Co-transcriptional splicing of constitutive and alternative exons (original) (raw)
Abstract
In metazoan organisms, pre-mRNA splicing is thought to occur during transcription, and it is postulated that these two processes are functionally coupled via still-unknown mechanisms. Current evidence supports co-transcriptional spliceosomal assembly, but there is little quantitative information on how much splicing is completed during RNA synthesis. Here we isolate nascent chromatin-associated RNA from free, nucleoplasmic RNA already released from the DNA template. Using a quantitative RT-PCR assay, we show that the majority of introns separating constitutive exons are already excised from the human c-Src and fibronectin pre-mRNAs that are still in the process of synthesis, and that these introns are removed in a general 5′-to-3′ order. Introns flanking alternative exons in these transcripts are also removed during synthesis, but show differences in excision efficiency between cell lines with different regulatory conditions. Our data suggest that skipping of an exon can induce a lag in splicing compared to intron removal under conditions of exon inclusion. Nevertheless, excision of the long intron encompassing the skipped exon is still completed prior to transcript release into the nucleoplasm. Thus, we demonstrate that the decision to include or skip an alternative exon is made during transcription and not post-transcriptionally.
Keywords: co-transcriptional splicing, alternative splicing, fibronectin, c-Src
INTRODUCTION
Transcription of messenger RNA precursors (pre-mRNAs) by RNA polymerase II (Pol II) and their splicing by the spliceosome are essential steps in gene expression. In vitro, these processes are often studied independently of one another, although there is substantial evidence that they occur simultaneously within cell nuclei. Initial data supporting co-transcriptional splicing came from electron micrographs of Drosophila melanogaster embryonic transcription units, which indicated that ribonucleoprotein complex formation on splice sites, and subsequent intron looping, can occur while the transcript remains tethered to the chromatin template (Beyer et al. 1981; Beyer and Osheim 1988). Later analyses of Balbiani ring 1 gene pre-mRNAs, obtained via microdissection of the polytene chromosomes of the Chironomus tentans salivary gland, demonstrated that over 90% of intron 3 excisions occurs during transcription (Bauren and Wieslander 1994). Similar analysis of the Balbiani Ring 3 gene indicated that splicing of nascent transcripts proceeds in a general, but not strict, 5′-to-3′ direction (Wetterberg et al. 1996). In mammalian cells, spliceosome components can be observed to accumulate at gene loci during transcription by immunofluorescence or chromatin immunoprecipitation (Huang and Spector 1991; Misteli et al. 1997; Neugebauer and Roth 1997; Listerman et al. 2006). This splicing factor accumulation occurs only on intron-containing transcripts (Huang and Spector 1996; Listerman et al. 2006) and requires Pol II with an intact C-terminal domain (CTD) (Misteli and Spector 1999). Recent in vitro assays of transcription and splicing within the same reaction have also shown that intron removal is more efficient when transcription is catalyzed by polymerase II (Pol II) rather than by phage RNA polymerases (Das et al. 2006; Hicks et al. 2006; Natalizio et al. 2009). These and other data led to the current hypothesis that splicing initiates prior to transcriptional termination and that the two processes can be functionally coupled.
The splicing reaction is highly regulated and, in mammalian cells, nearly all genes produce multiple mRNA products through the use of alternative exons and splice sites (Wang et al. 2008). Regulated splicing choices are mostly determined by non-splice-site sequences, located in alternative exons or neighboring introns, which recruit special regulatory proteins (Wang and Burge 2008). These RNA binding proteins, including members of the SR and hnRNP protein classes, can either facilitate or prevent spliceosome assembly at particular splice sites (Black 2003).
Two general models have been proposed for how the transcription apparatus might effect changes in alternative splicing patterns. One model for the functional coupling of transcription and splicing posits that the CTD acts as a “landing pad,” recruiting splicing factors to regions of ongoing transcription (Greenleaf 1993). Data indicate that SR-domain containing proteins can interact with Pol II (Yuryev et al. 1996; Misteli and Spector 1999). However, this interaction, if it occurs during transcription, is likely transient, as most SR proteins recruited to a gene locus depend on RNA for their gene association (Sapra et al. 2009). A second, less direct model for coupling has been described for a subset of alternative exons. This proposes that changes in Pol II elongation rate can modulate exon inclusion by altering the balance between competitive splice sites (de la Mata et al. 2003). In support of this “kinetic coupling” model, numerous effectors of transcription elongation rates including promoter identity (Cramer et al. 1997, 1999), chromatin-remodeling and modification (Batsche et al. 2006), and transcriptional activators (Dowhan et al. 2005) have been shown to affect the splicing patterns of specific alternative exons. The mechanistic basis for these models remains unresolved, but it is likely that processes postulated by both models are affecting alternative splicing choices.
Although, it is presumed that spliceosome assembly initiates during transcription, as supported by the detection of splicing factors accumulating at active gene loci, it is still not clear when the majority of splicing is completed. In Saccharomyces cerevisiae, snRNPs also accumulate on nascent transcripts as measured by chromatin immunoprecipitation. Pre-mRNAs with long second exons (>∼700 base pairs) are thought to be spliced co-transcriptionally and those containing short second exons post-transcriptionally, as judged by the different recruitment of spliceosome components to gene loci (Tardiff et al. 2006). In metazoans, there is little data regarding how much intron excision occurs prior to transcript release from the template. Analyses of endogenous and minigene-encoded intron-containing RNAs from isolates of mammalian total nuclear RNA indicate that intron removal will sometimes occur in a 5′-to-3′ fashion, consistent with a pattern of concurrent co-transcriptional intron removal (Hatzoglou et al. 1985; Lang and Spritz 1987; Neel et al. 1993). However, deviations from this pattern are also described (Gudas et al. 1990; Kessler et al. 1993). Moreover, it is not clear whether this intron excision occurs before or after transcript release from the template.
A greater understanding of how much splicing occurs during transcription is needed to resolve several questions regarding splicing regulation. For example, it is not known whether the excision efficiency of all the introns in the transcripts of a particular gene can be stimulated collectively over those in transcripts of other genes, as might be expected from some forms of transcriptional coupling. At the other extreme, it is not known whether some transcripts could be maintained in a pool of semispliced RNA after transcription is complete. Such a pool would allow the rapid processing of exons in response to particular cellular stimuli without additional transcription. There is evidence in yeast for pre-mRNAs containing meiotically spliced introns being maintained as unspliced pools in mitotic cells (Engebrecht et al. 1991; Moore et al. 2006). In mammalian cells, partially processed RNA can be detected in cell nuclei, but has not been quantified relative to fully spliced species or shown to exist in a pool separate from simple nascent RNA. Thus, we need more information on where and when the regulatory events of splicing occur relative to RNA synthesis.
Here, we systematically analyze the patterns of intron removal in the c-Src and fibronectin (FN) pre-mRNA transcripts. We compare the patterns seen in nascent chromatin-associated RNA with those found after release of the RNA into the nucleoplasm. We find that the vast majority of introns neighboring constitutive exons are excised co-transcriptionally. Introns flanking alternative exons are also removed during synthesis, but show differences in excision efficiency between cell lines with different regulatory conditions. Nevertheless, we find that the decision to include or skip an alternative exon is made during transcription and not post-transcriptionally.
RESULTS
Isolation and quantitative analysis of chromatin-associated pre-mRNA
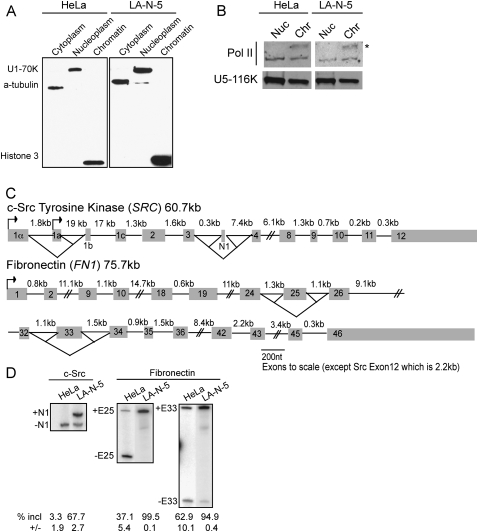
To examine intron excision levels in nascent RNA's associated with RNA Pol II and in RNA's released from the template; we separated chromatin-associated transcripts from free nucleoplasmic RNA. It was previously shown that isolation in urea and nonionic detergent removes loosely associated RNAs and proteins from the chromatin template, but does not disrupt ternary Pol II complexes (Wuarin and Schibler 1994). After cell lysis and separation of nuclei from the cytoplasmic fraction, we used this method to isolate chromatin-associated and soluble nucleoplasmic RNA fractions from both human HeLa and LA-N-5 neuroblastoma cells. The fractionation was assessed by immunoblot using antibodies to α-tubulin, U1-70K, and histone H3 proteins (Fig. 1A). Tubulin was found only in the cytoplasmic fraction and histone H3 solely in the chromatin fraction. The nucleoplasmic fraction should contain nucleoplasmic proteins and those components stripped off the chromatin. U1-70K, a component of the U1 snRNP that is easily removed from spliceosomes, is found only in the nucleoplasmic fraction under these conditions (Konarska and Sharp 1987; Konforti et al. 1993; Will and Lührmann 2006). In contrast, U5-116K, a subunit of the U5 snRNP whose association with the spliceosome is more stable than that of U1 (Konarska and Sharp 1987; Bessonov et al. 2008), is present in both nucleoplasmic and chromatin fractions, as is Pol II (Fig. 1B). Notably, the hyperphosphorylated form of Pol II, found in transcriptionally active complexes purifies exclusively with the chromatin, and constitutes 59% of the total Pol II in that fraction (Fig. 1B, asterisk; Egloff and Murphy 2008). Thus, this fractionation procedure cleanly separates different types of nuclear species. Components of the core transcriptional machinery, particularly the active Pol II complexes, and the catalytic spliceosome remain associated with the chromatin fraction.
FIGURE 1.
Purification of chromatin-associated pre-mRNA. (A) Chemiluminescent immunoblot of fractionated HeLa and LA-N-5 cell lines. Anti-α-tubulin, anti-U170K, and anti-Histone3 antibodies detect the cytoplasmic, nucleoplasmic, and chromatin fractions, respectively. Twenty-five micrograms of total protein from each fraction was loaded per lane. (B) Fluorescent immunoblot of HeLa and LA-N-5 chromatin and nucleoplasmic fractions. Twenty-five micrograms of total protein from each fraction was loaded per lane. An asterisk denotes hyperphosphorylated form of Pol II. (C) Diagrams of the c-Src and fibronectin gene structure. The 18 nt c-Src N1 exon is alternatively spliced and the 1A promoter is used in both HeLa and LA-N-5 cells. The fibronectin transcript contains exons 25 (EDII or EIIIB) and 33 (EDI or EIIIA), which are alternative. (D) Radiolabeled RT-PCR of cytoplasmic RNA showing inclusion levels of the c-Src N1, FN Exon 25 and 33 alternative exons in HeLa and LA-N-5 cell lines.
Poly(A) site cleavage is thought to occur prior to transcript release (West et al. 2008; Rigo and Martinson 2009). To assess how much RNA is stripped from the ternary complexes on the chromatin by the isolation procedure, we examined the amount of RNA uncleaved at the poly(A) site in the chromatin versus the nucleoplasmic fraction. We amplified uncleaved c-Src RNA from both nuclear fractions (Supplemental Fig. S1) by reverse transcription with a primer downstream from the cleavage site, followed by PCR amplification across exons 11 and 12. Interestingly, much of the RNA that is uncleaved at the poly(A) site is spliced across intron 11, and so both spliced and unspliced RNA were measured separately. The total c-Src RNA containing exon 11 was essentially the same in the two fractions. In contrast, the spliced RNA uncleaved at the poly(A) site is 7.0- and 5.4-fold more abundant in HeLa and LA-N-5 chromatin fractions compared to nucleoplasmic ones. In HeLa cells, a similar enrichment of RNA that is both uncleaved at the polyA site and unspliced at intron 11 is seen in the chromatin fraction. In LA-N-5 cells, there is more abundant unspliced RNA in the nucleoplasmic fraction. However, this is a small portion of the total nucleoplasmic c-Src RNA, which is almost entirely spliced in this fraction (see below). Thus, almost all nascent, uncleaved c-Src RNA remains attached to the template during the isolation procedure.
We chose two genes to study in detail. The c-Src and FN genes are well-characterized splicing templates that are ubiquitously expressed across cell types, and are of similar length (Kornblihtt et al. 1996; Chan and Black 1997). Fully spliced FN mRNAs have up to 46 exons depending on the inclusion of two regulated exons. Src has 15 exons when the alternative N1 exon is included (Fig. 1C). We examined a number of human cell lines (data not shown) and chose HeLa and LA-N-5 cells because they showed strong differences in splicing for the Src N1, FN 25 (also called EDII or EIIIB) and FN 33 alternative exons (EDI, EIIIA). As shown in Figure 1D, the c-Src exon N1 is completely excluded in HeLa cells, but shows ∼70% inclusion in LA-N-5 cells. FN exon 25 is predominantly skipped in HeLa, but is included in all LA-N-5 transcripts. FN exon 33 is included in 95% of the transcripts in LA-N-5 cells and 63% in HeLa cells. There is some culture-to-culture variation in exon 33 inclusion, which we attribute to changes in cell density.
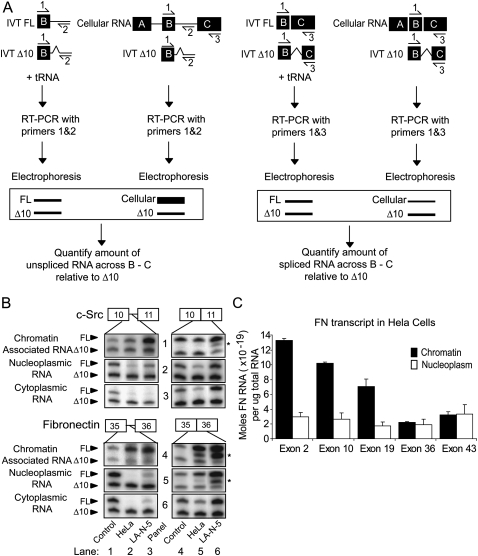
We next wanted to assess the structure and amount of RNA at different positions within the purified nascent transcripts. We employed a quantitative PCR protocol using in vitro transcribed (IVT) RNA's as internal normalization standards (Fig. 2A; Patel et al. 2002). To measure the abundance of a given intron, two sets of primers were designed. One pair flanked the 5′ exon–intron junction to detect pre-mRNA's containing that intron. A second pair of primers flanked the spliced exon–exon junction to measure the level of spliced RNA across the same exon–intron–exon region. For each intron analyzed, the exon–intron and exon–exon primer sets contained a common primer (Fig. 2A, primer 1), which was radiolabeled. For each set of primers, two IVT RNA controls were made. The first was the same size and sequence as the endogenous RNA. The second control RNA contained an internal 10 nucleotides (nt) deletion (Δ10). Equal quantities of the IVT FL and Δ10 internal standards were subjected to RT-PCR to confirm that the FL:Δ10 ratio was ∼1:1, indicating equally efficient amplification (labeled “control” in all figures). A defined amount (1 × 10−18 mol) of the Δ10 control RNA was added to each experimental sample of RNA, resulting in two bands per lane. One-tenth of each reverse transcription (RT) reaction was subsequently used in a PCR reaction, so that the upper experimental bands can be measured relative to 1 × 10−19 mol of RNA in the lower control band. The percentage of unspliced RNA remaining across each exon–intron–exon region was calculated by dividing the intron containing product by the sum of the intron containing and spliced products (Fig. 2A). RNA containing unspliced c-Src intron 10 and FN intron 35 is highly enriched in the chromatin-associated fraction (Fig. 2B, panels 1,4, lanes 2,3, upper bands), but not the cytoplasmic fraction (Fig. 2B, panels 3,6, lanes 2,3, upper bands). In contrast, RNA, where these introns have been excised, is abundant in all three fractions: chromatin-associated, nucleoplasmic, and cytoplasmic (Fig. 2B, all panels, lanes 5,6, upper bands). Each cellular fractionation and RT-PCR was repeated at least three times. In subsequent figures, values are an average of three experiments with error bars representing the standard error of mean (SEM).
FIGURE 2.
Exon abundance decreases with increasing proximity to 3′ end of the transcript within the chromatin fraction. (A) Depiction of quantitative RT-PCR protocol. HeLa and LA-N-5 chromatin-associated, nucleoplasmic, or cytoplasmic RNA was subjected to reverse transcription using a gene specific primer in the presence of 10−18 mol of in vitro transcribed (IVT) RNA internal controls. The amount of each analyzed exon–intron or exon–exon junction PCR product was calculated by comparison of band intensities between control and experimental samples. The fraction of each intron remaining in the three fractions was calculated by dividing the amount of the 5′ exon–intron junction product for each intron (the PCR product of primers 1 and 2) by the total product across the region (the sum of the intron product from primers 1 and 2 and the spliced product from primers 1 and 3). (B) Radiolabeled RT-PCR sample results for c-Src exons 10 and 11 and FN exon 35 and 36. Lanes 1,4 are control IVT RNA's as are the lower bands in lanes 2,3,5,6. Upper bands in lanes 2,5 (HeLa) and 3,6 (LA-N-5) represent the quantity of endogenous exon–intron or exon–exon junctions in each fraction of RNA. Asterisks denote nonspecific products. (C) Histogram representing the absolute amount of each analyzed constitutive FN exon in 1 μg of chromatin-associated or nucleoplasmic HeLa RNA. Error bars represent the standard error of mean (SEM).
The nascent, chromatin-associated RNAs should only be partially complete, whereas transcripts released into the nucleoplasm are expected to be fully transcribed and should have an equal quantity of 5′ and 3′ ends. To further validate and assess the purity of the chromatin-associated pre-mRNA pool, we measured the abundance of individual exons along the FN gene in the HeLa nuclear fractions. In the chromatin-associated fraction there was a decreasing 5′-to-3′ gradient of exon abundance, where the 5′ proximal exons were five- to sixfold more abundant than 3′ proximal ones (Fig. 2C). A similar gradient was seen in LA-N-5 cells (data not shown). Thus, the quantities of RNA in the chromatin fraction reflect the direction of transcription. In contrast, there was no difference in the abundance of the 5′ and 3′ proximal exons in the nucleoplasmic fraction, indicating that the majority of released, nucleoplasmic transcripts are complete.
The extent of intron excision decreases with increasing proximity to the 3′ end of the transcript
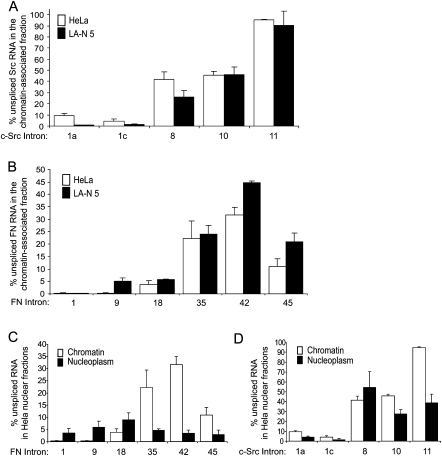
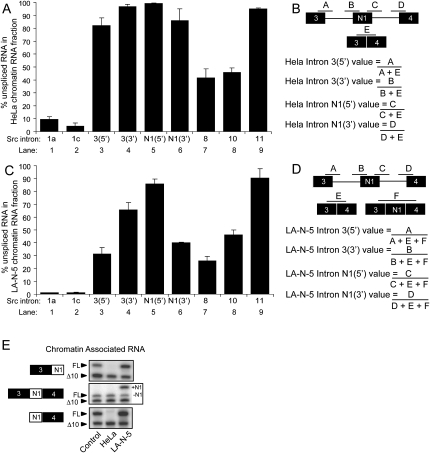
The removal of introns soon after they are transcribed would lead to the 5′ proximal region of a pre-mRNA being more completely spliced than the 3′ region. This idea is supported by observations from RNA and chromatin immunoprecipitation assays examining snRNP assembly along transcribed genes (Listerman et al. 2006), as well as EM experiments assaying the length of RNA extending from the polymerase as it progresses along a gene (Beyer et al. 1981). To test this directly, we measured the levels of representative exon–intron junctions at different positions along each gene. Within chromatin-associated c-Src and FN transcripts, in both cell lines, introns became more abundant with distance from the 5′ end, as predicted (Fig. 3A,B; representative raw data in Supplemental Fig. S2). Under these steady-state conditions, the initial introns at the 5′ end of Src (connecting exons 1a to 1b, and 1c to 2) are removed in over 90% of all transcripts. Similarly, FN introns 1, 9, and 18 are at least 94% spliced in both cell types. Indeed, FN intron 1 is barely detectable (Supplemental Fig. S2B), indicating that the intron is essentially completely removed in the chromatin fraction (i.e., fully co-transcriptionally excised). Extending this analysis toward the 3′ end of the transcript shows a steady increase in the level of unspliced introns for both genes. The only exception is the 3′ terminal FN intron 45, which is somewhat more fully spliced than the introns upstream (Fig. 3B). The percent of each intron retained is approximately equal in HeLa and LA-N-5 cells, suggesting that the efficiency of constitutive intron removal is similar between the two cell lines. Interestingly, all the analyzed introns, apart from the 3′ terminal Src intron 11, are removed in more than 50% of transcripts within the chromatin-associated RNA fraction. Thus, the majority of intron excision occurs co-transcriptionally, while the pre-mRNA is still attached to the DNA template. Note that nascent RNA's in which the intron has not been completely transcribed (i.e., the 5′ but not the 3′ splice site has been produced) will be detected as “unspliced” in this assay. Thus, the steady-state level of co-transcriptional intron excision may be higher than we can ascertain.
FIGURE 3.
Introns flanking constitutive exons in c-Src and FN are excised co-transcriptionally. (A) Histogram representing the percentage of unspliced RNA across c-Src introns 1a, 1c, 8, 10, and 11 in chromatin-associated pre-mRNAs from HeLa (white) and LA-N-5 (black) cells. (B) Histogram representing the percentage of unspliced RNA across FN introns 1, 9, 18, 35, 42, and 45 in chromatin-associated pre-mRNA from HeLa (white) and LA-N-5 (black) cell lines. (C) Histogram representing the percentage of unspliced RNA across FN introns 1, 9, 18, 35, 42, and 45 per microgram of HeLa chromatin-associated (white) and nucleoplasmic (black) RNA. (D) Same as (C) except data represent introns in the c-Src transcript from chromatin-associated and nucleoplasmic fractions. Error bars represent the standard error of mean (SEM).
We also measured the level of intron retention within transcripts from the nucleoplasmic fraction. These results were less consistent than with the chromatin-associated RNA fraction and varied with the gene and cell type examined. Most introns that were retained in the chromatin fraction showed increased excision in the nucleoplasm. This was particularly true for FN introns 35, 42, and 45 in HeLa cells (Fig. 3C), which do not show the 5′-to-3′ gradient of intron retention that was seen for the chromatin fraction. The 3′ most introns in the nucleoplasmic c-Src transcript continued to show higher levels of retention than those more 5′ proximal, although introns 10 and 11 were more completely excised than in the chromatin fraction. This was seen in both HeLa cells (Fig. 3D) and LA-N-5 cells (Supplemental Fig. S3B). In contrast, c-Src intron 8 was retained in nucleoplasmic transcripts at higher levels than within the chromatin fraction in both cell lines. It is tempting to speculate that these remaining introns will be excised in the nucleoplasm. However, from steady-state measurements we cannot be certain that all these transcripts will become mRNAs and are not aberrant products that will ultimately be degraded. The fact that these retained, nucleoplasmic introns do not always follow a 5′-to-3′ pattern of removal indicates that other processes are affecting their excision kinetics besides their position within the transcript.
In the HeLa chromatin-associated RNA fraction, FN intron 45 is excised in over 80% of transcripts, whereas intron 42 is removed in about 60% of chromatin-associated messages (Fig. 3B). A similar trend was seen in LA-N-5 cells. This is the only intron among the constitutive introns tested that was more extensively excised than those preceding it. We also assayed FN introns 43 and 44 (Supplemental Fig. S4), and found that exon 44 exhibited a higher level of splicing relative to the introns upstream. We further noted that the abundance of both FN intron 45 and the 3′ terminal Src intron 11 decreased by 50% or more between the chromatin-associated and nucleoplasmic fractions in both cell lines (Fig. 3C,D; Supplemental Fig. S3A,B). It has been suggested that splicing across terminal introns stimulates 3′-end processing and subsequent release of Pol II transcripts (Nesic and Maquat 1994; Rigo and Martinson 2008). Thus, the large increase in splicing between the chromatin and nucleoplasmic fractions for these terminal introns may result from their proximity to the poly(A) site.
Introns flanking alternative exons are excised co-transcriptionally
We next examined how alternative exons might differ in their patterns of intron excision. Although all the alternative exons in c-Src and FN show predominant inclusion in LA-N-5 cells and increased skipping in HeLa cells, they differ in their regulation. Src exon N1 and FN Exon 25 have both been described as targets for polypyrimidine tract binding (PTB, PTBP1) protein (Norton 1994; Chan and Black 1997) and Feminizing on X-2 (Fox2, RBM9) (Underwood et al. 2005), although they likely differ in their responses to other factors. In contrast, FN exon 33 splicing is controlled by SR proteins (Cramer et al. 1999; de la Mata and Kornblihtt 2006) and has been shown to increase in response to decreased transcription elongation rates (de la Mata et al. 2003).
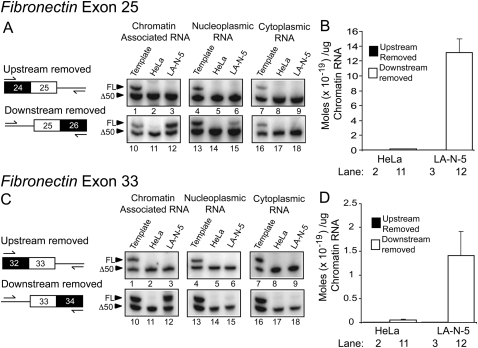
To assess the order of excision for the introns flanking these alternative exons, we measured the abundance of the two possible splicing intermediates retaining either only the upstream or only the downstream intron. For both FN exons 25 and 33, the product resulting from downstream intron excision and upstream intron retention is clearly detectable in the chromatin fraction of LA-N-5 cells (Fig. 4A,C, lane 12, upper band). These FN alternative splicing intermediates were, at best, only faintly detectible in HeLa cells, presumably due to lower FN RNA expression and reduced splicing of exons 25 and 33 in these cells. The reverse splicing intermediate, where the upstream, but not the downstream intron has been excised, was not detectable in either cell type for either FN exon 25 or 33 (Fig. 4A,C, lanes 2,3, upper band). Thus, both FN alternative exons are apparently spliced via a pathway where the downstream intron is removed prior to the upstream one.
FIGURE 4.
The introns downstream from FN alternative exons 25 and 33 are excised prior to the upstream ones. (A) Analysis of FN exon 25 splicing intermediates. The two possible splicing intermediates generated during alternative exon inclusion are diagrammed on the left. Denaturing PAGE analysis of RT-PCR splicing intermediate products is pictured on the right. The lower bands in all gels, as well as the upper bands in lanes 1,4,7,10,13,16 are internal IVT controls. Upper bands in lanes 2,3,11,12 represent splicing intermediate products from HeLa and LA-N-5 chromatin-associated RNA. Lanes 5,6,14,15 and lanes 8,9,17,18 represent the same information from the nucleoplasmic and cytoplasmic RNA fractions, respectively. Control reactions are shown in Supplemental Figure S5. (B) Quantification of lanes 2,3,11,12 in A. (C) Analysis of FN exon 33 splicing intermediates. The experimental setup is identical to A, except primers target exon 33. (D) Quantification of lanes 2,3,11,12 in C. Error bars represent the standard error of mean (SEM).
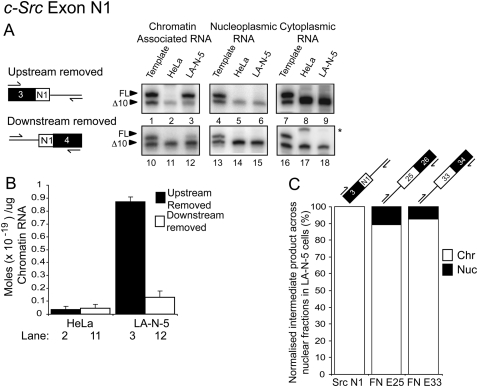
For the splicing of the c-Src exon N1, the opposite result was obtained. The product where the upstream, but not the downstream, intron is excised was readily detectable in the chromatin fraction of LA-N-5 cells (Fig. 5A, lane 3, upper band). The other intermediate is also observed, but is sevenfold less abundant than the major intermediate (Fig. 5A, lane 12, upper band). In HeLa cells, neither intermediate is detected because the N1 exon is entirely skipped (Fig. 5A, lanes 2,11).
FIGURE 5.
The intron upstream of c-Src exon N1 is excised prior to the downstream one. (A) Analysis of c-Src N1 splicing intermediates. The two possible splicing intermediates are diagrammed on the left. Denaturing PAGE analysis of RT-PCR splicing intermediate products is pictured on the right. The lower bands in all gels, as well as the upper bands in lanes 1,4,7,10,13,16 are internal controls. An asterisk denotes a nonspecific product. Upper bands in lanes 2,3,11,12 represent splicing intermediate products from HeLa and LA-N-5 chromatin-associated RNA. Lanes 5,6,14,15 and lanes 8,9,17,18 represent the same information except from the nucleoplasmic and cytoplasmic RNA fractions, respectively. Control reactions are shown in Supplemental Figure S5. (B) Quantification of lanes 2,3,11,12 in A. Asterisk indicates nonspecific product. (C) Stacked column graph showing percentage of the detectable splicing intermediate product found in LA-N-5 chromatin-associated and nucleoplasmic fractions normalized to the total amount of FN or c-Src RNA in each fraction. Error bars represent the standard error of mean (SEM).
Normalizing the amount of each intermediate to the total c-Src or FN RNA in the two fractions we find that 100% of the major c-Src splicing intermediate and over 85% of the major FN splicing intermediates purify with the chromatin-associated RNA (Fig. 5C). Thus, the majority of alternative splicing intermediates are generated, and then resolved, while the RNA is attached to the chromatin template.
We also compared the amount of intron retained in the alternatively spliced regions to that in constitutively spliced regions. Given that multiple splicing patterns are possible for each splice site flanking a regulated exon, we included all four exon–intron junctions (5′ and 3′ junctions of both upstream and downstream introns) in our analysis and plotted them for comparison with the constitutive introns.
The N1 exon splice junctions are essentially unspliced in the HeLa cell chromatin fraction, as expected from the skipping of this exon in these cells (Fig. 6A, lanes 4,5). However, the 5′ splice site of intron 3 and the 3′ splice site of intron N1 (Fig. 6A, lanes 3,6) are also over 80% retained in the HeLa cell chromatin fraction. This level of intron retention is twofold higher than for the downstream intron 8, indicating that splicing from exon 3 to exon 4 lags, relative to the flanking introns. Thus, the kinetics of excision when exon N1 is skipped is slower than the excision kinetics of constitutive introns. Nevertheless, the exon 3/exon 4 spliced product is present in the chromatin fraction indicating that splicing of exon 3 to 4 is still likely occurring on the chromatin (Fig. 6E, middle panel).
FIGURE 6.
Splicing across c-Src exon N1 is inefficient when N1 is skipped. (A) Histogram displaying the percentage of unspliced RNA across analyzed introns along the c-Src transcript in the HeLa chromatin-associated RNA fraction. Lanes 1,2,7–9 are the constitutive regions shown in Figure 3A. Lane 3 represents the percentage of unspliced RNA across the 5′ exon-intron junction of the intron upstream of N1 relative to the total spliced and unspliced RNA across exons 3–4. Lanes 4–6 indicate the amount of unspliced RNA across the other three exon-intron junctions. (B) Diagram of exon–intron junctions analyzed in A and method of quantification. (C) Same as in A, except analysis was done using LA-N-5 chromatin-associated RNA. (D) Same as in B, except analysis accounts for the situation where N1 is included. (E) RT-PCR analysis of spliced exon 3-N1-4 products in HeLa and LA-N-5 chromatin-associated RNA fraction indicating that spliced RNA is present in HeLa chromatin-associated fractions. Error bars represent the standard error of mean (SEM).
LA-N-5 cells carry out both N1 exon skipping and inclusion, with N1 being included ∼70% of the time (Fig. 1D). Both pathways contribute to the level of splicing observed at the intron 3 5′ splice site and the intron N1 3′ splice site (Fig. 6C, lanes 3,6). As expected, these splice sites show higher levels of splicing than the splice sites adjacent to N1 (Fig. 6C, lanes 4,5). Note that some of the RNA unspliced across the N1 exon may also derive from the already excised long intron, containing the skipped N1 exon. Again, the spliced products are readily detectable; indicating that splicing can be complete before release from the template (Fig. 6E, middle panel).
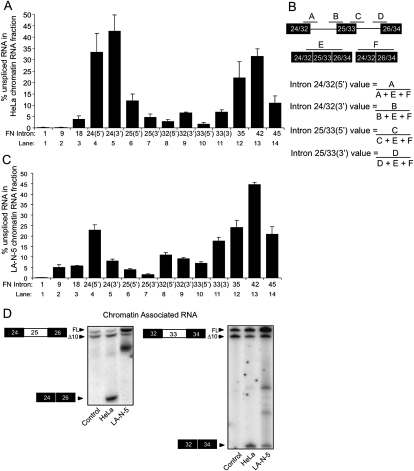
The same analysis on fibronectin gave somewhat different results. FN exons 25 and 33 are partially included in HeLa cells and almost completely so in LA-N-5 cells. In the HeLa chromatin fraction, intron 24 upstream of exon 25 shows less complete removal than intron 25 downstream, in agreement with the assays of the partially spliced products (Fig. 7A, lanes 4–7). Removal of this intron seems to lag behind excision of both the downstream intron 25 and the adjacent constitutive introns. This is also seen in LA-N-5 cells, although to a lesser extent. Although exon 25 is 100% included, the 5′ splice site of exon 24 is anomalously high relative to the other regulated sites (Fig. 7C, lane 4). The source of this is unknown, but it could result from transcriptional pausing in intron 24, between the 5′ and 3′ splice sites. In contrast to FN exon 25 and c-Src N1, FN exon 33 is very efficiently spliced. The introns flanking this exon show over 90% removal in HeLa chromatin-associated RNA and greater than 80% removal in the same LA-N-5 fraction (Fig. 7A,C, lanes 8–11). Once in the nucleoplasm, the amount of introns 32 and 33 remaining is at most, 2.5% (data not shown). This exon has been extensively studied for its response to transcription rates, and the observation that it is essentially fully spliced, while still associated with chromatin, is consistent with these analyses. The introns flanking the other two alternative exons, c-Src N1 and FN exon 25 are also greater than 50% spliced in the nucleoplasmic fraction (data not shown). However, their different retention levels may indicate a different kinetics of splicing and presumably different mechanism of regulation than exon 33.
FIGURE 7.
Splicing is efficient across the FN exon 25 and exon 33 alternative regions when the exons are completely included. (A) Histogram displaying the percentage of unspliced RNA across analyzed introns along the FN transcript in the HeLa chromatin-associated RNA fraction. Lanes 1–3,12–14 are the constitutive regions shown in Figure 3B. Lane 4 represents the percentage of unspliced RNA across the 5′ exon–intron junction of the intron upstream of exon 25 relative to the total amount RNA across exons 24–26. Lanes 5–7 indicate the amount of unspliced RNA across the other three exon–intron junctions. Similarly, lane 8 represents the percentage of unspliced RNA across the 5′ exon–intron junction of the intron upstream of exon 33 relative to the total amount RNA across exons 32–34. Lanes 9–11 indicate the amount of unspliced RNA across the other three exon–intron junctions. (B) Diagram of exon–intron junctions analyzed in A and C and method of quantification. (C) Same as in A, except analysis was done using LA-N-5 chromatin-associated RNA. (D) RT-PCR analysis of spliced exon 24–26 and 32–34 products in HeLa and LA-N-5 chromatin-associated RNA fraction. Error bars represent the standard error of mean (SEM).
DISCUSSION
Nearly all splicing occurs while pre-mRNA is attached to the chromatin template
In this study, we examined how introns vary in their co-transcriptional excision, with a goal of understanding how this process might by affected by regulatory inputs. To measure the amount of exon ligation occurring prior to transcript release from the chromatin template, we purified chromatin-associated RNAs and compared them to soluble nuclear ones. Since the contamination of chromatin-associated RNA with fully processed nucleoplasmic RNA would exaggerate the extent of co-transcriptional splicing, we confirmed that the chromatin-associated RNA exhibited several features expected of a pool of nascent transcripts. First, we detected a 5′-to-3′ decreasing gradient of exon abundance expected of the incompletely transcribed transcripts present in elongating ternary Pol II complexes (Fig. 2C). Second, this pool of RNA is enriched in messages that are uncleaved at the poly(A) site, consistent with data indicating that cleavage occurs prior to mRNA release (Supplemental Fig. S1; West et al. 2008). Last, the chromatin-associated RNA exhibits a rough 5′-to-3′ decreasing gradient of intron excision that would be expected for RNA undergoing co-transcriptional splicing (Fig. 3A,B). We do not observe these properties in the free nucleoplasmic fraction or in the total nuclear RNA preparations commonly studied as nascent RNA (Figs. 2C, 3C,D). Thus, chromatin-associated RNA provides an enriched source of truly nascent transcripts.
The ability to isolate chromatin-associated RNA is useful for a number of applications. An increase in the abundance of intronic RNA is sometimes used to verify that a rise in mRNA expression is due to increased synthesis rather than an alteration in mRNA degradation or other downstream process (Carey and Smale 2000). Such changes in pre-mRNA expression would be more sensitively measured in chromatin-associated RNA rather than RNA present in the usual nuclear preparation. Similarly, studies of splicing regulation in response to cellular stimuli are often hampered by the analysis of total or cytoplasmic mRNA levels, which change slowly relative to the stimulus. By assaying splicing in a true nascent RNA fraction, one might observe much more rapid responses to cell stimulation.
We find that the splicing of constitutive exons within the c-Src and FN genes does indeed proceed in a general 5′-to-3′ order (Fig. 3A,B). However, some introns did not follow this pattern; that is, they were more completely spliced than the preceding introns or less completely spliced than introns more 3′ proximal. The most anomalous examples were intron 44 and terminal intron 45 in FN, which were more efficiently spliced than the upstream intron 42 (Fig. 3B; Supplemental Fig. S4). There is evidence for functional interactions between terminal introns and the cleavage and polyadenylation machinery (Bauren et al. 1998; Vagner et al. 2000; Li et al. 2001; Rigo and Martinson 2008). Thus, it is tempting to speculate that the enhanced splicing of FN exon 45 is in some way related to its proximity to the poly(A) site, but this will require additional examination.
Nearly all the examined introns were over 50% excised in the chromatin fraction (Fig. 3A,B). FN intron 1 was virtually 100% excised in this pool, clearly indicating that its splicing is completed during RNA synthesis (Supplemental Fig. S2). The least efficiently excised intron was c-Src intron 11, which is retained in over 90% of chromatin-associated RNA transcripts (Fig. 3A). The anomalously high retention of this intron in nascent RNA may also be related to its location adjacent to the 3′ terminal exon. If terminal intron splicing is coupled to 3′ end formation, as suggested for FN, the rather long final c-Src exon may delay the splicing of intron 11 until transcription and poly-A site recognition are complete. In support of this, we find that intron 11 is over 60% removed in nucleoplasmic fractions (Fig. 3D; Supplemental Fig. S3B). Thus, although we observe the majority of splicing to occur in association with chromatin, RNAs containing unprocessed introns clearly can be released into the nucleoplasm.
We find that the pattern of retained introns seen in the nucleoplasm is quite different from that on the chromatin. This indicates that it will be difficult to determine the order of intron removal from studies of full-length or partially spliced pre-mRNAs in total nuclear RNA samples, as has been suggested (Hatzoglou et al. 1985; Lang and Spritz 1987; Shiels et al. 1987; Gudas et al. 1990; Kessler et al. 1993; Neel et al. 1993). While it is likely that some of these retained, nucleoplasmic introns are spliced post-transcriptionally, some nucleoplasmic transcripts may never mature and instead be degraded. The amount of nucleoplasmic splicing is difficult to assess without measurements of the competing rates of mature mRNA export and of “dead-end” transcript degradation.
Within the general decrease in splicing seen for more 3′ proximal introns on the chromatin, there is variability in the efficiency of intron excision. The source of this variability is not clear, but the level of splicing does not seem to correlate with intron length or splice site strength. For example, c-Src intron 1a is 19 kb long, but much more efficiently excised than intron 10, which is only about 200 nt long (Figs. 1C, 3A). FN introns 9, 42, and 44 all have similar maximum entropy scores for the strength of their splice sites, but show quite different levels of excision (Fig. 3B; Supplemental Fig. S6; Yeo and Burge 2004). Thus, along with the effect of transcriptional direction, other unidentified features, such as splicing factor abundance, RNA secondary structure, and transcription rate, must determine the relative efficiency of some of these splicing events.
Regulation of splicing during transcription
We examined three different alternative exons showing differential regulation between cell lines. To determine the order of upstream and downstream intron removal for each exon, we measured the relative abundance of the partially spliced intermediates. Both FN exons show exclusive removal of the downstream intron prior to the upstream one (Fig. 4). This partially spliced intermediate is highly enriched in the chromatin fraction indicating that the decision to include the exon is made during transcription. Unlike constitutive introns, these introns flanking the alternative FN exons are not being excised in the order of their transcription. Interestingly, although the pattern of flanking intron removal is identical, these two exons are thought to be controlled by different mechanisms. For exon 25, regulatory proteins like Fox2, which binds in the downstream intron, may promote its splicing. For exon 33, the “kinetic model” predicts that reduced elongation rates will promote splicing of the upstream intron prior to the synthesis of the complete downstream intron. It is not clear how to reconcile the earlier splicing of the downstream intron with the simplest forms of this model. However, the order of intron excision may not reflect the rates of partial spliceosome assembly at individual splice sites. For c-Src exon N1, both intermediates are detectible in LA-N-5 cells but, unlike the FN exons, excision of the upstream intron prior to the downstream predominates (Fig. 5A,B). These intermediates are undetectable in the nucleoplasm, again indicating that this exon is fully spliced by the time of transcript release. The introns flanking exon N1 are removed in a different order than FN exon 25, even though these exons are regulated by some of the same proteins. This may be due to additional features influencing the excision pathway of the flanking introns, such as a 10-fold differential in their length, or may be due to the PTB and Fox proteins acting differently in distinct exon settings.
We also examined these regulated introns under conditions of exon skipping, and relative to the constitutive introns in the same transcripts (Figs. 6, 7). In this case, c-Src N1 and FN exon 25 behave more similarly to each other than to FN exon 33. In HeLa cells, where these exons are predominantly or entirely skipped, the introns surrounding the regulated exons show high levels of retention relative to the adjacent constitutive introns both upstream and downstream. This is most striking for c-Src N1, which is entirely skipped in HeLa cells (Fig. 6A). In these cells, the intron encompassing the N1 exon is largely retained in the chromatin fraction. In LA-N-5 cells, there is a mixture of exon inclusion and exclusion, and the efficiency of splicing at the sites used for both splicing patterns is increased (the 5′ splice site of intron 3 and the 3′ splice site of intron N1) (Fig. 6C). For FN exon 25 in HeLa cells, the mixture of exon inclusion and exclusion again complicates the interpretation. Nevertheless, the level of unspliced RNA is increased under conditions of high exon skipping (Fig. 7A). For these exons, skipping of the exon apparently induces a lag in the splicing of the long intron encompassing it. It will be interesting to examine the role of exon-bound SR proteins in this lag, given results showing that the splicing apparatus has difficulty splicing over an SR protein bound within an intron (Ibrahim et al. 2005).
In contrast to c-Src exon N1 and FN exon 25, FN exon 33 and its flanking introns showed efficient splicing, and were more highly excised than the constitutive introns downstream. Both the exon 33 flanking introns, as well as the long intron derived from exon skipping, showed low levels of retention in the chromatin RNA (Fig. 7A,C) that were even lower than the nucleoplasm. This behavior, which is distinct from the c-Src N1 and FN exon 25, is in agreement with the idea that a different mechanism governs the regulation of this exon (Chan and Black 1997; Cramer et al. 1999; de la Mata et al. 2003; Underwood et al. 2005; de la Mata and Kornblihtt 2006).
The splicing of regulated cassette exons can be altered by a number of different mechanisms. However, the final outcome of all these mechanisms could be manifested at a later time, if splicing complexes assembled during transcription do not complete their assembly and catalysis until after transcriptional termination. For all three alternative exons examined here, very little unspliced RNA was observed in the nucleoplasm. Even for those exons showing increased unspliced RNA under exon skipping conditions, splicing seems to be completed prior to release from the chromatin template (Figs. 5C, 6E, 7D; data not shown). The very small pools of unspliced or partially spliced RNA in the nucleoplasm indicate that the regulation of splicing choice for alternative exons occurs during RNA synthesis.
We have drawn several conclusions regarding the level of excision for individual introns relative to each other and relative to transcript release. These experiments by necessity have examined steady-state levels of RNA. Thus, interpretations regarding the kinetics of these processes must be taken with caution, as some of the RNA species we observe may not be intermediates that will give rise to a mature mRNA. Developing assays to monitor the kinetics of in vivo splicing will be essential to further understanding the mechanisms behind transcriptionally coupled splicing.
MATERIALS AND METHODS
Cell culture
Human LA-N-5 neuroblastoma and HeLa cell lines were grown following standard tissue culture procedure, with guidelines provided by the American Type Culture Collection (http://www.atcc.org).
Antibodies
Anti-Histone3 was purchased from Abcam (ab1791), anti-α-tubulin from Calbiochem (CP-60), and Anti-Pol II from Santa Cruz Biotechnology (sc-899). The U5-116k antibody was a gift from R. Lührmann and anti-U170K was generated in the laboratory.
Constructs and primers
Primers were designed using Primer3 (http://frodo.wi.mit.edu/). T7 transcription templates for the quantitative full-length RNA controls were PCR amplified from human genomic DNA (for intron containing templates) or human cytoplasmic cDNA (for spliced templates). The T7 Δ10 transcription templates were obtained by PCR amplification of the full-length templates, using an extended version of the reverse primer designed to delete 10 internal nucleotides close to the 3′ end of the template, while preserving the original reverse primer binding sequence. All templates were then purified by agarose gel electrophoresis and quantified using a nanodrop-1000 spectrophotometer (Nanodrop Technologies) prior to in vitro transcription.
RNA transcription and quantification
A 25 μL reaction containing 250 ng of template DNA, 40 mM Tris-HCl at pH 7.9, 2 mM spermidine, 10 mM DTT, 12 mM MgCl2, 0.8 mM each NTP, 14U RNAguard (Amersham), and 10% (v/v) T7 RNA polymerase (homemade) was incubated at 37°C for 3 h. 10U RNase-free DNase (Roche) was then added for an additional 30 min. RNA was heated at 85°C for 5 min in RNA loading buffer (Ambion), loaded on a denaturing polyacrylamide gel and separated by electrophoresis. Bands were visualized by UV shadowing, extracted, and eluted overnight in 0.5 M C2H3NaO2, 0.2% SDS, 1 mM EDTA. Eluted RNA was precipitated, dissolved in TE (pH 7) and quantified (A260) using a Nanodrop-1000 spectrophotometer (Nanodrop Technologies). Molar RNA amounts were calculated, and the quantitative RNA controls were serially diluted to a final concentration of 4 × 10−19 mol/μL. Two and a half mcroliters of each FL or Δ10 RNA were added to each reverse transcription reaction, and 1/10 of the RT was added to each PCR. Prior to use, each IVT RNA was tested for template DNA contamination by RT-PCR lacking reverse transcriptase.
Cellular fractionation and RNA isolation
Briefly, 2 × 108 cells were gently trypsinized (HeLa) or removed from plate by pipetting (LA-N-5) and collected by centrifugation (adapted from Wuarin and Schibler [1994]). Cell pellets were rinsed in 1× PBS/1 mM EDTA and then the plasma membranes were lysed by resuspension in ice-cold NP-40 lysis buffer (10 mM Tris-HCl [pH 7.5], 0.05% [LA-N-5]–0.15% [HeLa] NP40 [Sigma], 150 mM NaCl), for 5 min. The lysate was then layered on top of 2.5 volumes of a chilled sucrose cushion (24% sucrose in lysis buffer) and centrifuged for 10 min, 4°C, 14,000 rpm in an Eppendorf 5415C microfuge. The supernatant (cytoplasmic fraction) was collected and treated with proteinase K for 1 h at 37°C, then phenol/chloroform extracted and ethanol precipitated. The nuclei pellet was gently rinsed with ice-cold 1× PBS/1 mM EDTA, then resuspended in a prechilled glycerol buffer (20 mM Tris-HCl [pH 7.9], 75 mM NaCl, 0.5 mM EDTA, 0.85 mM DTT, 0.125 mM PMSF, 50% glycerol) by gentle flicking of the tube. An equal volume of cold nuclei lysis buffer (10 mM HEPES [pH 7.6], 1 mM DTT, 7.5 mM MgCl2, 0.2 mM EDTA, 0.3 M NaCl, 1 M UREA, 1% NP-40) was added, the tube was gently vortexed for 2 × 2 sec, incubated for 2 min on ice, and then centrifuged for 2 min, 4°C, 14,000 rpm as above. The supernatant (soluble nuclear fraction) was treated with proteinase K for 1 h and 37°C, then phenol/chloroform extracted and ethanol precipitated. The chromatin pellet was gently rinsed with cold 1× PBS/1 mM EDTA and then dissolved in TRIzol (Invitrogen). Chromatin-associated RNA was purified according to the TRIzol protocol, but an additional phenol/chloroform extraction step was performed prior to precipitation. All RNA fractions were resuspended in TE (pH 7) and quantified using a Nanodrop-1000 spectrophotometer (Nanodrop Technologies) and tested for DNA contamination by RT-PCR lacking reverse transcriptase.
Reverse transcription and PCR
The experiment was performed as previously described (Patel et al. 2002) except 1 μg yeast tRNA was added to the control samples containing 1 × 10−18 mol of both the full-length and Δ10 IVT RNA fragments. Control reactions were performed alongside RT-PCR reactions assaying either 1 μg of chromatin-associated or nucleoplasmic RNA, or 2 μg of LA-N-5 or 4 μg of HeLa cytoplasmic RNA. Reverse transcription was performed according to the manufacturer's protocol (Superscript III, Invitrogen). PCR was performed using homemade Taq polymerase supplemented with homemade buffer (20 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2, 25 mM KCl, 0.05% Tween 20, 100 μg/mL BSA), 120 nM P32 labeled forward primer, 328 nM cold forward primer, 448 nM cold reverse primer, 200 μM dNTPs, 16% (v/v) cDNA, and 2% (v/v) Taq polymerase. Nineteen to 26 cycles of PCR were performed at an annealing temperature of 50°C for 45 sec with a 30 sec extension at 72°C. Amplicons were denatured in formamide buffer before being subjected to denaturing 8% PAGE. Gels were scanned using a Typhoon PhosphorImager (Molecular Dynamics) and quantified using Imagequant software (GE Healthcare). Quantification of intron abundance was performed by comparing the ratio of the control FL: Δ10 amplicons to the endogenous RNA:Δ10 amplicons after normalizing the Δ10 signal in the experimental sample to the Δ10 signal in the control.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Joan Steitz, Tim Nilsen, David Bentley, and members of the Black laboratory for helpful discussion and comments. We thank R. Lührmann and T. Achsel for the gift of the U5-116K antibody. A.P.-J. was supported in part by a fellowship from the American Association of University Women. This work was supported in part by NIH grant RO1GM49662 to D.L.B. D.L.B is an investigator of the Howards Hughes Medical Institute.
Footnotes
REFERENCES
- Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- Bauren G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- Bauren G, Belikov S, Wieslander L. Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′-end formation and excision of the 3′-terminal intron. Genes & Dev. 1998;12:2759–2769. doi: 10.1101/gad.12.17.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes & Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Beyer AL, Bouton AH, Miller OL., Jr Correlation of hnRNP structure and nascent transcript cleavage. Cell. 1981;26:155–165. doi: 10.1016/0092-8674(81)90299-3. [DOI] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Carey M, Smale ST. Transcriptional regulation in eukaryotes: Concepts, strategies, and techniques. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. [Google Scholar]
- Chan RC, Black DL. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol Cell Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Pesce CG, Baralle FE, Kornblihtt AR. Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Caceres JF, Cazalla D, Kadener S, Muro AF, Baralle FE, Kornblihtt AR. Coupling of transcription with alternative splicing: RNA Pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol Cell. 1999;4:251–258. doi: 10.1016/s1097-2765(00)80372-x. [DOI] [PubMed] [Google Scholar]
- Das R, Dufu K, Romney B, Feldt M, Elenko M, Reed R. Functional coupling of RNAP II transcription to spliceosome assembly. Genes & Dev. 2006;20:1100–1109. doi: 10.1101/gad.1397406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Kornblihtt AR. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol. 2006;13:973–980. doi: 10.1038/nsmb1155. [DOI] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, O'Malley BW. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERα and CAPERβ. Mol Cell. 2005;17:429–439. doi: 10.1016/j.molcel.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Engebrecht JA, Voelkel-Meiman K, Roeder GS. Meiosis-specific RNA splicing in yeast. Cell. 1991;66:1257–1268. doi: 10.1016/0092-8674(91)90047-3. [DOI] [PubMed] [Google Scholar]
- Greenleaf AL. Positive patches and negative noodles: Linking RNA processing to transcription? Trends Biochem Sci. 1993;18:117–119. doi: 10.1016/0968-0004(93)90016-g. [DOI] [PubMed] [Google Scholar]
- Gudas JM, Knight GB, Pardee AB. Ordered splicing of thymidine kinase pre-mRNA during the S phase of the cell cycle. Mol Cell Biol. 1990;10:5591–5595. doi: 10.1128/mcb.10.10.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzoglou M, Sekeris CE, Hanson RW. Processing of phosphoenolpyruvate carboxykinase (GTP) RNA in vivo. Proc Natl Acad Sci. 1985;82:4346–4350. doi: 10.1073/pnas.82.13.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks MJ, Yang CR, Kotlajich MV, Hertel KJ. Linking splicing to Pol II transcription stabilizes pre-mRNAs and influences splicing patterns. PLoS Biol. 2006;4:e147. doi: 10.1371/journal.pbio.0040147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Spector DL. Nascent pre-mRNA transcripts are associated with nuclear regions enriched in splicing factors. Genes & Dev. 1991;5:2288–2302. doi: 10.1101/gad.5.12a.2288. [DOI] [PubMed] [Google Scholar]
- Huang S, Spector DL. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J Cell Biol. 1996;133:719–732. doi: 10.1083/jcb.133.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim EC, Schaal TD, Hertel KJ, Reed R, Maniatis T. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc Natl Acad Sci. 2005;102:5002–5007. doi: 10.1073/pnas.0500543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler O, Jiang Y, Chasin LA. Order of intron removal during splicing of endogenous adenine phosphoribosyltransferase and dihydrofolate reductase pre-mRNA. Mol Cell Biol. 1993;13:6211–6222. doi: 10.1128/mcb.13.10.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Sharp PA. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987;49:763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- Konforti BB, Koziolkiewicz MJ, Konarska MM. Disruption of base pairing between the 5′ splice site and the 5′ end of U1 snRNA is required for spliceosome assembly. Cell. 1993;75:863–873. doi: 10.1016/0092-8674(93)90531-t. [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR, Pesce CG, Alonso CR, Cramer P, Srebrow A, Werbajh S, Muro AF. The fibronectin gene as a model for splicing and transcription studies. FASEB J. 1996;10:248–257. doi: 10.1096/fasebj.10.2.8641558. [DOI] [PubMed] [Google Scholar]
- Lang KM, Spritz RA. In vitro splicing pathways of pre-mRNAs containing multiple intervening sequences? Mol Cell Biol. 1987;7:3428–3437. doi: 10.1128/mcb.7.10.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chen ZY, Wang W, Baker CC, Krug RM. The 3′-end-processing factor CPSF is required for the splicing of single-intron pre-mRNAs in vivo. RNA. 2001;7:920–931. doi: 10.1017/s1355838201010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listerman I, Sapra AK, Neugebauer KM. Co-transcriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- Misteli T, Spector DL. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Schwartzfarb EM, Silver PA, Yu MC. Differential recruitment of the splicing machinery during transcription predicts genome-wide patterns of mRNA splicing. Mol Cell. 2006;24:903–915. doi: 10.1016/j.molcel.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Natalizio BJ, Robson-Dixon ND, Garcia-Blanco MA. The carboxyl-terminal domain of RNA polymerase II is not sufficient to enhance the efficiency of pre-mRNA capping or splicing in the context of a different polymerase. J Biol Chem. 2009;284:8692–8702. doi: 10.1074/jbc.M806919200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel H, Weil D, Giansante C, Dautry F. In vivo cooperation between introns during pre-mRNA processing. Genes & Dev. 1993;7:2194–2205. doi: 10.1101/gad.7.11.2194. [DOI] [PubMed] [Google Scholar]
- Nesic D, Maquat LE. Upstream introns influence the efficiency of final intron removal and RNA 3′-end formation. Genes & Dev. 1994;8:363–375. doi: 10.1101/gad.8.3.363. [DOI] [PubMed] [Google Scholar]
- Neugebauer KM, Roth MB. Distribution of pre-mRNA splicing factors at sites of RNA polymerase II transcription. Genes & Dev. 1997;11:1148–1159. doi: 10.1101/gad.11.9.1148. [DOI] [PubMed] [Google Scholar]
- Norton PA. Polypyrimidine tract sequences direct selection of alternative branch sites and influence protein binding. Nucleic Acids Res. 1994;22:3854–3860. doi: 10.1093/nar/22.19.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AA, McCarthy M, Steitz JA. The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J. 2002;21:3804–3815. doi: 10.1093/emboj/cdf297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigo F, Martinson HG. Functional coupling of last-intron splicing and 3′-end processing to transcription in vitro: The poly(A) signal couples to splicing before committing to cleavage. Mol Cell Biol. 2008;28:849–862. doi: 10.1128/MCB.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigo F, Martinson HG. Polyadenylation releases mRNA from RNA polymerase II in a process that is licensed by splicing. RNA. 2009;15:823–836. doi: 10.1261/rna.1409209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapra AK, Anko ML, Grishina I, Lorenz M, Pabis M, Poser I, Rollins J, Weiland EM, Neugebauer KM. SR protein family members display diverse activities in the formation of nascent and mature mRNPs in vivo. Mol Cell. 2009;34:179–190. doi: 10.1016/j.molcel.2009.02.031. [DOI] [PubMed] [Google Scholar]
- Shiels BR, Northemann W, Gehring MR, Fey GH. Modified nuclear processing of α 1-acid glycoprotein RNA during inflammation. J Biol Chem. 1987;262:12826–12831. [PubMed] [Google Scholar]
- Tardiff DF, Lacadie SA, Rosbash M. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly post-transcriptional. Mol Cell. 2006;24:917–929. doi: 10.1016/j.molcel.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologs of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S, Vagner C, Mattaj IW. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes & Dev. 2000;14:403–413. [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Burge CB. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Proudfoot NJ, Dye MJ. Molecular dissection of mammalian RNA polymerase II transcriptional termination. Mol Cell. 2008;29:600–610. doi: 10.1016/j.molcel.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetterberg I, Bauren G, Wieslander L. The intranuclear site of excision of each intron in Balbiani ring 3 pre-mRNA is influenced by the time remaining to transcription termination and different excision efficiencies for the various introns. RNA. 1996;2:641–651. [PMC free article] [PubMed] [Google Scholar]
- Will C, Lührmann R. The RNA world: The nature of modern RNA suggests a prebiotic RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. Spliceosome structure and function. [Google Scholar]
- Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: Evidence for co-transcriptional splicing. Mol Cell Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Yuryev A, Patturajan M, Litingtung Y, Joshi RV, Gentile C, Gebara M, Corden JL. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]