms. Two different approaches deal with this issue (Table 1). Some of the tools generate all of the designed probes that meet the selection criteria (HPD, OligoWiz, Oliz, Osprey, PROBEmer and YODA), while others let the user choose the number of probes per gene to be found (OligoFaktory, OligoPicker and PICKY). At the opposite extreme, Probesel automatically selects one probe per target gene. When multiple oligonucleotides are found in the software output, a visualization interface is available to help the user choose among them (OligoWiz, OligoFaktory, HPD, PROBEmer, Oligodb and MPrime). In this case, the user has to select each probe individually, which can be a very tedious task.
Sensitivity
Sensitivity is defined by the strength with which a probe binds to its target sequence. It influences the strength of the signal read from the microarray and the relevance of the obtained information. Modifying the oligonucleotide length is the first parameter that can influence sensitivity. We report in Table 2 the possibility to set oligonucleotide length in each evaluated program. Only one software candidate limits the design of probes to 50-mer (Oliz). In contrast, most of the tools allow the final oligonucleotide size to be set by the user, either exactly (CommOligo, GoArrays, HPD, Mprime, OliD, ArrayOligoSelector, Oligodb, OligoPicker, PROBEmer, ProbeSelect, ROSO, SEPON and YODA) or within a size range after optimization of other parameters (OligoArray, OligoFaktory, OligoWiz, Osprey, PICKY and Probesel). This latest solution can strengthen probe uniformity.
Table 2.
Description of the parameters involved in the oligonucleotide sensitivity and the way they are calculated
| Design program | Oligo length | _T_m calculation | GC percent | Secondary structure assessment |
|---|---|---|---|---|
| ArrayOligoSelector | Fixed by the user | Not a selection criteria | Filter out probes using user fixed threshold | Evaluated by self-complementarity |
| CommOligo | Fixed by the user from 10 to 128 mer | Range chosen by the user, _T_m calculated using NN model | Masking sequences using user defined range | Evaluated by self-complementarity |
| GoArrays | Fixed by the user | Range chosen by the user, _T_m calculated using NN model | Not a selection criteria | Using Mfold |
| HPD | Fixed by the user | Not a selection criteria | Filter out probes using user fixed threshold | Evaluated using self-folding energy (hairpin) |
| Mprime | Fixed by the user | Range chosen by the user, _T_m calculated using NN model | Filter out probes out of the user defined range | With scoring calculation from Kampke et al. (32) |
| OliD | Fixed by the user | Not available | Filter out probes using user fixed threshold | Using Mfold |
| OligoArray | Range chosen by the user from 15 to 75 mer | Range chosen by the user, _T_m calculated using NN model | Filter out probes out of the user defined range | Using a custom module similar to Mfold |
| Oligodb | Fixed by the user from 20 to 100 mer | Not a selection criteria, _T_m calculated using melting | Not a selection criteria | Using Mfold |
| OligoFaktory | Range chosen by the user | Range chosen by the user | Not a selection criteria | Not a selection criteria |
| OligoPicker | Fixed by the user from 20 to 100 mer | Range chosen by the user, _T_m calculated using a custom method | Not a selection criteria | Evaluated by self-complementarity |
| OligoWiz | Range chosen by the user, _T_m optimization | Range chosen by the user, _T_m calculated using NN model | Not a selection criteria | Evaluated using a folding energy algorithm |
| Oliz | Fixed at 50 mer | Fixed range around 76°C, _T_m calculated using prima (EMBOSS) | Range fixed between 45% and 50% | Not a selection criteria |
| Osprey | Range chosen by the user from 10 to 90 mer | Range chosen by the user, _T_m calculated using NN model | Not a selection criteria | With dimer and hairpin free energy calculation |
| PICKY | Range chosen by the user | _T_m optimisation, calculated using NN model | _T_m optimization apply user defined range | Include in the cross-hybridization screening |
| PROBEmer | Fixed by the user | Range chosen by the user | Filter out probes out of the user defined range | Evaluated by self-complementarity as in Primer3 |
| Probesel | Range chosen by the user | _T_m calculated using NN model | Not a selection criteria | Using Mfold |
| ProbeSelect | Fixed by the user | Not a selection criteria, _T_m calculated using a custom method | Not a selection criteria | Evaluated by self-complementarity |
| ROSO | Fixed by the user | Range chosen by the user, _T_m calculated using NN model | Preferred range between 40% and 65% | Hairpin and homoduplex free energy calculation |
| SEPON | Fixed by the user | _T_m calculated using melting | Using a penalty if GC content is far from 40% to 60% | Using Mfold |
| YODA | Fixed by the user | Range chosen by the user, _T_m calculated using NN model | Filter out probes out of the user defined range | Evaluated by self-complementarity |
Since the hybridization of all oligonucleotides with their targets occurs simultaneously on microarrays, the primary objective, in order to achieve maximum homogeneity of the oligo set, is to utilize probes that share a similar melting temperature (_T_m). Several methods are available to calculate the _T_m. The most frequently used method (Table 2) consists of applying the NN model with either the parameters from SantaLucia (23) or the parameters from Rychlik (27). This calculation can be performed directly by the program itself or by using an external software such as Melting (28) with Oligodb and SEPON, or prima, from the EMBOSS package (29), with Oliz. OligoPicker and ProbeSelect use a custom method. Although most of the programs allow the user to select a range in which the oligonucleotide _T_m must be found, some software (OligoArray, OligoPicker, PICKY and YODA) applies an optimization calculation to adapt other parameters (such as oligonucleotide size) in order to obtain the final narrowest _T_m range. For ArrayOligoSelector, Oligodb and ProbeSelect, _T_m is not taken into account, probably because it is assumed that oligonucleotides with uniform length and GC content share close _T_m values. Lastly, one has to know that all the formulas that are used calculate the _T_m for oligonucleotides free in solution, and not for attached DNA on a slide, as with microarrays. But we assume that the most important consideration is that the probes fit to the same _T_m range and not to a precise _T_m reference.
Closely related to the _T_m is the GC content of the oligonucleotide sequences. Eight of the software candidates (GoArrays, Oligodb, OligoFaktory, OligoPicker, OligoWiz, Osprey, Probesel and ProbeSelect) do not consider the GC content as a selective criterion during the oligonucleotide selection process (Table 2). As for the other tools, the designed probes that do not fulfil the GC percent range or threshold fixed by the user are filtered out from the final probe list (ArrayOligoSelector, CommOligo, HPD, Mprime, OliD, OligoArray, PROBEMer and YODA). Oliz sets oligonucleotide length (50-mer), _T_m optimum (76°C), and GC content (45–60%). In the same manner, ROSO uses a preferential range between 40% and 65% and SEPON applies a penalty score for oligonucleotide when the GC percent is outside of the 40–60% range. Finally, PICKY takes a GC percent that is defined by the user in order to perform a _T_m optimization and to select the best probe candidates. This method can be useful when dealing with sequences from genomes with very high or low GC percents.
In order to achieve maximal sensitivity, optimal probe design requires the exclusion of oligonucleotides that are able to form homo-dimers or stable intra-molecular secondary structures like hairpins or stem–loops. The goal of the secondary structure assessment step (Table 2) is to avoid the formation at the hybridization temperature of such structures that prevent stable target hybridization. Two methods take secondary structure formation into account. The first one deals with self-complementarity and uses alignments of the oligonucleotide with its reverse-complement sequence; the other one is based on thermodynamic calculations in order to determine the stability of potential secondary structures. The self-complementarity criterion used by ArrayOligoSelector, CommOligo, OligoPicker, PROBEmer, ProbeSelect and YODA assumes that a self-complementary oligonucleotide will form stable structures, such as dimers and hairpins. With PROBEmer, the self-complementarity computation uses the same methodology as the one found in Primer3 (30). In contrast, most of the programs that use thermodynamic calculation (GoArrays, OliD, OligoArray, Oligodb, Probesel and SEPON) rely on the Mfold tool (31) or similar derivatives that determine the stability of secondary structure. HPD, OligoWiz, Osprey and ROSO use an independent thermodynamic calculation, mostly because using the Mfold program is time-consuming. Mprime uses a different solution based on scoring calculation (32), while PICKY takes advantage of a suffix array data structure to perform the secondary structure stability search coupled with the cross-hybridization measurement.
Versatility and availability
Here we will go through some aspects of the software set up (Table 3). First, we will deal with the ability to design probes for a limited number of organisms. The vast majority of programs can design oligonucleotides for all organisms. Some programs only have a limitation on their sequence resource, as the NCBI database (OligoFaktory), or on their EST collections (SEPON). OligoWiz is more restricted, since it can work only for the organisms found on its server; however, all the classical model organisms are available, and other resources may be added upon request. In contrast, Mprime designs oligonucleotides only for rat, mouse, human, drosophila and zebra fish, while Oligodb works only for human. The choice of a particular organism permits specifying a precise database to perform the cross-hybridization analysis (Table 3). Most of the programs allow loading a FASTA file of all the sequences that we want to use as the ‘specificity bank’ (ArrayOligoSelector, GoArrays, OligoArray, OligoPicker, Osprey, PICKY, PROBEmer, ROSO and YODA), but other programs help the user to build the ‘specificity bank’ by retrieving an already existing one from RefSeq (Mprime), NCBI repository (OligoFaktory) or EST collection (SEPON). It is even easier when the organism is available and the ‘specificity bank’ is already formatted (Mprime, Oligodb, OligoFaktory and OligoWiz), even if the problem of database updating has to then be taken into account. On the other hand, the ‘specificity bank’ used by Probesel is the target database itself. This may be a real drawback for microarrays dedicated to gene subsets.
Table 3.
Description of the availability and flexibility of the oligonucleotide design software
| Design program | Organism | Specificity bank | Accessibility (free for academics) | User interface | Programming language | Running time |
|---|---|---|---|---|---|---|
| ArrayOligoSelector | No limitation | Fasta file | Downloadable from a website | Command line (L) | Python | 52 min |
| CommOligo | No limitation | Fasta file | Available upon request | Standalone GUI (W) | C++ | 1156 min |
| GoArrays | No limitation | Fasta file | Downloadable from a website | Standalone GUI (L) | Java | ND |
| HPD | Not available | Not available | Downloadable from a website | Standalone GUI (W) | Object Pascal | ND |
| Mprime | Rat, mouse, human, drosophila and zebrafish | RefSeq database for the organism | Web interface only | Web interface | C++ and Perl | 31 min |
| OliD | No limitation | Genome sequence | Available upon request | Command line (L) | Python | ND |
| OligoArray | No limitation | Fasta file | Downloadable from a website | Command Line (L) and Standalone GUI (L, W, M) | Java | 141 min |
| Oligodb | Human | All human cDNA transcripts in ENSEMBL | Web interface only | Web interface | Unknown | ND |
| OligoFaktory | All organisms with an NCBI database | A predefined or custom NCBI database | Web interface and downloadable program | Web interface and MacOS standalone GUI | Unknown | 43 min |
| OligoPicker | No limitation | Fasta file | Downloadable from a website | Command line (L) | Perl | 30 min |
| OligoWiz | All organisms found on the server | Located on the server | Client program downloadable | Client GUI (L, W, M) | Java client, Perl server | 26 min |
| Oliz | No limitation | Not available | Downloadable from a website | Command line (L) | Perl | ND |
| Osprey | No limitation | Fasta file | Web interface and source available upon request | Web interface | C and Perl | ND |
| PICKY | No limitation | Fasta file | Available upon request | Standalone GUI (L, W, M) | C++ | 11 min |
| PROBEmer | No limitation | Fasta file | Web interface only | Web interface | C | ND |
| Probesel | No limitation | Same as target sequences | Available upon request | Command line (L) | C++ | ND |
| ProbeSelect | No limitation | Proprietary format | Available upon request | Command line (L) | C++ | ND |
| ROSO | No limitation | Fasta file | Web interface and standalone program available upon request | Web interface and standalone GUI (W) | C | 418 min |
| SEPON | All organisms with an EST collection | A source EST collection | Available upon request | Command line (L) | Perl | ND |
| YODA | No limitation | Fasta file | Downloadable from a website | Command line (L, W, M) | Java | 3 min |
Only tools freely available for academics were considered in this overview. In terms of accessibility (Table 3), although a lot of tools are downloadable from their creators’ websites (ArrayOligoSelector, GoArrays, HPD, OligoArray, OligoFaktory, OligoPicker, Oliz and YODA), some are available from the authors only upon request (CommOligo, OliD, Osprey, PICKY, Probesel, ProbeSelect, ROSO and SEPON), which could potentially create accessibility limitations in the future. We were, for example, unable to obtain OliD. Mprime, Oligodb, OligoWiz, OligoFaktory, ROSO and PROBEmer are accessible through a server application. In addition, OligoFaktory and ROSO are also available using a standalone application.
The last parameter we consider here is the user interface (Table 3). Mprime, Oligodb, OligoFaktory, Osprey, PROBEmer and ROSO give access to oligonucleotide design through a web interface. Since it does not require any local software installation, this solution is the easiest one, but it can often hinder the design of a large number of oligonucleotides. In addition, parameter customization is often less transparent using such interfaces. For users who would like to perform the oligonucleotide design on their own computers (for confidentiality or speed reasons), CommOligo, GoArrays, HPD, OligoArray, OligoFaktory, PICKY, ROSO and YODA offer a stand-alone program with a graphical user interface (GUI). Most of these tools work on Linux/Unix systems, except for CommOligo and HPD, which work on Windows, and OligoFaktory, which works with Mac OS. In addition, Java-based programs (OligoArray, PICKY and YODA) are multi-platform and work on all the previous operating systems. A hybrid solution is found using OligoWiz, as this program has two components, a Java client with a GUI that works on Linux, Windows and Mac OS, and a server that performs thermodynamic calculations. This server can be installed locally, but this solution is only commercially available. Lastly, ArrayOligoSelector, OliD, OligoPicker, Oliz, Probesel, ProbeSelect and SEPON must be used with command lines and often require a local installation of other programs such as BLAST or Mfold. We provide the programming language used for each design software in Table 3.
TILING OLIGONUCLEOTIDE DESIGN
Oligonucleotide design for tiling encounters the same constraints as described above in terms of the sensitivity and specificity of each selected probe. In addition, tiling design needs to take the tiling path into account, which includes probe coverage and distribution. For the design of oligonucleotides in each sequence window, the program has to choose between position and hybridization quality. We selected five tools for our comparison, based on the same criteria as the custom oligonucleotide design. The first one, called MAMMOT, is dedicated to creating a tiling path in small regions across the genome. MAMMOT relies on the Primer3 program (30) in order to create a complete set of PCR primers along the region that is used as the tiling path. However, this method handles probe sensitivity and low complexity but does not deal with cross-hybridization management. OligoTiler and the Lipson et al. algorithm also focus on tiling path optimization in order to obtain a more uniform distribution of probes. In addition, OligoTiler is able to find tiling oligonucleotides even in repeat regions but offers limited control over sensitivity and specificity. Only low complexity (overlapping repeats) and secondary structures (inverted repeats) are evaluated. The Lipson et al. method works on a subset of high-quality oligonucleotides. Using this set, the algorithm selects the oligonucleotides that ensure the most homogenous distribution of the tiling path. With both solutions, the main constraint arises from the oligonucleotide position and not from its quality. On the other hand, ArrayDesign tries to select the oligonucleotide of better quality in each window of the tiling path, whereas Tileomatic tries to optimize both position and quality using an implementation of the shortest-path algorithm. All solutions using the BLAST program are ruled out due to the fact that the cross-hybridization assessment has to be done on a larger number of candidate oligonucleotides for tiling arrays than for the expression microarrays. The evaluation of specificity is performed using either a suffix array approach (Tileomatic) or a uniqueness score calculation (ArrayDesign) based on minimum unique prefix count for each oligonucleotide. Both methods allow the design of oligonucleotides in repeat regions if a large unique overlapping probe can be found. Both use a filter for GC content and _T_m temperature, but the precise information on quality probe measurement is not available from the Tileomatic publication.
DESIGN OF A MOUSE NERVOUS SYSTEM DEVELOPMENT OLIGO SET
For our custom test case, we chose a gene collection that focused on genes that are known to be involved in the development of the mouse nervous system. This set consists of 1421 genes, each of them associated with one unique Entrez Gene identifier. We selected half of the 20 design programs we evaluated previously for the oligonucleotide design. We first discarded nine of them because they did not fulfil all our needs. GoArrays has been developed to design a pair of short oligonucleotides linked together with a short spacer. HPD finds common or discriminant oligonucleotides of conserved genes. Oligodb is dedicated to human design only. Oliz designs oligonucleotides in the 3′-UTR using ESTs. SEPON works only with non-annotated sequences. PROBESEL uses the same specificity bank as the target one, which is a potential problem for the design of probes for a gene subset. Osprey is a web-server tool and cannot deal with a target DNA sequence file bigger than 10 Mb. ProbeSelect's file format does not fit wide genome organisms. Finally, ProbeMer's web server was not available and we did not obtain any response to our request for the OliD program.
We attempted to design one oligonucleotide for each of the 1421 genes with the programs we retained using the parameters described in the ‘Material and Methods’ section and in Supplementary Table 2. Only ArrayOligoSelector, OligoWiz2 and YODA succeeded in selecting 100% of the expected oligonucleotides with our design settings (Table 4). Commoligo, OligoArray and Mprime reached more than 90%. OligoPicker, ROSO and PICKY designed between 73% and 88% of our targets. Lastly, OligoFaktory was unable to design more than half of the oligonucleotides we expected.
Table 4.
Property for each oligonucleotide set created for the custom mouse array
| ArrayOligoSelector | CommOligo | Mprime | OligoArray | OligoFacktory | OligoPicker | OligoWiz | PICKY | ROSO | YODA | |
|---|---|---|---|---|---|---|---|---|---|---|
| Probe number | 1421 | 1392 | 1299 | 1383 | 580 | 1256 | 1421 | 1042 | 1163 | 1420 |
| Probe size | 50.0 ± 0 | 50.0 ± 0 | 50.0 ± 0 | 50.2 ± 0.46 | 51.5 ± 0.74 | 50.0 ± 0 | 49.9 ± 3.71 | 51.0 ± 0.84 | 50.0 ± 0 | 50.0 ± 0 |
| Specificity (%) | 94.23 | 81.82 | 78.06 | 82.21 | 80.34 | 98.89 | 83.11 | 98.46 | 78.59 | 81.83 |
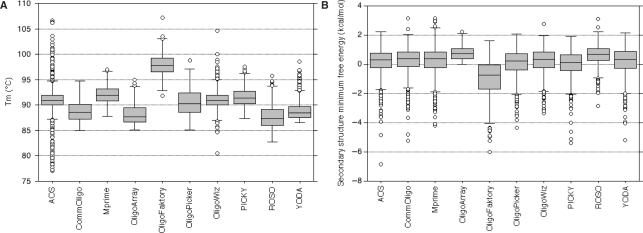
Homogeneous _T_m values are ideal to increase sensitivity during the hybridization process (Figure 1A). When we look at the _T_m mean and the interquartile range (IQR), ArrayOligoSelector (_T_m mean of 90.94°C, IQR of 1.9) and OligoWiz (_T_m mean of 91.09°C, IQR of 2) show the narrowest _T_m distribution associated with greater _T_m means. OligoFaktory also provides a high _T_m mean (97.84°C), but this result must be considered with caution, as the design was successful with only half of the expected oligonucleotides. The percentage of GC distribution (Supplementary Figure 1) is closely linked to _T_m values, since this parameter is required in the NN model used for _T_m calculation. Thus, the overall distribution of GC percent across all oligo sets follows the distribution of _T_m values. ArrayOligoSelector applies a strong constraint on this parameter (set by the user as 50% here), as the IQR around the fixed GC percent mean is equal to 0. The acceptable GC percent is usually set between 40% and 60%, and all oligo sets fit in this range except for OligoFaktory (63.9%). Sensitivity can also be adjusted by adapting the oligonucleotide size to fit within the _T_m and GC percent set up by the software. OligoArray, OligoPicker, ArrayOligoSelector, ROSO, YODA, CommOligo and Mprime design all the oligonucleotides with the same size (50 bp). The other software programs adapt the oligonucleotide size to optimize _T_m distribution (Table 4). This size distribution can be very wide, since OligoWiz designed oligonucleotides from 45-mer to 55-mer. The advantage of such a choice is not clear when looking at the _T_m distribution (Figure 1A), as OligoWiz does not have either a higher _T_m mean or a narrow IQR compared to programs that apply a strict constraint on the oligonucleotide size, such as ArrayOligoSelector or Mprime. The last parameter that can interfere with oligonucleotide sensitivity is the ability of oligonucleotides to form a stable secondary structure. We evaluated the self-hairpin free energy of designed oligonucleotides; as shown in Figure 1B, all box plots are very similar, except for OligoFaktory. The OligoFaktory profile can be explained by the higher GC percent of the oligonucleotides in this set, as GC-rich regions favour better stability of hairpin structures (33).
Figure 1.
Comparison of the sensitivity of the oligonucleotides designed for the custom mouse array. For each oligonucleotide set created we plot the distribution for all oligonucleotides in the set of _T_m (A) and free energies of the most probable secondary structure (B). The name of the software used for design is displayed on the _x_-axis. AOS stands for ArrayOligoSelector.
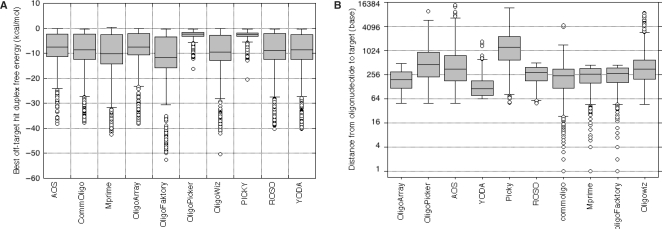
Oligonucleotide cross-hybridization (specificity) can be evaluated in several ways. One of them uses the first Kane parameter. For each designed probe, we counted the number of BLAST hits that had 75% or more identity on a full-size alignment along the whole probe. To avoid cross-hybridization, a specific oligonucleotide must not be able to bind to any other target than its dedicated one. This means that only one hit is supposed to be found after applying Kane's filter on BLAST output. We report in Table 4 the percentage of probes with only one hit according to these parameters. ArrayOligoSelector, OligoPicker and PICKY achieve the best specificity, with more than 94% of the designed probes without cross-hybridization according to Kane's first parameter. All the other software programs exhibit a quite narrow specificity, from 78% to 83% of probes with unique hits. In order to take sequence composition into account, we also computed the duplex free energy between the oligonucleotide and the best off-target hit, as determined using the BLAST program (Figure 2A). OligoPicker and PICKY clearly got the highest and narrowest free energy distributions in comparison with all other programs that did not show great differences. These observations demonstrate first that the results obtained using thermodynamic calculations are correlated with the one obtained with Kane's parameter. Indeed, we obtained the best measures with PICKY and OligoPicker. Second, these data do not point out any correlation between the estimated specificity and the way it is calculated. In fact, OligoPicker estimates specificity using BLAST, whereas PICKY uses suffix array to solve Kane's specifications and thermodynamic calculations.
Figure 2.
Evaluation of custom oligonucleotide specificity. (A) Duplex free energies between oligonucleotides and their best off-target hit. (B) Distribution of the distance between the 5′ of the oligonucleotide and the 3′ of the target gene sequence for each designed oligoset. The name of the software used for design is displayed on the _x_-axis. AOS stands for ArrayOligoSelector.
In addition, we looked at the distance between the oligonucleotide and the 3′-end of the target (Figure 2B). Oligonucleotide specificity does not take this measurement into account. Localization close to the 3′-end ensures the best chance to get a signal for small sample material. Limiting the distance to the target 3′-end is therefore useful and all the programs, except YODA, are able to design oligonucleotides at more than 4 kb from the 3′-end. Note that PICKY has the largest spread of oligonucleotide distance, which could be a potential drawback with amplified samples.
TILING OLIGO SET AGAINST A SMALL FUNGAL GENOME
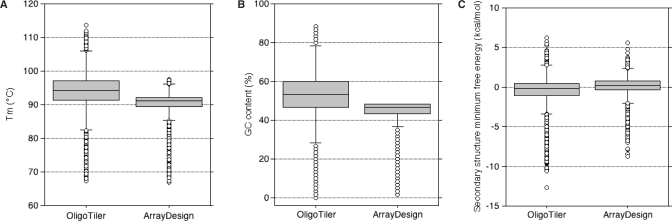
We also tested an oligonucleotide tiling design on the 34 Mbp T. reesei fungal genome (34). Our goal was to obtain coverage of one oligonucleotide per 150-bp window all along the genome. We used only ArrayDesign and OligoTiler, since MAMMOT is not dedicated to whole-genome design and neither the Tileomatic nor the Lipson et al. algorithms were available. Using the parameters described in the ‘Material and Methods’ section, ArrayDesign created 236 185 oligonucleotides and OligoTiler 222 778. The ArrayDesign oligo set exhibits the narrowest distribution of Tm and GC percent values (Figure 3A and B), which could point to better oligonucleotide sensitivity, even if the _T_m mean is lower than the OligoTiler one (93.91°C for OligoTiler and 90.16°C for ArrayDesign). ArrayDesign applies a range for _T_m selection that is represented by the upper cut-off on the _T_m distribution and GC percent. Oligos designed using OligoTiler may form more self-hairpin secondary structures than the ones designed with ArrayDesign (Figure 3C), although OligoTiler specifically used an inverted repeat filter.
Figure 3.
Comparison of the sensitivity of the oligonucleotides designed for tiling array. For each oligonucleotide set created we plot the distribution for all oligonucleotides in the set of _T_m (A), GC percent (B) and free energies of the most probable secondary structure (C).
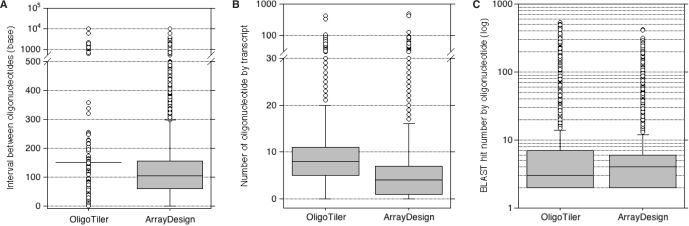
The critical step for tiling array design is the tiling path. Thus, for the same coverage (number of oligonucleotides on the genome), a uniform distribution of oligonucleotides provides greater detection of individual gene features. This can be measured using the interval between adjacent oligonucleotides. Figure 4A shows that the median of the interval designed by OligoTiler reflects our expectations (150 bp) with only a small variation (±27.8 bp), whereas the distribution of ArrayDesign's intervals has a median of 103 bp (±172.3 bp). The larger distribution of intervals between oligonucleotides with ArrayDesign may be a direct consequence of the ‘specificity’ optimization that the program performs, with a design mainly focused on conserved regions such as exons. We calculated the number of oligonucleotides designed for each transcript on the genome. Figure 4B shows that OligoTiler supplies a uniform distribution of oligonucleotides, and therefore achieves better coverage of transcripts than ArrayDesign. Indeed, OligoTiler designs eight probes per transcript, while ArrayDesign finds only four probes for each coding sequence. Finally, we evaluated oligonucleotide specificity using the first Kane parameter. For each designed oligonucleotide, we counted the number of BLAST hits that had an identity percentage ≥75% on a full-size alignment (60 bp). The number of oligonucleotides with only one hit is slightly greater (97%) using OligoTiler than ArrayDesign (96%). However, considering only the oligonucleotides with more than one hit (Figure 4C), the median hit number by oligonucleotide is four for ArrayDesign and three for OligoTiler. This comparison points out that these two different approaches achieve quite the same efficiency in terms of specificity based on BLAST hit calculation according to the first Kane parameter.
Figure 4.
Evaluation of tiling oligonucleotide specificity. (A) Distribution of the distance in base pair between oligonucleotide that follows each other on the tiling path. (B) Distribution of the number of oligonucleotide by transcript. (C) Distribution of the number of BLAST hits by oligonucleotide using the parameters described in the ‘Material and methods’ section. The _y_-axis is log scaled. To clearly display these distributions we removed all oligonucleotides with only one hit.
CONCLUSION
We reported the oligonucleotide properties of probe sets we obtained using several design programs to answer the same biological question. In light of these results, we wondered if it was possible to find common properties of design algorithms to select better probes. _T_m calculation is often described as the most important parameter of empirical experiments, even if this point is still controversial (35,36). Determining _T_m thresholds is a hard task. Probes with a high _T_m may cause saturating signals after scanning, whereas low _T_m probes may lead to weak signals that are undetectable from the background. This detection sensitivity is influenced by a lot of parameters, such as the scanner settings, the organism used or the quality of the samples. This is the reason why selecting absolute minimal and maximal _T_m values is almost impossible. Since hybridization occurs at the same temperature for all probes during the microarray experiment, we suggest obtaining the narrowest _T_m distribution in order to avoid low or high probe detection. ArrayOligoSelector and OligoWiz offer two different approaches to reach this goal (Figure 1A). If OligoWiz sets a _T_m range and estimates probe _T_m using the NN model, ArrayOligoSelector does not apply any constraint on _T_m values and filters probes using GC content only. In addition, it has been suggested that varying the length of oligonucleotides leads to a better isothermal probe design (36). From our data we do not see any differences in the _T_m value distributions between OligoWiz, which uses an isothermal design approach, and ArrayOligoSelector, which uses a fixed probe size.
Secondary-structure formation can also greatly influence the strength of the probe's signal. In previous studies, probes with a self-folding energy lower than –1 kcal/mol show decreasing detection signals (35). OligoArray and ROSO are the two design programs whose probes’ self-folding energy distribution is higher than this –1 kcal/mol threshold. Both methods use thermodynamic calculations to estimate secondary structure, whereas a large majority of other programs are based on self-complementarity evaluation. In addition, OligoArray uses Mfold to perform these calculations. This leads to less stable secondary structures and therefore to a better detection signal. In contrast, OligoFaktory does not take account of secondary-structure formation for probe selection. Oligonucleotides designed with this program show the lowest self-folding free energies and therefore the more stable secondary structure. Such structures avoid target hybridization and thus provide correct detection.
Most design programs claim that their goal is to reach the best oligonucleotide specificity. We show here that OligoPicker and PICKY are the best programs to perform a specificity estimation using either similarity search (Kane's parameters) or thermodynamic calculation. These two methods lead to similar results. In addition, it has been demonstrated in previous works that the impact of sequence similarity on hybridization signal intensities is not significant in comparison with other oligonucleotide properties (36), and that similarity may decrease detected signal only for probes with a high number of hits on the genome. It appears also that defining a precise cut-off for specificity is not appropriate, since an optimal design depends on a lot of other parameters, such as: microarray type, location of the similarity on the oligonucleotide sequence, genome type and so on (35). Finally, since OligoPicker uses BLAST for specificity calculation, whereas PICKY works with suffix arrays, both methods can be used as well.
As for probes, the selection of the ‘best’ oligonucleotide design program is a hard task. However, based on the results we obtained from our tests, we can recommend a few software programs that fit the design needs. We split these recommendations in two user categories: biologists with little computer support, and more experienced computer scientists. For the first category of users, OligoWiz is a good choice. This program finds 100% of the expected probes with correct _T_m homogeneity and low-energy secondary structure. OligoWiz does not reach the best specificity score, but this program offers biologists a complete graphical interface and a detailed tutorial. CommOligo may also be considered as an alternative. This design program creates fixed size probes, whereas OligoWiz selects oligonucleotides with variable sizes. CommOligo designs almost all of the expected probes with the correct parameters. Its major drawback is that it may need a very long running time. For scientists who work with high-resolution microarray, OligoTiler gives access through a web interface to a user-friendly tiling design algorithm. For computer scientists, ArrayOligoSelector is one of the best solutions. This program shows 100% of the expected probes designed, 94% specificity and a very narrow _T_m distribution, and therefore appears to be able to design probes in almost all cases. YODA must also be considered as an interesting design program. Almost all of the probes that YODA creates are located close to the 3′ end, and the design takes a very short time. But because of its low specificity score and a low median _T_m, YODA cannot be considered to be better than ArrayOligoSelector. Lastly, OligoArray is also a design program that has to be taken into account. OligoArray finds more than 97% of the expected probes designed with variable size length, and offers a lower secondary-structure energy in the oligonucleotide set than YODA and AligoArraySelector. However, OligoArray encounters the same specificity and _T_m drawbacks as YODA.
With custom microarrays, the selection of an oligonucleotide set is a key step. Several software solutions are available to help solve probe design problems, and each of them has its own advantages and drawbacks. The oligonucleotide design is an optimization problem among all the various parameters that influence the interaction between the probe and the sample. The selection of design programs must be done according to the objective the scientist wants to achieve, depending on the organism used, the number of oligonucleotides selected and their localization on the target sequence. The present work provides insights that will help users to select the most relevant software, according to these parameters and the nature of their projects.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Réseau National Génopôle (RNG); Institut national de la santé et de la recherche médicale (INSERM); Agence nationale de la recherche (ANR); Association pour la recherche sur le cancer (ARC); and Association Française contre les Myopathies (AFM). Funding for open access charge: Institut national de la santé et de la recherche médicale (INSERM).
Conflict of interest statement. None declared.
Supplementary Material
[Supplementary Data]
ACKNOWLEDGEMENTS
We wish to thank the oligonucleotide design software developers for their help in installation and program running. We are very grateful to Thomas Portnoy for his careful reading of the manuscript. We also thank the ENS transcriptome platform staff and collaborators for providing test data.
REFERENCES
- 1.Blick RJ, Revel AT, Hansen EJ. FindGDPs: identification of primers for labeling microbial transcriptomes for DNA microarray analysis. Bioinformatics. 2003;19:1718–1719. doi: 10.1093/bioinformatics/btg218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pozhitkov AE, Tautz D. An algorithm and program for finding sequence specific oligonucleotide probes for species identification. BMC Bioinformatics. 2002;3:9. doi: 10.1186/1471-2105-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, Li KB, Sung WK. G-PRIMER: greedy algorithm for selecting minimal primer set. Bioinformatics. 2004;20:2473–2475. doi: 10.1093/bioinformatics/bth259. [DOI] [PubMed] [Google Scholar]
- 4.Raddatz G, Dehio M, Meyer TF, Dehio C. PrimeArray: genome-scale primer design for DNA-microarray construction. Bioinformatics. 2001;17:98–99. doi: 10.1093/bioinformatics/17.1.98. [DOI] [PubMed] [Google Scholar]
- 5.Thareau V, Dehais P, Serizet C, Hilson P, Rouze P, Aubourg S. Automatic design of gene-specific sequence tags for genome-wide functional studies. Bioinformatics. 2003;19:2191–2198. doi: 10.1093/bioinformatics/btg286. [DOI] [PubMed] [Google Scholar]
- 6.Xu D, Li G, Wu L, Zhou J, Xu Y. PRIMEGENS: robust and efficient design of gene-specific probes for microarray analysis. Bioinformatics. 2002;18:1432–1437. doi: 10.1093/bioinformatics/18.11.1432. [DOI] [PubMed] [Google Scholar]
- 7.Herold KE, Rasooly A. Oligo Design: a computer program for development of probes for oligonucleotide microarrays. Biotechniques. 2003;35:1216–1221. doi: 10.2144/03356bc02. [DOI] [PubMed] [Google Scholar]
- 8.Relogio A, Schwager C, Richter A, Ansorge W, Valcarcel J. Optimization of oligonucleotide-based DNA microarrays. Nucleic Acids Res. 2002;30:e51. doi: 10.1093/nar/30.11.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahmann S. Fast large scale oligonucleotide selection using the longest common factor approach. J. Bioinform. Comput. Biol. 2003;1:343–361. doi: 10.1142/s0219720003000125. [DOI] [PubMed] [Google Scholar]
- 10.Tolstrup N, Nielsen PS, Kolberg JG, Frankel AM, Vissing H, Kauppinen S. OligoDesign: optimal design of LNA (locked nucleic acid) oligonucleotide capture probes for gene expression profiling. Nucleic Acids Res. 2003;31:3758–3762. doi: 10.1093/nar/gkg580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng J, Svensson JT, Madishetty K, Close TJ, Jiang T, Lonardi S. OligoSpawn: a software tool for the design of overgo probes from large unigene datasets. BMC Bioinformatics. 2006;7:7. doi: 10.1186/1471-2105-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertone P, Trifonov V, Rozowsky JS, Schubert F, Emanuelsson O, Karro J, Kao MY, Snyder M, Gerstein M. Design optimization methods for genomic DNA tiling arrays. Genome Res. 2006;16:271–281. doi: 10.1101/gr.4452906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graf S, Nielsen FG, Kurtz S, Huynen MA, Birney E, Stunnenberg H, Flicek P. Optimized design and assessment of whole genome tiling arrays. Bioinformatics. 2007;23:i195–i204. doi: 10.1093/bioinformatics/btm200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lipson D, Yakhini Z, Aumann Y. Optimization of probe coverage for high-resolution oligonucleotide aCGH. Bioinformatics. 2007;23:e77–e83. doi: 10.1093/bioinformatics/btl316. [DOI] [PubMed] [Google Scholar]
- 15.Ryder E, Jackson R, Ferguson-Smith A, Russell S. MAMMOT–a set of tools for the design, management and visualization of genomic tiling arrays. Bioinformatics. 2006;22:883–884. doi: 10.1093/bioinformatics/btl031. [DOI] [PubMed] [Google Scholar]
- 16.Schliep A, Krause R. Algorithms in Bioinformatics. Vol. 4645. Berlin/Heidelberg, Heidelberg: Springer; 2007. pp. 383–394. [Google Scholar]
- 17.Kane MD, Jatkoe TA, Stumpf CR, Lu J, Thomas JD, Madore SJ. Assessment of the sensitivity and specificity of oligonucleotide (50mer) microarrays. Nucleic Acids Res. 2000;28:4552–4557. doi: 10.1093/nar/28.22.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang HY, Malek RL, Kwitek AE, Greene AS, Luu TV, Behbahani B, Frank B, Quackenbush J, Lee NH. Assessing unmodified 70-mer oligonucleotide probe performance on glass-slide microarrays. Genome Biol. 2003;4:R5. doi: 10.1186/gb-2003-4-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol. Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 21.Chao KM, Pearson WR, Miller W. Aligning 2 Sequences within a Specified Diagonal Band. Comput. Appl. Biosci. 1992;8:481–487. doi: 10.1093/bioinformatics/8.5.481. [DOI] [PubMed] [Google Scholar]
- 22.Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 23.SantaLucia J., Jr. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manber U, Myers G. Suffix Arrays – a New Method for Online String Searches. SIAM J. Comput. 1993;22:935–948. [Google Scholar]
- 25.Hancock JM, Armstrong JS. SIMPLE34: an improved and enhanced implementation for VAX and Sun computers of the SIMPLE algorithm for analysis of clustered repetitive motifs in nucleotide sequences. Comput. Appl. Biosci. 1994;10:67–70. doi: 10.1093/bioinformatics/10.1.67. [DOI] [PubMed] [Google Scholar]
- 26.Tomiuk S, Hofmann K. Microarray probe selection strategies. Brief Bioinform. 2001;2:329–340. doi: 10.1093/bib/2.4.329. [DOI] [PubMed] [Google Scholar]
- 27.Rychlik W, Spencer WJ, Rhoads RE. Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res. 1990;18:6409–6412. doi: 10.1093/nar/18.21.6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Novere N. MELTING, computing the melting temperature of nucleic acid duplex. Bioinformatics. 2001;17:1226–1227. doi: 10.1093/bioinformatics/17.12.1226. [DOI] [PubMed] [Google Scholar]
- 29.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 30.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 31.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kampke T, Kieninger M, Mecklenburg M. Efficient primer design algorithms. Bioinformatics. 2001;17:214–225. doi: 10.1093/bioinformatics/17.3.214. [DOI] [PubMed] [Google Scholar]
- 33.Antao VP, Tinoco I., Jr. Thermodynamic parameters for loop formation in RNA and DNA hairpin tetraloops. Nucleic Acids Res. 1992;20:819–824. doi: 10.1093/nar/20.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM, Cullen D, et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat. Biotechnol. 2008;26:553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- 35.Flibotte S, Moerman DG. Experimental analysis of oligonucleotide microarray design criteria to detect deletions by comparative genomic hybridization. BMC Genomics. 2008;9:497. doi: 10.1186/1471-2164-9-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei H, Kuan PF, Tian S, Yang C, Nie J, Sengupta S, Ruotti V, Jonsdottir GA, Keles S, Thomson JA, et al. A study of the relationships between oligonucleotide properties and hybridization signal intensities from NimbleGen microarray datasets. Nucleic Acids Res. 2008;36:2926–2938. doi: 10.1093/nar/gkn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bozdech Z, Zhu J, Joachimiak MP, Cohen FE, Pulliam B, DeRisi JL. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 2003;4:R9. doi: 10.1186/gb-2003-4-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, He Z, Zhou J. Selection of optimal oligonucleotide probes for microarrays using multiple criteria, global alignment and parameter estimation. Nucleic Acids Res. 2005;33:6114–6123. doi: 10.1093/nar/gki914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rimour S, Hill D, Militon C, Peyret P. GoArrays: highly dynamic and efficient microarray probe design. Bioinformatics. 2005;21:1094–1103. doi: 10.1093/bioinformatics/bti112. [DOI] [PubMed] [Google Scholar]
- 40.Chung WH, Rhee SK, Wan XF, Bae JW, Quan ZX, Park YH. Design of long oligonucleotide probes for functional gene detection in a microbial community. Bioinformatics. 2005;21:4092–4100. doi: 10.1093/bioinformatics/bti673. [DOI] [PubMed] [Google Scholar]
- 41.Rouchka EC, Khalyfa A, Cooper NG. MPrime: efficient large scale multiple primer and oligonucleotide design for customized gene microarrays. BMC Bioinformatics. 2005;6:175. doi: 10.1186/1471-2105-6-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talla E, Tekaia F, Brino L, Dujon B. A novel design of whole-genome microarray probes for Saccharomyces cerevisiae which minimizes cross-hybridization. BMC Genomics. 2003;4:38. doi: 10.1186/1471-2164-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rouillard JM, Zuker M, Gulari E. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 2003;31:3057–3062. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mrowka R, Schuchhardt J, Gille C. Oligodb–interactive design of oligo DNA for transcription profiling of human genes. Bioinformatics. 2002;18:1686–1687. doi: 10.1093/bioinformatics/18.12.1686. [DOI] [PubMed] [Google Scholar]
- 45.Schretter C, Milinkovitch MC. OligoFaktory: a visual tool for interactive oligonucleotide design. Bioinformatics. 2006;22:115–116. doi: 10.1093/bioinformatics/bti728. [DOI] [PubMed] [Google Scholar]
- 46.Wang X, Seed B. Selection of oligonucleotide probes for protein coding sequences. Bioinformatics. 2003;19:796–802. doi: 10.1093/bioinformatics/btg086. [DOI] [PubMed] [Google Scholar]
- 47.Wernersson R, Nielsen HB. OligoWiz 2.0–integrating sequence feature annotation into the design of microarray probes. Nucleic Acids Res. 2005;33:W611–W615. doi: 10.1093/nar/gki399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen H, Sharp BM. Oliz, a suite of Perl scripts that assist in the design of microarrays using 50mer oligonucleotides from the 3′ untranslated region. BMC Bioinformatics. 2002;3:27. doi: 10.1186/1471-2105-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gordon PM, Sensen CW. Osprey: a comprehensive tool employing novel methods for the design of oligonucleotides for DNA sequencing and microarrays. Nucleic Acids Res. 2004;32:e133. doi: 10.1093/nar/gnh127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chou HH, Hsia AP, Mooney DL, Schnable PS. Picky: oligo microarray design for large genomes. Bioinformatics. 2004;20:2893–2902. doi: 10.1093/bioinformatics/bth347. [DOI] [PubMed] [Google Scholar]
- 51.Emrich SJ, Lowe M, Delcher AL. PROBEmer: a web-based software tool for selecting optimal DNA oligos. Nucleic Acids Res. 2003;31:3746–3750. doi: 10.1093/nar/gkg569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaderali L, Schliep A. Selecting signature oligonucleotides to identify organisms using DNA arrays. Bioinformatics. 2002;18:1340–1349. doi: 10.1093/bioinformatics/18.10.1340. [DOI] [PubMed] [Google Scholar]
- 53.Li F, Stormo GD. Selection of optimal DNA oligos for gene expression arrays. Bioinformatics. 2001;17:1067–1076. doi: 10.1093/bioinformatics/17.11.1067. [DOI] [PubMed] [Google Scholar]
- 54.Reymond N, Charles H, Duret L, Calevro F, Beslon G, Fayard JM. ROSO: optimizing oligonucleotide probes for microarrays. Bioinformatics. 2004;20:271–273. doi: 10.1093/bioinformatics/btg401. [DOI] [PubMed] [Google Scholar]
- 55.Hornshoj H, Stengaard H, Panitz F, Bendixen C. SEPON, a Selection and Evaluation Pipeline for OligoNucleotides based on ESTs with a non-target Tm algorithm for reducing cross-hybridization in microarray gene expression experiments. Bioinformatics. 2004;20:428–429. doi: 10.1093/bioinformatics/btg434. [DOI] [PubMed] [Google Scholar]
- 56.Nordberg EK. YODA: selecting signature oligonucleotides. Bioinformatics. 2005;21:1365–1370. doi: 10.1093/bioinformatics/bti182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]