Digital Disease Detection — Harnessing the Web for Public Health Surveillance (original) (raw)
. Author manuscript; available in PMC: 2010 Aug 5.
Published in final edited form as: N Engl J Med. 2009 May 7;360(21):2153–2157. doi: 10.1056/NEJMp0900702
The Internet has become a critical medium for clinicians, public health practitioners, and lay-people seeking health information. Data about diseases and outbreaks are disseminated not only through online announcements by government agencies but also through informal channels, ranging from press reports to blogs to chat rooms to analyses of Web searches (see box). Collectively, these sources provide a view of global health that is fundamentally different from that yielded by the disease reporting of the traditional public health infrastructure.1
Over the past 15 years, Internet technology has become integral to public health surveillance. Systems using informal electronic information have been credited with reducing the time to recognition of an outbreak, preventing governments from suppressing outbreak information, and facilitating public health responses to outbreaks and emerging diseases. Because Web-based sources frequently contain data not captured through traditional government communication channels, they are useful to public health agencies, including the Global Outbreak Alert and Response Network of the World Health Organization (WHO), which relies on such sources for daily surveillance activities.
Early efforts in this area were made by the International Society for Infectious Diseases' Program for Monitoring Emerging Diseases, or ProMED-mail, which was founded in 1994 and has grown into a large, publicly available reporting system, with more than 45,000 subscribers in 188 countries.2 ProMED uses the Internet to disseminate information on outbreaks by e-mailing and posting case reports, including many gleaned from readers, along with expert commentary. In 1997, the Public Health Agency of Canada, in collaboration with the WHO, created the Global Public Health Intelligence Network (GPHIN), whose software retrieves relevant articles from news aggregators every 15 minutes, using extensive search queries. ProMED and GPHIN played critical roles in informing public health officials of the outbreak of SARS, or severe acute respiratory syndrome, in Guangdong, China, as early as November 2002, by identifying informal reports on the Web through news media and chat-room discussions.
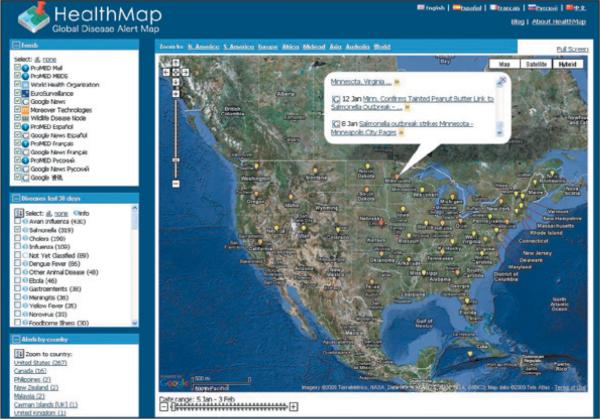
More recently, the advent of openly available news aggregators and visualization tools has spawned a new generation of disease-surveillance “mashups” (Web application hybrids) that can mine, categorize, filter, and visualize online intelligence about epidemics in real time. For instance, Health-Map (see image) is an openly available public health intelligence system that uses data from disparate sources to produce a global view of ongoing infectious disease threats. It has between 1000 and 150,000 users per day, including public health officials, clinicians, and international travelers. Other similar systems include MediSys, Argus, EpiSPIDER, BioCaster, and the Wildlife Disease Information Node. Automated analysis of online video materials and radio broadcasts will soon provide additional sources for early detection.
Screen Shot of HealthMap during the Recent Salmonella Typhimurium Outbreak.
HealthMap displays 319 articles about the outbreak that has affected 38 U.S. states.
The ease of use of blogs, mailing lists, RSS (Really Simple Syndication) feeds, and freely available mapping technology has meant that even an individual expert can create an important global resource. For instance, Declan Butler, a reporter at Nature, took aggregated data from various sources to provide a view of the spread of H5N1 avian influenza on a Google Earth interface. Similarly, Claudinne Roe of the Office of the Director of National Intelligence produces the Avian Influenza Daily Digest and blog, a collection of unclassified information about confirmed and suspected human and animal cases of H5N1 influenza.
Although news media represent an important adjunct to the public health infrastructure, the information they report pales in comparison to the potential collective intelligence that can be garnered from the public. An estimated 37 to 52% of Americans seek health-related information on the Internet each year, generally using search engines to find advice on conditions, symptoms, and treatments. Logs of users' chosen keywords and location information encoded in their computers' IP (Internet Protocol) addresses can be analyzed to provide a low-cost data stream yielding important insights into current disease trends.3 The power of these data has been demonstrated by studies of search engines provided by Google4 and Yahoo,5 in which data on searches using influenza-related keywords were used to generate an epidemic curve that closely matched that generated by traditional surveillance for influenza-related illness, deaths, and laboratory results. Google Flu Trends now provides a prospective view of current influenza search patterns throughout the United States. By making the information freely available to public health officials, clinicians, and ordinary citizens, such tools could help to guide medical decision making and underscore the importance of vaccination and other preventive measures.
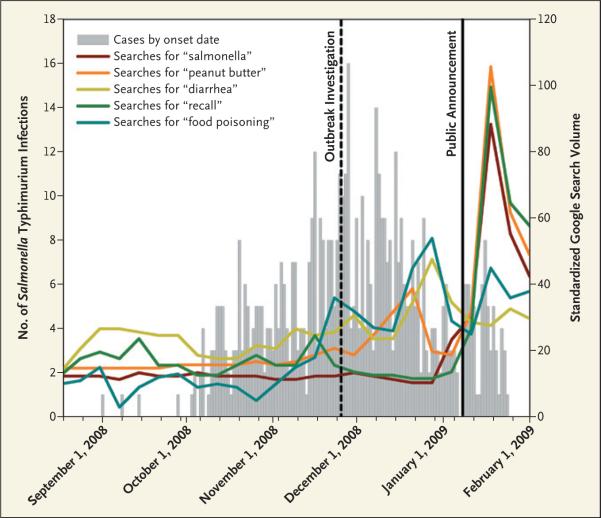
An example of the power of search-term surveillance can be found in an examination of the recent peanut-butter-associated outbreak of Salmonella enterica sero-type Typhimurium. Using Google Insights for Search, a search-volume reporting tool from Google, we compared the epidemic curve of onset dates for confirmed infections with trends in the volume of Internet searches on related terms in the United States (see graph). Search terms included “diarrhea,” “peanut butter,” “food poisoning,” “recall,” and “salmonella,” and search volumes were compared with the corresponding volumes from the previous year. The initial public report of salmonella was released on January 7, 2009, triggering an increase in searches for “salmonella,” “recall,” and “peanut butter,” but we saw earlier peaks in searches for “diarrhea” and “food poisoning.” Admittedly, these data provide only preliminary evidence of an emerging problem and require further study, but they highlight possibilities for early disease detection.
Infections with the Outbreak Strain of Salmonella Typhimurium, as reported by the CDC as of February 8, 2009.
Lines show data from Google Insights for Search, representing a portion of Web searches based in the United States across all Google domains relative to the total number of searches done on Google over time and scaled to a maximum value of 100. The data have been standardized by subtracting the mean volume from the previous 12 months for each term.
Though mining the Web is a valuable new direction (see sidebar on the H1N1 influenza epidemic), these sources cannot replace the efforts of public health practitioners and clinicians. The Internet is also providing new opportunities for connecting experts who identify and report outbreaks. Information technologies such as wikis, social networks, and Web-based portals can facilitate communication and collaboration to accelerate the dissemination of reports of infectious diseases and aid in mobilizing a response. Some scientific societies are now leveraging technologies for distributed data exchange, analysis, and visualization. For instance, the International Society for Disease Surveillance has created the Distributed Surveillance Taskforce for Real-Time Influenza Burden Tracking and Evaluation (DiSTRIBuTE), a group of state and local health departments that use the Web to share, integrate, and analyze health data across large regions. And the International Society of Travel Medicine, in collaboration with the Centers for Disease Control and Prevention (CDC), has created the GeoSentinel project, which brings together travel and tropical-medicine clinics in an electronic network for surveillance of travel-related illnesses. Similarly, the Emerging Infections Network, administered by the Infectious Diseases Society of America in collaboration with the CDC, is a Web-based network of more than 1000 infectious disease specialists that is geared toward finding cases during outbreaks and detecting new or unusual clinical events.
Broader Web-based networks are also proving useful for surveillance. Social-networking sites for clinicians, patients, and the general public hold potential for harnessing the collective wisdom of the masses for disease detection. Given the continued deployment of personally controlled electronic health records, we expect that patients' contributions to disease surveillance will increase. Eventually, mobile-phone technology, enabled by global positioning systems and coupled with short-message-service messaging (texting) and “microblogging” (with Twitter), might also come into play. For instance, an organization called Innovative Support to Emergencies, Diseases, and Disasters (InSTEDD) has developed open-source technology to permit seamless cross-border communication between mobile devices for early warning and response in resource-constrained settings.
These Internet-based systems are quickly becoming dominant sources of information on emerging diseases, though their effects on public health measures remain uncertain. Information overload, false reports, lack of specificity of signals, and sensitivity to external forces such as media interest may limit the realization of their potential for public health practice and clinical decision making. Sources such as analyses of search-term use and news media may also face difficulties with verification and follow-up. Though they hold promise, these new technologies require careful evaluation. Ultimately, the Internet provides a powerful communications channel, but it is health care professionals and the public who will best determine how to use this channel for surveillance, prevention, and control of emerging diseases.
Digital Resources for Disease Detection.
Sample Web-based data sources
ProMED-mail, www.promedmail.org
Global Public Health Intelligence Network (GPHIN), www.phac-aspc.gc.ca/media/nr-rp/2004/2004_gphin-rmispbk-eng.php
HealthMap, www.healthmap.org
MediSys, http://medusa.jrc.it
EpiSPIDER, www.epispider.org
BioCaster, http://biocaster.nii.ac.jp
Wildlife Disease Information Node, http://wildlifedisease.nbii.gov
H5N1 Google Earth mashup, www.nature.com/avianflu/google-earth
Avian Influenza Daily Digest and blog, www.aidailydigest.blogspot.com
Google Flu Trends, www.google.org/flutrends
Google Insights for Search, www.google.com/insights/search
DiSTRIBuTE, www.syndromic.org/projects/DiSTRIBuTE.htm
GeoSentinel, www.istm.org/geosentinel/main.html
Emerging Infections Network, http://ein.idsociety.org
Argus, http://biodefense.georgetown.edu
Sample health-related social-networking sites
Physicians, www.sermo.com
Patients, www.patientslikeme.com
Everyone, www.healthysocial.org
Acknowledgments
Dr. Brownstein Mr. Freifeld and Dr. Madoff report receiving grant support from Google.org.
Footnotes
No other potential conflict of interest relevant to this article was reported.
References
- 1.Brownstein JS, Freifeld CC, Reis BY, Mandl KD. Surveillance Sans Frontières: Internet-based emerging infectious disease intelligence and the HealthMap Project. PLoS Med. 2008;5(7):e151. doi: 10.1371/journal.pmed.0050151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madoff LC. ProMED-mail: an early warning system for emerging diseases. Clin Infect Dis. 2004;39:227–32. doi: 10.1086/422003. [DOI] [PubMed] [Google Scholar]
- 3.Eysenbach G. Infodemiology: tracking flu-related searches on the web for syndromic surveillance. AMIA Annu Symp Proc. 2006:244–8. [PMC free article] [PubMed] [Google Scholar]
- 4.Ginsberg J, Mohebbi MH, Patel RS, Brammer L, Smolinski MS, Brilliant L. Detecting influenza epidemics using search engine query data. Nature. 2009;457:1012–4. doi: 10.1038/nature07634. [DOI] [PubMed] [Google Scholar]
- 5.Polgreen PM, Chen Y, Pennock DM, Nelson FD. Using Internet searches for influenza surveillance. Clin Infect Dis. 2008;47:1443–8. doi: 10.1086/593098. [DOI] [PubMed] [Google Scholar]