The β3 subunit of the Na+,K+-ATPase mediates variable nociceptive sensitivity in the formalin test (original) (raw)
. Author manuscript; available in PMC: 2010 Aug 1.
Abstract
It is widely appreciated that there is significant inter-individual variability in pain sensitivity, yet only a handful of contributing genetic variants have been identified. Computational genetic mapping and quantitative trait locus analysis suggested that variation within the gene coding for the β3 subunit of the Na+,K+-ATPase pump (Atp1b3) contributes to inter-strain differences in the early phase formalin pain behavior. Significant strain differences in Atp1b3 gene expression, β3 protein expression, and biophysical properties of the Na+,K+ pump in dorsal root ganglia neurons from resistant (A/J) and sensitive (C57BL/6J) mouse strains supported the genetic prediction. Furthermore, in vivo siRNA knockdown of the β3 subunit produced strain-specific changes in the early phase pain response, completely rescuing the strain difference. These findings indicate that the β3 subunit of the Na+,K+-ATPase is a novel determinant of nociceptive sensitivity and further supports the notion that pain variability genes can have very selective effects on individual pain modalities.
Keywords: genetics, nociception, gene mapping, sodium-potassium pump, formalin
1. Introduction
Identification and characterization of genetic factors responsible for interindividual variability in pain sensitivity is of critical significance for improving our understanding of pain mechanisms and for developing novel therapies. A handful of genetic variants affecting sensitivity to noxious stimuli in mice and humans, under controlled laboratory conditions, have been identified within genes encoding proteins such as calcitonin gene-related peptide [38], catechol-_O_-methyltransferase [60], fatty acid hydrolase [21], GTP cyclohydrolase [51], the melanocortin-1 receptor [40,42], the δ-opioid receptor [21,39], the μ-opioid receptor [14], the transient receptor potential A subtype 1 (TRPA1) channel [21], and the transient receptor potential V subtype 1 (TPRV1) channel [22] (see ref. [24] for review). Some of these genes were obvious candidates and were directly tested in human genetic association studies, whereas others were first identified by genetic analysis of animal models. There are several reasons for the continuing importance of genetic animal models of pain. First, the inability to control many key variables and to assemble sufficiently powered cohorts has led to failures to demonstrate genetic association with pain phenotypes in humans for many seemingly obvious gene candidates [5]. Second, human genetic association studies have often generated controversial results; different studies examining related pain phenotypes have reported either no association, association with different haplotypes, or directly opposite allelic effects on the pain response [9,20,21,29]. Third, the large number of documented pain-related differences between transgenic knockout mice and their wildtype controls [23] suggests that many more relevant genes have yet to be identified. As of this writing, null mutations in at least 279 murine genes have been associated with an altered pain phenotype, and there are known genetic variants within the human homologues of most of these murine genes.
The formalin test of chemical/inflammatory pain produces well-characterized, biphasic recuperative behaviors in rodents [13]. Acute/early phase behaviors are due to direct chemical stimulation of peripheral nociceptors whereas tonic/late phase behaviors are thought to be due to central sensitization and/or ongoing inflammatory input [52]. This preclinical assay is one of the most common in current use [41], and one of the few existing models of spontaneous (non-evoked) pain, the most prevalent and bothersome symptom of human clinical pain [37]. A previous quantitative trait locus (QTL) analysis of intercross progeny from a resistant (A/J) and a sensitive (C57BL/6) strain identified a region on mouse chromosome 9 (called Nociq1; LOD = 5.2; 44−68 cM) that robustly affected pain sensitivity on the formalin test [56]. One or more genes within this genomic interval affect pain response during the early but not the late phase of the formalin test. We report here that the Atp1b3 gene underlies this early phase-specific QTL. We provide genetic, immunohistochemical and electrophysiological data supporting the unexpected candidacy of the β3 subunit of the Na+,K+-ATPase in the mediation of nociception.
2. Methods
2.1. Subjects
Subjects in all experiments were naïve, adult (6−12 weeks old) mice of both sexes [36]. Mice were purchased from The Jackson Laboratory (Bar Harbor, ME), or bred in-house from breeders so obtained. Upon weaning (at 18−21 d) or immediately after arrival, mice were housed in standard shoebox cages of 2−4 with same-sex littermates in a temperature-controlled (20 ±1°C) environment (14 h:10 h light/dark cycle; lights on at 07:00 h), and with ad lib access to food (Harlan Teklad 8604) and tap water. Purchased mice were habituated to the laboratory for at least one week before any behavioral testing commenced.
2.2. Formalin test
Mice were habituated for 30 min in individual transparent Plexiglas cylinders prior to a subcutaneous injection of 5% formalin into the plantar right hindpaw (20 μl volume) and were digitally videotaped for 60 min. Video files were later sampled for 5 sec at 1-min intervals, and the presence or absence of right hindpaw licking/biting in that 5-sec period was scored using Observer software (Noldus, Leesburg, VA). The early (acute) phase of the formalin test was defined as 0−5 min post-injection, and the late (tonic) phase as 10−60 min post-injection. We chose to define the early phase conservatively (0−5 min, instead of the more common 0−10 min) to ensure that we were measuring only early phase behavior and not interphase or beginning late phase behavior. Across the strains shown in Fig. 1A, the correlation between 0−5 min licking/biting and 0−10 min licking/biting (not shown) is _r_=0.78 (p<0.005), and haplotype analysis performed using 0−10 min strain means yields essentially the same list as that shown in Fig. 1C. Data are presented in most cases as the percentage of samples in which licking/biting was detected [1,6]. In some experiments, the total time spent licking in the early phase was assessed by continuous scoring of video files instead of sampling, although data were similar using either method. Hindpaw edema was quantified as done previously [56].
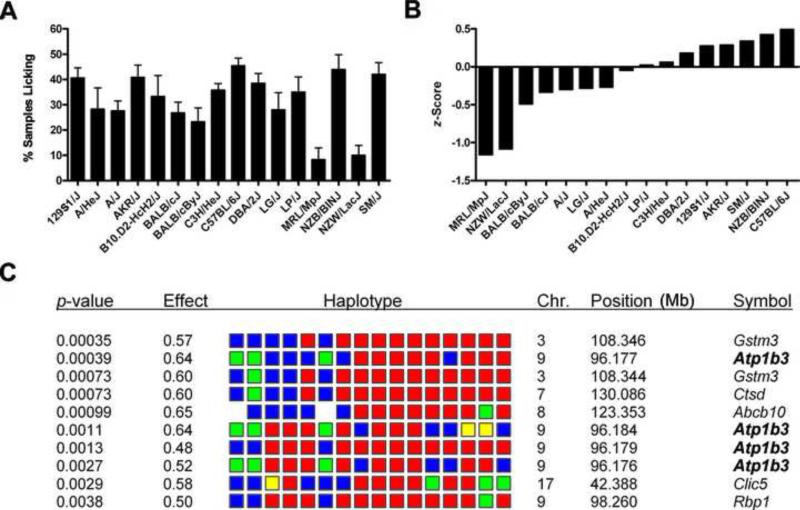
Figure 1.
Haplotype mapping implicates Atp1b3 as a candidate gene underlying variable sensitivity to formalin pain. A, Sensitivity of 16 inbred mouse strains in the early phase of the formalin test (5%, 20 μl). Bars represent mean ± SEM percentage of samples featuring licking/biting behavior from 0−5 min post-injection (_n_=10−38/strain).
B, Standardized scores (_z_-scores) of data in panel a, arranged from least sensitive (left) to most sensitive (right). C, Top 10 genome-wide correlations (arranged by _p_-value) between 5,694 SNP haplotype blocks and formalin test _z_-scores. Colored blocks represent the haplotype of the 16 strains, in the left-to-right order shown in graph B (most common haplotype in red, 2nd most common in blue, 3rd most common in green, least common in yellow). The effect score represents the proportion of the observed inter-strain phenotypic difference that could be explained by genetic variation within that haplotype block.
2.3. Computational haplotype-based genetic mapping
SNPs were organized into haplotype blocks; only a limited number of haplotypes—typically 2, 3 or 4—are present within a haplotype block. The haplotype-based computational analysis identifies haplotype blocks in which the haplotypic strain grouping within the block correlates with the distribution of phenotypic data among the inbred strains analyzed. To do this, a _p_-value that assesses the likelihood that genetic variation within each block could underlie the observed distribution of phenotypes among the inbred strains was calculated. The haplotype blocks were then ranked based upon the calculated _p_-value. When this computational analysis was performed, the haplotype map had 5,694 haplotype blocks generated from 215,155 SNPs characterized across 19 inbred strains covering 2,609 genes (see http://mouseSNP.roche.com). HapMapper computation analysis of the early phase response phenotypes proceeded as described previously [28,55].
2.4. qPCR experiments
We quantified basal expression of Atp1b1, Atp1b2, and Atp1b3 in A/J and C57BL/6J mice in the dorsal root ganglion (DRG). All experiments were conducted in triplicate. Lumbar DRGs (L4-L6; n_=6−12/mouse) were harvested and total RNA was isolated and reverse transcribed using standard protocols. Gene expression was quantified using the 7700 Sequence Detector (TaqMan) and the SYBR Green PCR Core Reagent Kit, as described in the manufacturer's manual (Applied Biosystems, Foster City, CA). In a first PCR reaction the glyceraldehyde-3-phosphate dehydrogenase (Gapdh) content of each cDNA sample was quantified to control for variations in cDNA amounts and diluted accordingly. The values were averaged in each direction and normalized by referring mean values of the target gene to mean values of Gapdh. The specificity of the PCR products was verified by gel electrophoresis. TaqMan-PCR reactions were performed in 25-μl volumes with a final primer concentration of 900 nM at 95°C for 30 s and 60°C for 60 s for 40 cycles. Primer and probe sets for Atp1b1 (Mm00437614_m1), Atp1b2_ (Mm01337151_m1), Gapdh, and Atp1b3 forward 5′- tctgagccacagacttacaaaaagt -3′ and Atp1b3 reverse 5′- cttgtgaggttcttctgttcttcca -3′ with probe 5′-6FAM- tttcctaaagccatattctg -TAMRA-3′ were obtained from Applied Biosystems.
2.5. Immunohistochemistry
The β3 subunit of the Na+,K+-ATPase was labeled in isolated L4-L5 DRG tissue using rabbit polyclonal antibody raised against an oligopeptide corresponding to the N-terminal sequence of the rat β3 isoform plus a terminal cysteine residue [2] (a generous gift of Dr. Kathleen J. Sweadner of Massachusetts General Hospital, Charlestown, MA). Staining for the β3 Na+,K+-ATPase in paraffin sections was conducted as described previously [10]. Isolated L4-L5 DRGs with short segments of attached spinal nerve and root were rinsed in phosphate-buffered saline (PBS; pH = 7.4) and immediately immersed in fixative (4% formaldehyde in PBS) for 4−7 days at 4 °C. After fixation, tissue samples were dehydrated in graded alcohol solutions, cleared in xylene, and embedded in paraffin for cross sectioning. Five-μm thick sections were cut, and the largest sections from the middle third of the ganglia were collected and mounted on slides. Slides with sections of DRGs from both strains of mice were further processed in parallel under equivalent conditions (immunohistochemical procedures and light intensity) during image capturing.
Staining for β3 Na+,K+-ATPase in paraffin sections was conducted as described previously [10,46]. Briefly, tissue sections were deparaffinized with xylene and rehydrated with graded alcohols and water. After high-temperature antigen retrieval in citrate buffer (pH 6), endogenous peroxidase and non-specific antibody binding sites were blocked by incubating the sections with hydrogen peroxide (3% in methanol for 10 min at room temperature) and heat-inactivated 10% donkey serum (30 min at 37 °C), respectively. Following overnight incubation with primary antibody (dilution 1:1000 in PBS containing 1% donkey serum, 4 °C), sections were incubated with secondary biotinylated antibody (donkey anti-rabbit antibodies; 1:600 in PBS/10% mouse Fab fragment; Jackson ImmunoResearch Laboratories, West Grove, PA), peroxidase-conjugated streptavidin label (1:400 in PBS), and finally with 3,3′-diaminobenzidine chromogen solution (Vector Laboratories, Burlingame, CA). Development of the staining was monitored under a microscope and stopped by washing with PBS. In negative control experiments, the incubations with primary antibody were substituted with an equivalent time of incubation in 1% donkey serum in PBS. Stained sections, dehydrated through 95% and 100% ethanol, cleared with xylene and coverslipped with Permount medium (Fisher Scientific, Fair Lawn, NJ).
Images were analyzed using Image-Pro 5.1 (Media Cybernetics, Silver Spring, MA) and custom “Clock-scan” protocol software [11]. Cell diameters were determined in cells in which the nucleus was readily determined by calculating the average of major and minor axes of an ellipse automatically best fit to the cell outline [46]. Average intensities of cytoplasmic and background pixels were subtracted yielding the value of cytoplasmic labelling with minimal (if any) dependence on within and between sections variations in staining or section thickness and illumination intensities [11].
2.6. siRNA synthesis and validation
Five Dicer-substrate siRNA duplexes (siRNAs) [19,47] were designed to be specific for the mouse Atp1b3 gene (NM_007502) using established criteria [45]. RNA duplexes were obtained from Integrated DNA Technologies (IDT, Coralville, IA). All RNA duplexes were HPLC-purified, verified for identity by electrospray mass spectrometry, and were verified at >90% purity as assessed by analytical HPLC. Duplexes were validated for functional potency in NIH3T3 cells in vitro prior to use in vivo. NIH3T3 cells were obtained from ATCC and maintained under standard culture conditions in high glucose DMEM containing 10% newborn calf serum and 1% penicillin/streptomycin. Cells were plated at 3.5× 104/well in 24-well plates the day before transfection in 0.5 ml complete media without antibiotics. Transfections were done in triplicate for each sample using 0.5 μl per well of siLentFect cationic lipid (Bio-Rad, Hercules, CA) and 5 pmol or 0.05 pmol of siRNA duplex to give a final concentration of either 10 nM or 0.1 nM. As a control, the TriFECTa DS Scrambled Negative DsiRNA duplex (Integrated DNA Technologies) was transfected in parallel at 10 nM. Cells were incubated with the transfection mixture for 24 h. Total RNA was isolated using the Promega SV96 Total RNA Isolation System (Promega, Madison, WI) as per the manufacturer's protocol. cDNA was synthesized using 150 ng total RNA the 30U SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA).
Quantitative real-time PCR was done using a 7900HT Real Time PCR System (Applied Biosystems, Foster City, CA) in 10 μl volume in 384-well format. Reaction mixes employed Immolase DNA Polymerase (Bioline, Randolph, MA), 200 nM primers and probe, and 10 ng cDNA. Reactions were cycled using the following program: 95 °C 10 min followed by 95 °C for 15 s, 60°C for 60 s for 40 cycles. All reactions were performed in triplicate. Primers employed were: Atp1b3 forward 5′-cagtttccagtctccttgcttg -3′ and Atp1b3 reverse 5′-gatatttgtggatatccgtctggg-3′ with probe 5′-6FAM-tccaaaggacagccttgcatccttgtga -IBFQ-3′. The fluorophore was 6-carboxyfluorescein (6FAM) and the quencher was Iowa Black FQ (IBFQ). Expression data were normalized to an internal control run in parallel for the Mus musculus ribosomal protein L23 gene, Rpl23 (NM_022891) using the ΔΔCt method. Primers and probes were Rpl23 forward: 5′- ctgtgaagggaatcaaggga -3′, Rpl23 reverse 5′- tgtcgaattaccactgctgg -3′, and probe 5′-6FAM-ctgaacagacttcctgctgctggtg -IBFQ-3′. Final expression data were normalized using the negative control DsiRNA transfections as 100%.
All five of the Atp1b3 siRNAs tested reduced the target gene expression by >90% at 10 nM. At low dose, potency differences were detectable between the reagents and duplexes 3.1 and 3.5 were most potent, suppressing target expression by 82% and 63% respectively at 0.1 nM (see Supplementary Figure 1).
_2.7. siRNA studies_- in vivo knockdown
In vivo delivery of siRNA molecules was performed similar to a previously published protocol [30]. The siRNA (2 μg/μl) was combined with i-Fect transfection reagent (Neuromics, Edina, MN) at a ratio of 1:5 (weight/volume) and the combination was allowed to complex for 5 min at room temperature. The siRNA/i-Fect complex was then further diluted in sterile physiological saline to give a final dose of 0.5 μg of siRNA per injection. A total of 5 μl of this solution was delivered via i.t. injection to the lumbar region using a 20-μl Hamilton microsyringe with a 30-gauge needle (Hamilton Company, Reno NV). This injection paradigm was repeated approximately 24 h and 48 h later (total of three injections).
2.8. Patch clamp electrophysiology
Under deep halothane/oxygen anesthesia, the spinal column was hemisected and dorsal root ganglia (DRG) from the lumbar region of either A/J or C57BL/6J mice were extracted and placed into Ca2+- and Mg2+-free DMEM (Invitrogen, Carlsbad, CA), then transferred at 37 °C and Ca2+- and Mg2+-free DMEM containing 1 mg/ml papain and 2 mg/ml collagenase (Sigma Aldrich, Saint Louis, MO). After enzymatic digestion, the DRGs were rinsed three times in DMEM containing 10% certified fetal bovine serum (FBS) and 2 mM L-glutamine. Mechanical trituration with a fire-polished Pasteur pipette was used to dissociate the DRG neurons, which were then rinsed three times in culturing media. Prior to plating, the DRG neurons were passed through a 100 μm nylon cell strainer (BD Biosciences, San Jose, CA). The DRG neurons were then plated onto 35-mm dishes coated with laminin (BD Biosciences) and poly-D-lysine (Sigma Aldrich), and incubated at 37 °C in 5% CO2.
Dissociated DRG neurons were cultured in F-12 media supplemented with 10% certified FBS, 2 mM L-glutamine, and 100 U/ml penicillin and streptomycin (Invitrogen). The external solution (ECS) used during patch-clamp recordings contained (in mM): 145 NaCl, 5.4 KCl, 1 MgCl2, 10 HEPES, 0.2 CdCl2, 2 BaCl2, and 5.1 D-glucose. Suppression of K+ and Ca2+ currents was achieved with the addition of Cd2+ and Ba2+, and the absence of Ca2+. NaOH was used to adjust the ECS to a pH of ∼7.4. The pipette solution or intracellular solution (ICS) contained (in mM): 60 Na-gluconate, 70 Cs-gluconate, 10 CsCl, 5 sodium phosphocreatine, 10 HEPES, 5 EGTA, 10 TEA-Cl, 4 Mg-ATP and 0.3 GTP. High concentrations of Na+ as well as ATP, creatine phosphate, TEA and pyruvate were used to enhance pump activation while further suppressing K+ currents. The ICS (osmolarity of ∼290 mOsm) was adjusted to a pH of ∼7.2.
Dorsal root ganglion neurons were cultured for 24 h prior to experiments. Currents were recorded in the whole-cell patch clamp mode using an Axopatch 200B amplifier with onboard lowpass Bessel filter at 5 kHz and digitized through a Digidata 3200A at a sampling rate of 2 kHz. Recording electrodes (borosilicate glass , P-97 puller, Sutter Instruments, Novato, CA), were fire-polished to a tip resistance of 3−6 MΩ (MP-830 microforge, Narishige, East Meadow, NY). During patch-clamp recording, cells were perfused with ECS at a rate of 1 ml/min. Fast solution exchange was achieved using a three barrel perfusion system (SF-77B; Warner Instruments, Hamden, CT). Small cells were verified to be neurons by eliciting voltage-dependent Na+ current through gradual depolarization above −60mV in whole cell configuration. Neuronal current activities or holding currents (Ih) were continuously recorded under condition of holding potential (Vh) clamped at −40 mV. Membrane capacitance (Cm) and series resistance (Rs) were measured through the peak amplitude and decay constant of transients induced by repetitive depolarizing pulses of 10 mV.
To evaluate the ability of A/J and C57BL/6 DRG neurons to sustain repetitive firing, they were stimulated by trains of depolarizing currents set at 2 times their action potential (AP) threshold at a frequency of 10 Hz for 1 second, repeated 50 times with 5 second intervals between stimulations. The AP threshold of each neuron was determined by applying 500 ms-long depolarizing current steps by increments of 10 pA. The acquisition frequency was 50 kHz to fully capture the AP spikes and the highest amplitude of response to each stimulus train was used as index of the gradual decay of cellular response over the 50 trials, with or without treatment with 10 μM ouabain.
2.9. Rotarod test
Motor ataxia was determined using a standard mouse rotarod (Ugo Basile, Verese, Italy) which provided a accelerating rotational speed from 22 rpm at the start of the test to a maximum speed of 50 rpm. Mice were baseline tested 2−4 times prior to any experiments. The latency to fall off of the rotarod was determined automatically by a timer that measured to the nearest second. A cutoff latency of 300 sec was used for all rotarod assessments.
2.10. Statistics
Data were analyzed by ANOVA followed by Student's _t_-test or Bonferroni multiple comparison post hoc test, as appropriate. The criterionα=0.05 was used throughout.
3. Results
3.1. Haplotype-based computational genetic mapping identifies Atp1b3 as a candidate gene
The early phase response to formalin pain was evaluated in 16 inbred mouse strains. There were robust inter-strain differences (p<0.001) in recuperative licking/biting behavior (Fig. 1A), and the data were subjected to computational haplotype-based genetic analysis (HapMapper). This method identifies genomic regions where the pattern of genetic variation (strain groupings within a SNP haplotype block) correlates with the distribution of phenotypic responses among the inbred strains analyzed. Among the 10 genomic regions that best correlated with the standardized strain means (Fig. 1B), four were associated with the Atp1b3 gene (Fig. 1C). This gene was of particular interest since it is located well within the interval on chromosome 9 (96.142 Mb; 51.0 cM) that was previously identified by QTL analysis [56]. Two strains, MRL/MpJ and NZW/LacJ, were significantly less sensitive to formalin-induced pain than all others tested (p<0.05). The 14 remaining strains appeared to be normally distributed, with C57BL/6J the most sensitive and A/J among the most resistant. The 74 SNPs within the Atp1b3 gene divided the 16 inbred strains analyzed into three major haplotypic groupings, one of which consisted of the MRL/MpJ, NZW/LacJ and LG/J strains. Of importance, there is a non-synonymous polymorphism (Ser127Asn) within exon 4 of the Atp1b3 gene, and these three strains have the Asn127 allele. This non-conserved amino acid difference could contribute (at least in part) to the reduced pain response in MRL/MpJ and NZW/LacJ mice, whereas other genetic loci could modify the response of LG/J mice. Although non-synonymous SNPs distinguishing A/J from C57BL/6J mice were not present, three polymorphisms were clustered at positions −1285, −1371 and −1447 bases upstream of the Atp1b3 transcription start site. Each haplotype utilizes a different allelic combination of these three polymorphisms, which could produce a difference in Atp1b3 mRNA expression between A/J and C57BL/6J mice.
3.2. Differences in β3 subunit mRNA and protein expression are strain-specific
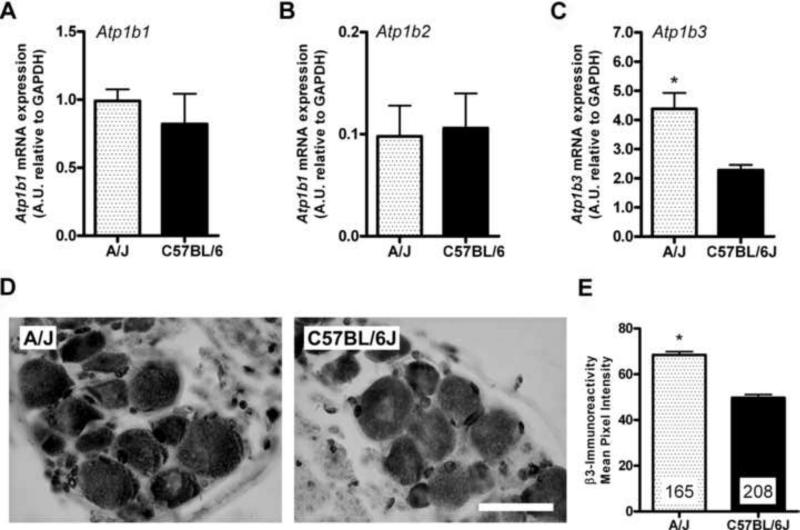
We next determined whether there was a difference in the basal level of expression of Atp1b3 in C57BL/6J and A/J mice, which might explain the observed phenotypic difference between these mouse strains. Microarray gene expression profiling suggests that 5% formalin is not a sufficient stimulus to regulate a large number of genes (unpublished data; also see ref. [3]). It is also highly questionable as to whether strain differences, which are obvious even in the first two minutes of the formalin test, could be secondary to formalin-induced gene expression changes. Thus, basal strain differences in gene expression are likely responsible for phenotypic strain differences in this trait. We observed that there was significantly higher (p<0.05) Atp1b3 mRNA expression in L4-L6 dorsal root ganglia (DRG) isolated from A/J mice relative to those of C57BL/6J mice. By contrast, there was no difference in the level of Atp1b1 or Atp1b2 mRNA in DRG between these two strains (Fig. 2A-C). There was also no strain difference in Atp1b3 expression in whole-brain tissue (data not shown). We also observed a significantly higher (p<0.05) level of β3-like immunoreactivity in A/J DRG soma (Fig. 2D,E) that was distributed equally among neurons of differing diameters (data not shown). These findings demonstrate that higher Atp1b3 mRNA and protein expression is associated with lower early phase formalin pain behavior.
Figure 2.
Expression of β subunits in dorsal root ganglion tissue. qPCR analysis of A, Atp1b1, B, Atp1b2, and C, Atp1b3 mRNA expression in lumbar DRG tissue. Bars represent the mean ± SEM ratios for _n_=3 technical replicates performed on pooled DRG tissue from _n_=5−6 mice/strain. D, Representative photomicrographs of DRG sections from A/J (left) and C57BL/6J (middle) mice stained with an antibody against β3 protein. Scale bar = 30 μm. E, Quantification of β3-like immunoreactivity in DRG neurons from A/J and C57BL/6J mice. Bars represent the mean ± SEM pixel intensity; numbers represent the number of individual neuronal soma in DRG sections from _n_=4−5 mice/strain. *p<0.05 by Student's _t_-test.
3.3. Strain-specific differences in DRG neuron excitability
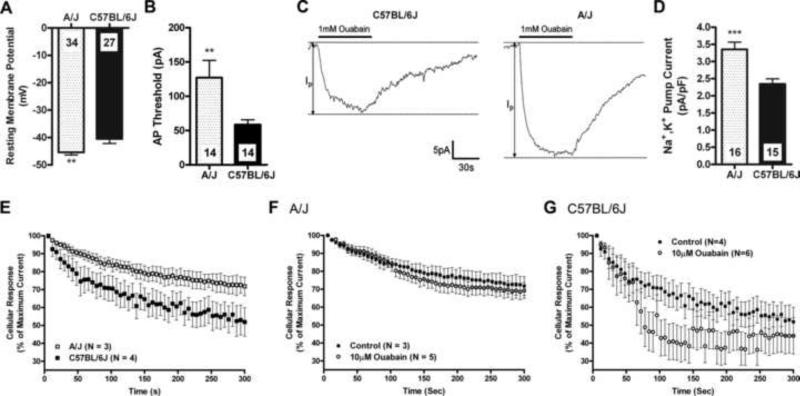
The Atp1b3 gene encodes the non-catalytic β3 subunit [32] of the sodium-potassium Na+,K+-ATPase (Na+,K+ pump), a complex of α- and β-subunits that maintains the ionic gradient responsible for the neuronal resting potential via the active uptake of K+ ions and efflux of Na+ ions [49,50]. The β-subunits of the Na+,K+-ATPase are recognized as having a critical role in determining intrinsic ion transport properties, and are responsible for proper trafficking and insertion of nascent pumps into the cell membrane [16,17]. Thus, increased Atp1b3 expression in A/J mice could increase the number of functional Na+,K+ pumps on the cell surface. To investigate this possibility, patch clamp electrophysiology was used to measure several biophysical parameters in dissociated DRG neurons obtained from C57BL/6J and A/J mice. DRG neurons from A/J mice had −4.9 mV more negative resting membrane potentials than those from C57BL/6 mice (p<0.01; Fig. 3A). Furthermore, DRG neurons from A/J mice exhibited a significantly (p<0.01) higher threshold to elicit an action potential as compared to those from C57BL/6J mice (Fig. 3B). We also observed that DRG neurons from A/J mice had significantly (p<0.001) higher Na+,K+ pump current density than DRG neurons from C57BL/6J mice (Fig. 3C,D).
Figure 3.
DRG neurons from A/J mice have greater Na+,K+ pump currents and are ouabain insensitive compared to C57BL/6J mice. A, Measurement of the resting membrane potential and B, the stimulus threshold required to elicit an action potential in dissociated DRG neurons from A/J and C57BL/6J mice (mean ± SEM ; _n_=14−34 neurons/strain). **p<0.01 by Student's _t_-test. C, Representative traces of the Na+,K+ pump current, defined as the ouabain (1 mM)-sensitive fraction of the holding current, in dissociated DRG neurons from A/J and C57BL/6J mice. D, Quantification of Na+,K+ pump current density in A/J and C57BL/6J DRG neurons (mean ± SEM ; _n_=15−16 neurons/strain). ***p<0.001 by Student's _t_-test. Measurement of the ability of neurons to maintain repetitive firing using a 10Hz stimulation train over 300 s. The maximal action potential amplitude of A/J and C57BL/6J DRG neurons was recorded at in the presence of no drug (E) and 10 μM ouabain (F,G). There was a significant difference between strains (_F_1,400 = 149.1, p<0.001) in panel E and between C57BL/6J drug treatment groups (_F_1,400 = 56.38, p<0.001) in panel G as determined by ANOVA.
We next sought to determine whether this basal increase in pump density would confer a higher ability to sustain repetitive neuronal firing. We observed that DRG neurons from A/J mice had significantly less attenuation of action potential amplitude compared to DRG neurons from C57BL/6J mice in response to repetitive 10-Hz stimulus trains delivered over 300 s (Fig. 3E). Furthermore, the addition of 10 μM ouabain had no discernible effect on repetitive stimulus-induced decrease in action potential amplitude in A/J neurons (Fig. 3F), but drastically exacerbated this decrease in C57BL6/J neurons (Fig. 3G). Thus, despite their lower excitability, DRG neurons from A/J mice have a considerably increased ability to support repetitive neuronal firing, and that this property is related to the increased pools of Na+,K+ pumps that these neurons possess. These findings suggest also that the inter-strain behavioral differences observed during the early phase of the formalin test are more linked to the excitability level of DRG neurons (action potential threshold, resting membrane potential) than to their ability to sustain repetitive neuronal firing.
3.4. siRNA-mediated knockdown of Atp1b3 produces phase- and strain-specific effects on formalin pain
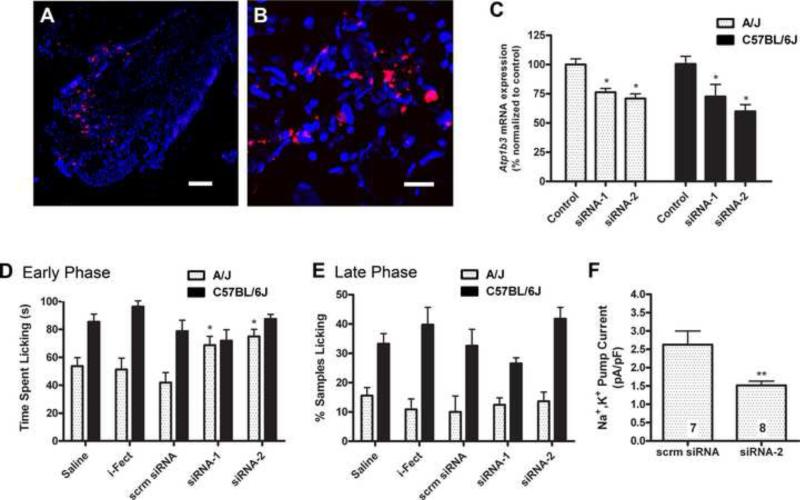
Based on the expression and electrophysiological data, we predicted that manipulations that decrease Atp1b3 expression and/or pump functioning would increase pain sensitivity, especially in strains (like A/J) with a high level of Atp1b3 expression. Although in vivo pharmacological experiments with ouabain, a Na+,K+-ATPase inhibitor, were possible, ouabain is not selective to β3 subunits, and pilot experiments revealed that effects on formalin behaviors were obtained only at doses that also produces ataxia as measured on the rotarod test (data not shown). To produce a more selective knockdown, we turned to an siRNA strategy. Two unique Dicer-substrate small interfering RNAs (siRNAs) that selectively target Atp1b3 were synthesized and shown to potently suppress Atp1b3 mRNA expression in vitro (Supplementary Fig.1A). Wide-field and confocal microscopy revealed that the cationic lipid-encapsulated siRNAs were evenly distributed throughout the lumbar DRGs (Fig. 4A,B) and spinal cord (Supplementary Fig.1D,E). We also confirmed that the intrathecal (i.t.) injection of the siRNAs significantly (p<0.05) decreased the expression of Atp1b3 mRNA in the DRG of A/J and C57BL/6J mice (Fig. 4C). Our finding that i.t. injection permits efficient delivery of oligonucleotides to the DRG is consistent with previous reports [25,27]. There were no significant changes in the expression of either Atp1b1 or Atp1b2 mRNA following siRNA treatment (data not shown), confirming the specificity of the siRNAs.
Figure 4.
Rescue of the strain difference in early phase formalin pain behavior by intrathecal siRNA against Atp1b3. A, Wide field and B, confocal microscopy of 20-μm sections of lumbar DRG tissue from mice treated with three daily i.t injections of i-Fect + 0.5 μg/day of Cy3-labeled scrambled siRNA (red). Nuclei were counterstained with DAPI (blue). Scale bars = 100 μm (wide field) and 20 μμm (confocal).C, qPCR analysis of Atp1b3 mRNA expression in lumbar DRG tissue from C57BL/6J and A/J mice treated with i-Fect transfection reagent alone (control) or i-Fect plus one of two siRNAs targeting Atp1b3 (siRNA-1 and siRNA-2). Bars represent the percent of Atp1b3 mRNA expression as compared to the respective control groups ± SEM on pooled DRG tissue from _n_=6 mice/strain. *p<0.05 by Bonferroni post hoc test compared to same-strain control group. D, Early phase (0−5 min) and E, late phase (5−60 min) formalin-induced licking/biting in A/J and C57BL/6J mice treated with i-Fect transfection reagent alone (i-Fect), i-Fect plus a scrambled siRNA (scrm siRNA), or i-Fect plus one of two unique siRNAs targeting Atp1b3 (siRNA-1 and siRNA-2). Bars represent mean ± SEM total time spent licking (in panel D) or the percentage of sampled intervals showing licking behavior (in panel E) (_n_=8−13 mice/strain/treatment). *p<0.05 by Bonferroni post hoc test compared to same-strain saline group. F, Quantification of Na+,K+ pump current density in DRG neurons from A/J mice previous treated with i-Fect plus a scrambled siRNA (scrm siRNA) or i-Fect plus siRNA-2 for three days prior to measurement (mean ± SEM ; _n_=7−8 neurons/group). **p<0.01 by Student's _t_-test.
The siRNAs were administered i.t. to mice of both strains once daily for three consecutive days prior to measurement of the formalin responses. This injection regimen produced transient ataxia on the rotarod test (Supplementary Fig.1B), which fully resolved by the time of formalin testing (Supplementary Fig.1C). Both siRNAs significantly (p<0.05) and selectively increased early phase formalin licking behavior in A/J mice, but had no effect in C57BL/6J mice (Fig. 4D). None of the controls produced any significant effects. The siRNA-mediated effect on the early phase response fully rescued the strain difference between A/J and C57BL6 mice. In contrast, none of the siRNAs produced a significant effect on the late phase formalin licking in either strain, and the strain difference in late phase behavior was maintained (Fig. 4E). In order to determine whether siRNA knockdown of Atp1b3 caused alterations in Na+,K+-ATPase activity we measured the Na+,K+ pump current in DRG neurons obtained from siRNA-treated A/J mice. We observed a significant decrease in pump current in the siRNA-2 treated mice compared the scrambled siRNA control group (Fig. 4F).
4. Discussion
Computational genetic mapping and QTL analysis indicated that genetic variation within the gene encoding the β3 subunit of the Na+,K+-ATPase contributes to inter-strain differences in the early phase of the formalin test. Consistent with this genetic prediction, we observed significant strain differences in Atp1b3 gene expression, β3 protein expression, and in Na+,K+ pump current in DRG neurons of resistant (A/J) and sensitive (C57BL/6) mouse strains. I_n vivo_ siRNA knockdown of the β3 subunit produced strain-specific changes in the early phase licking behavior, completely rescuing the original strain difference between A/J and C57BL/6J mice. These facts suggest that Atp1b3 represents the sole gene underlying the originally identified QTL on mouse chromosome 9 [56]. It is likely not the only gene contributing to variability in the early phase of the formalin test, however, and the candidacy of Gstm3 (glutathione S-transferase, mu 3) (see Fig. 1C) is presently being evaluated.
4.1. The Na+,K+-ATPase and pain
It is well known that the Na+,K+-ATPase is found throughout the nociceptive axis [48] and is absolutely critical to neuronal function [26]. However, only a very limited number of studies—with somewhat disparate findings—have examined the role of the Na+,K+-ATPase in modulating pain. Spinal administration of ouabain in rats was shown to produce dose-dependent analgesia to noxious heat [59]. Ouabain was clinically evaluated as a peripheral nerve block agent for pain management [15], but is not currently used in pain pharmacotherapy. Analgesic properties of ouabain are partly supported by our data showing its inhibitory effect on the ability of DRG neurons to sustain action potential generation. However, ouabain blocks the function of the Na+,K+ pump at all isoforms, and therefore cannot be used in vivo to selectively evaluate the effects of the β3 subunit-specific polymorphism. It was recently shown that Na+ 3,K+ pump activity in the dorsal horn of the spinal cord is markedly increased following acute peripheral inflammation produced by a burn injury [8]. Furthermore, several studies assaying global gene expression changes using high density oligonucleotide microarrays have in fact observed significant regulation of Atp1b3 expression in both neuropathic [54,57] and inflammatory [3,58] pain states.
4.2. Role of the β subunit of Na+,K+-ATPase
Analysis of mice that are heterozygous for a null mutation of the α-isoform provided the first whole-animal evidence of behavioral defects resulting from genetic knockdown of a Na+,K+-ATPase subunit [43], but these mice were not tested for pain. Although the α subunits are responsible for the catalytic transport of Na+ and K+ ions across the membrane, the importance of the β isoforms is evidenced by the fact that defects in β subunits result in a structurally and functionally deficient Na+,K+-ATPase in vitro [17]. Recent studies have also demonstrated that the β subunits of the Na+,K+-ATPase may have actions independent of ion transport, including the activation of the phosphatidylinositol 3-kinase cell signaling cascade [4] and cell-cell adhesion [53], raising the possibility that the β subunits may be able modulate nociceptive sensitivity through intracellular signaling mechanisms as well.
The β subunit of Na+,K+-ATPase is required for assembly and delivery to the plasma membrane of [16,17], and thus availability of β subunits is a critical determinant of the cell's capacity to maintain low intracellular Na+ under a wide range of conditions [7,34]. Well in accord with this concept, our data suggest that the diminished Na+,K+ pump current and capacity of C57BL/6J DRG neurons to maintain Na+ gradient and action potential amplitude during continuing frequency discharge is a result of decreased β3 subunit pool in these neurons. In addition to the control of Na+ gradient, neuronal Na+,K+-ATPase also controls accumulation of extracellular K+ and is responsible for the electrogenic component of the cell resting membrane potential and after-discharges (see ref. [12]). Thus, two other consequences of the limited pool of β3 subunits and associated decrease of Na+,K+-ATPase activity in C57BL/6J nociceptive afferents were also observed, namely a slightly depolarized resting membrane potential and a decrease in the action potential excitability threshold. This, we believe, is the most likely explanation of the relatively high behavioral sensitivity of C57BL/6J mice to formalin. It is also possible, of course, that central neurons in the spinal circuits involved in formalin pain processing show β3-dependent strain differences in excitability.
As compared to A/J neurons having stronger Na+,K+-ATPase activity, C57BL/6J nociceptors should also be more sensitive to the action of ouabain. Indeed, in agreement with this prediction, exposure of A/J neurons to ouaba (10 μM)did not affect their ability to sustain repetitive firing. However, in the same test ouabain treatment resulted in a dramatic acceleration of the decline and instability of action potential amplitude in C57BL/6J neurons.
4.3. Implications of these findings
It is of particular interest that both the QTL effect and the manipulations of Atp1b3 (as the gene likely responsible for the QTL effect), displayed specificity to the early phase of the formalin test. The early phase of the formalin test is admittedly not likely as clinically relevant as the late phase, and we are not suggesting that Atp1b3 is an appropriate target for drug development. However, the fact that the genetic mediation of pain can be so exquisitely specific has important scientific implications. First, it suggests that the total number of pain-relevant genes is likely to be larger than currently expected, since at least some genes will have effects limited to certain modalities. Second, it adds to the ongoing debate over whether scores from multiple pain tests of different modalities should be averaged into composite pain scores in genetic association study designs [18,44].
In conclusion, we have demonstrated that the Atp1b3 gene is a novel determinant of pain variability. Beyond demonstrating that the Na+,K+ pump modulates nociception, our findings provide the first description of a role in a systems-level biological process for any βsubunit of the Na+,K+-ATPase. It is of particular interest that genetic variability within a regulatory rather than a catalytic subunit of this protein affects pain. More broadly, this finding illustrates that genetic variables affecting biomedically relevant traits may not always be found within the obvious candidate genes, and supports the use of genome-wide approaches for genetic discovery.
Supplementary Material
01
Sequences of Atp1b3 DsiRNAs
| Name | Target accession | Sequence |
|---|---|---|
| 5′ pGGGAUACAAUGGUACCUUGCCAAca 3′ | ||
| siRNA 3.1 | NM_007502 | |
| 3′ AUCCCUAUGUUACCAUGGAACGGUUGU 5′ | ||
| 5′ pCGCCCAUCAUGACGAAGACUGAGaa 3′ | ||
| siRNA 3.5 | NM_007502 | |
| 3′ UGGCGGGUAGUACUGCUUCUGACUCUU 5′ |
Acknowledgments
This work was supported by NIH grants NS41670 (J.S.M.), DA021332 (D.J.C.), and DK067284 (M.D.) and CIHR grant MOP-14718 (P.S.). The authors wish to thank Michael Collingwood for expert technical assistance. M.A.B. is employed by Integrated DNA Technologies, Inc. (IDT), which offers oligonucleotides for sale similar to some of the compounds described in the manuscript. IDT is not a publicly traded company and M.A.B. does not own any shares/equity in IDT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abbott FV, Ocvirk R, Najafee R, Franklin KBJ. Improving the efficiency of the formalin test. Pain. 1999;83:561–569. doi: 10.1016/S0304-3959(99)00168-2. [DOI] [PubMed] [Google Scholar]
- 2.Arystarkhova E, Sweadner KJ. Tissue-specific expression of the Na,K-ATPase beta3 subunit. The presence of beta3 in lung and liver addresses the problem of the missing subunit. J Biol Chem. 1997;272:22405–22408. doi: 10.1074/jbc.272.36.22405. [DOI] [PubMed] [Google Scholar]
- 3.Barr GA, Gao P, Wang S, Cheng J, Qin J, Sibille EL, Pavlidis P. Microarray analysis of gene expression following the formalin test in the infant rat. Pain. 2005;117:6–18. doi: 10.1016/j.pain.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 4.Barwe SP, Anilkumar G, Moon SY, Zheng Y, Whitelegge JP, Rajasekaran SA, Rajasekaran AK. Novel role for Na,K-ATPase in phosphatidylinositol 3-kinase signaling and suppression of cell motility. Mol Biol Cell. 2005;16:1082–1094. doi: 10.1091/mbc.E04-05-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belfer I, Wu T, Kingman A, Krishnaraju RK, Goldman D, Max MB. Candidate gene studies of human pain mechanisms: a method for optimizing choice of polymorphisms and sample size. Anesthesiology. 2004;100:1562–1572. doi: 10.1097/00000542-200406000-00032. [DOI] [PubMed] [Google Scholar]
- 6.Chesler EJ, Ritchie J, Kokayeff A, Lariviere WR, Wilson SG, Mogil JS. Genotype-dependence of gabapentin and pregabalin sensitivity: the pharmacogenetic mediation of analgesia is specific to the type of pain being inhibited. Pain. 2003;106:325–335. doi: 10.1016/S0304-3959(03)00330-0. [DOI] [PubMed] [Google Scholar]
- 7.Chow DC, Forte JG. Functional significance of the beta-subunit for heterodimeric P-type ATPases. J Exp Biol. 1995;198:1–17. doi: 10.1242/jeb.198.1.1. [DOI] [PubMed] [Google Scholar]
- 8.Czaplinski M, Abad C, Eblen-Zajjur A. Normal expression and inflammation-induced changes of Na and Na/K ATPase activity in spinal dorsal horn of the rat. Neurosci Lett. 2005;374:147–151. doi: 10.1016/j.neulet.2004.10.046. [DOI] [PubMed] [Google Scholar]
- 9.Diatchenko L, Slade GD, Nackley AG, Bhalang K, Sigurdsson A, Belfer I, Goldman D, Xu K, Shabalina SA, Shagin D, Max MB, Makarov SS, Maixner W. Genetic basis for individual variations in pain perception and the development of a chronic pain condition. Hum Mol Genet. 2005;14:135–143. doi: 10.1093/hmg/ddi013. [DOI] [PubMed] [Google Scholar]
- 10.Dobretsov M, Hastings SL, Sims TJ, Stimers JR, Romanovsky D. Stretch receptor-associated expression of alpha 3 isoform of the Na+, K+-ATPase in rat peripheral nervous system. Neuroscience. 2003;116:1069–1080. doi: 10.1016/s0306-4522(02)00922-3. [DOI] [PubMed] [Google Scholar]
- 11.Dobretsov M, Romanovsky D. “Clock-scan” protocol for image analysis. Am J Physiol Cell Physiol. 2006;291:C869–879. doi: 10.1152/ajpcell.00182.2006. [DOI] [PubMed] [Google Scholar]
- 12.Dobretsov M, Stimers JR. Neuronal function and alpha3 isoform of the Na/K-ATPase. Front Biosci. 2005;10:2373–2396. doi: 10.2741/1704. [DOI] [PubMed] [Google Scholar]
- 13.Dubuisson D, Dennis SG. The formalin test: a quantitative study of the analgesic effects of morphine, meperidine, and brain stem stimulation in rats and cats. Pain. 1977;4:161–174. doi: 10.1016/0304-3959(77)90130-0. [DOI] [PubMed] [Google Scholar]
- 14.Fillingim RB, Kaplan L, Staud R, Ness TJ, Glover TL, Campbell CM, Mogil JS, Wallace MR. The A118G single nucleotide polymorphism of the μ-opioid receptor gene (OPRM1) is associated with pressure pain sensitivity in humans. J Pain. 2005;6:159–167. doi: 10.1016/j.jpain.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Fink BR, Cairns AM. A new approach to differential peripheral nerve fiber block: Na+,K+-ATPase inhibition. Anesthesiology. 1983;59:127–131. doi: 10.1097/00000542-198308000-00011. [DOI] [PubMed] [Google Scholar]
- 16.Geering K. The functional role of beta subunits in oligomeric P-type ATPases. J Bioenerg Biomembr. 2001;33:425–438. doi: 10.1023/a:1010623724749. [DOI] [PubMed] [Google Scholar]
- 17.Hasler U, Wang X, Crambert G, Beguin P, Jaisser F, Horisberger JD, Geering K. Role of beta-subunit domains in the assembly, stable expression, intracellular routing, and functional properties of Na,K-ATPase. J Biol Chem. 1998;273:30826–30835. doi: 10.1074/jbc.273.46.30826. [DOI] [PubMed] [Google Scholar]
- 18.Hastie BA, Riley JL, III, Robinson ME, Glover T, Campbell CM, Staud R, Fillingim RB. Cluster analysis of multiple experimental pain modalities. Pain. 2005;116:227–237. doi: 10.1016/j.pain.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 19.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nature Biotech. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 20.Kim H, Dionne RA. Lack of influence of GTP cyclohydrolase gene (GCH1) variations on pain sensitivity in humans. Mol Pain. 2007;3:6. doi: 10.1186/1744-8069-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H, Mittal DP, Iadarola MJ, Dionne RA. Genetic predictors for acute experimental cold and heat pain sensitivity in humans. J Med Genet. 2006;43:e40. doi: 10.1136/jmg.2005.036079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H, Neubert JK, San Miguel A, Xu K, Krishnaraju RK, Iadarola MJ, Goldman D, Dionne RA. Genetic influence on variability in human acute experimental pain sensitivity associated with gender, ethnicity and psychological temperament. Pain. 2004;109:488–496. doi: 10.1016/j.pain.2004.02.027. [DOI] [PubMed] [Google Scholar]
- 23.LaCroix-Fralish ML, Ledoux JB, Mogil JS. The Pain Genes Database: an interactive web browser of pain-related transgenic knockout studies. Pain. 2007;131:3.e1–3.e4. doi: 10.1016/j.pain.2007.04.041. [DOI] [PubMed] [Google Scholar]
- 24.LaCroix-Fralish ML, Mogil JS. Progress in genetic studies of pain and analgesia. Annu Rev Pharmacol Toxicol. 2009;49:97–121. doi: 10.1146/annurev-pharmtox-061008-103222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai J, Hunter JC, Ossipov MH, Porreca F. Blockade of neuropathic pain by antisense targeting of tetrodotoxin-resistant sodium channels in sensory neurons. Meth Enzymol. 2000;314:201–213. doi: 10.1016/s0076-6879(99)14104-1. [DOI] [PubMed] [Google Scholar]
- 26.Lees GJ. Inhibition of sodium-potassium-ATPase: a potentially ubiquitous mechanism contributing to central nervous system neuropathology. Brain Res Rev. 1991;16:283–300. doi: 10.1016/0165-0173(91)90011-v. [DOI] [PubMed] [Google Scholar]
- 27.Li CY, Song YH, Higuera ES, Luo ZD. Spinal dorsal horn calcium channel alpha2delta-1 subunit upregulation contributes to peripheral nerve injury-induced tactile allodynia. J Neurosci. 2004;24:8494–8499. doi: 10.1523/JNEUROSCI.2982-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao G, Wang J, Guo J, Allard J, Cheng J, Ng A, Shafer S, Puech A, McPherson JD, Foernzler D, Peltz G, Usuka J. In silico genetics: identification of a functional element regulating H2-Eα gene expression. Science. 2004;306:690–695. doi: 10.1126/science.1100636. [DOI] [PubMed] [Google Scholar]
- 29.Liem EB, Joiner TV, Tsueda K, Sessler DI. Increased sensitivity to thermal pain and reduced subcutaneous lidocaine efficacy in redheads. Anesthesiology. 2005;102:509–514. doi: 10.1097/00000542-200503000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo MC, Zhang DQ, Ma SW, Huang YY, Shuster SJ, Porreca F, Lai J. An efficient intrathecal delivery of small interfering RNA to the spinal cord and peripheral neurons. Mol Pain. 2005;1:29. doi: 10.1186/1744-8069-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macpherson LJ, Xiao B, Kwan KY, Petrus MJ, Dubin AE, Hwang S, Cravatt B, Corey DP, Patapoutian A. An ion channel essential for sensing chemical damage. J Neurosci. 2007;27:11412–11415. doi: 10.1523/JNEUROSCI.3600-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malik N, Canfield VA, Beckers MC, Gros P, Levenson R. Identification of the mammalian Na,K-ATPase β3 subunit. J Biol Chem. 1996;271:22754–22758. doi: 10.1074/jbc.271.37.22754. [DOI] [PubMed] [Google Scholar]
- 33.Masocha W, Horvath G, Agil A, Ocana M, Del Pozo E, Szikszay M, Baeyens JM. Role of Na(+), K(+)-ATPase in morphine-induced antinociception. J Pharmacol Exp Ther. 2003;306:1122–1128. doi: 10.1124/jpet.103.052977. [DOI] [PubMed] [Google Scholar]
- 34.McDonough AA, Azuma KK, Lescale-Matys L, Tang MJ, Nakhoul F, Hensley CB, Komatsu Y. Physiologic rationale for multiple sodium pump isoforms. Differential regulation of alpha 1 vs alpha 2 by ionic stimuli. Ann N Y Acad Sci. 1992;671:156–168. doi: 10.1111/j.1749-6632.1992.tb43793.x. [DOI] [PubMed] [Google Scholar]
- 35.McNamara CR, Mandel-Brehm J, Bautista DM, Siemens J, Deranian KL, Zhao M, Hayward NJ, Chong JA, Julius D, Moran MM, Fanger CM. TRPA1 mediates formalin-induced pain. Proc Natl Acad Sci USA. 2007;104:13525–13530. doi: 10.1073/pnas.0705924104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mogil JS, Chanda ML. The case for the inclusion of female subjects in basic science studies of pain. Pain. 2005;117:1–5. doi: 10.1016/j.pain.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 37.Mogil JS, Crager SE. What should we be measuring in behavioral studies of chronic pain in animals? Pain. 2004;112:12–15. doi: 10.1016/j.pain.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 38.Mogil JS, Meirmeister F, Seifert F, Strasburg K, Zimmermann K, Reinold H, Austin J-S, Bernardini N, Chesler EJ, Hoffman HA, Hordo C, Messlinger K, Nemmani KVS, Rankin AL, Ritchie J, Siegling A, Smith SB, Sotocinal SB, Vater A, Lehto SG, Klussmann S, Quirion R, Michaelis M, Devor M, Reeh PW. Variable sensitivity to noxious heat is mediated by differential expression of the CGRP gene. Proc Natl Acad Sci USA. 2005;102:12938–12943. doi: 10.1073/pnas.0503264102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mogil JS, Richards SP, O'Toole LA, Helms ML, Mitchell SR, Belknap JK. Genetic sensitivity to hot-plate nociception in DBA/2J and C57BL/6J inbred mouse strains: possible sex-specific mediation by α2-opioid receptors. Pain. 1997;70:267–277. doi: 10.1016/s0304-3959(97)03333-2. [DOI] [PubMed] [Google Scholar]
- 40.Mogil JS, Ritchie J, Smith SB, Strasburg K, Kaplan L, Wallace MR, Romberg RR, Bijl H, Sarton EY, Fillingim RB, Dahan A. Melanocortin-1 receptor gene variants affect pain and μ-opioid analgesia in mice and humans. J Med Genet. 2005;42:583–587. doi: 10.1136/jmg.2004.027698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mogil JS, Ritchie J, Sotocinal SG, Smith SB, Croteau S, Levitin DJ, Naumova AK. Screening for pain phenotypes: analysis of three congenic mouse strains on a battery of nine nociceptive assays. Pain. 2006;126:24–34. doi: 10.1016/j.pain.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 42.Mogil JS, Wilson SG, Chesler EJ, Rankin AL, Nemmani KVS, Lariviere WR, Groce MK, Wallace MR, Kaplan L, Staud R, Ness TJ, Glover TL, Stankova M, Mayorov A, Hruby VJ, Grisel JE, Fillingim RB. The melanocortin-1 receptor gene mediates female-specific mechanisms of analgesia in mice and humans. Proc Natl Acad Sci USA. 2003;100:4867–4872. doi: 10.1073/pnas.0730053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moseley AE, Williams MT, Schaefer TL, Bohanan CS, Neumann JC, Behbehani MM, Vorhees CV, Lingrel JB. Deficiency in Na,K-ATPase alpha isoform genes alters spatial learning, motor activity, and anxiety in mice. J Neurosci. 2007;27:616–626. doi: 10.1523/JNEUROSCI.4464-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neddermeyer TJ, Fluhr K, Lotsch J. Principle components analysis of pain thresholds to thermal, electrical, and mechanical stimuli suggests a predominant common source of variance. Pain. 2008;138:286–291. doi: 10.1016/j.pain.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 45.Peek AS, Behlke MA. Design of active small interfering RNAs. Curr Opin Mol Ther. 2007;9:110–118. [PubMed] [Google Scholar]
- 46.Romanovsky D, Light KE, Walker J, Dobretsov M. Target-determined expression of alpha3 isoform of the Na+,K+-ATPase in the somatic nervous system of rat. J Comp Neurol. 2005;483:114–123. doi: 10.1002/cne.20401. [DOI] [PubMed] [Google Scholar]
- 47.Rose SD, Kim DH, Amarzguioui M, Heidel JD, Collingwood MA, Davis ME, Rossi JJ, Behlke MA. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33:4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sayers ST, Khan T, Shahid R, Dauzvardis MF, Siegel GJ. Distribution of alpha 1 subunit isoform of (Na,K)-ATPase in the rat spinal cord. Neurochem Res. 1994;19:597–602. doi: 10.1007/BF00971336. [DOI] [PubMed] [Google Scholar]
- 49.Skou JC. The influence of some cations on an adenosine triphosphatase from peripheral nerves. Biochim Biophys Acta. 1957;23:394–401. doi: 10.1016/0006-3002(57)90343-8. [DOI] [PubMed] [Google Scholar]
- 50.Skou JC, Esmann M. The Na,K-ATPase. J Bioenerg Biomembr. 1992;24:249–261. doi: 10.1007/BF00768846. [DOI] [PubMed] [Google Scholar]
- 51.Tegeder I, Costigan M, Griffin RS, Abele A, Belfer I, Schmidt H, Ehnert C, Nejim J, Marian C, Scholz J, Wu T, Allchorne A, Diatchenko L, Binshtok AM, Goldman D, Adolph J, Sama S, Atlas SJ, Carlezon WA, Parsegian A, Lotsch J, Fillingim RB, Maixner W, Geisslinger G, Max MB, Woolf CJ. GTP cyclohydrolase and tetrahydrobiopterin regulate pain sensitivity and persistence. Nature Med. 2006;12:1269–1277. doi: 10.1038/nm1490. [DOI] [PubMed] [Google Scholar]
- 52.Tjolsen A, Berge O-G, Hunskaar S, Rosland JH, Hole K. The formalin test: an evaluation of the method. Pain. 1992;51:5–17. doi: 10.1016/0304-3959(92)90003-T. [DOI] [PubMed] [Google Scholar]
- 53.Vagin O, Tokhtaeva E, Sachs G. The role of the beta1 subunit of the Na,K-ATPase and its glycosylation in cell-cell adhesion. J Biol Chem. 2006;281:39573–39587. doi: 10.1074/jbc.M606507200. [DOI] [PubMed] [Google Scholar]
- 54.Valder CR, Liu J-J, Song Y-H, Luo ZD. Coupling gene chip analyses and rat genetic variances in identifying potential target genes that may contribute to neuropathic allodynia development. J Neurochem. 2003;87:560–573. doi: 10.1046/j.1471-4159.2003.02016.x. [DOI] [PubMed] [Google Scholar]
- 55.Wang J, Peltz G. Peltz G, editor. Haplotype-based computational genetic analysis in mice. Book Title|, Vol. Volume|. City|: Publisher|, Year|. p.^pp. Pages|.
- 56.Wilson SG, Chesler EJ, Hain HS, Rankin AL, Schwarz JZ, Call SB, Murray MR, West EE, Teuscher C, Rodriquez-Zas S, Belknap JK, Mogil JS. Identification of quantitative trait loci for chemical/inflammatory nociception in mice. Pain. 2002;96:385–391. doi: 10.1016/S0304-3959(01)00489-4. [DOI] [PubMed] [Google Scholar]
- 57.Yang L, Zhang FX, Huang F, Lu YJ, Li GD, Bao L, Xiao HS, Zhang X. Peripheral nerve injury induces trans-synaptic modification of channels, receptors and signal pathways in rat dorsal spinal cord. Eur J Neurosci. 2004;19:871–883. doi: 10.1111/j.0953-816x.2004.03121.x. [DOI] [PubMed] [Google Scholar]
- 58.Yukhananov R, Kissin I. Persistent changes in spinal cord gene expression after recovery from inflammatory hyperalgesia: a preliminary study on pain memory. BMC Neurosci. 2008;9:32. doi: 10.1186/1471-2202-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeng W, Dohi S, Shimonaka H, Asano T. Spinal antinociceptive action of Na+, K+ pump inhibitor ouabain and its interaction with morphine and lidocaine in rats. Anesthesiology. 1999;90:500–508. doi: 10.1097/00000542-199902000-00026. [DOI] [PubMed] [Google Scholar]
- 60.Zubieta J-K, Heitzeg MM, Smith YR, Bueller JA, Xu K, Xu Y, Koeppe RA, Stohler CS, Goldman D. COMT val158met genotype affects μ-opioid neurotransmitter responses to a pain stressor. Science. 2003;299:1240–1243. doi: 10.1126/science.1078546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01